Abstract
Influenza viruses of the N1 neuraminidase (NA) subtype affecting both animals and humans caused the 2009 pandemic. Anti-influenza virus NA inhibitors are crucial early in a pandemic, when specific influenza vaccines are unavailable. Thus, it is urgent to confirm the antiviral susceptibility of the avian viruses, a potential source of a pandemic virus. We evaluated the NA inhibitor susceptibilities of viruses of the N1 subtype isolated from wild waterbirds, swine, and humans. Most avian viruses were highly or moderately susceptible to oseltamivir (50% inhibitory concentration [IC50], <5.1 to 50 nM). Of 91 avian isolates, 7 (7.7%) had reduced susceptibility (IC50, >50 nM) but were sensitive to the NA inhibitors zanamivir and peramivir. Oseltamivir susceptibility ranged more widely among the waterbird viruses (IC50, 0.5 to 154.43 nM) than among swine and human viruses (IC50, 0.33 to 2.56 nM). Swine viruses were sensitive to oseltamivir, compared to human seasonal H1N1 isolated before 2007 (mean IC50, 1.4 nM). Avian viruses from 2007 to 2008 were sensitive to oseltamivir, in contrast to the emergence of resistant H1N1 in humans. Susceptibility remained high to moderate over time among influenza viruses. Sequence analysis of the outliers did not detect molecular markers of drug-resistance (e.g., H275Y NA mutation [N1 numbering]) but revealed mutations outside the NA active site. In particular, V267I, N307D, and V321I residue changes were found, and structural analyses suggest that these mutations distort hydrophobic pockets and affect residues in the NA active site. We determined that natural oseltamivir resistance among swine and wild waterbirds is rare. Minor naturally occurring variants in NA can affect antiviral susceptibility.
In little more than 1 decade, there have been three remarkable events involving the emergence and control of seasonal, prepandemic, and pandemic influenza viruses of the N1 neuraminidase (NA) subtype. The first was the emergence and transmission of a highly pathogenic H5N1 influenza virus from waterbirds to domestic poultry with inefficient transmission to humans that was detected in 1997 (10, 45). Over the next 13 years, the H5N1 virus evolved into more than 10 phylogenetically distinct hemagglutinin (HA) clades, directly or indirectly killed hundreds of millions of gallinaceous birds, and spread to many countries in Eurasia, infecting 442 people and killing 262 (43, 47, 48, 50). These highly pathogenic viruses are continuing to evolve in multiple epicenters, including China, Indonesia, and Egypt (3, 7, 30, 47, 53). The second remarkable event occurred in seasonal influenza in 2007 when resistance to the anti-influenza virus drug oseltamivir was detected in Norway in the absence of drug selection pressure (22, 49, 52). The naturally occurring oseltamivir-resistant H1N1 influenza viruses have been surprisingly fit and had spread globally in humans by early 2009 (13, 24). The third event was the emergence of the pandemic H1N1 2009 influenza virus that was antigenically distinct from the 2007-2008 seasonal H1N1 viruses (16, 40, 51). The pandemic H1N1 influenza viruses also possess the N1 NA from avian sources (8, 16). Unlike previously circulating viruses, the pandemic H1N1 viruses contained a complex of influenza virus genes of Eurasian and North American swine influenza virus origin (40) which were previously derived from reassorted genes of human, swine, and avian lineages. This novel reassortment of HA and NA genes resulted in a virus that was effectively transmitted in humans. Immunocompromised patients, children under the age of 10 years, pregnant women, and people with underlying medical conditions, including obesity, have been particularly affected (16, 25, 26, 36). Efficacious vaccines have been prepared and are being administered. At this time approximately 99% of pandemic H1N1 2009 viruses tested are sensitive to NA inhibitors (NAIs) but are resistant to the adamantanes (49).
A common feature among these influenza virus events is the possession of an N1 subtype of NA. Although each of these N1s is antigenically and phylogenetically distinct, each emerged at different times in the past from the wild waterbird reservoir. While vaccination remains the primary option for the prevention and control of both seasonal and pandemic influenza, anti-influenza virus drugs are being recognized as the immediate option for treatment and control, especially during the 6 months or more needed for vaccine preparation and testing (18). The inherent problem with the use of monovalent chemotherapy for the treatment of influenza is that at the onset of a pandemic influenza virus outbreak, rapid vaccine production methods or novel prophylactic vaccines cannot be introduced fast enough. A good example of this was the emergence of pandemic H1N1 2009 viruses and the lack of a novel vaccine. Oseltamivir-resistant seasonal H1N1 influenza viruses emerged in 2007 and were naturally occurring drug-resistant viruses without drug selection pressure. The surprising aspect of the emergence of the resistant seasonal virus was that the resistance appeared in Norway, where little oseltamivir has been used (22, 49). Oseltamivir-resistant strains of H5N1 and pandemic H1N1 have been detected (6, 11), but to date these resistant viruses have not spread consistently and are sensitive to zanamivir, a drug which is more closely fitted to the structure of the NA active site (5, 6, 18, 20, 21, 42).
Close monitoring of clinical isolates has detected drug-resistant influenza viruses from humans, but limited information or monitoring is available concerning the sensitivities of influenza viruses to NAIs in their natural avian reservoir or species other than humans. Our hypothesis was that influenza viruses with reduced susceptibility to anti-influenza virus drugs may exist within the natural reservoir of wild waterbirds and domestic swine. These influenza viruses with reduced susceptibility or resistance to anti-influenza virus drugs could potentially be transmitted to humans. We examined the susceptibilities of N1 NAs from wild waterbirds and domestic swine to the NAIs in a phenotypic NA enzyme inhibition assay and sequenced the NAs of the identified outliers.
MATERIALS AND METHODS
Compounds.
Oseltamivir carboxylate (oseltamivir; GS4071; [3R,4R,5S]-4-acetamido-5-amino-3-[1-ethylpropoxy]-1-cyclohexane-1 carboxylic acid) was provided by Hoffmann-La Roche (Basel, Switzerland). Zanamivir (GG167; 4-guanidino-Neu5Ac2en) was provided by GlaxoSmithKline (Research Triangle Park, NC). Peramivir ([1S,2S,3R,4R,1′S]-3-[1′-acetylamino-2′-ethyl]butyl-4-[(aminoimino)-methyl]amino-2-hydroxycyclopentane-1-carboxylic acid; BCX-1812) was provided by BioCryst Pharmaceuticals (Birmingham, AL). The compounds were dissolved in distilled water, and aliquots of stock were stored at −20°C until used.
Viruses and cells.
The avian and swine influenza viruses out of 123 of the N1 NA subtype are shown in Table 1. Most isolates were from North America. Viruses obtained from avian sources were all from apparently healthy wild waterbirds and were isolated from fecal samples or from tracheal/oropharyngeal or cloacal swabs obtained from Anseriformes (ducks), Anas platyrhynchos (mallard), Anas acuta (pintail), Anas discors (blue-winged teal), Anas carolinensis (green-winged teal), Spatula clypeata (shoveler), Aythya americana (redhead), and Aythya valisineria (canvasback) and from Charadriiformes (shorebirds), Arenaria interpres (ruddy turnstone), Larus atricilla (laughing gull), and Larus argentatus (herring gull). The influenza viruses from pigs were from animals with respiratory disease. Viruses were isolated from ducks from 1983 to 2008 and from shorebirds/gulls from 1979 to 2007. Swine isolates were obtained from 2005 to 2009, and human isolates were from 1976 to 2009.
TABLE 1.
Susceptibility of avian influenza A viruses of N1 NA subtype to the NA inhibitor oseltamivir
| Source and name of N1 NA virus | Subtype | Yr isolated | Origin | Mean IC50 ± SD (nM)b |
|---|---|---|---|---|
| Ducksa | ||||
| A/Mallard/Alberta/743/83 | H9N1 | 1983 | Canada | 20.40 ± 2.69 |
| A/Blue-Winged Teal/Alberta/212/84 | H1N1 | 1984 | Canada | 9.01 ± 1.58 |
| A/Mallard/TN/11464/85 | H1N1 | 1985 | USA | 39.75 ± 10.00 |
| A/Blue-Winged Teal/LA/B228/86 | H1N1 | 1986 | USA | 5.98 ± 2.02 |
| A/Mallard/Alberta/323/88 | H2N1 | 1988 | USA | 24.88 ± 11.16 |
| A/Mallard/Alberta/253/90 | H6N1 | 1990 | Canada | 1.28 ± 0.34 |
| A/Mallard/Alberta/107/91 | H3N1 | 1991 | USA | 12.11 ± 3.60 |
| A/Mallard/Alberta/196/92 | H3N1 | 1992 | Canada | 22.75 ± 0.00 |
| A/Pintail/Alberta/129/93 | H7N1 | 1993 | Canada | 0.81 ± 0.36 |
| A/Mallard/Alberta/5/95 | H10N1 | 1995 | Canada | 1.82 ± 0.55 |
| A/Mallard/Alberta/267/96 | H1N1 | 1996 | Canada | 56.70 ± 11.82 |
| A/Mallard/Alberta/119/98 | H1N1 | 1998 | Canada | 4.35 ± 0.00 |
| A/Mallard/Alberta/201/98 | H1N1 | 1998 | Canada | 13.40 ± 0.00 |
| A/Mallard/Alberta/34/2001 | H7N1 | 2001 | Canada | 102.25 ± 3.80 |
| A/Pintail/Alberta/210/2002 | H1N1 | 2002 | Canada | 8.50 ± 10.25 |
| A/Mallard/Alberta/130/2003 | H4N1 | 2003 | Canada | 8.99 ± 6.40 |
| A/Mallard/Alberta/88/2004 | H1N1 | 2004 | Canada | 99.17 ± 61.85 |
| A/Mallard/Alberta/226/2004 | H3N1 | 2004 | Canada | 51.61 ± 14.43 |
| A/Pintail/Alberta/68/2005 | H1N1 | 2005 | Canada | 40.57 ± 8.54 |
| A/Pintail/Alberta/69/2005 | H1N1 | 2005 | Canada | 11.52 ± 3.65 |
| A/Canvasback/Alberta/276/2005 | H1N1 | 2005 | Canada | 39.30 ± 8.81 |
| A/Mallard/PA/454069-9/2006 | H5N1 | 2006 | USA | 2.33 ± 0.96 |
| A/Mallard/MN/SG-000220/2007 | H6N1 | 2007 | USA | 7.36 ± 3.96 |
| A/Green-Winged Teal/LA/SG-00090/2007 | H1N1 | 2007 | USA | 18.03 ± 8.36 |
| A/Mallard/MN/AI07-3018/2007 | H1N1 | 2007 | USA | 29.61 ± 20.09 |
| A/Mallard/MN/AI07-3019/2007 | H3N1 | 2007 | USA | 12.97 ± 2.85 |
| A/Mallard/MN/AI07-3099/2007 | H3N1 | 2007 | USA | 12.83 ± 4.18 |
| A/Mallard/MN/AI07-3100/2007 | H1N1 | 2007 | USA | 10.54 ± 2.97 |
| A/Mallard/OH/510306-4/2007 | H5N1 | 2007 | USA | 6.55 ± 2.33 |
| A/Mallard/MI/463796-7/2007 | H5N1 | 2007 | USA | 4.48 ± 1.17 |
| A/Mallard/MN/AI07-3108/2007 | H3N1 | 2007 | USA | 6.39 ± 2.10 |
| A/Mallard/MN/AI07-3124/2007 | H3N1 | 2007 | USA | 8.39 ± 2.45 |
| A/Mallard/MN/AI07-3127/2007 | H1N1 | 2007 | USA | 5.11 ± 1.89 |
| A/Mallard/MN/AI07-3136/2007 | H1N1 | 2007 | USA | 6.41 ± 3.39 |
| A/Mallard/MN/AI07-3140/2007 | H1N1 | 2007 | USA | 7.50 ± 2.36 |
| A/Mallard/MN/AI07-3189/2007 | H10N1 | 2007 | USA | 6.74 ± 4.38 |
| A/Mallard/MN/SG-000220/2007 | H6N1 | 2007 | USA | 5.69 ± 3.12 |
| A/Mallard/MN/SG-000223/2007 | H6N1 | 2007 | USA | 3.97 ± 2.68 |
| A/Mallard/MN/SG-000170/2007 | H6N1 | 2007 | USA | 12.53 ± 3.84 |
| A/Mallard/MN/SG-000214/2007 | H6N1 | 2007 | USA | 6.18 ± 1.81 |
| A/Mallard/MN/SG-000104/2007 | H6N1 | 2007 | USA | 7.62 ± 3.82 |
| A/Mallard/MN/SG-000105/2007 | H6N1 | 2007 | USA | 7.02 ± 2.56 |
| A/Mallard/MN/SG-000121/2007 | H1N1 | 2007 | USA | 12.41 ± 2.43 |
| A/Red Headed Duck/MN/SG-000123/2007 | H1N1 | 2007 | USA | 10.49 ± 2.25 |
| A/Blue-Winged Teal/TX/SG-00087/2007 | H4N1 | 2007 | USA | 6.03 ± 2.11 |
| A/Mallard/MI/463796-7/2007 | H5N1 | 2007 | USA | 4.48 ± 1.17 |
| A/Mallard/OH/510306-4/2007 | H5N1 | 2007 | USA | 6.55 ± 2.33 |
| A/Mallard/OH/4809/2008 | H1N1 | 2008 | USA | 8.58 ± 1.47 |
| A/Mallard/MN/SG-00572/2008 | H3N1 | 2008 | USA | 8.83 ± 1.96 |
| A/Mallard/MN/SG-00627/2008 | H1N1 | 2008 | USA | 6.95 ± 3.18 |
| A/Mallard/MN/SG-00628/2008 | H1N1 | 2008 | USA | 4.91 ± 1.63 |
| A/Northern Shoveler/MN/SG-00651/2008 | H1N1 | 2008 | USA | 8.80 ± 2.81 |
| A/Northern Shoveler/MN/SG-00655/2008 | H1N1 | 2008 | USA | 9.39 ± 4.22 |
| A/Northern Shoveler/MN/SG-00661/2008 | H3N1 | 2008 | USA | 8.15 ± 2.41 |
| A/Northern Shoveler/MN/SG-00665/2008 | H3N1 | 2008 | USA | 11.40 ± 4.95 |
| A/Mallard/MN/AI08-3825/2008 | H5N1 | 2008 | USA | 9.56 ± 4.80 |
| A/Mallard/MN/AI08-4507/2008 | H3N1 | 2008 | USA | 7.56 ± 4.44 |
| A/Green-Winged Teal/AI08-4655/2008 | H5N1 | 2008 | USA | 1.87 ± 0.73 |
| A/Mallard/MN/AI08-5384/2008 | H5N1 | 2008 | USA | 12.00 ± 1.14 |
| Shorebirds/gullsa | ||||
| A/Gull/Kazakhstan/870/79 | H1N1 | 1979 | USSR | 3.07 ± 0.91 |
| A/Ruddy Turnstone/NJ/61/85 | H11N1 | 1985 | USA | 23.97 ± 6.89 |
| A/Herring Gull/DE/698/88 | H2N1 | 1988 | USA | 10.85 ± 0.14 |
| A/Ruddy Turnstone/DE/34/93 | H2N1 | 1993 | USA | 41.13 ± 15.51 |
| A/Ruddy Turnstone/DE/78/93 | H11N1 | 1993 | USA | 10.05 ± 0.85 |
| A/Ruddy Turnstone/DE/8/93 | H2N1 | 1993 | USA | 14.22 ± 10.33 |
| A/Laughing Gull/DE/254/93 | H13N1 | 1993 | USA | 0.50 ± 0.21 |
| A/Ruddy Turnstone/DE/81/93 | H2N1 | 1993 | USA | 23.97 ± 6.89 |
| A/Ruddy Turnstone/DE/170/94 | H3N1 | 1994 | USA | 0.98 ± 0.46 |
| A/Ruddy Turnstone/DE/185/94 | H3N1 | 1994 | USA | 3.59 ± 1.41 |
| A/Ruddy Turnstone/DE/183/94 | H3N1 | 1994 | USA | 1.12 ± 0.39 |
| A/Shorebird/DE/39/95 | H3N1 | 1995 | USA | 3.23 ± 1.33 |
| A/Shorebird/DE/288/95 | H3N1 | 1995 | USA | 0.92 ± 0.33 |
| A/Shorebird/DE/24/96 | H11N1 | 1996 | USA | 35.25 ± 3.06 |
| A/Ruddy Turnstone/DE/125/96 | H12N1 | 1996 | USA | 45.82 ± 2.44 |
| A/Shorebird/DE/111/97 | H2N1 | 1997 | USA | 39.00 ± 2.84 |
| A/Shorebird/DE/138/97 | H2N1 | 1997 | USA | 2.68 ± 0.21 |
| A/Shorebird/DE/182/97 | H2N1 | 1997 | USA | 154.43 ± 38.53 |
| A/Shorebird/DE/24/98 | H2N1 | 1998 | USA | 111.07 ± 18.61 |
| A/Shorebird/DE/95/2003 | H9N1 | 2003 | USA | 67.00 ± 11.16 |
| A/Shorebird/DE/68/2003 | H9N1 | 2003 | USA | 52.85 ± 13.95 |
| A/Laughing Gull/DE/5/2003 | H9N1 | 2003 | USA | 65.80 ± 17.88 |
| A/Shorebird/DE/65/2003 | H9N1 | 2003 | USA | 5.63 ± 2.63 |
| A/Ruddy Turnstone/NJ/AI07-69/2007 | H5N1 | 2007 | USA | 5.79 ± 1.36 |
| A/Ruddy Turnstone/NJ/AI07-72/2007 | H9N1 | 2007 | USA | 6.46 ± 1.57 |
| A/Ruddy Turnstone/NJ/AI07-283/2007 | H9N1 | 2007 | USA | 5.59 ± 1.61 |
| A/Ruddy Turnstone/NJ/AI07-296/2007 | H6N1 | 2007 | USA | 7.36 ± 3.96 |
| A/Ruddy Turnstone/NJ/AI07-699/2007 | H5N1 | 2007 | USA | 4.60 ± 1.44 |
| A/Ruddy Turnstone/NJ/AI07-839/2007 | H12N1 | 2007 | USA | 6.60 ± 2.22 |
| Swinec | ||||
| A/Swine/NC/38448-1/2005 | H1N1 | 2005 | USA | 2.11 ± 0.91 |
| A/Swine/KS/029170/2007 | H1N1 | 2007 | USA | 0.33 ± 0.12 |
| A/Swine/OK/038826/2007 | H1N1 | 2007 | USA | 1.82 ± 0.42 |
| A/Swine/NE/047330/2007 | H1N1 | 2007 | USA | 0.90 ± 0.49 |
| A/Swine/OH/004880/2008 | H1N1 | 2008 | USA | 3.31 ± 1.52 |
| A/Swine/NC/007270/2008 | H1N1 | 2008 | USA | 1.12 ± 0.65 |
| A/Swine/KY/012454/2008 | H1N1 | 2008 | USA | 1.44 ± 0.18 |
| A/Swine/MN/016245/2008 | H1N1 | 2008 | USA | 0.66 ± 0.43 |
| A/Swine/WI/018247-2/2008 | H1N1 | 2008 | USA | 1.27 ± 0.48 |
| A/Swine/IL/020968/2008 | H1N1 | 2008 | USA | 1.90 ± 0.66 |
| A/Swine/MO/044329/2008 | H1N1 | 2008 | USA | 0.65 ± 0.25 |
| A/Swine/MN/055403-3/2008 | H1N1 | 2008 | USA | 0.79 ± 0.27 |
| A/Swine/IA/056944/2008 | H1N1 | 2008 | USA | 1.77 ± 0.79 |
| A/Swine/IL/060530/2008 | H1N1 | 2008 | USA | 0.92 ± 0.12 |
| A/Swine/IA/003479/2009 | H1N1 | 2009 | USA | 2.56 ± 1.50 |
Influenza viruses obtained from avian sources were all from apparently healthy wild birds and were isolated from fecal samples or from tracheal/oropharyngeal or cloacal swabs. Stock viruses of avian isolates were made by passaging in 10-day-old embryonated chicken eggs. Two-letter abbreviations in virus names correspond to standard postal abbreviations for states.
The NA inhibition assay used MUNANA as substrate (final concentration, 100 μM). Values are the means of at least three independent determinations.
Influenza viruses from pigs were from animals with respiratory disease and were isolated from bronchial swabs, nasal swabs, or lung tissues.
Avian viruses were obtained from the St. Jude Children's Research Hospital influenza repository, USDA-APHIS, and the University of Georgia. Swine viruses were obtained from the University of Minnesota. Human isolates were also obtained from the St. Jude Children's Research Hospital influenza virus repository and were used for comparison in this study. Stocks were made for each virus by passaging in 10-day-old embryonated chicken eggs (avian influenza viruses) or in Madin-Darby canine kidney cells (MDCK; for human and swine influenza viruses). These virus stocks were frozen at −80°C until used. MDCK cells were obtained from the American Type Culture Collection (Manassas, VA) and were maintained as described previously (54).
NA activity and NA inhibition assays.
NA activity of the virus was measured in a fluorescent assay by using the fluorigenic substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid sodium salt hydrate (MUNANA; Sigma, St. Louis, MO) as the substrate, based on the method of Potier et al. (37). The susceptibility of viruses was tested in an NA enzyme inhibition assay (4). Fluorimetric determinations were quantified with a Fluoroskan II (Labsystems, Helsinki, Finland) or Synergy 2 (BioTek Instruments, Winooski, VT) fluorimeter using an excitation wavelength of 360 nm and an emission wavelength of 460 nm and measuring fluorescence of the released 4-methyl-umbelliferone.
Viruses were standardized to equivalent NA enzyme activity in the linear range of the curve and incubated with NA inhibitor at concentrations of 0.00005 to 100 μM. For NA inhibition assays, 10 μl of drug and 10 μl of diluted virus were mixed and preincubated for 30 min at 37°C. Next, 30 μl of 100 μM MUNANA in 325 mM 2-(N-morpholino)ethanesulfonic acid (MES; pH 6.5; Sigma, St. Louis, MO) buffer containing 10 mM CaCl2 was added. After 30 min at 37°C, the reaction was stopped by addition of 150 μl of freshly prepared stop solution (25% ethanol and 12.5% glycine; Fisher Scientific, Rochester, NY) in distilled water. The concentration of NA inhibitor that reduced NA activity by 50% relative to a control mixture with no inhibitor (IC50) was determined by plotting the percent inhibition of NA activity as a function of the compound concentrations calculated. IC50s were calculated on a Microsoft Excel spreadsheet (Microsoft Corp., Redmond, WA), and values were transferred to a GraphPad Prism (GraphPad Software, La Jolla, CA). IC50s are reported as the means of three independent determinations.
Statistics.
Data were heteroscedastic (Fligner-Killeen; P < 0.0001), and so the Kruskal-Wallis (nonparametric test, equivalent to an analysis of variance) and Dunn's multiple comparison tests were used to analyze differences in the mean IC50s among the four groups (ducks, shorebirds, swine, and humans) and year of virus isolation.
Sequencing.
The RNeasy kit (Qiagen, Chatsworth, CA) was used to extract viral RNA, and a one-step reverse transcription-PCR kit (Qiagen, Chatsworth, CA) and universal primers to the NA and M2 genes were used for amplification. The sequences were determined by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital by using a BigDye Terminator (version 3) cycle sequencing kit and synthetic oligonucleotides. Samples were analyzed on Applied Biosystems 3700 DNA analyzers (Foster City, CA). Analysis of amino acid residues was based on the N1 numbering system.
Three-dimensional macromolecular structural modeling and visualization.
The PyMOL molecular visualization system (DeLano Scientific LLC, Palo Alto, CA) was used to produce high-quality three-dimensional images based on the crystal structure of the influenza virus NA of the N1 subtype (PDB code 3cl0) from the RCSB protein structure database. Analysis of three-dimensional structure was based on the N1 numbering system.
RESULTS
Oseltamivir susceptibility among avian species.
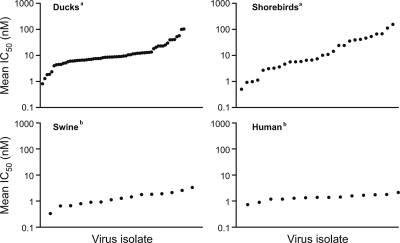
Migratory waterfowl have been accepted as the primordial source of influenza viruses that transmit to other species, including humans. Although humans can be infected with this primordial avian source of influenza virus, little attention has been given to the sensitivity of the viruses to anti-influenza virus drugs. Influenza viruses from 62 ducks (Anseriformes), 25 shorebirds (Charadriiformes), and 4 gulls (Laridae), listed in Table 1, were examined for antiviral susceptibility in the phenotypic NA enzyme inhibition assay. Currently there is no guidance on how to scale susceptibility and analyze influenza viruses with different IC50s. However, the Neuraminidase Inhibitor Susceptibility Network (NISN) provides a panel of reference viruses for the standardization of IC50s, and these were used as a basis for our analysis. The two resistant viruses, A/Fukui/45/2004 (H3N2) carrying an E119V mutation and A/Mississippi/3/2001 (H1N1) carrying an H274Y NA mutation, exhibited a range of IC50s for oseltamivir (48 to 413 nM; fluorescence-based NA enzyme inhibition assays). We chose an IC50 of >50 nM as representing reduced susceptibility. Although resistant human viruses exhibited an IC50 of 2,700 nM (Table 2), none of the avian isolates tested approached this number. Thus, we classified our avian viruses as having “reduced susceptibility” instead of being “resistant.” The “highly susceptible” isolates possessed IC50s (<5 nM) comparable with the susceptible reference viruses from the NISN panel, A/Fukui/20/2004 (H3N2) and A/Mississippi/3/2001 (H1N1) (histidine at 275), which exhibit a range of IC50 values for oseltamivir of 0.2 to 3.0 nM. The higher IC50 cutoff was used instead of 3.0 nM because 5 nM represents a 10-fold change from the reduced susceptibility cutoff of 50 nM. Table 2 shows the levels of susceptibility among avian, swine, and human influenza viruses. Importantly, we did not detect any NAI-resistant viruses among the N1 NA avian influenza virus strains. Approximately 16% of influenza viruses isolated from ducks were highly susceptible to oseltamivir (mean IC50, 0.81 to 4.91 nM), 77% were moderately susceptible (mean IC50, 5.11 to 40.57 nM), and only 6.5% had reduced susceptibility (mean IC50, 51.61 to 102.25 nM) (Table 2). Among combined shorebird and gull influenza isolates, 31% were highly susceptible (mean IC50, 0.5 to 4.6 nM), 52% were moderately susceptible (mean IC50, 5.59 to 45.82 nM), and 17% had reduced susceptibility (mean IC50, 52.85 to 154.43 nM). The avian viral isolates from wild waterbirds showed similar levels of sensitivity to oseltamivir in all ranges of susceptibility, whereas human and swine influenza viruses had comparable mean IC50s that were highly susceptible (Table 2). Furthermore, the variation in the waterbirds is higher than that of the mammalian samples (Fligner-Killeen test; P < 0.0001). In addition, within each level of susceptibility among wild waterbirds, a range in values of oseltamivir susceptibility was observed. Duck and shorebird isolates ranged mainly from ≤5 nM to <50 nM (highly susceptible to moderately susceptible), which is in contrast to the values observed for swine and human isolates. Of the avian viruses (combining ducks, shorebirds, and gulls), 19 were highly susceptible to oseltamivir (mean IC50, 0.5 to 5.0 nM) and 63 were moderately susceptible (mean IC50, 5.1 to 50 nM) (Table 2). Seven avian isolates (three from ducks, three from shorebirds, and one from gulls) had reduced susceptibility to oseltamivir (mean IC50, >50 nM). Four avian isolates had an IC50 between 50 and 100 nM, and three others had an IC50 of >100 nM. Therefore, approximately 21% of all wild avian influenza virus isolates were highly susceptible, 69% were moderately susceptible, and only ∼10% had reduced susceptibility. Overall, the avian isolates analyzed had variations in their susceptibilities to oseltamivir. The range of IC50s that was observed among the avian isolates was then categorized into baseline values of susceptibility. The influenza viruses of the N1 NA subtype isolated from shorebirds had higher IC50s (mean IC50 of 26 nM) and therefore were less susceptible to oseltamivir in vitro than were the susceptible human isolates. The overall mean IC50 for influenza viruses isolated from ducks was 16.1 nM. Figure 1 shows distribution plots that further illustrate that most of the avian influenza virus isolates fell within the range of moderate susceptibility (middle portion of the distribution graph). Ducks and shorebirds/gulls had comparable distribution patterns of antiviral susceptibility. Swine and human seasonal influenza virus isolates (except 2001 to 2008 viruses) showed a difference in their distribution pattern of antiviral susceptibility from those seen in wild waterbirds.
TABLE 2.
Susceptibilities of the avian, swine, and human influenza viruses of the N1 NA subtype to the NA inhibitor oseltamivir
| Species | Susceptibilitya | IC50 range (nM)b | No. of isolates (% of total) | Mean IC50 ± SD (nM) |
|---|---|---|---|---|
| Ducks | High | 0.81-4.91 | 12 (18.5) | 2.97 ± 1.49 |
| Moderate | 5.11-40.57 | 50 (77) | 12.85 ± 8.82 | |
| Reduced | 51.61-102.25 | 4 (6) | 77.43 ± 26.99 | |
| Shorebirds/gulls | High | 0.5-4.6 | 9 (31) | 2.80 ± 1.34 |
| Moderate | 5.59-45.82 | 15 (52) | 13.57 ± 9.62 | |
| Reduced | 52.85-154.43 | 5 (17) | 92.76 ± 36.59 | |
| Swine | High | 0.33-2.56 | 15 (100) | 1.44 ± 0.81 |
| Moderate | NAc | 0 | NA | |
| Reduced | NA | 0 | NA | |
| Humans | Sensitive | 0.89-2.12 | 14 (82) | 1.40 ± 0.36 |
| Resistant | 2,588.92-2,887.63 | 3 (17) | 2,725.1 ± 151.1 |
Susceptibilities of influenza viruses to oseltamivir were recorded as high (mean IC50, 0.5 to 5.0 nM), moderate (mean IC50, 5.1 to 50.0 nM), or reduced (mean IC50, 50 to ≥100 nM).
The NA inhibition assay was performed with viruses standardized to equivalent NA activity and incubated with NAIs at concentrations of 0.00005 to 100 μM with MUNANA as a substrate. The IC50 was determined by plotting the dose-response curve of inhibition of NA activity as a function of the compound concentration. Values are from at least three independent determinations.
NA, not applicable.
FIG. 1.
Plots showing the IC50 (in nM) ranges of oseltamivir for avian, swine, and human influenza viruses of the N1 NA subtype. Isolates are ranked in order by the IC50. Sixty-two duck, 25 shorebird, 4 gull, and 15 swine isolates and 14 human seasonal influenza viruses were analyzed. Mean IC50s between the four types of hosts were analyzed using a Kruskal-Wallis test and Dunn's multiple comparison post hoc test. The groups designated with the same letter (either a or b) did not differ significantly (P > 0.05). The groups designated with different letters differed significantly (P < 0.05).
Oseltamivir susceptibility among human and swine influenza virus isolates.
Swine and human influenza viruses had mean IC50s that were much lower (1.4 nM) than those of ducks and shorebirds/gulls, which ranged from as low as 0.5 nM to as high as 154.43 nM. In addition, influenza viruses that were resistant to oseltamivir were found only in the seasonal human influenza viruses, such as A/Georgia/20/2006, whose resistance is well established (39). We determined that the mean IC50s differed among the four groups analyzed (ducks, shorebirds, swine, and humans; P < 0.0001). Dunn's multiple comparison test of the mean IC50s between the pairs of different groups revealed the following P values: (i) duck versus shorebirds, P > 0.05; (ii) duck versus swine, P < 0.05; (iii) duck versus human, P < 0.05; (iv) swine versus human, P > 0.05; (v) shorebirds versus swine, P < 0.05; (vi) shorebirds versus humans, P < 0.05. Thus, there is no significant difference in the mean IC50s between ducks and shorebirds or between swine and humans, but the avian (ducks and shorebirds) values were significantly higher than the mammalian (swine and humans) isolates.
Analysis of susceptibility over time.
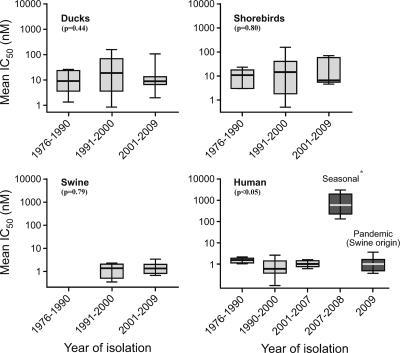
Of particular interest was whether the data over time would reveal either a significant increase in the number of isolates with an IC50 outside the quantile range or, alternatively, whether a significant increase in mean IC50 over time would be evident. Viruses originating primarily in North America were also analyzed by year of isolation to determine whether NAI susceptibility had varied over 10-year periods between 1976 and 2009 (Fig. 2). All avian and swine influenza viruses remained susceptible to oseltamivir over time. The differences between years of isolation did not show evidence of a clear linear upward trend but remained fairly stable from year to year among the avian species over the period studied. This is quite different from the dramatic increase in the number of human influenza viruses that were resistant to oseltamivir in 2007 and 2008. Interestingly, human pandemic H1N1 influenza viruses isolated in 2009 possessed similar susceptibility to oseltamivir as the sensitive seasonal H1N1 Brisbane-like viruses from previous years (Fig. 2). The surveillance of circulating human H1N1 influenza viruses by the World Health Organization, Centers for Disease Control and Prevention, and others revealed the same trend of oseltamivir resistance among human influenza virus isolates from 2007 to 2009 (13, 22, 39). Statistical analysis revealed that there was no significant difference between isolation years within a species and their susceptibility to oseltamivir with the exception of human influenza viruses (Fig. 2). A clear upward trend of the mean IC50s was observed among human isolates (P = 0.0010). This was due to seasonal influenza virus that was resistant to oseltamivir collected from 2007 to 2008, which is contrast to isolates collected from avian and swine hosts (P > 0.05).
FIG. 2.
Quantile box plots illustrating the log10 mean IC50s for oseltamivir for each species from which virus was isolated. The range of isolation years for each species that virus was isolated is shown on the x axis. Viruses collected were all from various wild birds, mainly from the United States and Canada. Some isolates and their respective IC50s are from published data (5, 21, 33,22, 28, 29). Results within a graph were analyzed using a Kruskal-Wallis test and Dunn's multiple comparison post hoc test; P values are included on the graphs. Data marked with an * are statistically different from data for other years within a graph.
Hemagglutinin subtype and oseltamivir susceptibility.
All influenza A virus subtypes, H1 to H16, have been isolated from wild aquatic birds (35, 46), and their function is related to the HA/NA balance (27). In this study, avian isolates were examined by HA subtype to determine if different combinations of HA would have an effect on susceptibility to oseltamivir. We found that different combinations of HA and NA had no correlation to oseltamivir susceptibility (data not shown).
Characterization of minor outliers.
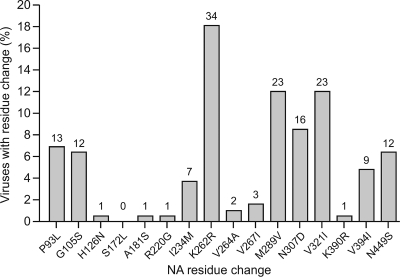
Seven of the 123 viruses tested (three isolates from ducks, three from shorebirds, and one from gull) had IC50s greater than any other viruses tested (Table 3). The three that had the highest IC50s (>100 nM) were from A/Shorebird/DE/182/97 (H2N1), A/Shorebird/DE/24/98 (H2N1), and A/Mallard/Alberta/34/2001 (H7N1). One avian isolate with a value that was near 100 nM was A/Mallard/Alberta/88/2004 (H1N1). The A/Laughing Gull/DE/5/2003 (H9N1) and A/Shorebird/DE/95/2003 (H9N1) isolates had IC50s of >50 nM but <100 nM. The NA genes of the seven avian outliers were sequenced and were aligned with the NA amino acid sequences from 188 other North American avian N1 NA isolates available in the public domain (from online databases, Influenza Sequence NCBI Influenza Virus Resource, and GenBank) to determine the consensus sequence. Sequence analysis revealed a number of NA amino acids that differed between the isolates with reduced oseltamivir susceptibility and the consensus sequence (Fig. 3 and 4). Furthermore, these mutations were also found at varying frequencies in the 188 other N1 NA sequences (Fig. 4). Significantly, none of the sequences had any of the known NA mutations, such as H275Y, that confer oseltamivir resistance (9, 34).
TABLE 3.
Characterization of minor outliers among avian influenza viruses of the N1 NA subtype
| Species | Virus strain | Subtype | Mean IC50 ± SD (nM)a |
NA mutation(s)b | ||
|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | Peramivir | ||||
| Ducks | A/Mallard/Alberta/34/2001 | H7N1 | 102.25 ± 3.80 | 0.48 ± 0.11 | 2.76 ± 2.00 | N307D |
| A/Mallard/Alberta/88/2004 | H1N1 | 99.17 ± 61.85 | 8.72 ± 5.48 | 8.18 ± 2.30 | K262R; V321I | |
| A/Mallard/TN/11464/85 | H1N1 | 39.75 ± 10.0 | 6.21 ± 2.84 | 3.51 ± 2.65 | G105S; H126N; I234M; M289V;V394I; N449S | |
| Shorebirds/Gulls | A/Shorebird/DE/182/97 | H2N1 | 154.43 ± 38.53 | 7.22 ± 4.11 | 4.25 ± 1.97 | V267I; N307D |
| A/Shorebird/DE/24/98 | H2N1 | 111.07 ± 18.61 | 6.00 ± 4.86 | 8.67 ± 1.51 | S172L; V267I; N307D | |
| A/Laughing Gull/DE/5/2003 | H9N1 | 65.80 ± 17.88 | 3.93 ± 0.87 | 4.92 ± 1.57 | P93L;V264A; K390R; | |
| A/Shorebird/DE/95/2003 | H9N1 | 67.00 ± 11.16 | 5.50 ± 0.40 | 4.76 ± 1.38 | P93L; A181S; R220G; 264A | |
The NA inhibition assay was performed with viruses standardized to equivalent NA activity and incubated with NAIs at concentrations of 0.00005 to 100 μM with MUNANA as a substrate. The IC50 was determined by plotting the dose-response curve of inhibition of NA activity as a function of the compound concentration. Values are from at least three independent determinations.
RNA was isolated directly from virus-containing allantoic or cultural fluids. Amino acid numbering is based on N1 NA.
FIG. 3.
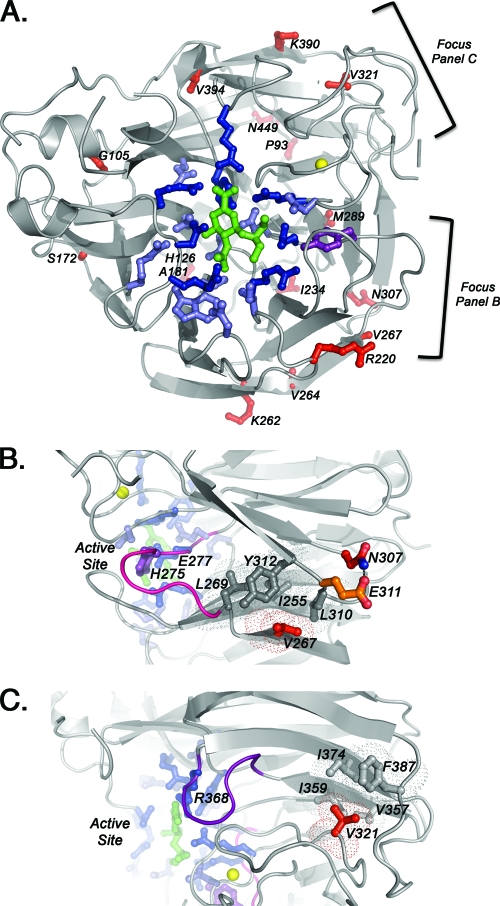
Structure of the complex between N1 influenza virus NA and oseltamivir (PDB code 3cl0) and the amino acid substitutions that were found in seven avian isolates with reduced susceptibility. Amino acid analysis was based on N1 numbering system. (A) Locations of NA residue changes found in seven avian isolates with reduced susceptibility (red). Also shown are catalytic residues (dark blue and purple; 8 residues), framework residues (light blue; 11 residues), oseltamivir (green), and calcium ion (yellow). The H275Y (N1 numbering; H274Y is in N2 numbering). The NA mutation is shown in purple. (B) Two residues, V267 and N307, may be responsible for reduced susceptibility to oseltamivir. Mutation may destabilize the hydrophobic patch (dark gray; four residues), which in turn may destabilize the nearby loop (pink) containing oseltamivir-interacting residues H275 and E277. (C) Residue V321, which may be responsible for reduced susceptibility to oseltamivir. The mutation may destabilize the hydrophobic patch (light gray; four residues), which in turn may destabilize the nearby loop (purple) containing oseltamivir-interacting residue R368. All residues are labeled using N1 numbering.
FIG. 4.
Frequencies of the NA mutations identified in the seven outliers across 188 influenza viruses isolated from North American waterfowl (from the online databases, Influenza Sequence NCBI Influenza Virus Resource, GenBank). The numbers above each bar represent the number of virus isolates exhibiting the indicated mutation.
NA active site residues that directly interact with the substrate are the catalytic residues, and the framework residues provide a structural scaffold for the catalytic residues (9, 19). Figure 3A shows these regions within a subunit (monomer) of the NA tetramer in which these important residues are conserved. Interestingly, analysis of the residue changes observed among avian isolates with reduced oseltamivir susceptibility revealed that none of the changes was interacting directly with catalytic or framework residues (Fig. 3A). These data suggest that binding of the oseltamivir may be affected by NA amino acid residues outside the active site, and additional studies are required to test this hypothesis. One particular residue change that appeared most frequently (in approximately 8.5% [16/188] of all avian isolates) was N307D. This residue change also appeared in three of the seven avian influenza viruses with reduced susceptibility. Furthermore, V267I appeared in two of the seven of these avian influenza viruses. Residues V267 and N307 are near a hydrophobic patch (Fig. 3B). Another strain of avian influenza virus with reduced susceptibility had NA mutations K262R and V321I. Residue K262 lies on the surface of NA. In contrast, residue V321 lies within another hydrophobic patch (Fig. 3C). The three mutations (V267I, N307D, and V321I) that are potentially responsible for reduced oseltamivir susceptibility were not present in moderately or highly susceptible isolates tested in this study. Lastly, a few viral isolates had several NA mutations, making it difficult to identify the mutation that may be contributing to the reduced oseltamivir susceptibility (Fig. 4). All seven influenza viruses with reduced oseltamivir susceptibility were sensitive to other NAIs, zanamivir and peramivir (Table 3).
Sequence analysis of M2 protein.
The matrix protein residues that were examined are conserved regions of the protein. These regions are shared among all avian influenza viruses and are known to confer amantadine resistance (e.g., M2 residue at positions 26, 27, 30, 31, and 34). Only the absence of known amino acid residues conferring resistance to adamantanes was analyzed in the study. Sequence alignment and analysis of the seven avian influenza virus outliers did not reveal any mutations in their M2 protein, which confers amantadine resistance (data not shown). In addition, sequence alignment and analysis of at least 10 additional avian influenza viruses of the N1 NA subtype did not reveal any residue changes at positions 26, 27, 30, 31, or 34 in the transmembrane region of the M2 protein.
DISCUSSION
There is particular importance for confirming the activity of NAIs against avian and swine influenza viruses which may infect humans and could potentially initiate a new influenza pandemic. We found that natural oseltamivir resistance among wild aquatic avian and swine influenza viruses of the N1 NA subtype is rare; most (90%) avian isolates from 1979 to 2008 were highly (21%) or moderately (69%) susceptible to oseltamivir (mean IC50 range, 5 to 50 nM). Out of 91 avian isolates, seven isolates (7.7%) had reduced susceptibility to oseltamivir (IC50, >50 nM). However, those seven outliers were susceptible to zanamivir and peramivir. All swine influenza viruses examined (15/15) were susceptible to oseltamivir in vitro and had IC50s of 1.4 nM, which is comparable to those of seasonal H1N1 human viruses isolated prior to 2007. We suggest that possible molecular markers of reduced oseltamivir susceptibility in avian N1 NA viruses are amino acid changes located outside the active site (V267I, N307D, and V321I) that potentially distort hydrophobic pockets and indirectly affect the NA catalytic and framework residues. This study is the first to our knowledge to define the levels of susceptibility to NAIs among the influenza viruses isolated from wild waterbirds and swine and focus on the susceptibility of influenza viruses in their natural reservoir.
For all influenza virus subtypes there is the potential for the virus to become resistant to adamantanes or NAIs by a mutation(s), usually in response to treatment with the drug. Positive selective pressures caused by anti-influenza virus prophylaxis, such as the NAIs that are used to treat influenza virus infections in humans, do not occur among wild waterbirds or swine in nature. We hypothesize that antiviral resistance and reduced susceptibility can occur in avian influenza viruses by three different mechanisms. First, continuous evolution of influenza viruses could result in the random acquisition of specific mutations in NA that reduce NAI susceptibility or even lead to drug resistance. In addition, mutations in other viral genes could improve viral fitness and transmissibility and thus result in dissemination of these variants. A striking example of this possibility is the emergence and widespread use of oseltamivir-resistant variants with the well-established H275Y NA amino acid substitution among seasonal H1N1 influenza viruses of the A/Solomon Islands/3/2006 and A/Brisbane/2007 lineages circulating in 2007-2008 (1, 12, 29, 31, 49). It is still not well understood why oseltamivir-resistant H1N1 viruses circulating in 2007-2008 were highly fit in humans.
Second, the resistance could occur through gene reassortment with human and swine viruses carrying drug-resistant mutations. Direct transmission of whole avian influenza virus to mammals, or incorporation of avian gene segments into mammalian strains, e.g., H3N2 triple reassortants in pigs in the United States, has been documented (8, 10, 44). These viruses obtained their NA from avian viruses that do not likely have drug resistance mutations. To date, there is no evidence of drug-resistant avian influenza virus strains being transferred to humans or other species or their contribution to a reassortant event.
Finally, with low probability, resistance could occur in the avian species through drug pressure when oseltamivir is released in water or sewage. Recent studies have shown that oseltamivir carboxylate (the active metabolite of oseltamivir) is not degraded in aquatic environments and is present at detectable levels in river water and sewage discharge (17, 41). Although oseltamivir carboxylate is not orally bioavailable in humans, there is the potential that wild waterbirds and gallinaceous birds, or their excretions carrying influenza viruses, may encounter water contaminated with the drug (15). This is notable because migratory waterfowl can travel to these areas and become exposed to oseltamivir. Therefore, potential exposure of influenza viruses from wild waterbirds to oseltamivir could promote the evolution of viral resistance in nature. However, in our study, no resistance was detected among N1 NA influenza viruses isolated from wild waterbirds.
Our study revealed that duck and shorebird/gull influenza viruses have a wider range of susceptibility to oseltamivir (IC50 range, 0.5 to 154.43 nM) than swine or human isolates (IC50 range, 0.33 to 2.56 nM). The mean IC50 for both swine and humans is 1.4 nM, which indicates that the oseltamivir susceptibility of the swine NA is more “human-like.” Oseltamivir susceptibility among wild avian and swine influenza viruses remained stable over a 10-year period except for the seven outliers. Interestingly, four out of these seven outliers were isolated in the same span of time (2001 to 2004) in which oseltamivir was put into use (1999). Other investigators have determined baseline levels of susceptibility to oseltamivir, but only among human influenza isolates after NAIs were put into clinical use (14, 20, 23, 24, 32, 33, 39). We found small differences in the mean IC50s for the inhibition of avian and swine influenza virus N1 NAs for isolates from 1979 to 2008. Similarly, it was reported for human isolates of N1 and N2 NA subtypes that susceptibility to NAIs is stable over time (23, 31). In addition, no correlation to oseltamivir susceptibility was detected with different HA/NA combinations.
Analysis of the NA sequences of the isolates that had reduced oseltamivir susceptibility revealed a number of mutations outside the catalytic and framework residues. Structural analysis suggests that these mutations may indirectly affect the stability of the active site residues and their interaction with oseltamivir. It has been documented that residues far away from an enzymatic active site can affect the enzymatic activity indirectly, through “energy channels” throughout the protein (2, 38), and this may be the case for the NA enzyme of influenza virus as well. Interestingly, a number of residue changes, such as V267I and N307D, appeared in two of three of the avian isolates with reduced oseltamivir susceptibility. One isolate carried a V321I substitution. Residues V267 and N307 are near a hydrophobic patch that may be important for stabilizing active site residues H274 and E276 (Fig. 3B). Residue V267 resides in the hydrophobic patch, and mutation to a bulkier isoleucine may distort this region. Residue N307 forms a hydrogen bond with E311, and mutation of N307 to negatively charged aspartate would likely disrupt this hydrogen bond and cause repulsion. Repulsion of E311 would disrupt hydrophobic patch residues, which again might affect the stabilization of active site residues H274 and E276. Another strain of avian influenza virus with reduced susceptibility had NA mutations K262R and V321I. Residue K262 lies on the surface of NA and is unlikely to affect oseltamivir binding upon mutation to the similar residue arginine. Taken together, these data suggest that residues outside the NA active site, such as V267I, N307D, and V321I, may distort hydrophobic pockets and indirectly affect the NA catalytic and framework residues. These residues may be new potential markers for reduced susceptibility to oseltamivir. It is also possible that two or more mutations could be acting in concert. Analysis of 188 sequences of avian influenza viruses of N1 NA subtype available in the public domain revealed that changes in the NA residues determined in our study apparently occur among the wild waterbird species but at a relatively low frequency. Thus, these NA mutations do not appear to have a selective advantage and probably would not be maintained in wild birds. It is also possible that a combination of mutations located outside the NA active site act together and is the basis for the reduced susceptibility to oseltamivir in some isolates. One report identified three NA mutations in avian H5N1 viruses (K150N, I222L, and S246N) that had decreased susceptibility to oseltamivir (4).
From our study, we can conclude that reduced susceptibility and resistance to oseltamivir can occur in influenza viruses in the wild waterbird reservoir, but these naturally occurring variants probably have no evolutionary advantage in nature. The biological significance of reduced NA susceptibility and the role of particular mutations on oseltamivir binding warrant testing by appropriate mutagenesis and binding experiments before considering the effect of a mutation on binding affinity.
Acknowledgments
This study was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, contract no. HHSN266200700005C, and by the American Lebanese Syrian Associated Charities.
We thank Subrata Barman, John Franks, and Patrick Seiler for technical assistance and helpful advice, David Galloway for scientific editing, and James Knowles for manuscript preparation.
Footnotes
Published ahead of print on 21 July 2010.
REFERENCES
- 1.Abed, Y., B. Nehmé, M. Baz, and G. Boivin. 2008. Activity of the neuraminidase inhibitor A-315675 against oseltamivir-resistant influenza neuraminidases of N1 and N2 subtypes. Antiviral Res. 77:163-166. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, P. K. 2006. Enzymes: an integrated view of structure, dynamics and function. Microb. Cell Fact. 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Azemi, A., J. Bahl, S. Al-Zenki, Y. Al-Shayji, S. Al-Amad, H. Chen, Y. Guan, J. S. M. Peiris, and G. J. D. Smith. 2008. Avian influenza A virus (H5N1) outbreaks, Kuwait, 2007. Emerg. Infect. Dis. 14:958-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boltz, D. A., B. Douangngeun, P. Phommachanh, S. Sinthasak, R. Mondry, C. A. Obert, P. Seiler, R. Keating, Y. Suzuki, H. Hiramatsu, E. Govorkova, and R. G. Webster. 2009. Emergence of H5N1 avian influenza viruses with reduced sensitivity to neuraminidase inhibitors and novel reassortants in Lao People's Democratic Republic. J. Gen. Virol. 91:949-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2009. Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis—North Carolina 2009. MMWR Morb. Mortal. Wkly. Rep. 58:969-972. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2009. Three reports of oseltamivir-resistant novel influenza A (H1N1) viruses. Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/ h1n1flu/HAN/070909.htm.
- 7.Chen, H., G. Deng, Z. Li, G. Tian, Y. Li, P. Jiao, L. Zhang, Z. Liu, R. G. Webster, and K. Yu. 2004. The evolution of H5N1 influenza viruses in ducks in southern China. Proc. Natl. Acad. Sci. U. S. A. 101:10452-10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, J. 2009. New details on virus's promiscuous past. Science 324:1127. [DOI] [PubMed] [Google Scholar]
- 9.Collins, P. J., L. F. Hairea, Y. P. Lina, J. Liua, R. J. Russell, P. A. Walkera, S. R. Martina, R. S. Danielsa, V. Gregorya, J. J. Skehel, S. J. Gamblina, and A. J. Hay. 2009. Structural basis for oseltamivir resistance of influenza viruses. Vaccine 27:6317-6323. [DOI] [PubMed] [Google Scholar]
- 10.de Jong, J. C., E. C. J. Claas, A. D. M. E. Osterhaus, R. G. Webster, and W. L. Lim. 1997. A pandemic warning? Nature 389:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Jong, M. D., T. T. Tran, H. K. Truong, M. H. Vo, G. J. D. Smith, V. C. Nguyen, V. C. Cam, T. Q. Phan, Q. H. Do, Y. Guan, J. S. M. Peiris, T. H. Tran, and J. Farrar. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353:2667-2672. [DOI] [PubMed] [Google Scholar]
- 12.Democratis, J., M. Pareek, and I. Stephenson. 2006. Use of neuraminidase inhibitors to combat pandemic influenza. J. Antimicrob. Chemother. 58:911-915. [DOI] [PubMed] [Google Scholar]
- 13.Dharan, N. J., L. V. Gubareva, J. J. Meyer, M. Okomo-Adhiambo, R. C. McClinton, S. A. Marshall, K. St. George, S. Epperson, L. Brammer, A. I. Klimov, J. S. Bresee, and A. M. Fry for the Oseltamivir-Resistance Working Group. 2009. Infections with oseltamivir-resistant influenza A (H1N1) virus in the United States. JAMA 301:1034-1041. [DOI] [PubMed] [Google Scholar]
- 14.Ferraris, O., N. Kessler, and B. Lina. 2005. Sensitivity of influenza viruses to zanamivir and oseltamivir: a study performed on viruses circulating in France prior to the introduction of neuraminidase inhibitors in clinical practice. Antiviral Res. 68:43-48. [DOI] [PubMed] [Google Scholar]
- 15.Fick, J., R. H. Lindberg, M. Tysklind, P. D. Haemig, J. Waldenström, A. Wallensten, and B. Olsen. 2007. Antiviral oseltamivir is not removed or degraded in normal sewage water treatment: implications for development of resistance by influenza A virus. PLoS One 2:e986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. M. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. López-Gatell, H. Olivera, I. López, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St. George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 2009. Antigenic and genetic characteristics of swine-origin 2009 A (H1N1) influenza viruses circulating in humans. Science 325:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh, G. C., N. Nakada, N. Yamashita, and H. Tanaka. 2010. Oseltamivir carboxylate, the active metabolite of oseltamivir phosphate (Tamiflu), detected in sewage discharge and river water in Japan. Environ. Health Perspect. 118:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govorkova, E. A., I. A. Leneva, O. G. Goloubeva, K. Bush, and R. G. Webster. 2001. Comparison of efficacies of RWJ-270201, zanamivir, and oseltamivir against H5N1, H9N2, and other avian influenza viruses. Antimicrob. Agents Chemother. 45:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gubareva, L. V., M. J. Robinson, R. C. Bethell, and R. G. Webster. 1997. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J. Virol. 71:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gubareva, L. V., R. G. Webster, and G. Hayden. 2001. Comparison of the activities of zanamivir, oseltamivir, and RWJ-270201 against clinical isolates of influenza virus and neuraminidase inhibitor-resistant variants. Antimicrob. Agents Chemother. 45:3403-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall, R. J., M. P. Peacey, J. C. Ralston, J. Bocacao, M. Ziki, W. Gunn, A. Quirk, and Q. S. Huang. 2009. Pandemic influenza A (H1N1) viruses currently circulating in New Zealand are sensitive to oseltamivir. Eurosurveillance 14:1-2. [DOI] [PubMed] [Google Scholar]
- 22.Hauge, S. H., S. Dudman, K. Borgen, A. Lackenby, and O. Hungnes. 2009. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08. Emerg. Infect. Dis. 15:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurt, A. C., I. G. Barr, C. J. Durrant, R. P. Shaw, H. M. Sjorgen, and A. W. Hampson. 2003. Surveillance for neuraminidase inhibitor resistance in influenza viruses from Australia. Commun. Dis. Intell. 27:542-547. [PubMed] [Google Scholar]
- 24.Hurt, A. C., J. Ernest, Y-M. Deng, P. Iannello, T. G. Besselaar, C. Birch, P. Buchy, M. Chittaganpitch, S-C. Chiu, D. Dwyer, A. Guigoni, B. Harrower, I. P. Kei, T. Kok, C. Lin, K. McPhie, A. Mohd, R. Olveda, T. Panayotou, W. Rawlinson, L. Scott, D. Smith, H. D'Souza, N. Komadina, R. Shaw, A. Kelso, and I. G. Barr. 2009. Emergence and spread of oseltamivir-resistant A (H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res. 83:90-93. [DOI] [PubMed] [Google Scholar]
- 25.Jain, S., L. Kamimoto, A. M. Bramley, A. M. Schmitz, S. R. Benoit, J. Louie, D. E. Sugerman, J. K. Druckenmiller, K. A. Ritger, R. Chugh, S. Jasuja, M. Deutscher, S. Chen, J. D. Walker, J. S. Duchin, S. Lett., S. Soliva, E. V. Wells, D. Swerdlow, T. M. Uyeki, A. E. Fiore, S. J. Olsen, A. M. Fry, C. B. Bridges, and L. Finelli. 2009. The 2009 pandemic influenza A (H1N1) virus hospitalizations investigation. Hospitalized patients with 2009 H1N1 influenza in the United States, April-June 2009. N. Engl. J. Med. 361:1935-1944. [DOI] [PubMed] [Google Scholar]
- 26.Jamieson, D. J., M. A. Honein, S. A. Rasmussen, J. L. Williams, D. L. Swerdlow, M. S. Biggerstaff, S. Lindstrom, J. K. Louie, C. M. Christ, S. R. Bohm, V. P. Fonseca, K. A. Ritger, D. J. Kuhles, P. Eggers, H. Bruce, H. A. Davidson, E. Lutterloh, M. L. Harris, C. Burke, N. Cocoros, L. Finelli, K. F. MacFarlane, B. Shu, and S. J. Olsen for the Novel Influenza A (H1N1) Pregnancy Working Group. 2009. H1N1 2009 influenza virus infection during pregnancy in the USA. Lancet 374:451-458. [DOI] [PubMed] [Google Scholar]
- 27.Kaverin, N. V., A. S. Gambaryan, N. V. Bovin, I. A. Rudneva, A. A. Shilov, O. M. Khodova, N. L. Varich, B. V. Sinitsin, N. V. Makarova, and E. A. Kropotkina. 1998. Postreassortment changes in influenza A virus hemagglutinin restoring HA-NA functional match. Virology 244:315-321. [DOI] [PubMed] [Google Scholar]
- 28.Kawakami, C., M. Obuchi, M. Saikusa, Y. Noguchi, M. Ujike, T. Odagirir, M. Iwata, M. Takahiro, T. Toyozawa, and M. Tashiro. 2009. Isolation of oseltamivir-resistant influenza A/H1N1 virus of different origins in Yokohama City, Japan, during the 2007-2008 influenza season. Jpn. J. Infect. Dis. 62:83-86. [PubMed] [Google Scholar]
- 29.Lackenby, A., O. Hungnes, S. G. Dudman, A. Meijer, W. J. Paget, A. J. Hay, and M. C. Zambon. 2008. Emergence of resistance to oseltamivir among influenza A (H1N1) viruses in Europe. Eurosurveillance 13:1-3. [DOI] [PubMed] [Google Scholar]
- 30.Li, K. S., Y. Guan, J. Wang, G. J. D. Smith, K. M. Xu, L. Duan, A. P. Rahardjo, P. Puthavathana, C. Buranathai, T. D. Nguyen, A. T. S. Estoepangestie, A. Chaisingh, P. Auewarakul, H. T. Long, N. T. H. Hanh, R. J. Webby, L. L. M. Poon, H. Chen, K. F. Shortridge, K. Y. Yuen, R. G. Webster, and J. S. M. Peiris. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209-213. [DOI] [PubMed] [Google Scholar]
- 31.McKimm-Breshkin, J., T. Trivedi, A. Hampson, A. Hay, A. Klimov, M. Tashiro, F. Hayden, and M. Zambon. 2003. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antivir. Ther. 47:2264-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meijer A., A. Lackenby, O. Hungnes, B. Lina, S. van der Werf, B. Schweiger, M. Opp, J. Paget, J. van de Kassteele, A. Hay, and M. Zambon for the European Influenza Surveillance Scheme. 2009. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerg. Infect. Dis. 15:552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monto, A. S., J. L. McKimm-Breschkin, C. Macken, A. W. Hampson, A. Hay, A. Klimov, M. Tashiro, R. G. Webster, M. Aymard, F. G. Hayden, and M. Zambon. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 50:2395-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moscona, A. 2005. Oseltamivir resistance: disabling our influenza defenses. N. Engl. J. Med. 353:2633-2636. [DOI] [PubMed] [Google Scholar]
- 35.Munster, V. J., A. Wallensten, C. Baas, G. F. Rimmelzwaan, M. Schutten, B. Olsen, A. D. Osterhaus, and R. A. Fouchier. 2005. Mallards and highly pathogenic avian influenza ancestral viruses, northern Europe. Emerg. Infect. Dis. 11:1545-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peiris, J. S., L. L. M. Poon, and Y. Guan. 2009. Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J. Clin. Virol. 45:169-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potier, M., L. Mameli, M. Belisle, L. Dallaire, and S. B. Melancon. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-N-acetylneuraminate) substrate. Anal. Biochem. 94:287-296. [DOI] [PubMed] [Google Scholar]
- 38.Rameix-Welti, M. A. Enouf, V. Cuvelier, F. Jeannin, and S. van der Werf. 2008. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog. 4:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheu, T., V. M. Deyde, M. Okomo-Adhiambo, R. J. Garten, X. Xu, R. A. Bright, E. N. Butler, T. R. Wallis, A. I. Klimov, and L. V. Gubareva. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide in 2004-2008. Antimicrob. Agents Chemother. 52:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, G. J. D., V. Dhanasekaran, J. Bahl, S. J. Lycett, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, S. Bhatt, J. S. M. Peiris, Y. Guan, and A. Rambaut. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122-1125. [DOI] [PubMed] [Google Scholar]
- 41.Söderström, H., J. D. J.ärhult, B. Olsen, R. H. Lindberg, H. Tanaka, and J. Fick. 2009. Detection of the antiviral drug oseltamivir in aquatic environments. PLoS One 4:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stittelaara, K. J., M. Tisdale, G. van Amerongena, R. F. van Lavierena, F. Pistoora, J. Simona, and A. D. M. E. Osterhaus. 2008. Evaluation of intravenous zanamivir against experimental influenza A (H5N1) virus infection in cynomolgus macaques. Antiviral Res. 80:225-228. [DOI] [PubMed] [Google Scholar]
- 43.Taubenberger, J. K., and D. M. Morens. 2009. Pandemic influenza, including a risk assessment of H5N1. Rev. Sci. Tech. 28:187-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taubenberger, J. K., A. H. Reid, R. M. Lourens, R. Wang, G. Jin, and T. G. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889-893. [DOI] [PubMed] [Google Scholar]
- 45.Webster, R. 1998. Influenza: an emerging disease. Emerg. Infect. Dis. 4:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster, R., S. Krauss, D. Hulse-Post, and K. Sturm-Ramirez. 2007. Evolution of influenza A viruses in wild birds. J. Wildl. Dis. 43:1-6.17347388 [Google Scholar]
- 47.Webster, R. G., and E. A. Govorkova. 2006. H5N1 influenza: continuing evolution and spread. N. Engl. J. Med. 355:2174-2177. [DOI] [PubMed] [Google Scholar]
- 48.World Health Organization. 2010. Continuing progress towards a unified nomenclature system for the highly pathogenic H5N1 avian influenza viruses. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/avian_influenza/guidelines/nomenclature/en/.
- 49.World Health Organization. 2007. Influenza in the world September 2006-August 2007. Wkly. Epidemiol. Rec. 82:357-360.17933085 [Google Scholar]
- 50.World Health Organization Global Influenza Program Surveillance Network. 2005. Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 11:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.World Health Organization. 2009. Margaret Chan, Director, general statement. World now at the start of 2009 influenza pandemic. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/index.html.
- 52.World Health Organization. 2009. Influenza A (H1N1) virus resistance to oseltamivir. Last quarter 2007 to 7 February 2008. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/influenza/oseltamivir_summary/en/.
- 53.Wu, W. 2009. Control of avian influenza A H5N1 in China. Lancet Infect. Dis. 9:460-461. [DOI] [PubMed] [Google Scholar]
- 54.Yen, H.-L., E. Hoffmann, G. Taylor, C. Scholtissek, A. S. Monto, R. G. Webster, and E. A. Govorkova. 2006. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J. Virol. 80:8787-8795. [DOI] [PMC free article] [PubMed] [Google Scholar]