Abstract
The nsp2 replicase protein of porcine reproductive and respiratory syndrome virus (PRRSV) was recently demonstrated to be processed from its precursor by the PL2 protease at or near the G1196|G1197 dipeptide in transfected CHO cells. Here the proteolytic cleavage of PRRSV nsp2 was further investigated in virally infected MARC-145 cells by using two recombinant PRRSVs expressing epitope-tagged nsp2. The data revealed that PRRSV nsp2 exists as different isoforms, termed nsp2a, nsp2b, nsp2c, nsp2d, nsp2e, and nsp2f, during PRRSV infection. Moreover, on the basis of deletion mutagenesis and antibody probing, these nsp2 species appeared to share the same N terminus but to differ in their C termini. The largest protein, nsp2a, corresponded to the nsp2 product identified in transfected CHO cells. nsp2b and nsp2c were processed within or near the transmembrane (TM) region, presumably at or near the conserved sites G981|G982 and G828|G829|G830, respectively. The C termini for nsp2d, -e, and -f were mapped within the nsp2 middle hypervariable region, but no conserved cleavage sites could be definitively predicted. The larger nsp2 species emerged almost simultaneously in the early stage of PRRSV infection. Pulse-chase analysis revealed that all six nsp2 species were relatively stable and had low turnover rates. Deletion mutagenesis revealed that the smaller nsp2 species (e.g., nsp2d, nsp2e, and nsp2f) were not essential for viral replication in cell culture. Lastly, we identified a cellular chaperone, named heat shock 70-kDa protein 5 (HSPA5), that was strongly associated with nsp2, which may have important implications for PRRSV replication. Overall, these findings indicate that PRRSV nsp2 is increasingly emerging as a multifunctional protein and may have a profound impact on PRRSV replication and viral pathogenesis.
Replicase polyprotein maturation is a highly orchestrated and precisely regulated process, which plays a very important role in the life cycle of positive-stranded RNA viruses. Its products are critical for the downstream assembly of viral replication complexes and are often antagonists of host innate immunity. The proteolytic cleavage of these replicase polyproteins is usually carried out by viral proteases, with occasional action by cellular proteases (10, 27). The focus of this report is the maturation of porcine reproductive and respiratory syndrome virus (PRRSV) replicase protein. PRRSV causes reproductive failure (e.g., abortions, mummies, stillbirths) in sows and respiratory distress (e.g., interstitial pneumonia) in young pigs, leading to millions of dollars of losses every year in North America (25) and even more in other regions (e.g., Southeast Asia) in recent years (36, 41). PRRSV is a positive-stranded RNA virus with a genome of about 15.4 kb and is a member of the family Arteriviridae in the order Nidovirales (3). PRRSV replication generates two replicase polyprotein precursors, pp1a and pp1ab, specified by ORF1a and ORF1a/b, respectively (28, 38). pp1a is made directly from ORF1a, whereas pp1ab stems from the translation of ORF1a/b via frameshift reading of ORF1 (28, 38). These polyproteins, either cotranslationally or subsequently, undergo extensive proteolytic maturation; at least 14 mature replicase subunits have been predicted according to studies of equine arteritis virus (EAV) (37, 42), the prototype of the family Arteriviridae. The proteolytic processing is thought to involve four virally encoded proteases within ORF1a, including two papain-like cysteine proteases (PCP1α and PCP1β), a cysteine protease (PL2), a picornavirus 3C-like serine protease (3CLpro), and possibly cellular proteases (8, 30-32, 42).
The studies of EAV support a general model for PRRSV replicase maturation: nsp1 is released from the precursor by PCP1α and PCP1β; PL2 of nsp2 then cleaves the site at the nsp2-nsp3 (nsp2/3) junction; and the maturation of the remaining pp1a and pp1ab is executed by a 3C-like serine protease contained in nsp4. Lines of evidence have shown that nsp1 is processed in vitro into nsp1α and nsp1β by cis-active PCP1α and PCP1β, respectively, and that this processing occurs cotranslationally and rapidly (8, 30, 34). The nsp2 protein of PRRSV strain VR-2332, situated immediately downstream of nsp1, is a large multidomain protein of about 1,197 amino acids (aa) containing an N-terminal PL2 cysteine protease, a 500- to 700-aa middle hypervariable region, a putative transmembrane (TM) domain, and a C-terminal tail (13, 15). The PL2 protease is active both in cis and in trans and mediates the processing of nsp2 into one predominant product in CHO cells (14). In vitro mutagenesis studies of strain VR-2332 have revealed that the nsp2/3 cleavage is sensitive to mutations at the G1196|G1197 site (14). For example, even a conserved G1197A substitution abolishes nsp2/3 proteolysis in CHO cells. Similar mutations also abolished the production of PRRSV progeny virions (14). These studies have pointed to the conclusion that the PL2-induced nsp2/3 cleavage most likely takes place at or near the G1196|G1197 dipeptide. In contrast to that in transfected CHO cells, nsp2 protein processing in PRRSV-infected cells is not yet understood. The goal of the experiments described here was to further assess the proteolytic products of nsp2 in the context of infection with the PRRSV type 2 strain VR-2332.
A major hindrance to the understanding of PRRSV replicase maturation has been the lack of antibodies of good quality. Here, by utilizing the genetically flexible nature of nsp2, two recombinant PRRSVs expressing nsp2 derivatives tagged with foreign epitopes were constructed and then utilized to further investigate the proteolytic processing of nsp2 in PRRSV-infected MARC-145 cells. We showed that nsp2 existed as several isoforms with apparently different C termini during PRRSV infection. Total proteolysis of PRRSV nsp2 likely involved both the PL2 protease and other, unknown viral or cellular proteases. The processing was rapid, and the cleaved products were relatively stable and finely balanced. Additionally, a cellular protein chaperone named HSPA5 was found to interact with the nsp2 protein and could be specifically coimmunoprecipitated by anti-nsp2 antibodies.
MATERIALS AND METHODS
Plasmids and antibodies.
The plasmids used in this study, including pNsp2-3, pNsp2-3 C55A, pNsp2-3 G1197P, and pPL2, have been described previously (14). The genes specifying nsp2 polypeptides comprising aa 12 to 813, aa 12 to 981, and aa 12 to 1196 were amplified from plasmid pNsp2-3 and cloned into the site between BamHI and XbaI in plasmid pcDNA/HA-FLAG (GenBank accession number FJ524378) to generate the new plasmids pNsp2(12-813), pNsp2(12-981), and pNsp2(12-1196), respectively.
The antibodies used in this study include anti-c-myc monoclonal antibody 9E10 (Developmental Studies Hybridoma Bank at the University of Iowa), rabbit polyclonal anti-c-myc antibodies (Abcam), mouse anti-hemagglutinin (anti-HA) antibodies (Covance), mouse anti-FLAG antibodies (M2; Sigma), horseradish peroxidase (HRP)-conjugated anti-mouse IgG or anti-rabbit IgG secondary antibodies (Southern Biotechnology, Inc.), and a rabbit anti-HSPA5 antibody (Santa Cruz Biotechnology). Mouse monoclonal antibodies D3A4 and E5F8 are gifts from Hanchun Yang (China Agricultural University) (40). The rabbit polyclonal antibody V was raised against a peptide containing PRRSV strain VR-2332 nsp2 aa 1078 to 1094 (SEKPIAFAQLDEKKITA) (Covance) and has been described in a previous report (14).
Construction of PRRSV deletion mutants.
Plasmid pV7-nsp2Δ324-434-GFP has been reported previously (13). To generate a recombinant virus tagged with the c-myc epitope, the green fluorescent protein (GFP)-encoding gene was replaced by 3 c-myc epitopes (ASEQKLISEEDLEQKLISEEDLEQKLISEED) to produce plasmid pV7-nsp2Δ324-434-myc (pV7-myc) by overlapping PCR as described previously (GenBank accession number FJ524377) (13). To generate the HA-c-myc double-tagged virus pV7-HA-myc (GenBank accession number FJ524376), two copies of the influenza A virus HA epitope tag (YPYDVPDYA) replaced nsp2 aa 12 to 24 in plasmid pV7-myc. A similar strategy was applied to create nsp2 deletion viruses based on plasmid pV7-myc, and the new plasmids were designated pV7-myc-nsp2Δ543-632, pV7-myc-nsp2Δ633-726, and pV7-myc-nsp2Δ727-813. The sequences of the primers used for the construction of these viruses are given in Table 1.
TABLE 1.
Primers used in this study
| Primera | Genome positionb | Sequencec |
|---|---|---|
| Construction of pV7-myc and pV7-HA-myc | ||
| VR-1051U27/ | 1051-1077 | 5′ TCGCCATGCTAACCAATTTGGCTATC |
| /VR-2430L24 | 2407-2430 | 5′ TTGGCATGAGCCCATATTCTTCTC |
| /VR-1859L33 | 1827-1859 | 5′ ATGCTCGAGTTATCAGCTAGGCAGGTGCATCAC |
| VR-1824U24/ | 1824-1847 | 5′ TTGACCGGCTGGCTGAGGTGATGC |
| /VR-3349L26 | 3324-3349 | 5′ GCGTAGCAGGGTCATCAAGCTTAGTC |
| dVR-67U22/ | 2167-2187 | 5′ CGCCCGCCACGCGTAATCGACA |
| /Nsp2Δ324-434-myc 2306L64 | c-myc | 5′ GATCTTCTTCTGAAATCAACTTTTGTTCCAGATCTTCTTCAGAGATGAGTTTCTGCTCGCTAGC |
| Nsp2Δ324-434-myc 2344U64/ | c-myc | 5′ ACAAAAGTTGATTTCAGAAGAAGATCTGGAACAGAAGCTCATCTCTGAGGAAGATCTGCCTAGG |
| Nsp2Δ324-434-myc 2387U43/ | c-myc | 5′ TCTGAGGAAGATCTGCCTAGGCCAAAAGTTCAGCCTCGAAAAA |
| VR-HA2 1387U65/ | HA | 5′ TCCAGATTACGCTTACCCATACGATGTCCCTGATTACGCAGTTCGTGAAACCCGGCAGGCCAAG |
| /VR-HA2 1350L65 | HA | 5′ GACATCGTATGGGTAAGCGTAATCTGGAACATCGTATGGGTAACAAGAGCGTGCTTTTCTTGCTC |
| Construction of nsp2 truncation mutants and cloning of HSPA5 | ||
| Nsp2 PL2 1U37/ | 1373-1387 | 5′ AGCTAAGCTTGCCACCATGGTGGCGACTGCTACAGTC |
| /VR-3755L33 | 3732-3755 | 5′ TACTCTAGAAGAGCCGGCGCCACCTGTGCCTGCC |
| /VR-4258L31 | 4237-4258 | 5′ CTATCTAGAGCCCAGTAACCTGCCAAGAATGG |
| /VR-4898L33 | 4875-4898 | 5′ CCCTCTAGATGAAGGCTTGGAAATTTGCCTGATT |
| /VR-5583L35 | 5558-5583 | 5′ CTATCTAGAAGACCCAAGCTGGGACGGGGTAAACAA |
| HSPA5 1U74/ | HSPA5 | 5′ CCCAAGCTTGCCACCATGGGAGAACAAAAACTCATCTCAGAAGAGGATCTGAAGCTCTCCCTGGTGGCCGCGAT |
| /HSPA5 1988L38 | HSPA5 | 5′ ATGCTCGAGTTACTACAACTCATCTTTTTCTGCTGTAT |
Forward primers are indicated by a slash after the designation; reverse primer designations are preceded by a slash.
Based on GenBank accession number U87392 (VR-2332).
Boldface italics indicate engineered restriction sites, and boldface indicates the engineered start codon for nsp2 expression.
Immunostaining of nsp2 protein.
V7-myc was monitored for nsp2 protein expression using antibodies to c-myc. At 20 h postinfection, V7-myc-infected MARC-145 cells at passage 3 were labeled using monoclonal antibody 9E10 against a c-myc epitope and an Alexa 568-conjugated secondary antibody (red) (Molecular Probes). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue).
Transient expression.
MARC-145 (ATCC) and CHO cells (Invitrogen) were maintained in Eagle's minimal essential medium (EMEM) (SAFC Biosciences) supplemented with 10% fetal bovine serum (FBS) at 37°C under 5% CO2. CHO cells were transiently transfected by using Lipofectamine 2000 (Invitrogen) as described previously (14). RNA transfection of MARC-145 cells was performed as described previously (13).
Viral growth assays.
MARC-145 cells in T25 flasks were infected at a multiplicity of infection (MOI) of 0.1 with either parental or mutant viruses at passage 3. After 1 h of attachment at room temperature with gentle mixing, unbound viruses were removed; the monolayers were washed three times with serum-free EMEM; and the medium was replaced with 7 ml complete medium. Samples were collected from the medium at different time points after infection and were titrated by viral plaque assays on MARC-145 cells.
Immunoprecipitation and Western blotting.
Transfected CHO cells or infected MARC-145 cells were rinsed twice with cold phosphate-buffered saline (PBS) (0.14 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.5 mM KH2PO4) and lysed with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.5% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, and 100 μl/ml protease inhibitor cocktail [P8340; Sigma]) on a platform shaker for 30 min at 4°C. The cell debris was removed by centrifugation at 13,000 rpm for 20 min. The supernatants were precleared by protein G agarose or protein A agarose and were then incubated with selected antibodies as well as protein G PLUS-agarose (Santa Cruz Biotechnology) or protein A agarose (Roche) at 4°C overnight. The immunocomplexes were washed twice with cold RIPA buffer, once with 0.1% sodium dodecyl sulfate (SDS) in RIPA buffer, and once with PBS. After being heated for 5 min at 100°C, the proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and were then electrophoretically transferred to a nitrocellulose membrane. For Western blotting, the membrane was blocked with 5% milk powder in PBST (0.14 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.5 mM KH2PO4, 0.1% Tween 20) for 1 h and was then incubated with the appropriate primary antibodies diluted in PBST-5% milk overnight at 4°C. After three washes with PBST for 30 min, the blot was incubated with an appropriate secondary antibody diluted in PBST for 1 h. The membrane was again washed and was then developed with the ECL Western blot analysis system (Pierce).
Radiolabeling and pulse-chase analysis of the nsp2 protein.
MARC-145 cells in 60-mm-diameter petri dishes were infected with V7-myc (MOI, 0.1) at passage 3 and were incubated in 5 ml EMEM with 10% FBS at 37°C. For pulse-chase analysis, at 12 to 18 h postinfection, MARC-145 cells were washed with Dulbecco's modified essential medium (DMEM) deficient in methionine and cysteine (Met/Cys) and were starved for 30 min. The cells were then labeled with 4 ml of DMEM with 100 μCi/ml [35S]Met-Cys. After 5 to 6 h of labeling, MARC-145 cells were washed twice with DMEM; then they were incubated with 5 ml DMEM with 10% FBS. At various times (0, 15, 30, 60, 120, 180, and 240 min), one dish was removed from the incubator; the cells were washed with cold PBS and were then lysed in RIPA buffer. Alternatively, MARC-145 cells were infected with PRRSV at an MOI of 50. At 13.5 h postinfection, 60-mm-diameter dishes were each labeled with 350 μCi/ml [35S]Met-Cys in 1 ml DMEM for 20 min; the plates were washed; and DMEM with excess amounts of unlabeled methionine and cysteine (2 mM each) was added. Individual cell monolayers were lysed with RIPA buffer at 0, 5, 10, 15, 20, 25, 30, 60, 90, and 120 min. The cell lysates were cleared by centrifugation and were immunoprecipitated using suitable antibodies.
Tandem affinity purification and mass spectrometry.
MARC-145 cells were infected with the recombinant PRRSV V7-myc (passage 3) at an MOI of 0.1 and were then incubated in EMEM with 10% FBS at 37°C. The cells were harvested at 36 h postinfection and were lysed in RIPA buffer. nsp2-associated complexes were purified by two rounds of immunoprecipitation with mouse anti-c-myc monoclonal antibodies and rabbit polyclonal antibody V. The complexes were washed twice with RIPA buffer, once with RIPA buffer with 0.1% SDS, and once with PBS and were then eluted by boiling in SDS-PAGE loading buffer with 5% 2-mercaptoethanol. The complexes were separated by SDS-PAGE on a 4 to 12% NuPage gel (Invitrogen) and were visualized with Coomassie blue or Sypro Ruby (Invitrogen). Sypro Ruby-stained bands were excised, trypsin digested, extracted, and analyzed by liquid chromatography and tandem mass spectrometry (LC-MS-MS) at the University of Minnesota Mass Spectrometry Center.
RESULTS
Construction and recovery of c-myc- and HA-tagged PRRSV.
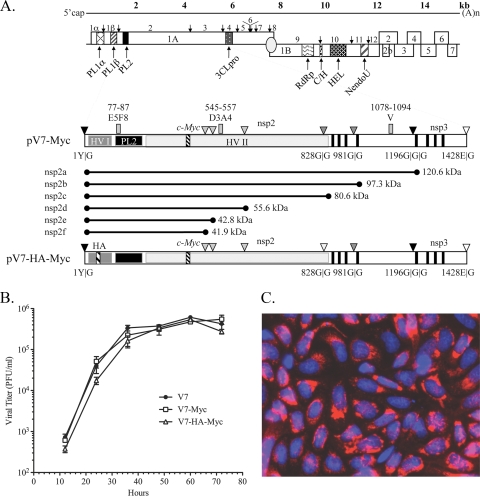
The nsp2 regions comprising aa 12 to 35 and aa 324 to 434 of PRRSV strain VR-2332 have been shown to be dispensable for viral replication in cell culture, and a recombinant PRRSV with a GFP-encoding gene in place of nsp2 aa 324 to 434 is viable (13). We accordingly replaced the deletions of PRRSV nsp2 aa 12 to 35 and aa 324 to 432 with foreign epitope tags in order to facilitate analysis of the nsp2 product(s) in PRRSV-infected MARC-145 cells utilizing high-quality commercial antibodies. To generate a c-myc-tagged virus, the GFP-encoding gene in the PRRSV infectious clone pV7-nsp2Δ324-434-GFP was replaced with 3 consecutive c-myc epitopes to generate the new plasmid pV7-nsp2Δ323-434-myc (pV7-myc) (Fig. 1 A). To produce a double-tagged virus, two copies of an HA epitope were engineered into plasmid pV7-myc to replace nsp2 aa 12 to 24, and the new plasmid was designated pV7-nsp2Δ12-24-HA-nsp2Δ323-434-myc (pV7-HA-myc) (Fig. 1A). The plasmids were linearized and were transcribed in vitro. The RNA transcripts were transfected into MARC-145 cells. A virus-induced cytopathic effect (CPE), characterized by cell rounding, clustering, and detachment, was readily detected 4 to 5 days posttransfection (data not shown). The two mutants displayed growth kinetics indistinguishable from that of the parental virus VR-V7 in MARC-145 cells (Fig. 1B), suggesting that the insertion of one to two small foreign epitopes into the nsp2 region did not affect PRRSV replication. Mutant V7-myc was stable for at least 10 passages, as confirmed by sequence analysis of the nsp2-coding region (data not shown). Immunostaining with anti-c-myc antibodies revealed a typical perinuclear localization pattern of nsp2 (Fig. 1C), as reported previously (11, 13, 40). Mutant V7-HA-myc was stable for at least 3 passages, as confirmed by sequence analysis of the nsp2-coding region (data not shown). However, anti-HA antibodies failed to stain nsp2 in V7-HA-myc-infected cells, perhaps due to inaccessibility of the HA epitope under native conditions.
FIG. 1.
Characterization of foreign-epitope-tagged PRRSV. (A) PRRSV genome annotation. The genome of PRRSV is shown with all identified open reading frames. ORF1A and ORF1B are posttranslationally cleaved by virally encoded papain-like proteases (PCP1α and PCP1β), a cysteine protease (PL2), and a poliovirus 3C-like serine protease (3CL). The following polymerase signature regions are indicated: RNA-dependent RNA polymerase (RdRp), cysteine and histidine rich (C/H), helicase (HEL), and nidovirus uridylate-specific endoribonuclease (NendoU). Below the genome schematic are expanded diagrams of nsp2-3 and the construction of recombinant PRRSVs expressing c-myc- or c-myc- and HA-tagged nsp2. nsp2 contains an N-terminal small hypervariable region (HV-I), a PL2 protease domain, a middle hypervariable region (HV-II), a putative transmembrane domain (filled vertical bars), and a C-terminal domain. The nsp2 PL2 protease is predicted to cleave at G|G dipeptides; there are 10 such dipeptides in type 2 strain VR-2332, some of which are not conserved in other strains. The major conserved predicted cleavage sites discussed in this report are represented by dark shaded triangles; potential cleavage sites that are not conserved are shown as light shaded triangles; and the cleavage sites of nsp1|nsp2 and nsp3|nsp4 are shown as filled and open triangles, respectively. Three consecutive c-myc epitopes were inserted in place of nsp2 aa 324 to 434, based on strain VR-2332 full-length cDNA clone pVR-V7, to generate mutant pV7-myc. In the bottom construct, an HA epitope replaced nsp2 aa 12 to 24 of pV7-myc to generate the double-tagged mutant pV7-HA-myc. (B) Growth kinetics of mutants V7-myc and V7-HA-myc and of parental virus VR-V7 at passage 3. The viruses were used to infect MARC-145 cells in T25 flasks at an MOI of 0.1. The virus-infected cell supernatants were collected every 12 h and were titrated by a viral plaque assay. The mean results were plotted; error bars indicate standard deviations. (C) Immunostaining of nsp2 protein. At 20 h postinfection, the nsp2 protein in V7-myc-infected MARC-145 cells was labeled using monoclonal antibody 9E10 against a c-myc epitope and an Alexa 568-conjugated secondary antibody (red) (Molecular Probes). Nuclei were stained with DAPI (blue). The fields were merged using Photoshop, version 8.
Detection of nsp2 products in PRRSV-infected MARC-145 cells.
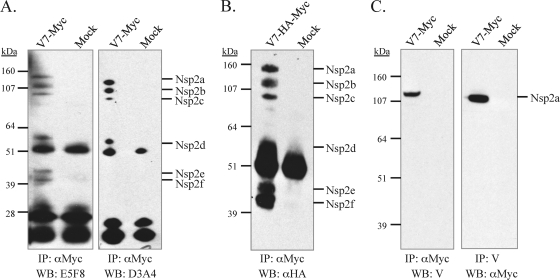
To detect nsp2 products, MARC-145 cells were infected with V7-myc at an MOI of 0.1. The cells were lysed with RIPA buffer at 24 to 36 h postinfection and were cleared by centrifugation. Following immunoprecipitation with mouse monoclonal antibodies (9E10) against the c-myc epitope and separation by SDS-PAGE, the nsp2 protein was probed with mouse monoclonal antibody E5F8, which recognizes the nsp2 region comprising aa 77 to 87 (40). Six specific products, with estimated sizes of 120, 100, 80, 51, 43, and 41 kDa, were detected and designated nsp2a, nsp2b, nsp2c, nsp2d, nsp2e, and nsp2f, respectively (Fig. 2A, first lane). To rule out the possibility that the nsp2 products were being generated during sample preparation or during immunoprecipitation, V7-myc-infected cells were pelleted after being washed with cold PBS three times and were then boiled in SDS-PAGE sample buffer. The proteins were separated, transferred to a nitrocellulose membrane, and probed with anti-c-myc antibodies. Again, we detected all 6 nsp2 species (data not shown). A similar processing pattern was also observed in V7-HA-myc-infected MARC-145 cells probed with anti-HA antibodies after immunoprecipitation with anti-c-myc antibody 9E10 (Fig. 2B, first lane). Since the HA epitope is in place of nsp2 aa 12 to 24 and the recognition site of antibody E5F8 is very near the N terminus of nsp2 (Fig. 1A), we conclude that these products most likely have the same N terminus.
FIG. 2.
Identification of nsp2 products in PRRSV-infected MARC-145 cells. (A) MARC-145 cells either were mock infected or were infected with V7-myc (MOI, 0.1) at passage 3. At 24 to 36 h postinfection, the cells were lysed, and nsp2 proteins were immunoprecipitated (IP) by mouse monoclonal antibody 9E10 recognizing the c-myc epitope. The samples were analyzed by reducing SDS-PAGE on a 4 to 12% polyacrylamide gel followed by Western blotting (WB) with the nsp2-specific antibody E5F8 or D3A4. (B) Analysis of nsp2-associated products in V7-HA-myc-infected MARC-145 cells. The nsp2 products were first immunoprecipitated by anti-c-myc antibody 9E10, then separated by SDS-PAGE, and finally analyzed by WB with a mouse monoclonal antibody to the HA epitope. (C) V7-myc-infected cell lysates were immunoprecipitated with anti-c-myc antibody 9E10 or rabbit antibody V, recognizing a peptide near the C terminus of nsp2; then they were separated by SDS-PAGE and analyzed by immunoblotting with antibody V or rabbit anti-c-myc polyclonal antibodies. The use of different antibody combinations for IP and WB frequently detected nonspecific proteins (25 kDa and 50 kDa, respectively), which are likely immunoglobulin light and heavy chains, respectively, that reacted with the secondary antibody.
Mapping of the C termini of the nsp2 species.
To differentiate the C termini of the nsp2 products, we initially attempted to use antibody probing to distinguish the nsp2 isoforms. Mouse monoclonal antibody D3A4 recognizes the nsp2 peptide comprising aa 545 to 557 (40) and reacted with nsp2 segments a, b, c, and d but not with segments e and f (Fig. 2A, third lane). Since the proteins had initially been precipitated with an anti-c-myc monoclonal antibody recognizing the epitope located just before nsp2 aa 434, the antibody recognition pattern signified that the C termini of nsp2e and nsp2f are located between nsp2 aa 434 and aa 545 (Fig. 2A, third lane).
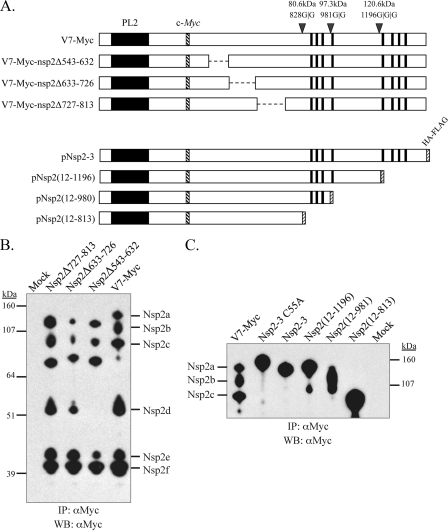
The fact that nsp2d reacted with antibody D3A4 suggests that nsp2d is cleaved after residue 557. To further map the cleavage site, three nsp2 deletion mutants based on V7-myc (V7-myc-nsp2Δ543-632, V7-myc-nsp2Δ633-726, and V7-myc-nsp2Δ727-813) were generated (Fig. 3 A). The mutants were viable and showed growth titers comparable to those of V7-myc and the parental virus VR-V7 (data not shown). By using these mutants in analyses similar to those described above, nsp2d could be distinguished from nsp2c and nsp2e by the deletion of the nsp2 hypervariable region comprising aa 543 to 632, because no nsp2d product was detected after the region was deleted (Fig. 3B), suggesting that the cleavage site of nsp2d is located between nsp2 aa 557 and aa 632.
FIG. 3.
Mapping of the relative positions of the nsp2 isoforms. (A) V7-myc is shown in schematic form with PL2 cleavage sites and the calculated molecular sizes of predicted proteins that included the nsp2 N terminus. V7-myc nsp2 regions comprising aa 543 to 632, aa 633 to 726, or aa 727 to 813 were deleted to generate new full-length infectious cDNA clone mutants pV7-myc-nsp2Δ543-632, pV7-myc-nsp2Δ633-726, and pV7-myc-nsp2Δ727-813, respectively. Polypeptides corresponding to nsp2 aa 12 to 813, aa 12 to 981, and aa 12 to 1196 were cloned into pcDNA3 to generate plasmid constructs pNsp2(12-813), pNsp2(12-981), and pNsp2(12-1196). The HA-FLAG epitope was attached to the C terminus of each polypeptide. The predicted molecular sizes for the corresponding shortened polypeptides were calculated and are given in the text. (B) MARC-145 cells were infected with nsp2 deletion mutant V7-myc-nsp2Δ543-632, V7-myc-nsp2Δ633-726, or V7-myc-nsp2Δ727-813. At 24 to 36 h postinfection, the cells were lysed and immunoprecipitated (IP) with the anti-c-myc monoclonal antibody 9E10, separated by SDS-PAGE on a 4 to 12% NuPage gel, and subjected to Western blotting (WB) with anti-c-myc rabbit polyclonal antibodies. (C) CHO cells were transfected with plasmids expressing either nsp2-3, nsp2-3(C55A), which does not undergo PL2 proteolysis (14), or one of the nsp2 truncation mutants. The cells were lysed after 48 h of transfection; then they were immunoprecipitated with anti-c-myc monoclonal antibody 9E10 and analyzed by Western blotting with an anti-c-myc rabbit polyclonal antibody. The nsp2 proteins immunoprecipitated from V7-myc-infected MARC-145 cells served as a control. The extra bands in the lanes for truncation mutants Nsp2(12-1196) and Nsp2(12-981) may represent degraded products, possibly due to cellular proteases.
In contrast to that of nsp2d, the detection of nsp2c was not affected by the deletion of nsp2 aa 543 to 632, aa 633 to 726, or aa 727 to 813 (Fig. 3B). Thus, the cleavage site of nsp2c lies downstream of residue Gly-813. nsp2b and nsp2c could not be differentiated by deletion mutagenesis, since further deletion of the putative transmembrane domains (aa 876 to 898, 911 to 930, 963 to 979, and 989 to 1009) or their upstream flanking sequence (aa 814 to 845) is lethal to the virus (13). To assess the relative sizes of nsp2b and nsp2c, peptides corresponding to nsp2 aa 12 to 813 and nsp2 aa 12 to 981 were in vitro expressed in CHO cells by cloning the respective nsp2 fragment into pcDNA3/HA-FLAG. The nsp2 C-terminal glycine residues (aa 813 and 981) were chosen because they resided at or near potential alternative cleavage sites for PL2, which has been shown to prefer G|G dipeptides (14, 31). As shown in Fig. 3C, nsp2c and nsp2b showed comparable migration rates with nsp2(12-813) (82 kDa) and nsp2(12-981) (100.3 kDa), respectively, suggesting that they have different C termini.
Rabbit antipeptide antibody V recognizes the nsp2 region comprising aa 1078 to 1094 (14). As shown in Fig. 2C, antibody V detected nsp2a but not other nsp2 species, consistent with the previous result that antibody V recognized the processed nsp2 in CHO cells (14). In Fig. 3C, nsp2a had a migration rate similar to that of nsp2 processed from transfected construct expressing nsp2-3 that was cleaved at or around the G1196|G1197 site in CHO cells, as demonstrated by site-directed mutagenesis, but migrated faster than the uncleaved precursor nsp2-3(C55A) (14). In addition, nsp2a had a migration rate similar to that of the in vitro-expressed polypeptide corresponding to nsp2 aa 12 to 1196 (123.6 kDa). Furthermore, mutations at the G1196|G1197 site that block the nsp2-3 cleavage in vitro are lethal to PRRSV (14). Therefore, we conclude that nsp2a is the equivalent of the nsp2 product processed in CHO cells.
nsp2d, nsp2e, and nsp2f are not essential for viral replication in cell culture.
The experiments described above suggested that the cleavage sites of nsp2d, nsp2e, and nsp2f lie in the middle hypervariable region of nsp2. The deletion mutagenesis in this study, combined with the findings of a previous report (13), demonstrated that deletion of either nsp2 aa 324 to 525, containing putative cleavage sites for nsp2e and nsp2f, or nsp2 aa 543 to 632, containing the putative cleavage site for nsp2d, did not disrupt viral replication in cell culture. In addition, a 400-aa deletion (aa 324 to 726) of the nsp2 hypervariable region that contains the proposed cleavage sites of nsp2d, nsp2e, and nsp2f generated a virus that was viable but exhibited delayed growth kinetics, reduced cytotoxicity, and diminished plaque formation (13). Thus, consistent with the poor conservation of the respective possible G|G cleavage sites among PRRSV strains (Fig. 1A), nsp2d, nsp2e, and nsp2f are not essential for viral replication in cell culture.
We concluded previously that the cleavage of nsp2a most likely occurs at or near residues G1196|G1197 (14). By using reverse genetics, we had also shown that mutations that block the cleavage at this site are lethal to the virus (14). Thus, the proteolytic generation of nsp2a is critical for viral replication. The cleavage sites that generate nsp2b and nsp2c were mapped to the region around the transmembrane (TM) domains. We could not directly determine the importance of nsp2b and nsp2c at this point, since deletion of either the TM domain or its upstream (aa 814 to 845) or downstream sequence is lethal to PRRSV (13).
Accumulation, stability, and turnover of the nsp2 isoforms.
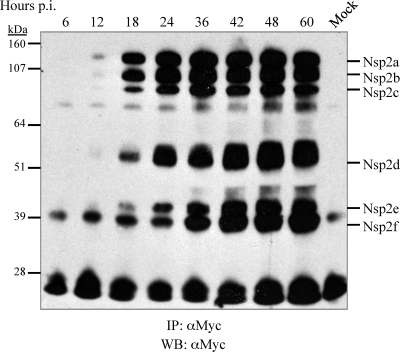
We wanted to study when and how the nsp2 species emerge during PRRSV infection. Accordingly, V7-myc was used to infect MARC-145 cells at a low MOI of 0.1 in order to observe the accumulation of nsp2 species. As shown in Fig. 4, the nsp2 proteins emerged almost simultaneously in the early stages of PRRSV infection, from 6 to 12 h, especially for the larger nsp2 isoforms, and accumulated to peak levels at 36 to 42 h postinfection, consistent with the viral growth curve (Fig. 1B). To investigate the processing kinetics of nsp2, radioimmunoprecipitation assays were carried out with anti-c-myc antibody 9E10 (Fig. 5 A and B) or the rabbit antipeptide antibody V (Fig. 5C and D). In line with the results described above, products with sizes similar to those of the six nsp2 species were recognized in the radioimmunoprecipitation assay by anti-c-myc antibodies (Fig. 5A). As expected, of the six nsp2 cleavage products, antibody V immunoprecipitated only nsp2a (Fig. 5C). Interestingly, one product of host or viral origin with an estimated size of 70 to 80 kDa was markedly coimmunoprecipitated by both anti-myc and V antibodies. Two specific bands with molecular masses of about 14 kDa and 39 kDa were immunoprecipitated only by antibody V. The origins of these bands were not clear, but they could represent downstream cleavage products from nsp2b and nsp2c proteolysis (Fig. 5C). In addition, the large band precipitated by antibody 9E10 only in mock-infected cells was of unknown origin (Fig. 5A).
FIG. 4.
Accumulation of the nsp2 isoforms during virus infection. MARC-145 cells in 60-mm-diameter petri dishes were infected with V7-myc at an MOI of 0.1. At different time points after infection, as indicated, the cells were lysed, immunoprecipitated (IP) with monoclonal antibody 9E10 against the c-myc epitope, separated by SDS-PAGE on a 4 to 12% NuPage gel with 5% β-mercaptoethanol, and subjected to Western blotting (WB) with rabbit polyclonal antibodies to c-myc.
FIG. 5.
Pulse-chase analysis of the nsp2 isoforms. (A to D) MARC-145 cells in 60-mm-diameter petri dishes were infected with V7-myc at an MOI of 0.1. At 20 h postinfection, the cells were labeled with [35S]methionine-cysteine. Cells were lysed and immunoprecipitated (IP) with either anti-c-myc antibody 9E10 (A) or rabbit antipeptide antibody V (C). For pulse-chase analysis, the cells were pulsed for 5 h and then chased for as long as 4 h. nsp2 proteins were immunoprecipitated with anti-c-myc monoclonal antibody 9E10 (B) or rabbit antipeptide antibody V (D). (E and F) In an effort to elucidate the precursor-product relationship, cells were pulsed for 20 min and chased for as long as 2 h. nsp2 proteins were then immunoprecipitated with anti-c-myc monoclonal antibody 9E10 (E) or rabbit antipeptide antibody V (F). The proteins were separated by 4 to 12% SDS-PAGE with 10% dithiothreitol. The gel was dried, and radiolabeled nsp2 proteins were detected by autoradiography.
The stability of nsp2-associated products was analyzed by pulse-chase assays. Initially, MARC-145 cells were infected with the V7-myc virus at an MOI of 0.1 and were metabolically labeled with [35S]methionine-cysteine for 5 h at 18 h postinfection. The nsp2 species accumulated to a maximal level after a pulse for 5 to 6 h, as revealed by immunoprecipitation with anti-c-myc antibodies (Fig. 5B, left). After a 5-h pulse, the 35S-labeled nsp2 species were chased for various periods and were immunoprecipitated either with anti-c-myc antibodies for as long as 240 min (Fig. 5B, right) or with antibody V for as long as 180 min (Fig. 5D). Bands consistent with all six nsp2 isoforms were identified when immunoprecipitation was performed with anti-c-myc antibodies. When antibody V was used for immunoprecipitation, nsp2a was easily detected, as well as the 14-kDa and 39-kDa PRRSV-specific products seen in Fig. 5C. The cleavage of nsp2 from polyprotein pp1a occurred rapidly; no obvious precursor proteins were detected (Fig. 5B and D). Additionally, the ratio among different nsp2 isoforms appeared to be relatively constant (Fig. 5B).
The lack of an obvious precursor-product relationship prompted us to examine the possibility that the amounts of the individual nsp2 products may have reached a plateau during the extended time of [35S]methionine labeling before the completion of the chase. To rule out that possibility, we performed infection with PRRSV V7-myc at a high MOI of 50, shortened the labeling time to 20 min, and then chased for various times. Immunoprecipitations were performed using an anti-c-myc antibody and antibody V (Fig. 5E and F). Again, we observed a similar processing pattern. The failure to establish a clear precursor-product relationship indicated that nsp2 appeared to be processed primarily in cis and that this processing occurred cotranslationally and rapidly.
Coimmunoprecipitation of heat shock 70-kDa protein 5 with the nsp2 replicase protein.
The assays described above also revealed that a protein band with an apparent molecular mass of 70 to 80 kDa was strongly coimmunoprecipitated with nsp2 by either anti-c-myc antibodies or antibody V against nsp2 (Fig. 5B and D) but was not detected when the assays were subsequently analyzed by Western blotting (Fig. 2 to 4). In order to identify the protein, we performed two rounds of immunoprecipitation, with anti-c-myc antibody 9E10 and antipeptide antibody V, respectively. The immunocomplexes were eluted and resolved by SDS-PAGE (Fig. 6 A). The 70- to 80-kDa specific band coimmunoprecipitating with nsp2 was visualized, excised, digested with trypsin, extracted, and subjected to LC-MS-MS analysis. The recovered peptides highly matched heat shock 70-kDa protein 5 (HSPA5, or GRP78; 72 kDa) (Fig. 6B). The coverage was approximately 32%, and a total of 19 unique peptides that matched the HSPA5 sequence of the rhesus monkey, a cousin to the African green monkey, from which the MARC-145 cell line is derived, were recovered (Fig. 6B). The identity of the protein as HSPA5 and its association with nsp2 were further confirmed by Western blotting with rabbit anti-HSPA5 specific antibodies after nsp2 immunoprecipitation with anti-c-myc antibodies (Fig. 6C). The Dana-Farber Cancer Institute Sus scrofa Gene Index (SsGI) (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gireport.pl?gudb=pig) was used to identify a protein sequence of swine GRP78 (TC395795) that was 98.9% identical to rhesus monkey HSPA5, illustrating that the association was not merely an aberrant result of PRRSV growth in nonhost cells (data not shown).
FIG. 6.
Coimmunoprecipitation of HSPA5 with nsp2. (A) MARC-145 cells were infected with V7-myc at an MOI of 0.1. At 24 to 36 h postinfection, the cells were lysed and subjected to two rounds of immunoprecipitation (IP), with anti-c-myc monoclonal antibody 9E10 and rabbit antipeptide antibody V, respectively. The proteins were resolved on a 4 to 12% NuPage reducing gel and were stained with Coomassie blue. (B) The separated proteins were then stained with Sypro Ruby, excised, digested with trypsin, extracted, and subjected to LC-MS-MS analysis. The HSPA5 (GRP78) amino acid sequence of Macaca mulatta (rhesus monkey; GenBank ID 109112231) is shown, with the recovered sequences, sometimes overlapping each other, highlighted in gray. (C) Immunoblot analysis of the immunoprecipitated proteins with a rabbit polyclonal antibody to HSPA5.
DISCUSSION
The experiments described in this report revealed new information regarding the processing of PRRSV nsp2 in the context of viral infection. This protein is often discussed as though it consisted of only one proteolytic product. There now exists clear evidence that type 2 PRRSV strain VR-2332 nsp2 is processed into multiple species. Specifically, the nsp2 species that were identified in this report share the same N terminus but differ in their truncated C termini. This finding is consistent with the observation that EAV nsp2, the counterpart of PRRSV nsp2, undergoes internal cleavage, but in a cell type-dependent manner (29). Whether this finding will apply to other PRRSV strains or cell types (macrophages) remains to be determined.
Functional implications of multiple nsp2 species.
The observation that PRRSV nsp2 has several isoforms has parallels in DNA and RNA viruses. For instance, herpes simplex virus always encodes full-length and truncated versions of the same protein that carry overlapping or distinct functions, although various mechanisms are employed to generate the isoforms, such as alternative splicing, proteolytic cleavage, or translation reinitiation (1, 21, 22, 26). The most salient examples are herpes simplex virus protein pairs UL26/UL26.5, ICP22/US1.5, and US3/US3.5 (21, 22, 26). For arteriviruses, besides EAV nsp2, it was recently reported that EAV nsp7 contains an internal cleavage site for 3CLpro (37). Mutational analysis with an infectious clone of EAV demonstrated that both isoforms are critical for viral replication (37). Additional representative examples from RNA viruses include measles virus V/P, infectious bursal disease virus VP2/VPX, and Sendai virus C/C′ proteins (2, 7, 39). By analogy, the PRRSV nsp2 isoforms may serve different functions in the replication cycle of PRRSV. One benefit of adopting such a strategy, for a positive-strand RNA virus such as PRRSV, is to help maximize coding capacity. Other reasons for generation of isoforms may be to dictate protein abundance, to regulate protein-protein interactions, or to control protein trafficking.
Potential cleavage sites of nsp2 species.
Our finding raises an important question regarding the mechanism for nsp2 cleavage. The PL2 protease displays a preference for a G|G dipeptide and mediates a single nsp2/3 cleavage at or around G1196|G1197 in transfected CHO cells (14). This preference resonates with the deubiquitinating activity of the PRRSV PL2 protease (12, 23, 35). It has been shown that several currently known viral deubiquitinating enzymes (DUBs) recognize and cleave after a G|G dipeptide in ubiquitin conjugates (33). Thus, the preference for a G|G dipeptide may represent a general feature of substrate recognition by the PRRSV PL2 protease.
We propose that G828|G829|G830 and G981|G982 may serve as potential cleavage sites for nsp2c and nsp2b, respectively, based on the substrate recognition property of the PL2 protease and the fact that these sites are fairly conserved among PRRSV strains (15). Additional indirect evidence that nsp2b is cleaved at or near G981|G982 is as follows: (i) an in vitro-expressed polypeptide corresponding to nsp2 aa 12 to 981 had a gel migration rate similar to that of nsp2b (Fig. 3C), and (ii) the G981A and G981P substitutions were lethal to the virus (data not shown). Similarly, there is additional indirect evidence to support G828|G829|G830 as the site for nsp2c cleavage: (i) nsp2c was processed after the residue Gly-813 and had a gel migration pattern similar to that of the polypeptide corresponding to nsp2 aa 12 to 813, expressed in CHO cells (Fig. 3C), and (ii) deletion of nsp2 aa 727 to 813 does not affect viral viability, while deletion of nsp2 aa 727 to 845 is lethal to PRRSV (13). The deleterious effect may be due to the elimination of the nsp2c processing site. Thus, the region comprising aa 813 to 845, containing the putative cleavage site G828|G829|G830, appears to be crucial for viral replication. It should be noted that G828|G829|G830 is highly conserved only in North American strains. Therefore, generation of nsp2c may be type or strain specific, as with hepatitis C virus, in which the internal cleavage of HCV NS3 takes place only in group 2a strains (19). The puzzling fact that the PL2 protease mainly recognizes the nsp2a cleavage site (G1196|G1197) but not other sites (G828|G829|G830 and G981|G982) in CHO cells, in contrast to its behavior in PRRSV-infected cells, suggests that the processing of nsp2b and nsp2c species may need additional cofactors of either host or viral origin. Cofactor binding may alter the overall folding of the nsp2 precursor so that the cleavage sites would be exposed due to the conformational change, or it may change the substrate binding specificity.
The nsp2 region comprising aa 324 to 632, which contains the cleavage sites of nsp2d, nsp2e, and nsp2f, is highly heterogeneous among PRRSV strains (15). From an evolutionary point of view, it is unlikely that the virus would choose such a highly variable region for its substrate. In addition, no dipeptides in VR-2332 nsp2 aa 324 to 632 could be identified as conserved among PRRSV strains and thus able to serve as potential sites that would satisfy the cleavage properties of the PL2 cysteine protease. However, there are several nonconserved G|G dipeptides that could serve as proteolytic substrates for the production of products of the estimated sizes of nsp2d to nsp2f in a strain-specific manner (Fig. 1A). Alternatively, these nsp2 species could be due to polypeptide degradation or translational attenuation, i.e., the ribosomes falling off the viral template during translation. However, we could not rule out the possibility that the PL2 protease recognizes residues other than the G|G dipeptide, or that other viral or host proteases may be involved in nsp2 cleavage.
Processing mechanism of nsp2.
Another important issue concerns the regulation of nsp2 processing. In our pulse-chase assays, we did not observe a gradual decrease in the level of larger nsp2 species and an increase in the level of small nsp2 products, such as would be assumed for a standard precursor-product relationship. Instead, the results revealed that the nsp2 species showed up rapidly and had low turnover (Fig. 5B, D, E, and F) and that the ratio among them, in particular for nsp2a, nsp2b, and nsp2c, remained relatively constant during 2 to 4 h of chase (Fig. 4B and E). Therefore, we propose that the nsp2 species are likely processed in cis instead of trans and that this processing occurs cotranslationally and rapidly, in a manner similar to that of nsp1 (8). This hypothesis also makes sense for explaining the processing of nsp2a and nsp2b, the cleavage sites of which are predicted to be located on opposite sides (cytoplasmic versus lumen) of intracellular membranes. In addition, the well-established ratio balance among nsp2 species indicates that the processing of nsp2 is highly regulated and finely tuned in such a way that additional cleavage sites may not be accessible to the PL2 protease once they are processed.
Potential role of HSPA5.
HSPA5 (also known as GRP78, or BiP) is a member of the heat shock 70-kDa protein family and is constitutively expressed in the lumen of the endoplasmic reticulum (ER) (6). This cellular protein is known to be associated with a variety of folding and assembly intermediates of cellular or viral membrane proteins (4, 5, 9, 24). In this report, we unexpectedly found that HSPA5 strongly coimmunoprecipitated with the nsp2 protein (Fig. 5 and 6). Although the details of the interaction need to be further analyzed, the observation has several important implications, as follows. (i) HSPA5 may play an important part in assisting nsp2 folding for proteolysis by either PL2 or host cell proteases, or both. It has been reported that the proteolytic activity of bovine viral diarrhea virus (BVDV) NS2 protein depends on a cellular cofactor, namely, a chaperone protein termed Jiv (J-domain protein interacting with viral protein) or its 90-aa fragment Jiv90 (20). It is possible that the interaction of HSPA5 with nsp2 could change the conformation of nsp2 and expose additional cleavage sites to PL2 protease activity. (ii) HSPA5 may be involved in the regulation of viral replication. The production of viral progeny depends on the successful recruitment of host cellular components for viral replication, protein synthesis, and virion assembly. During viral replication, a massive amount of proteins is synthesized in a relatively short time, and protein folding can thus become a limiting step (24). The assembly of viral replication complexes on intracellular membranes involves large numbers of viral and/or host proteins. The recruitment of HSPA5 to viral replication complexes may facilitate the folding of the PRRSV macromolecular replication complex. (iii) HSPA5 may be involved in immune evasion. Sequestration of molecular chaperones may lead to unfolded protein responses (UPS) or ER stress (16), leaving many host proteins incorrectly folded. The unfolded proteins, including those involved in host antiviral responses, are sent to the ubiquitin-proteosomal system (UPS) for degradation (16-18). In addition, ER stress also causes attenuation of host translation (16-18). Future work may be directed toward dissecting the detailed interaction between PRRSV nsp2 and HSPA5 and toward defining the role of HSPA5 in the PRRSV replication cycle.
In summary, we report the presence of nsp2 isoforms with apparently different C termini in PRRSV-infected MARC-145 cells and the interaction of nsp2 with the host chaperone HSPA5. These findings indicate that PRRSV nsp2 is increasingly emerging as a multifunctional protein and may have important roles in viral replication and pathogenesis.
Acknowledgments
We thank Bruce Witthuhn of the University of Minnesota Mass Spectrometry Center for technical help with the mass spectrometry analysis. We also thank Hanchun Yang (China Agricultural University) for kindly providing monoclonal antibodies to PRRSV nsp2.
This study was funded by NRI-CSREES 2006-01598.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 28 July 2010.
REFERENCES
- 1.Carter, K. L., and B. Roizman. 1996. The promoter and transcriptional unit of a novel herpes simplex virus 1 alpha gene are contained in, and encode a protein in frame with, the open reading frame of the alpha 22 gene. J. Virol. 70:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castón, J. R., J. L. Martinez-Torrecuadrada, A. Maraver, E. Lombardo, J. F. Rodriguez, J. I. Casal, and J. L. Carrascosa. 2001. C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J. Virol. 75:10815-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 4.Cho, D. Y., G. H. Yang, C. J. Ryu, and H. J. Hong. 2003. Molecular chaperone GRP78/BiP interacts with the large surface protein of hepatitis B virus in vitro and in vivo. J. Virol. 77:2784-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choukhi, A., S. Ung, C. Wychowski, and J. Dubuisson. 1998. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J. Virol. 72:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daugaard, M., M. Rohde, and M. Jaattela. 2007. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581:3702-3710. [DOI] [PubMed] [Google Scholar]
- 7.de Breyne, S., R. Stalder, and J. Curran. 2005. Intracellular processing of the Sendai virus C′ protein leads to the generation of a Y protein module: structure-functional implications. FEBS Lett. 579:5685-5690. [DOI] [PubMed] [Google Scholar]
- 8.den Boon, J. A., K. S. Faaberg, J. J. Meulenberg, A. L. Wassenaar, P. G. Plagemann, A. E. Gorbalenya, and E. J. Snijder. 1995. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J. Virol. 69:4500-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doms, R. W., R. A. Lamb, J. K. Rose, and A. Helenius. 1993. Folding and assembly of viral membrane proteins. Virology 193:545-562. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang, Y., R. R. Rowland, M. Roof, J. K. Lunney, J. Christopher-Hennings, and E. A. Nelson. 2006. A full-length cDNA infectious clone of North American type 1 porcine reproductive and respiratory syndrome virus: expression of green fluorescent protein in the Nsp2 region. J. Virol. 80:11447-11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frias-Staheli, N., N. V. Giannakopoulos, M. Kikkert, S. L. Taylor, A. Bridgen, J. Paragas, J. A. Richt, R. R. Rowland, C. S. Schmaljohn, D. J. Lenschow, E. J. Snijder, A. Garcia-Sastre, and H. W. Virgin IV. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, J., G. Liu, Y. Wang, and K. S. Faaberg. 2007. Identification of nonessential regions of the nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J. Virol. 81:9878-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, J., M. S. Rutherford, and K. S. Faaberg. 2009. The porcine reproductive and respiratory syndrome virus nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities. J. Virol. 83:9449-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, J., Y. Wang, and K. S. Faaberg. 2006. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 122:175-182. [DOI] [PubMed] [Google Scholar]
- 16.He, B. 2006. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 13:393-403. [DOI] [PubMed] [Google Scholar]
- 17.Kim, R., M. Emi, K. Tanabe, and S. Murakami. 2006. Role of the unfolded protein response in cell death. Apoptosis 11:5-13. [DOI] [PubMed] [Google Scholar]
- 18.Kleizen, B., and I. Braakman. 2004. Protein folding and quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 16:343-349. [DOI] [PubMed] [Google Scholar]
- 19.Kou, Y. H., M. F. Chang, Y. M. Wang, T. M. Hung, and S. C. Chang. 2007. Differential requirements of NS4A for internal NS3 cleavage and polyprotein processing of hepatitis C virus. J. Virol. 81:7999-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lackner, T., H. J. Thiel, and N. Tautz. 2006. Dissection of a viral autoprotease elucidates a function of a cellular chaperone in proteolysis. Proc. Natl. Acad. Sci. U. S. A. 103:1510-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, F. Y., and B. Roizman. 1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 65:5149-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, F. Y., and B. Roizman. 1991. The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J. Virol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarova, K. S., L. Aravind, and E. V. Koonin. 2000. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 25:50-52. [DOI] [PubMed] [Google Scholar]
- 24.Mayer, M. P. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 153:1-46. [DOI] [PubMed] [Google Scholar]
- 25.Neumann, E. J., J. B. Kliebenstein, C. D. Johnson, J. W. Mabry, E. J. Bush, A. H. Seitzinger, A. L. Green, and J. J. Zimmerman. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 227:385-392. [DOI] [PubMed] [Google Scholar]
- 26.Poon, A. P., L. Benetti, and B. Roizman. 2006. U(S)3 and U(S)3.5 protein kinases of herpes simplex virus 1 differ with respect to their functions in blocking apoptosis and in virion maturation and egress. J. Virol. 80:3752-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan, M. D., S. Monaghan, and M. Flint. 1998. Virus-encoded proteinases of the Flaviviridae. J. Gen. Virol. 79(Pt. 5):947-959. [DOI] [PubMed] [Google Scholar]
- 28.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 29.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 30.Snijder, E. J., A. L. Wassenaar, and W. J. Spaan. 1992. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J. Virol. 66:7040-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snijder, E. J., A. L. Wassenaar, W. J. Spaan, and A. E. Gorbalenya. 1995. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 270:16671-16676. [DOI] [PubMed] [Google Scholar]
- 32.Snijder, E. J., A. L. Wassenaar, L. C. van Dinten, W. J. Spaan, and A. E. Gorbalenya. 1996. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J. Biol. Chem. 271:4864-4871. [DOI] [PubMed] [Google Scholar]
- 33.Sulea, T., H. A. Lindner, and R. Menard. 2006. Structural aspects of recently discovered viral deubiquitinating activities. Biol. Chem. 387:853-862. [DOI] [PubMed] [Google Scholar]
- 34.Sun, Y., F. Xue, Y. Guo, M. Ma, N. Hao, X. C. Zhang, Z. Lou, X. Li, and Z. Rao. 2009. Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1α. J. Virol. 83:10931-10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun, Z., Z. Chen, S. Lawson, and Y. Fang. 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 84:7832-7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian, K., X. Yu, T. Zhao, Y. Feng, Z. Cao, C. Wang, Y. Hu, X. Chen, D. Hu, X. Tian, D. Liu, S. Zhang, X. Deng, Y. Ding, L. Yang, Y. Zhang, H. Xiao, M. Qiao, B. Wang, L. Hou, X. Wang, X. Yang, L. Kang, M. Sun, P. Jin, S. Wang, Y. Kitamura, J. Yan, and G. F. Gao. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Aken, D., J. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 87:3473-3482. [DOI] [PubMed] [Google Scholar]
- 38.van Hemert, M. J., and E. J. Snijder. 2007. The arterivirus replicase, p. 83-101. In S. Perlman, T. Gallagher, and E. J. Snijder (ed.), Nidoviruses. ASM Press, Washington, DC.
- 39.Vidal, S., J. Curran, and D. Kolakofsky. 1990. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J. Virol. 64:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan, Y., X. Guo, X. Ge, Y. Chen, Z. Cha, and H. Yang. 2007. Monoclonal antibody and porcine antisera recognized B-cell epitopes of Nsp2 protein of a Chinese strain of porcine reproductive and respiratory syndrome virus. Virus Res. 126:207-215. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, Y. J., X. F. Hao, Z. J. Tian, G. Z. Tong, D. Yoo, T. Q. An, T. Zhou, G. X. Li, H. J. Qiu, T. C. Wei, and X. F. Yuan. 2008. Highly virulent porcine reproductive and respiratory syndrome virus emerged in China. Transbound. Emerg. Dis. 55:152-164. [DOI] [PubMed] [Google Scholar]
- 42.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]