Abstract
The switch from Epstein-Barr virus (EBV) latent infection to lytic replication is governed by two viral transactivators, Zta and Rta. We previously reported that the EBV protein LF2 binds Rta, inhibits Rta promoter activation, and blocks EBV replication in cells. In addition, LF2 induces SUMO2/3 modification of Rta. We now show that this modification occurs at four lysines within the Rta activation domain (426, 446, 517, and 530) and that sumoylation of Rta is not essential for its repression. Coexpression studies demonstrated that Rta is sequestered to the extranuclear cytoskeleton in the presence of LF2. We mapped the LF2 binding site to Rta amino acids (aa) 476 to 519 and showed that LF2 binding is critical for Rta relocalization and repression. The core of this binding site, Rta aa 500 to 526, confers LF2-mediated relocalization and repression onto the artificial transcription factor GAL4-VP16. Mutational analysis of LF2 provided further evidence that Rta redistribution is essential for repression. Rta localization changes during replication of the LF2-positive P3HR1 genome, but not during replication of the LF2-negative B95-8 genome. BLRF2 protein expression was decreased and delayed in P3HR1 cells compared with B95-8 cells, consistent with reduced Rta activity. By contrast, BMRF1 expression, regulated primarily by Zta, did not differ significantly between the two cell lines. Our results support a model in which LF2 regulates EBV replication by binding to Rta and redistributing it out of the nucleus.
Epstein-Barr virus (EBV), the prototypical gammaherpesvirus, causes infectious mononucleosis in healthy individuals, B-cell lymphoproliferative disease in immunosuppressed individuals, and rarely, B-cell lymphomas, Hodgkin lymphoma, and nasopharyngeal carcinoma in otherwise-healthy persons (45, 65). Gammaherpesviruses, including EBV and Kaposi's sarcoma-associated herpesvirus (KSHV), differ from other viruses because their associated diseases are not the consequence of virus replication. Instead, EBV-associated malignancies are a by-product of the growth and survival signals triggered by limited viral gene expression that allows EBV to persist in a latent state in infected cells, and hence the human population (44). Because viral replication is not occurring in most EBV-infected cells, inhibitors of replication are not efficacious in treating infectious mononucleosis or EBV-associated malignancies. On the contrary, activation of EBV replication has been suggested as therapy, because virus replication can directly kill EBV-infected tumor cells, sensitize them to nucleoside analogues, and stimulate immune-mediated killing via increased virus antigen expression in tumor cells (21, 22, 55).
Entry into replication is regulated by the EBV genes BZLF1 and BRLF1, encoding the transcriptional activators Z (Zta) and R (Rta), respectively (44, 53). Zta and Rta must act in concert for EBV replication to occur: deletion of either BZLF1 or BRLF1 renders the virus incompetent for DNA replication and virion production (20). Some EBV lytic genes are activated primarily by Rta, others primarily by Zta, and some are synergistically activated by the combined actions of Rta and Zta (13, 20, 24, 27, 37, 43, 48, 49, 53, 60, 64). A subset of EBV genes is activated by Rta but repressed by Zta (18, 49, 64). In most EBV-positive cell lines, expression of either Zta or Rta induces the expression of the other protein and disrupts latency (62, 73). Rta activation of some lytic promoters can be enhanced by coexpression of the BRRF1 gene product (Na) (35). Rta and Zta expression can be induced by B-cell receptor cross-linking, phorbol esters, butyrate, and ionophores, but the physiological signals responsible for inducing EBV replication are not well defined (12, 52). In latently infected cells, lytic gene expression is suppressed by extensive methylation of the genome (3, 19, 40, 54). Preferential binding of Zta to methylated DNA is thought to be important for initiating replication from this epigenetic repressed state (3, 16, 23).
Rta is a 605-amino-acid (aa) member of the gammaherpesvirus ORF50 family of transcriptional activators, which have no known homology to cell transcription factors. The Rta N terminus contains overlapping dimerization (aa 1 to 232) and DNA binding (aa 1 to 280) domains (50). The C-terminal activation domain is comprised of an essential acidic activation domain (aa 520 to 605) and an accessory activation domain (aa 416 to 519), which adds activation potential (33, 50). This accessory domain is required for transactivation in B cells but not in epithelial cells (33). Rta activates many EBV promoters, including the BALF2, BMRF1, and BMLF1 promoters, through a direct mechanism by binding to Rta response elements (RREs) that conform to the consensus GNCCN9GGNG (11, 28-30). Other promoters lacking RREs are activated through indirect mechanisms that may involve direct promoter targeting through interactions with cell transcription factors or activation of signaling pathways in the cytoplasm (1, 24, 46, 47, 63, 68). Rta predominantly localizes to the nucleus (13, 14), and an Rta nuclear localization signal mutant was defective for activation of the BGLF5 promoter and induction of BALF2 expression (36).
We discovered that EBV LF2, a gene deleted from the B95-8 reference strain (59, 61), encodes an Rta binding protein (6). LF2 was also identified as an IRF7 antagonist in a screen for EBV-encoded inhibitors of interferon signaling (72). In EBV-infected cells, LF2 associated with Rta and abrogated Rta's ability to induce viral DNA replication (5). We further demonstrated that LF2 impairs Rta lytic promoter activation and inhibited Rta DNA binding in electrophoretic mobility shift assays. However, the repressive effects of LF2 persisted even when Rta was fused to a heterologous GAL4 DNA binding domain that retained DNA binding activity in the presence of LF2. This suggested that LF2's repressive effects could also act directly on the activation domain. The observation that the small ubiquitin-like modifer (SUMO) is covalently linked to Rta in the presence of LF2 provides a plausible mechanism by which LF2 could modulate Rta activity. SUMO modification of transcription factors frequently results in their repression, through recruitment of histone deacetylases (HDACs) and other repressive complexes (2, 25, 26, 31, 70). Although sumoylation of Rta lysines 19, 213, and 517 previously had been reported to be essential for activation (10), we demonstrated that mutation of these residues to arginines, which cannot serve as SUMO acceptors, does not impair activation (5). Instead, the observed loss of activation seen in the earlier study appears to be due to the K213A mutation, which in addition to preventing sumoylation at that residue disrupts DNA binding (9). Thus, SUMO modification does not appear to contribute to Rta activation but could mediate LF2 represssion of Rta. In this study we sought to determine the mechanism by which LF2 inhibits Rta activity, beginning with an examination of the role of SUMO modification.
MATERIALS AND METHODS
Cell culture.
293T cells are from a human embryonic kidney cell line. The B95-8 Z-HT (B95/Z-HT) and P3HR1 Z-HT (P3/Z-HT) cells have been described elsewhere (39, 71). B95/Z-HT pCEP4-GFP and pCEP4-Flag-LF2 cells were derived from B95/Z-HT cells transfected with pCEP4-GFP or pCEP4-Flag-LF2, followed by continuous hygromycin selection. All cell lines were cultured in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY) or RPMI 1640 medium (Gibco) supplemented with l-glutamine (Gibco), penicillin-streptomycin (Gibco), and 10% fetalplex (Gemini Bio-Products, West Sacramento, CA).
Plasmids.
pcDNA3-Rta, dsRed-HA-Rta, pFlag-LF2, pGL3-BALF2, pGL3-BMLF1, and pGL3-BMRF1 have been described previously (5, 6). The BZLF1 (−221/+13) and BMRF1 (−299/+59) promoters were amplified from B95-8 genomic DNA and cloned into the pGL3 promoter vector (Promega, Madison, WI). pCEP4-GFP was described previously (51), and pCEP4-Flag-LF2 was constructed by cloning 3×Flag-LF2 from pFlag-LF2 into the BamHI/SnaBI sites of pCEP4 (Invitrogen, Carlsbad, CA). pcDNA3-His-SUMO3 was constructed by inserting a double-stranded oligo into the KpnI/BamHI sites of pcDNA3-HA-SUMO3 (5), which replaced the hemagglutinin (HA) tag with the 6×His tag-containing sequence GGTACCATGGGCAGCAGCCATCATCATCATCATCACGGATCC. pcDNA3-Rta 4KR and pcDNA3-Rta 4KR-AD were constructed from pcDNA3-Rta with QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) using oligos containing the appropriate point mutations. Single lysine revertant mutants K530, K517, K446, and K426 were constructed from pcDNA3-Rta 4KR-AD. pcDNA3-Rta-V5 was constructed by cloning an oligo encoding the V5 epitope (GTACGGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGCGTACCGGTTAGTAATGATCTAG) downstream of the Rta open reading frame in pcDNA3-Rta. pcDNA3-Rta-d476-519-V5 was produced by generating the 476-519 deletion by site overlap extension and cloning it into pcDNA3-Rta-V5. pGAL4-VP16 was constructed by PCR amplifying the VP16 activation domain from pVP16 (Clontech, Mountain View, CA) and cloning it into the MluI and XbaI site of pM (Clontech). GAL4-Rta480-501-VP16, GAL4-Rta500-526-VP16, and GAL4-Rta527-550-VP16 were produced by amplifying the appropriate Rta residues by PCR and inserting the fragments into the EcoRI and MluI sites of pGAL4-VP16. The LF2 deletion series was constructed by site overlap extension using external primers containing gateway recombination sequences (shown in capital letters; GGGGACAACTTTGTACAAAAAAGTTGGCatggccgaagcttaccccgga and GGGGACAACTTTGTACAAGAAAGTTGGcagagtgccctcggaggctac) and internal primers containing each desired deletion. Final PCR products were cloned into the pDONR223 gateway entry vector by BP recombinase reaction and sequenced. Each mutant was subsequently cloned by LR recombinase into the destination vectors pN-2×HA and pDEST-myc-eGFP, which have been described previously (6).
Antibodies.
The following antibodies were used for Western blotting, immunoprecipitations, and immunofluorescence: mouse monoclonal antibodies against HA (16B12; Covance, Emeryville, CA), Flag (M2; Sigma, St. Louis, MO), V5 (V5-10; Sigma), VP16 (1-21; Santa Cruz Biotechnology, Santa Cruz, CA), EBV Rta (8C12; Argene, Varilhes, France), EBV BMRF1 (R3; Millipore, Bedford, MA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 6C5; Millipore), and α-tubulin (B-5-1-2; Sigma); goat polyclonal anti-lamin B (C-20; Santa Cruz); rabbit polyclonal anti-BRG1 (H-88; Santa Cruz), and EBV BLRF2 (SLO25-1; a generous gift from George Miller, Yale University School of Medicine, New Haven, CT). A polyclonal anti-LF2 rabbit serum was raised against LF2 aa 26 to 44 and affinity purified (Open Biosystems, Huntsville, AL).
Western blot analysis.
Total cell lysates or immunoprecipitated proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, blotted onto nitrocellulose membranes, and probed with appropriate antibodies. After extensive washing, horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were applied, and the membrane was washed again and then developed with a chemiluminescence reagent (Perkin-Elmer, Waltham, MA) and visualized on a Kodak Image Station 4000R (Kodak Molecular Imaging Systems, Rochester, NY).
Immunoprecipitation.
Transfected 293T cells were lysed in high-salt NP-40 lysis buffer (1% [vol/vol] Igepal CA-630, 40 mM Tris-HCl [pH 7.5], 300 mM NaCl, and 10 mM MgCl2) containing EDTA-free Complete protease inhibitor (Roche, Mannheim, Germany) and cleared by centrifugation at 10,000 × g for 10 min. Supernatants were diluted 1:1 with 1% (vol/vol) NP-40 and precleared with Sepharose (Sigma) for 2 h at 4°C before incubation with anti-Flag M2 agarose (Sigma) overnight at 4°C. The beads were washed extensively with NP-40 lysis buffer (1% [vol/vol] Igepal CA-630, 20 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 5 mM MgCl2) and eluted with 0.4 mg/ml Flag peptide (Sigma) in NP-40 lysis buffer. The eluted proteins were analyzed by Western blotting.
Sumoylation assays.
A total of 1.5 μg of DNA was transfected with Effectene (Qiagen, Valencia, CA) into 293T cells at 50% confluence in six-well plates. The sumoylation assay was performed as described elsewhere (38, 69). Briefly, after 48 h, the cells were washed and resuspended with ice-cold phosphate-buffered saline (PBS). A fraction was taken and prepared as total cell lysate (input). Remaining cells were lysed in lysis buffer (6 M guanidinium-HCl, 100 mM sodium phosphate buffer [pH 8.0], 10 mM 2-mercaptoethanol) containing 5 mM imidazole and sonicated. His-SUMO-modified proteins were purified by nickel affinity chromatography using nickel-nitrilotriacetic acid (NTA) agarose (Qiagen). The agarose was incubated with the lysates for 2 h at 25°C, washed with lysis buffer and 8 M urea wash buffers (8 M urea, 10 mM Tris, 0.1 M sodium phosphate buffer [pH 8.0], 10 mM 2-mercaptoethanol, and 8 M urea, 10 mM Tris, 0.1 M sodium phosphate buffer [pH 6.3], 10 mM 2-mercaptoethanol with 0 to 0.2% [vol/vol] Triton X-100) and eluted with elution buffer (250 mM imidazole, 150 mM Tris [pH 6.8]) for 30 min at 25°C. The input samples and eluates were analyzed by Western blotting.
Reporter assays.
A total of 1.3 μg of DNA was transfected with Effectene (Qiagen) into 293T cells. After 48 h, cells were lysed in reporter lysis buffer (Promega) and clarified by centrifugation. Luciferase (Luciferase assay system; Promega) and β-galactosidase (Galacto-Light Plus system; Applied Biosystems, Bedford, MA) activities were measured using an Optocomp I luminometer (MGM Instruments, Hamden, CT). Luciferase assay results were corrected for transfection efficiency based on β-galactosidase activity.
Immunofluorescence analysis.
293T cells were grown on glass coverslips coated with poly-d-lysine (BD Biosciences, San Jose, CA), and 0.5 to 1.0 μg of total DNA was transfected with Effectene. After 48 h, cells were subjected to Draq5 (Biostatus Limited, Shepshed, United Kingdom) DNA staining, fixed with 1% (wt/vol) paraformaldehyde in PBS, and permeabilized with 0.1% (vol/vol) Triton X-100 in PBS containing 1 mM glycine. Cells were blocked with 5% serum in PBS and incubated with primary antibodies diluted in PBS plus 1% (wt/vol) bovine serum albumin (BSA). Secondary antibodies were goat Alexa Fluor 488 and Alexa Fluor 568 (Molecular Probes, Eugene, OR) and diluted in PBS plus 1% (wt/vol) BSA. The coverslips were mounted with ProLong Gold antifade reagent (Molecular Probes). Images were taken with a laser-scanning Zeiss Axioskop PCM 2000 microscope with a plan-apochromatic 1.40 numerical-aperture 63× objective (Zeiss, Germany) and are single confocal slices.
Subcellular fractionation.
A 0.5-μg aliquot of LF2 and 0.5 μg of Rta expression plasmids were transfected with Effectene into 293T cells. For crude cytoplasmic nuclear fractionations (17), cells were washed and gently dislodged with ice-cold PBS. Cells were swollen in hypotonic buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin) for 1 h on ice, homogenized by 20 dounce strokes using a tight pestle, and pelleted at 350 × g. One percent of the supernatant and pellet was analyzed by Western blotting. The pellet fraction was lysed with isotonic NP-40 buffer (1% [vol/vol] Igepal CA-630, 20 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 5 mM MgCl2) containing Complete protease inhibitor (Roche). Two percent of the NP-40-soluble lysate and of the NP-40-insoluble pellet were analyzed by Western blotting. Subcellular fractions were also generated using a commercially available sequential extraction procedure (Pierce, Rockford, IL) according to the manufacturer's instructions. Each of the five final fractions were analyzed by Western blotting.
RESULTS
Sumoylation of Rta is not required for LF2 repression.
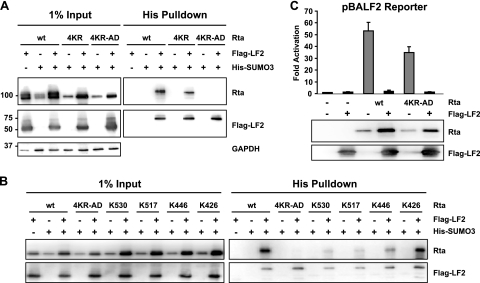
To determine whether LF2-induced sumoylation of Rta is required for inhibition of transactivation, Rta residues that become SUMO modified in the presence of LF2 were identified by using a candidate lysine approach. We previously demonstrated that lysines 19, 213, and 517, which are reported to be SUMO modified (10), are not essential for LF2 repression. The SUMOplot algorithm (Abgent, San Diego, CA) predicts lysine 316 as the most probable SUMO acceptor site within Rta. To address the possibility that Rta might be SUMO modified at any or all of these residues, lysine 316 was mutated to an arginine in combination with lysines 19, 213, and 517 (Rta 4KR). SUMO modification assays were performed by transfecting Rta or Rta mutants into 293T cells along with His-tagged SUMO3, with or without Flag-tagged LF2. SUMO-modified proteins were purified by performing His pull-down assays under denaturing conditions, which abrogated noncovalent protein interactions and inactivated SUMO proteases (69). When LF2 was cotransfected with His-SUMO3 and Rta or Rta 4KR, higher-molecular-weight Rta species were specifically pulled down (Fig. 1 A). These modified Rta forms were dependent on the presence of both His-SUMO3 and LF2, consistent with SUMO3 modification of Rta and Rta 4KR. This confirms the previously published result that LF2 promotes sumoylation of Rta (5) and demonstrates that this sumoylation is not significantly reduced by the 4KR mutations.
FIG. 1.
LF2-dependent sumoylation of Rta is not essential for repression of Rta by LF2. (A) Sumoylation assay to detect SUMO3 modification of Rta (wild type [wt]), Rta K19R K213R K316R K517R (4KR), or Rta K426R K446R K517R K530R (4KR-AD) in the presence of Flag-LF2. Western blotting with anti-Rta antibodies (upper panels) demonstrated the levels of SUMO3-modified Rta or Rta mutants in the presence of Flag-LF2. Input lysates (1%) for pull-down assays are shown in the left half of each panel. Western blotting using anti-Flag antibodies revealed bands corresponding to SUMO3-modified LF2 (lower right panel). (B) Sumoylation assay to detect SUMO3 modification of wt Rta, Rta 4KR-AD, Rta K426R K446R K517R (K530), Rta K426R K446R K530R (K517), Rta K426R K517R K530R (K446), or Rta K446R K517R K530R (K426). Western blotting with anti-Rta antibodies (upper panels) demonstrated the level of SUMO3-modified wt Rta or Rta mutant in the presence of Flag-LF2. Input lysates (1%) for pull-down assays are shown in the left half of each panel. (C) Reporter assay results from 293T cells transfected with a BALF2 promoter-driven luciferase reporter construct with or without Rta or Rta 4KR-AD in the presence or absence of Flag-LF2. Luciferase activities are shown as fold activation over reporter alone and were normalized for transfection efficiency as determined by β-galactosidase activity. Data are averages for six transfections from three independent experiments. Error bars indicate standard deviations. Western blot assays of the cell lysates with anti-Rta and anti-Flag antibodies demonstrated Rta and LF2 protein expression levels.
The above results argue that LF2-induced Rta sumoylation must occur at lysines not previously reported or predicted to be SUMO modified. Of the 31 lysines within Rta, only 4 are present within the activation domain. We considered these to be likely candidates for SUMO modification because the Rta activation domain fused to the GAL4 DNA binding domain is susceptible to LF2 repression (5). Indeed, mutation of these four lysine residues, 426, 446, 517, and 530, to arginines (Rta 4KR-AD) resulted in the complete loss of Rta sumoylation (Fig. 1A). Flag-LF2 was pulled down with His-SUMO3, which confirmed that LF2 itself is SUMO modified under these conditions and served as an internal control to demonstrate the consistency among His pull-down assays.
To determine which of the four lysine residues is targeted for sumoylation by LF2, four single lysine revertant mutants of Rta 4KR-AD were made (K426, K446, K517, and K530) and tested for sumoylation as described above (Fig. 1B). These assays revealed that any of these four lysine residues could serve as a SUMO acceptor site but that lysine 426 appeared to be the preferred residue for sumoylation. The limited ability of lysine 517 to serve as a SUMO acceptor is consistent with the insignificant decrease in sumoylation seen with the original Rta 4KR mutant.
In luciferase reporter gene assays, Rta 4KR-AD activated the BALF2 promoter about 35-fold, comparable to the 50-fold activation seen with wild-type Rta (Fig. 1C). LF2 cotransfection completely abrogated the ability of Rta as well as that of Rta 4KR-AD to transactivate the BALF2 promoter. Because Rta 4KR-AD is not SUMO modified in the presence of LF2 but is repressed to the same extent as wild-type Rta under the conditions tested, we conclude that Rta sumoylation is not essential for LF2-mediated repression.
LF2 redistributes Rta out of the nucleus to the cytoskeleton.
A notable feature of the LF2-Rta interaction is that Rta protein levels become substantially increased in the presence of LF2, despite reduced Rta activity. This does not appear to be due to LF2 transcriptional activation of the cytomegalovirus (CMV) promoter, because LF2 does not exert this effect on CMV-driven reporter genes or on other proteins expressed from the same pcDNA3 vector used to express Rta (data not shown). Although SUMO modification may enhance Rta protein stability, the Rta 4KR-AD experiments described above argue that sumoylation is not required for Rta protein levels to increase in the presence of LF2 (Fig. 1C). Rather, the accumulation of Rta induced by LF2 appears to correlate with Rta repression. Indeed, transcription factor degradation and activation are reported to be functionally linked (56, 66). For example, when transcription factors are sequestered into nuclear domain 10 (PML) bodies, their protein levels are frequently increased (57, 74). We sought to determine if LF2 relocalization of Rta into one of these transcriptionally inactive sites might be the cause of Rta repression and protein accumulation.
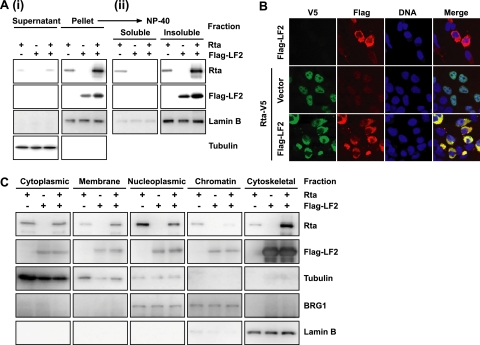
To assess whether LF2 affects Rta localization, we expressed Rta and Flag-tagged LF2 alone and in combination in 293T cells and performed crude cytoplasmic-nuclear fractionations based on the method of Dignam et al. (17). Western blotting confirmed that α-tubulin, a cytoplasmic protein, was predominantly in the supernatant and that lamin B, a nuclear protein, localized to the pellet (Fig. 2 A, panel i). Western blotting revealed that the majorities of Rta and LF2 were in the pellet. Therefore, this crude nuclear fraction was further extracted into NP-40-soluble and -insoluble fractions. As expected, lamin B, a component of the nuclear matrix, was predominantly found in the NP-40-insoluble fraction (Fig. 2A, panel ii). Western blotting for Flag demonstrated that LF2 was always found in the NP-40-insoluble component of the crude nuclear fraction. Rta, when expressed alone, was distributed approximately equally in the NP-40-soluble and -insoluble fractions, with a small amount present in the cytoplasm. When cotransfected with LF2, however, Rta was completely redistributed into the NP-40-insoluble fraction, suggesting that LF2 sequesters Rta into an NP-40-insoluble compartment, such as the nuclear matrix.
FIG. 2.
Coexpression of LF2 results in redistribution of Rta out of the nucleus to the cytoskeleton. (A) Crude cytoplasmic nuclear fractionation and subsequent NP-40 extraction of the nuclear fraction. 293T cells were transfected with Rta, Flag-LF2, or both, harvested 48 h after transfection, and fractionated. (i) One percent of the cytoplasmic and crude nuclear fractions was analyzed by Western blotting using anti-Rta and anti-Flag antibodies. (ii) The crude nuclear fraction was lysed in 1% NP-40 buffer and fractionated into soluble and insoluble components by centrifugation. Two percent of the NP-40 lysate and pellet were analyzed by Western blotting. (B) Immunofluorescence analysis of 293T cells transfected with Flag-LF2, Rta-V5, or both. Cells were subjected to DNA (blue) staining, fixation with 1% paraformaldehyde, indirect immunofluorescence using anti-Flag (red) and anti-V5 (green) antibodies, and confocal microscopy. Single-channel and merged images are shown. (C) Subcellular fractionation of 293T cells transfected with Rta, Flag-LF2, or both. Transfected cells were fractionated into cytoplasm, membrane, nucleoplasm, chromatin-bound, and cytoskeleton fractions and analyzed by Western blotting using anti-Rta and anti-Flag antibodies. Purity of the fractions was monitored by blotting for α-tubulin, BRG1, and lamin B.
The subcellular localization of Rta in the presence of LF2 was also examined by immunofluorescence microscopy using V5-tagged Rta and Flag-tagged LF2 expressed alone or in combination in 293T cells. Surprisingly, LF2 was detected exclusively in the perinuclear cytoplasm (Fig. 2B). The cytoplasmic localization was confirmed for untagged LF2 by immunofluorescence (data not shown) and green fluorescent protein (GFP)-tagged LF2 (see Fig. S1 and S2 in the supplemental material). Rta expressed alone was predominantly nuclear. Coexpression of LF2 resulted in a dramatic relocalization of Rta to the cytoplasm (Fig. 2B). These results confirmed that Rta relocalizes in the presence of LF2 and suggested that Rta exclusion from the nucleus may mediate LF2's repressive effects on Rta.
Because nuclei prepared by the Dignam method frequently have cytoplasmic contamination, we sought to more precisely define the subcellular localization of the LF2-Rta complex. Rta and Flag-LF2 were again expressed alone and in combination in 293T cells. The transfected cells were fractionated into cytoplasm, membrane, nucleoplasm, chromatin-bound, and cytoskeleton fractions by a sequential extraction procedure. Fractions were monitored using control antibodies for proteins of known localizations: α-tubulin (cytoplasm), BRG1 (nucleoplasm and chromatin), and lamin B (nuclear matrix/cytoskeleton). LF2 was predominantly detected in the cytoskeletal fraction and was not affected by Rta cotransfection (Fig. 2C). By contrast, Rta was found mostly in the nucleoplasm when expressed alone but, in the presence of coexpressed LF2, was enriched in the cytoskeletal fraction and depleted from the nucleoplasm. These data and the fluorescence microscopy analysis (Fig. 2A and B) (see Fig. S2 in the supplemental material) suggest that exogenously expressed LF2 localizes to a cytoskeletal compartment that appears to be juxtaposed to the nucleus. In the presence of LF2, Rta is substantially excluded from the nucleoplasm and colocalizes with LF2.
LF2 binding to Rta is required for repression and redistribution.
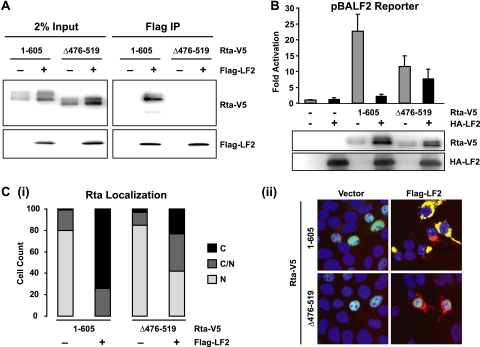
To facilitate testing of whether LF2-mediated relocalization is required for Rta repression, the LF2 binding site within Rta was mapped. Our previous data had suggested that the C-terminal 149 aa of the Rta activation domain are required for interaction with LF2 (5). Because this region includes the epitope detected by the 8C12 Rta monoclonal antibody, Rta deletion mutants were constructed with a C-terminal V5 tag to permit Western blotting. As expected, Rta-V5 strongly interacted with Flag-LF2. In contrast, Rta-Δ476-519-V5 was not detected in the Flag immunoprecipitation reaction (Fig. 3 A). Because stringent extraction conditions (1% NP-40 buffer containing 300 mM NaCl) were required to solubilize LF2-Rta complexes, we cannot exclude the possibility of weak residual binding between LF2 and Rta-Δ476-519-V5, as is suggested by the accumulation of this Rta deletion mutant in the presence of LF2. Nevertheless, our data demonstrate that Rta with aa 476 to 519 deleted is highly deficient for LF2 binding.
FIG. 3.
LF2 binding to Rta is required for repression and redistribution of Rta. (A) Coimmunoprecipitation assay using 293T cells transfected with V5-tagged full-length Rta (1-605) or Rta with aa 476 to 519 deleted (Δ476-519) in the presence or absence of Flag-LF2. Proteins immunoprecipitated by anti-Flag agarose from high-salt 1% NP-40 lysates were identified by Western blotting (right panels) with anti-V5 and anti-Flag antibodies. In each case, 2% of the input lysate is shown for comparison (left panels). (B) Reporter assay results from 293T cells transfected with a BALF2 promoter-driven luciferase reporter with or without Rta-V5 1-605 and Δ476-519 in the presence or absence of HA-LF2 are shown. Luciferase activities are shown as fold activation compared to reporter alone and were normalized for transfection efficiency as determined by β-galactosidase activity. Data are averages for six transfections from three independent experiments. Error bars indicate standard deviations. Western blot results of the cell lysates with anti-V5 and anti-LF2 antibodies demonstrate Rta and LF2 expression levels. (C) Immunofluorescence analysis of 293T cells transfected with Rta-V5 1-605 or Δ476-519 in the presence or absence of Flag-LF2. After DNA staining and fixation, the cells were subjected to indirect immunofluorescence using anti-V5 and anti-Flag antibodies and confocal microscopy. (i) Number of cells observed with exclusively nuclear, exclusively cytoplasmic, and nuclear and cytoplasmic V5 staining in the presence or absence of Flag-LF2. (ii) Merged representative images of the green (anti-V5), red (anti-Flag), and blue (DNA) channels.
To determine whether LF2 binding is required for LF2 repression of Rta transactivation, full-length Rta and Rta Δ476-519 were assessed for BALF2 promoter activation in the absence and presence of LF2. Consistent with previous reports that Rta aa 416 to 519 serve as an accessory activation domain (33), Rta Δ476-519 activated less than the full-length protein (Fig. 3B). Wild-type Rta activation was reduced in the presence of LF2 to background levels, but Rta Δ476-519 activation was only moderately reduced (about 33%), retaining activation at levels considerably higher than the background.
The effect of LF2 binding on Rta localization was assessed by expressing Rta or Rta Δ476-519 in the presence or absence of Flag-LF2 in 293T cells and determining their localization by immunofluorescence. In the absence of LF2, both Rta and Rta Δ476-519 were almost exclusively nuclear (Fig. 3C). Wild-type Rta was substantially relocalized out of the nucleus in the presence of LF2, whereas Rta Δ476-519 showed only a modest decrease in nuclear staining with LF2 coexpression (Fig. 3C). To quantify the effect of LF2 on Rta localization under each condition, at least 100 cells were observed and classified as demonstrating exclusively cytoplasmic, exclusively nuclear, or cytoplasmic and nuclear Rta staining. When LF2 and wild-type Rta were coexpressed, cells exhibiting exclusively nuclear staining were not observed, and only 26% of cells showed any nuclear Rta staining. By contrast, in the presence of LF2, Rta Δ476-519 nuclear localization was only modestly reduced: 77% of cells showed partial or complete nuclear Rta localization. Thus, an Rta mutant with 44 aa deleted within the accessory activation domain is markedly impaired for LF2 binding, is relocalized to the cytoplasm with substantially lower efficiency, and is resistant to LF2 repression.
Rta aa 500 to 526 are sufficient to confer LF2-mediated redistribution and repression onto an artificial transcription factor.
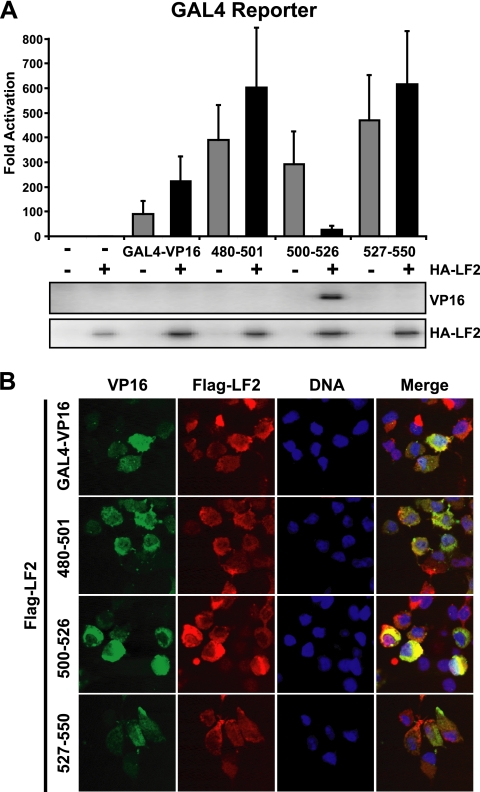
Because Rta Δ476-519 may have some residual capacity to interact with LF2, we investigated whether this Rta domain is sufficient to mediate LF2 interaction. The ability of Rta residues to confer LF2 repression onto the artificial transcription factor GAL4-VP16, in which the GAL4 DNA binding domain is fused to the VP16 activation domain, was tested. Consistent with our previous report (5), GAL4-VP16 was not repressed by LF2 coexpresssion (Fig. 4 A). Additionally, when Rta aa 480 to 501 or aa 527 to 550 were fused in frame between the GAL4-VP16 DNA binding and activation domains, no LF2 repressive effects were observed. In contrast, fusion of Rta aa 500 to 526 to GAL4-VP16 resulted in a transcription factor whose activity was inhibited by 90% when LF2 was cotransfected. Although the GAL4-VP16 fusion proteins could be detected by immunofluorescence (Fig. 4B), their expression could not be demonstrated by Western blotting despite robust activity in reporter assays. This is consistent with other reports using GAL4-VP16 fusion proteins (67). Only the LF2-repressed GAL4-500-526-VP16 accumulated to detectible levels in the presence of LF2, providing further evidence that LF2-mediated repression results in accumulation of the inhibited transactivator.
FIG. 4.
Rta aa 500 to 526 are sufficient to confer LF2-dependent repression and redistribution onto GAL4-VP16. (A) Reporter assay results from 293T cells transfected with luciferase GAL4 reporter constructs along with GAL4-VP16 or with Rta aa 480 to 501, aa 500 to 526, or aa 527 to 550 fused between the GAL4 DNA binding domain and the VP16 activation domain (GAL4-VP16, 480-501, 500-526, and 527-550) in the presence or absence of HA-LF2 are shown. Luciferase activities are shown as fold activation over reporter alone and were normalized for transfection efficiency as determined by β-galactosidase activity. Data are averages for six transfections from three independent experiments. Error bars indicate standard errors of the means. Western blots of the cell lysates with anti-VP16 and anti-HA antibodies are shown below the graph. (B) Indirect immunofluorescence analysis of 293T cells transfected with GAL4-VP16, 480-501, 500-526, or 527-550 in the presence or absence of Flag-LF2. Single-channel (green, anti-VP16; red, anti-Flag; blue, DNA) and merged images are shown.
To investigate whether the LF2 repression was due to relocalization, GAL4-VP16 by itself or fused to Rta aa 480 to 501, 500 to 526, or 527 to 550 was transiently expressed along with Flag-LF2. Immunofluorescence was performed using VP16 and Flag antibodies. Like Rta, GAL4-500-526-VP16 was cytoplasmic in the presence of Flag-LF2. By contrast, GAL4-VP16 by itself or fused to Rta aa 480 to 501 or 527 to 550 was detected in the nucleus when coexpressed with LF2 (Fig. 4B). These data suggest that Rta residues within aa 500 to 526 are sufficient to confer LF2-mediated transcriptional repression and redistribution out of the nucleus.
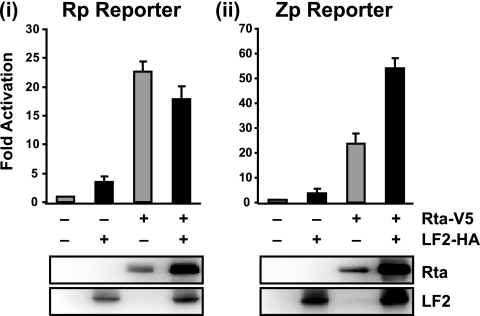
LF2 does not inhibit Rta activation of the BZLF1 or BRLF1 immediate-early gene promoters.
Our previous data and the above experiments suggest that LF2 inhibits Rta transactivation of lytic promoters that contain RREs, such as the BALF2 promoter (5). In order to test whether LF2 can affect activation of promoters that lack RREs, reporter assays with the BRLF1 and BZLF1 promoters were performed. In the absence of LF2, Rta activated the BRLF1 promoter (Rp) approximately 20-fold, and coexpression of LF2 did not significantly impair this activation (Fig. 5 i). The BZLF1 promoter (Zp) was activated approximately 25-fold by Rta expression, and coexpression of LF2 resulted in 2-fold further activation (Fig.5ii). Thus, LF2 relocalization of Rta out of the nucleus does not impair Rp or Zp activation, consistent with induction of cytoplasmic signaling as an important mechanism of Rta upregulation of these immediate-early promoters.
FIG. 5.
LF2 does not inhibit Rta activation of the BRLF1 or BZLF1 promoter. Reporter assay results from 293T cells transfected with luciferase reporter constructs from the BLRF1 (Rp) (i) or BZLF1 (Zp) promoter (ii) with or without Rta-V5 and LF2-HA are shown. Luciferase activities are shown as fold activation over reporter alone and were normalized for transfection efficiency as determined by β-galactosidase activity. Data are averages for six transfections from three independent experiments. Error bars indicate standard deviations. Western blots of the cell lysates with anti-Rta and anti-LF2 antibodies demonstrated Rta and LF2 protein expression levels.
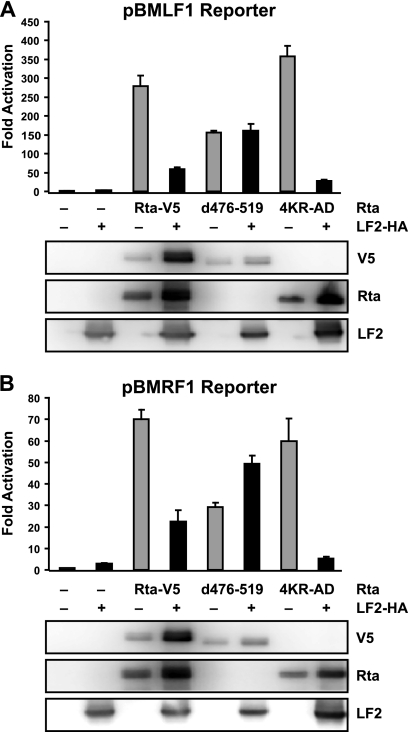
LF2 inhibition of RRE-containing promoters requires direct binding of LF2 to Rta but not Rta sumoylation.
In order to test whether effects of the LF2 binding-deficient and sumoylation-deficient Rta mutants observed on BALF2 promoter activation were characteristic of RRE-containing promoters in general, the effects of these mutants were tested on the BMLF1 and BMRF1 promoters. As with the BALF2 promoter, Rta Δ476-519 activation of the BMLF1 and BMRF1 promoters was approximately half that seen with wild-type Rta and was not decreased by LF2 coexpression (Fig. 6). Further, Rta Δ476-519 was expressed at levels comparable to wild-type Rta but accumulated to a lesser extent in the presence of LF2. By contrast, the sumoylation-deficient Rta 4KR-AD mutant was indistinguishable from wild-type Rta in its activation of the BMLF1 and BMRF1 promoters, and this activation was inhibited by LF2 comparably to that seen with wild-type Rta. These data suggest that results obtained with the BALF2 promoter were representative of RRE promoters in general: LF2 binding is essential for inhibition of Rta activation, but sumoylation is not.
FIG. 6.
LF2 binding is required for inhibition of Rta activation of RRE-containing lytic promoters, but Rta sumoylation is not. Reporter assay results from 293T cells transfected with luciferase reporter constructs from the BMLF1 (A) or BMRF1 (B) promoter with or without Rta-V5 1-605, Rta-V5 Δ476-519, or Rta 4KR-AD in the presence or absence of LF2-HA. Luciferase activities are shown as fold activation over reporter alone and were normalized for transfection efficiency as determined by β-galactosidase activity. Data are averages for six transfections from three independent experiments. Error bars indicate standard deviations. Western blots of the cell lysates with anti-V5, anti-Rta, and anti-LF2 antibodies demonstrate Rta and LF2 protein expression levels.
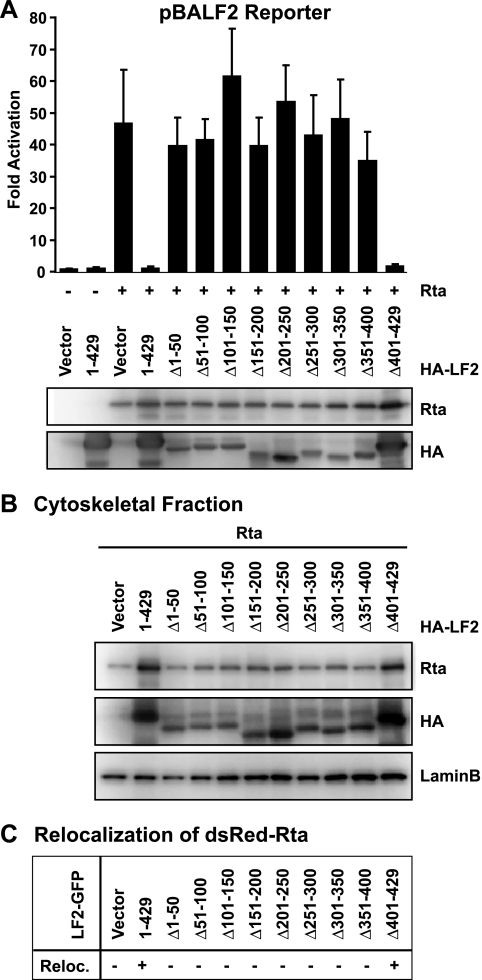
LF2 mutants defective for Rta redistribution cannot repress Rta.
To define the LF2 domains that confer Rta repression and cytoskeletal localization, a series of eight 50-aa deletions (Δ1-50 to Δ351-400) and one C-terminal 29-aa deletion (Δ401-429) were constructed. The resulting nine LF2 deletion mutants and full-length LF2 (1-429) were tested for their ability to prevent Rta activation of the BALF2 promoter in reporter gene assays. Only LF2 Δ401-429 reduced Rta transactivation to the same extent as full-length LF2 (Fig. 7 A). None of the other LF2 deletion mutants significantly affected Rta-induced activation of the BALF2 promoter, despite the fact that these deletions were expressed.
FIG. 7.
LF2 mutants defective for Rta redistribution cannot repress Rta. (A) Reporter assay results from 293T cells transfected with a BALF2 promoter-driven luciferase reporter with or without Rta in the presence or absence of HA-tagged full-length LF2 (1-429) or the indicated LF2 deletion mutants. Luciferase activities are shown as fold activation over reporter alone and were normalized for transfection efficiency as determined by β-galactosidase activity. Data are averages for six transfections from three independent experiments. Error bars indicate standard deviations. Western blots of the cell lysates with anti-Rta and anti-HA antibodies demonstrated Rta and LF2 protein expression levels. (B) 293T cells were transfected with Rta in the presence or absence of HA-tagged full-length LF2 or the indicated LF2 deletion constructs and fractionated. The cytoskeletal fraction was analyzed by Western blotting using anti-Rta and anti-HA antibodies. Detection of lamin B was used to demonstrate relative purity of the cytoskeletal fraction in comparison to other fractions (not shown) and served as a loading control. (C) Summary of fluorescence microscopy analysis (see Fig. S1) of 293T cells transfected with dsRed-tagged Rta in the presence or absence of GFP-tagged LF2 (1-429) or LF2 deletion constructs (Δ1-50, Δ51-100, Δ101-150, Δ151-200, Δ201-250, Δ251-300, Δ301-350, Δ351-400, Δ401-429). Transfected cells were observed by confocal microscopy and scored for the cytoplasmic redistribution of dsRed signal.
Subcellular fractionation of 293T cells cotransfected with the HA-tagged LF2 deletions and Rta demonstrated that only LF2 Δ401-429 resulted in accumulation of Rta in the cytoskeletal fraction, as seen with full-length LF2 (Fig. 7B). To confirm this correlation of Rta repression and relocalization, dsRed-tagged Rta was cotransfected with GFP-tagged LF2 deletions, and the subcellular localizations of Rta and LF2 were examined by fluorescence microscopy. Coexpression of LF2 1-429 or Δ401-429 resulted in the redistribution of otherwise-nuclear dsRed-Rta (see Fig. S1 in the supplemental material). The redistributed dsRed-Rta localized to a perinuclear ring, while dsRed-Rta in the presence of other LF2 deletion mutants remained exclusively nuclear. Interestingly, although most of the LF2 mutants were defective for repression, they all showed the same juxtanuclear distribution as wild-type LF2 (see Fig. S1 and S2 in the supplemental material), suggesting that they are not grossly misfolded. Thus, the C-terminal 29 aa of LF2 are dispensable for Rta repression and relocalization, but the Rta binding domain could not be further defined within the first 400 aa of LF2 based on this panel of deletion mutants.
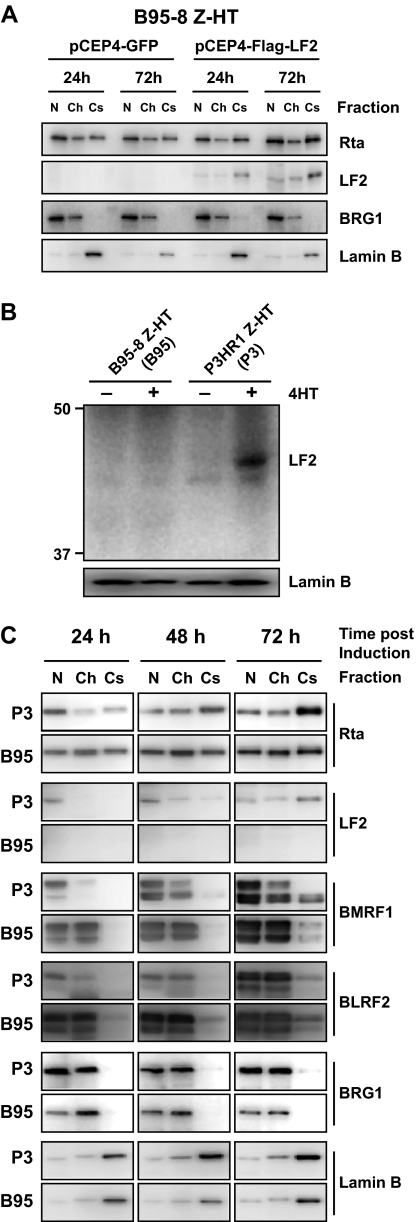
Rta relocalizes to the cytoskeleton during replication of an LF2-positive EBV genome.
In order to assess LF2 effects on Rta during replication of the EBV genome, we compared Rta localization in B95-8 cells, which lack LF2, to B95-8 cells stably transfected with LF2 and to P3HR1 cells, which contain an intact LF2 gene. To allow efficient induction of lytic replication, Zta fused to the hydroxytamoxifen-responsive estrogen receptor hormone binding domain (Z-HT) was stably expressed in B95-8 (B95/Z-HT) and P3HR1 (P3/Z-HT) cells. Addition of 4-hydroxy-tamoxifen (4HT) results in nuclear translocation of the otherwise cytoplasmic Z-HT and induces the lytic cascade.
For stable expression in B95/Z-HT cells, the episomal pCEP4 vector containing Flag-LF2 or GFP was used. After hygromycin selection, B95/Z-HT cells expressing Flag-LF2 or GFP were induced for lytic replication and subjected to subcellular fractionation at 24 and 72 h after addition of 4HT. LF2 was found predominantly in the cytoskeletal fraction (Fig. 8A), and its expression increased over time, possibly due to activation of the pCEP4 CMV promoter during EBV replication. Compared to GFP-expressing controls, cytoskeletal-associated Rta increased when LF2 was expressed in B95/Z-HT, albeit to a lesser extent than seen in P3/Z-HT cells. Surprisingly, immunofluorescence microscopy demonstrated that only about 20% of B95/Z-HT cells expressed detectable Flag-LF2, despite continuous culture in hygromycin (see Fig. S3 in the supplemental material). This suggests that the degree of Rta redistribution in response to LF2 may be much greater than that observed, since 80% of fractionated cells did not contain detectable levels of LF2.
FIG. 8.
Rta relocalizes from the nucleoplasm to the cytoskeleton during replication of an LF2-positive EBV genome. (A) B95-8 cells that stably express Zta-HT and GFP or Flag-LF2 were induced for replication by treatment with 4HT for 24 or 72 h and fractionated into cytoplasm, membrane, nucleoplasm, chromatin-bound, and cytoskeleton fractions. Equal relative amounts of the nucleoplasm (N), chromatin-bound (Ch), and cytoskeleton (Cs) fractions were analyzed by Western blotting for Rta and LF2. Fraction purity was assessed by detection of nucleoplasmic and chromatin-associated BRG1 protein and the nuclear matrix/cytoskeleton component lamin B. (B) LF2 Western blot results with anti-LF2 polyclonal rabbit sera. B95-8 or P3HR1 cells that stably express Zta-HT (P3 or B95) were treated with 4HT for 72 h and lysed in SDS loading buffer. Detection of lamin B served as a loading control. (C) P3 and B95 cells were treated with 4HT for 24, 48, and 72 h and fractionated as described for panel A. Equal relative amounts of the N, Ch, and Cs fractions were analyzed by Western blotting for the indicated EBV proteins. Fraction purity was assessed by detection of the subcellular markers BRG1 and lamin B.
Although profiling experiments have demonstrated that LF2 mRNA levels are detectible by 8 h and peak within 12 h of lytic induction, LF2 protein expression levels during replication have never been reported. Preliminary experiments using LF2-specific rabbit sera (see Fig. S4 in the supplemental material) detected a 45-kDa band corresponding to the predicted size of LF2 in P3/Z-HT cells within 48 h of 4-HT addition (Fig. 8B and C and unpublished observations). This band was not detected in B95/Z-HT cells. These experiments demonstrated that LF2 is expressed in P3HR1 cells during lytic replication and that LF2 protein levels are increasing during the first 72 h postinduction. We therefore induced P3/Z-HT and B95/Z-HT cells for 24, 48, and 72 h and collected nucleoplasmic, chromatin, and cytoskeletal fractions (Fig. 8C). As previously found, detection of nuclear/chromatin (BRG1) and cytoskeletal (lamin B) markers was used to monitor subcellular fraction purity. Western blotting revealed that LF2 was enriched in the cytoskeletal fraction of P3/Z-HT cells at 72 h and to a lesser degree at 48 h. LF2 appeared to be predominantly nucleoplasmic at 24 h in P3/Z-HT cells.
Rta protein was detected primarily in the nucleoplasm of P3/Z-HT cells at 24 h. With time, nucleoplasmic Rta levels decreased, and Rta accumulated in the cytoskeleton. By 72 h, Rta was mostly cytoskeletal in the P3/Z-HT cells. Thus, the Rta distribution correlates with LF2 protein expression in P3/Z-HT cells in a temporal and spatial fashion. By contrast, in B95/Z-HT cells, Rta was detected equally in the nucleoplasmic, chromatin, and cytoskeletal fractions, and this distribution did not change over time. LF2 was absent in all three B95/Z-HT fractions at any time point.
To identify differences in lytic cascade activation, expression of the protein products of the Zta-responsive BMRF1 gene and the Rta-responsive BLRF2 gene were examined. In both P3/Z-HT and B95/Z-HT cells, BMRF1 protein was detectable in the nucleoplasmic and chromatin fractions at 24 h and increased through the 72-h time point. There were no significant differences in the level or distribution of BMRF1 between the two cell lines. By contrast, BLRF2 expression was markedly delayed in P3/Z-HT cells compared to B95/Z-HT cells during replication. In B95/Z-HT cells, the BLRF2 protein was highly expressed in the nucleoplasm and chromatin fractions by 24 h and remained at elevated levels through 72 h. In P3/Z-HT cells, BLRF2 distribution was similar, but expression levels were markedly decreased at the earlier time points and this difference persisted to a lesser degree through 72 h. In summary, proteins encoded by early and late genes are expressed in the presence or absence of LF2. The observation that the Rta-responsive BLRF2 gene product can be expressed at lower levels with delayed kinetics in P3/Z-HT cells suggests that Rta activity is decreased in these cells compared to B95/Z-HT.
DISCUSSION
Data presented here demonstrate that Rta redistributes from the nucleoplasm to the cytoskeleton during EBV replication. This event is mediated by LF2, as coexpression of LF2 in the absence of other EBV proteins is sufficient for Rta redistribution. Moreover, Rta redistribution is not observed during EBV replication in LF2-deficient B95-8 cells but does occur when LF2 is exogenously expressed. Direct binding of LF2 to Rta appears to be critical, because an Rta mutant deficient for LF2 binding is not efficiently relocalized. Our experiments leave open the question of whether LF2 initially encounters Rta in the nucleus or the cytoplasm. At least three mechanisms of Rta nuclear exclusion can be imagined. LF2 may sequester Rta in the cytoplasm by tethering it to the extranuclear cytoskeleton, LF2 may compete with an Rta binding partner required for nuclear localization, or LF2 may induce an Rta modification that retains Rta outside of the nucleus. Many cell transcription factors are regulated by exclusion from the nucleus (76), and there are examples for all three mechanisms in the literature: the antioxidant response element transcription factor Nrf2 is sequestered in the cytoplasm by Keap1, which anchors Nrf2 to the actin cytoskeleton (41); NF-κB heterodimers are retained in the cytoplasm through masking of their nuclear localization signal by IκB proteins (34, 58); monoubiquitination promotes cytoplasmic localization of p53 (7, 8).
Our finding that the sumoylation-deficient Rta mutant 4KR-AD is efficiently repressed and relocalized in the presence of LF2 (Fig. 1C and data not shown) argues that sumoylation is not responsible for Rta redistribution. It is possible that LF2 induces modifications other than sumoylation that play a role in Rta localization. However, these modifications would have to occur within the 27-aa core of the LF2 binding site or be induced by this binding site in both Rta and the unrelated GAL4-VP16 sequence. The GAL4-VP16 experiment also suggests that nuclear localization signal masking is an unlikely mechanism. Thus, we favor a sequestration mechanism. Sequestration is supported by the observation that Rta accumulates in the same subcellular fraction as LF2. Further, sequestration best explains how the core of the LF2 binding site is sufficient to confer relocalization and repression onto the heterologous GAL4-VP16 transcription factor. Additionally, the inability of LF2 to inhibit activation of Zp and Rp, which can be activated without direct binding of Rta to RREs, is consistent with a sequestration mechanism. Although Rta exclusion from the nucleus is clearly sufficient for repression of RRE-containing promoters, an important outstanding question is whether Rta redistribution is essential. The strong correlation of Rta repression and redistribution observed for the LF2 deletion mutants suggests that it is. The proposed sequestration mechanism predicts that LF2 binds to a cell protein in order to tether the Rta-LF2 complex in the cytoskeleton.
Attempts to prove this sequestration mechanism by making specific mutations in LF2 have been hampered by the lack of an LF2 structure or defined subdomains. LF2 is thought to have evolved from the herpesvirus dUTPase gene, which encodes a protein that folds into a single globular structure (15). Current threading algorithms (42) using the EBV dUTPase structure as a basis predict folds for only about 25% of the LF2 residues. Consequently, we constructed 50-aa deletions spanning the LF2 protein in an effort to define an LF2 mutant competent for Rta binding that could not relocalize Rta. With the exception of LF2 with the C-terminal 29 aa deleted, all LF2 mutants studied failed to repress or relocalize Rta. While this supports a correlation between repression and relocalization, 50-aa deletions could disrupt the folding of a globular protein like LF2. On the other hand, the LF2 mutants exhibited the same localization as wild-type LF2 (see Fig. S1 in the supplemental material) and only slightly reduced expression levels (Fig. 7), suggesting that they are not grossly misfolded. Construction of LF2 mutants that are deficient for specific functions or protein interactions remains an important unresolved problem.
By contrast, efforts to map the LF2 binding domain within Rta were highly successful, identifying a 27-aa sequence that is a critical mediator of LF2 effects. This likely reflects the modular architecture of the Rta protein, which has well-delineated DNA binding and transcriptional activation domains. The newly identified LF2 binding site falls within the accessory activation domain and contains a cluster of residues that are highly conserved among Rta homologues of other lymphocryptoviruses (see Fig. S5 in the supplemental material). Secondary structure algorithms predict an α-helix within these conserved residues, and it is tempting to view this LF2 binding domain as a rod-like structure that is enveloped by the globular LF2 protein. The fact that the 8C12 monoclonal epitope overlaps with this binding site and can recognize Rta in its native conformation (e.g., immunofluorescence or immunoprecipitation) lends further support to the notion that this binding site is readily accessible on the surface of the Rta protein. LF2 binding has also been mapped to the central inhibitory association domain of IRF7 (aa 285 to 457) (72). Although limited regions of homology can be identified between these two binding sites, the IRF7 site will need to be more precisely mapped before it can be reliably judged whether LF2 binds to these proteins via the same mechanism.
Our results demonstrate that the Rta activation domain becomes SUMO modified in the presence of LF2. Rta lysine 426 is the predominant SUMO acceptor, but lysines 446, 517, and 530 can also be SUMO modified to a lesser degree. Rta SUMO modification is not required for activation or essential for LF2-mediated repression, but we cannot completely exclude a role for SUMO modification in LF2-mediated effects. For example, sumoylation may be an important means of Rta inactivation if LF2 levels are limiting or Rta sequestration is incomplete. Our studies of P3HR1 cells demonstrated that Rta persists in the nucleoplasm at 72 h, despite substantial redistribution to the cytoskeletal compartment. Whether nucleoplasmic Rta is further inhibited by LF2-induced sumoylation or inhibition of DNA binding, as we have previously observed, is unknown. Introduction of the Rta 4KR-AD mutant into a recombinant EBV genome may uncover a role for Rta sumoylation that is not apparent in transient assays. It is also possible that sumoylation of LF2 is what determines its localization and, indirectly, mediates Rta relocalization. Alternatively, if LF2 is a SUMO E3 ligase or recruits one, other proteins bound by LF2 may become sumolyated. IRF7 is a potential candidate; however, in preliminary experiments SUMO-modified IRF7 levels were not increased by LF2 coexpression.
The precise role of LF2 in EBV biology remains uncertain. However, our results may explain why Rta activates promoters through two distinct mechanisms. LF2 sequestration of Rta markedly impairs activation of RRE-containing promoters without significantly affecting activation of promoters that lack RREs. If LF2 is an EBV virion protein, as are its homologues in other gammaherpesviruses (4, 75), this differential regulation of Rta may play out in the newly infected cell. In this setting, in cells in which the immediate-early promoters are activated, LF2 would prevent EBV replication but allow activation of other promoters through mechanisms that do not involve Rta binding to RREs (32). LF2 is unlikely to accomplish this feat alone, as “abortive” lytic replication has been observed in experiments with LF2-negative B95-8 genomes (40). In those experiments, the unmethylated state of the newly delivered EBV genome was proposed to prevent activation of replication by Zta. It is also probable that LF2's ability to inhibit IRF7 signaling is most important during initial infection. Thus, the inclusion of LF2 into the EBV tegument could foster the establishment of latent infection and allow activation of a subset of Rta-responsive promoters, while simultaneously subverting the cell's innate immune response.
Our current results suggest an important role for LF2 in cells undergoing EBV replication as well. Although LF2 can abrogate EBV replication when transfected into EBV-positive cells (5), this activity must be regulated for EBV to be able to reproduce itself. Data presented here clearly demonstrate that despite LF2 expression and Rta relocalization in P3HR1 cells, both early and late gene expression proceeds. It is likely that the lower levels of LF2 expression directed by its endogenous promoter temper the ability of LF2 to block replication. LF2 levels may accumulate during replication to some threshold level and serve as a feedback mechanism to force the cell to exit replication and, potentially, to survive. LF2 could also define an upper limit for lytic gene product expression that might otherwise reach toxic levels. The EBV SM protein may also act in this fashion, by limiting the feed-forward activation of Rta on its own promoter (71). Alternatively, LF2 may be required to balance the expression of lytic genes controlled by Rta with those regulated by Zta. Our results show that BMRF1 expression, driven by the combined effects of ectopic Z-HT and endogenous Zta, is similar in induced B95/Z-HT and P3/Z-HT cells. In contrast, BLRF2 is markedly less well expressed in P3/Z-HT cells. Although Zta has been reported to repress BLRF2 expression, differences in Zta activity seem unlikely in view of the BMRF1 data. Instead, the decrease in BLRF2 levels in P3/Z-HT is probably due to reduced Rta activity. This did not appear to be due to differences in Rta expression, but we cannot exclude unmeasured effects, such as differences in BRRF1 (Na) coactivation of Rta. To control for these possibilities, an EBV genome selectively deleted for the LF2 open reading frame is being constructed which should allow the effects of LF2 to be studied in an isogenic background. Nevertheless, the observation that Rta is redistributed during EBV replication of the P3HR1 genome, but not the B95-8 genome, implicates LF2 in the observed differences in BLRF2 expression and, by extension, in regulation of lytic replication.
Supplementary Material
Acknowledgments
This work was supported by a Howard Hughes Medical Institute Early Career Award (to E.J.) and the exchange program between Harvard Medical School and the graduate training program 1071 (DFG-GK 1071) at the Friedrich-Alexander University Erlangen-Nuremberg, Germany (to A.M.F.H.).
We thank Amy Holthaus for helpful discussions and excellent technical support.
Footnotes
Published ahead of print on 14 July 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adamson, A. L., D. Darr, E. Holley-Guthrie, R. A. Johnson, A. Mauser, J. Swenson, and S. Kenney. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, A. L., and S. Kenney. 2001. Epstein-barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhende, P. M., W. T. Seaman, H. J. Delecluse, and S. C. Kenney. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 36:1099-1104. [DOI] [PubMed] [Google Scholar]
- 4.Boname, J. M., J. S. May, and P. G. Stevenson. 2005. Murine gammaherpesvirus 68 open reading frame 11 encodes a nonessential virion component. J. Virol. 79:3163-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderwood, M. A., A. M. Holthaus, and E. Johannsen. 2008. The Epstein-Barr virus LF2 protein inhibits viral replication. J. Virol. 82:8509-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calderwood, M. A., K. Venkatesan, L. Xing, M. R. Chase, A. Vazquez, A. M. Holthaus, A. E. Ewence, N. Li, T. Hirozane-Kishikawa, D. E. Hill, M. Vidal, E. Kieff, and E. Johannsen. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U. S. A. 104:7606-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, S., O. Bischof, A. Dejean, and K. H. Vousden. 2007. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 9:428-435. [DOI] [PubMed] [Google Scholar]
- 8.Carter, S., and K. H. Vousden. 2009. Modifications of p53: competing for the lysines. Curr. Opin. Genet. Dev. 19:18-24. [DOI] [PubMed] [Google Scholar]
- 9.Chang, L. K., J. Y. Chung, Y. R. Hong, T. Ichimura, M. Nakao, and S. T. Liu. 2005. Activation of Sp1-mediated transcription by Rta of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids Res. 33:6528-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, L. K., Y. H. Lee, T. S. Cheng, Y. R. Hong, P. J. Lu, J. J. Wang, W. H. Wang, C. W. Kuo, S. S. Li, and S. T. Liu. 2004. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 279:38803-38812. [DOI] [PubMed] [Google Scholar]
- 11.Chen, L. W., P. J. Chang, H. J. Delecluse, and G. Miller. 2005. Marked variation in response of consensus binding elements for the Rta protein of Epstein-Barr virus. J. Virol. 79:9635-9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Countryman, J., L. Gradoville, S. Bhaduri-McIntosh, J. Ye, L. Heston, S. Himmelfarb, D. Shedd, and G. Miller. 2009. Stimulus duration and response time independently influence the kinetics of lytic cycle reactivation of Epstein-Barr virus. J. Virol. 83:10694-10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox, M. A., J. Leahy, and J. M. Hardwick. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darr, C. D., A. Mauser, and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BRLF1 induces the lytic form of viral replication through a mechanism involving phosphatidylinositol-3 kinase activation. J. Virol. 75:6135-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davison, A. J., and N. D. Stow. 2005. New genes from old: redeployment of dUTPase by herpesviruses. J. Virol. 79:12880-12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickerson, S. J., Y. Xing, A. R. Robinson, W. T. Seaman, H. Gruffat, and S. C. Kenney. 2009. Methylation-dependent binding of the epstein-barr virus BZLF1 protein to viral promoters. PLoS Pathog. 5:e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Guindy, A. S., and G. Miller. 2004. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J. Virol. 78:7634-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernberg, I., K. Falk, J. Minarovits, P. Busson, T. Tursz, M. G. Masucci, and G. Klein. 1989. The role of methylation in the phenotype-dependent modulation of Epstein-Barr nuclear antigen 2 and latent membrane protein genes in cells latently infected with Epstein-Barr virus. J. Gen. Virol. 70:2989-3002. [DOI] [PubMed] [Google Scholar]
- 20.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng, W. H., G. Hong, H. J. Delecluse, and S. C. Kenney. 2004. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J. Virol. 78:1893-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng, W. H., E. Westphal, A. Mauser, N. Raab-Traub, M. L. Gulley, P. Busson, and S. C. Kenney. 2002. Use of adenovirus vectors expressing Epstein-Barr virus (EBV) immediate-early protein BZLF1 or BRLF1 to treat EBV-positive tumors. J. Virol. 76:10951-10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis, A. L., L. Gradoville, and G. Miller. 1997. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J. Virol. 71:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furnari, F. B., M. D. Adams, and J. S. Pagano. 1992. Regulation of the Epstein-Barr virus DNA polymerase gene. J. Virol. 66:2837-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Dominguez, M., and J. C. Reyes. 2009. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim. Biophys. Acta 1789:451-459. [DOI] [PubMed] [Google Scholar]
- 26.Gill, G. 2005. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15:536-541. [DOI] [PubMed] [Google Scholar]
- 27.Granato, M., A. Farina, R. Gonnella, R. Santarelli, L. Frati, A. Faggioni, and A. Angeloni. 2006. Regulation of the expression of the Epstein-Barr virus early gene BFRF1. Virology 347:109-116. [DOI] [PubMed] [Google Scholar]
- 28.Gruffat, H., N. Duran, M. Buisson, F. Wild, R. Buckland, and A. Sergeant. 1992. Characterization of an R-binding site mediating the R-induced activation of the Epstein-Barr virus BMLF1 promoter. J. Virol. 66:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruffat, H., E. Manet, A. Rigolet, and A. Sergeant. 1990. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 18:6835-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gruffat, H., and A. Sergeant. 1994. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 22:1172-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagemeier, S. R., S. J. Dickerson, Q. Meng, X. Yu, J. E. Mertz, and S. C. Kenney. 2010. Sumoylation of the Epstein-Barr virus BZLF1 protein inhibits its transcriptional activity and is regulated by the virus-encoded protein kinase. J. Virol. 84:4383-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halder, S., M. Murakami, S. C. Verma, P. Kumar, F. Yi, and E. S. Robertson. 2009. Early events associated with infection of Epstein-Barr virus infection of primary B-cells. PLoS One 4:e7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardwick, J. M., L. Tse, N. Applegren, J. Nicholas, and M. A. Veliuona. 1992. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J. Virol. 66:5500-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-κB. Genes Dev. 18:2195-2224. [DOI] [PubMed] [Google Scholar]
- 35.Hong, G. K., H. J. Delecluse, H. Gruffat, T. E. Morrison, W. H. Feng, A. Sergeant, and S. C. Kenney. 2004. The BRRF1 early gene of Epstein-Barr virus encodes a transcription factor that enhances induction of lytic infection by BRLF1. J. Virol. 78:4983-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu, T. Y., Y. Chang, P. W. Wang, M. Y. Liu, M. R. Chen, J. Y. Chen, and C. H. Tsai. 2005. Reactivation of Epstein-Barr virus can be triggered by an Rta protein mutated at the nuclear localization signal. J. Gen. Virol. 86:317-322. [DOI] [PubMed] [Google Scholar]
- 37.Hung, C. H., and S. T. Liu. 1999. Characterization of the Epstein-Barr virus BALF2 promoter. J. Gen. Virol. 80:2747-2750. [DOI] [PubMed] [Google Scholar]
- 38.Jaffray, E. G., and R. T. Hay. 2006. Detection of modification by ubiquitin-like proteins. Methods 38:35-38. [DOI] [PubMed] [Google Scholar]
- 39.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalla, M., A. Schmeinck, M. Bergbauer, D. Pich, and W. Hammerschmidt. 2010. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc. Natl. Acad. Sci. U. S. A. 107:850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang, M. I., A. Kobayashi, N. Wakabayashi, S. G. Kim, and M. Yamamoto. 2004. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. U. S. A. 101:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 43.Kenney, S., E. Holley-Guthrie, E. C. Mar, and M. Smith. 1989. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63:3878-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kieff, E., and A. B. Rickinson. 2007. Epstein-Barr virus and its replication, p. 2602-2654. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 45.Kutok, J. L., and F. Wang. 2006. Spectrum of Epstein-Barr virus-associated diseases. Annu. Rev. Pathol. 1:375-404. [DOI] [PubMed] [Google Scholar]
- 46.Lee, Y. H., Y. F. Chiu, W. H. Wang, L. K. Chang, and S. T. Liu. 2008. Activation of the ERK signal transduction pathway by Epstein-Barr virus immediate-early protein Rta. J. Gen. Virol. 89:2437-2446. [DOI] [PubMed] [Google Scholar]
- 47.Liu, C., N. D. Sista, and J. S. Pagano. 1996. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J. Virol. 70:2545-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, P., and S. H. Speck. 2003. Synergistic autoactivation of the Epstein-Barr virus immediate-early BRLF1 promoter by Rta and Zta. Virology 310:199-206. [DOI] [PubMed] [Google Scholar]
- 49.Lu, C. C., Y. Y. Jeng, C. H. Tsai, M. Y. Liu, S. W. Yeh, T. Y. Hsu, and M. R. Chen. 2006. Genome-wide transcription program and expression of the Rta responsive gene of Epstein-Barr virus. Virology 345:358-372. [DOI] [PubMed] [Google Scholar]
- 50.Manet, E., A. Rigolet, H. Gruffat, J. F. Giot, and A. Sergeant. 1991. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 19:2661-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maruo, S., E. Johannsen, D. Illanes, A. Cooper, and E. Kieff. 2003. Epstein-Barr virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J. Virol. 77:10437-10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller, G., A. El-Guindy, J. Countryman, J. Ye, and L. Gradoville. 2007. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv. Cancer Res. 97:81-109. [DOI] [PubMed] [Google Scholar]
- 53.Miller, I. G., Jr., and A. El-Guindy. 2002. Regulation of Epstein-Barr virus lytic cycle activation in malignant and nonmalignant disease. J. Natl. Cancer Inst. 94:1733-1735. [DOI] [PubMed] [Google Scholar]
- 54.Minarovits, J., S. Minarovits-Kormuta, B. Ehlin-Henriksson, K. Falk, G. Klein, and I. Ernberg. 1991. Host cell phenotype-dependent methylation patterns of Epstein-Barr virus DNA. J. Gen. Virol. 72:1591-1599. [DOI] [PubMed] [Google Scholar]
- 55.Moore, S. M., J. S. Cannon, Y. C. Tanhehco, F. M. Hamzeh, and R. F. Ambinder. 2001. Induction of Epstein-Barr virus kinases to sensitize tumor cells to nucleoside analogues. Antimicrob. Agents Chemother. 45:2082-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4:192-201. [DOI] [PubMed] [Google Scholar]
- 57.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 58.Palombella, V. J., O. J. Rando, A. L. Goldberg, and T. Maniatis. 1994. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell 78:773-785. [DOI] [PubMed] [Google Scholar]
- 59.Parker, B. D., A. Bankier, S. Satchwell, B. Barrell, and P. J. Farrell. 1990. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology 179:339-346. [DOI] [PubMed] [Google Scholar]
- 60.Quinlivan, E. B., E. A. Holley-Guthrie, M. Norris, D. Gutsch, S. L. Bachenheimer, and S. C. Kenney. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21:1999-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raab-Traub, N., T. Dambaugh, and E. Kieff. 1980. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell 22:257-267. [DOI] [PubMed] [Google Scholar]
- 62.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ragoczy, T., and G. Miller. 2001. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J. Virol. 75:5240-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ragoczy, T., and G. Miller. 1999. Role of the Epstein-Barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J. Virol. 73:9858-9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 66.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 67.Salghetti, S. E., M. Muratani, H. Wijnen, B. Futcher, and W. P. Tansey. 2000. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. U. S. A. 97:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 69.Tatham, M. H., M. S. Rodriguez, D. P. Xirodimas, and R. T. Hay. 2009. Detection of protein SUMOylation in vivo. Nat. Protoc. 4:1363-1371. [DOI] [PubMed] [Google Scholar]
- 70.Verger, A., J. Perdomo, and M. Crossley. 2003. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 4:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verma, D., C. Ling, E. Johannsen, T. Nagaraja, and S. Swaminathan. 2009. Negative autoregulation of Epstein-Barr virus (EBV) replicative gene expression by EBV SM protein. J. Virol. 83:8041-8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, L., E. Fossum, C. H. Joo, K. S. Inn, Y. C. Shin, E. Johannsen, L. M. Hutt-Fletcher, J. Hass, and J. U. Jung. 2009. Epstein-Barr virus LF2: an antagonist to type I interferon. J. Virol. 83:1140-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. U. S. A. 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]
- 75.Zhu, F. X., J. M. Chong, L. Wu, and Y. Yuan. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ziegler, E. C., and S. Ghosh. 2005. Regulating inducible transcription through controlled localization. Sci. STKE 2005:re6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.