Abstract
There is considerable HIV-1 variation in patients. The extent of the variation is due to the high rate of viral replication, the high viral load, and the errors made during viral replication. Mutations can arise from errors made either by host DNA-dependent RNA polymerase II or by HIV-1 reverse transcriptase (RT), but the relative contributions of these two enzymes to the mutation rate are unknown. In addition, mutations in RT can affect its fidelity, but the effect of mutations in RT on the nature of the mutations that arise in vivo is poorly understood. We have developed an efficient system, based on existing technology, to analyze the mutations that arise in an HIV-1 vector in a single cycle of replication. A lacZα reporter gene is used to identify viral DNAs that contain mutations which are analyzed by DNA sequencing. The forward mutation rate in this system is 1.4 × 10−5 mutations/bp/cycle, equivalent to the retroviral average. This rate is about 3-fold lower than previously reported for HIV-1 in vivo and is much lower than what has been reported for purified HIV-1 RT in vitro. Although the mutation rate was not affected by the orientation of lacZα, the sites favored for mutations (hot spots) in lacZα depended on which strand of lacZα was present in the viral RNA. The pattern of hot spots seen in lacZα in vivo did not match any of the published data obtained when purified RT was used to copy lacZα in vitro.
Human immunodeficiency virus type 1 (HIV-1) is genetically diverse both within and between infected individuals (6, 12, 66). The high rate of viral replication, the size of the viral population in individual patients (13, 25), and extensive recombination (28, 77) contribute to genetic variation (17, 18). However, the root cause of HIV-1 diversification is mutations that arise during retroviral replication. The ability to rapidly diversify allows HIV-1 to evade the host's immune system (51), alter its tropism, and develop resistance to antiretroviral drugs (31). Hence, the fidelity of replication contributes to the adaptability and fitness of the virus.
Mutations can occur at three stages of retroviral replication: (i) when viral RNA is transcribed from the provirus by host DNA-dependent RNA polymerase II (Pol II), (ii) when the single-stranded viral RNA genome is converted into double-stranded DNA (dsDNA) by viral reverse transcriptase (RT), or (iii) when the provirus is copied by the host DNA-dependent DNA polymerase when the infected cell replicates. Although the available in vitro data are incomplete, several groups have suggested that RT is the major cause of mutations made during HIV-1 replication (32, 46, 53, 59). RT lacks exonucleolytic proofreading activity, and in vitro studies have suggested that it is error-prone (discussed below) (4, 30, 59, 62). The cellular DNA replication machinery has a much higher fidelity, with a mutation rate between 1 × 10−9 and 1 × 10−12 mutations/bp/cycle, which implies that the host DNA polymerases make a negligible contribution to the errors that arise during active HIV-1 replication (36, 75). Although the contribution of the errors made by host RNA Pol II to the fidelity of HIV-1 replication is not known, it could be significant (19, 32). Experiments with a reporter gene inserted into a retroviral long terminal repeat (LTR) have shown that approximately two-thirds of mutations occur either during RNA transcription or during first (minus)-strand viral DNA synthesis (32, 53). However, these data do not differentiate between the errors made by RNA Pol II and those made by RT. Host factors can also contribute to the overall error rate of HIV-1. In the absence of HIV-1 Vif, APOBEC3G (A3G) can cause extensive G-to-A mutations by deaminating cytidines in the minus strand of the viral DNA (27). It is also possible that host DNA repair enzymes cause mutations in the newly synthesized viral DNA (7, 60).
A number of groups have measured the fidelity of HIV-1 RT in vitro by copying a DNA or an RNA template encoding a target in which mutations can easily be identified, most commonly the α-complementing peptide of Escherichia coli β- galactosidase (lacZα). These assays have identified RT residues involved in nucleotide discrimination, binding, and incorporation (4, 9, 39, 65, 68). In vitro, HIV-1 RT has a high rate of nucleotide misincorporation, in the range of 5.9 × 10−4 to 5.3 × 10−5 mutations/bp/cycle (reviewed in references 61 and 69). Despite the fact that many groups have used the same mutagenic target (lacZα), different error rates have been reported, and there has been little agreement on the proportions and frequencies of the specific types of mutations RT makes (4, 8, 9, 30, 68). A part of the problem is that HIV-1 RT fidelity in vitro depends both on the recombinant RT used and on the assay conditions (39, 48). It has also been reported that recombinant HIV-1 RT is more error-prone than the RTs of other retroviruses (1, 2, 63, 68, 70). Together, these data have led to proposals that the low fidelity of RT makes HIV-1 replication particularly error-prone (50, 59).
In vivo (cell culture) systems that include a number of the viral and host components that contribute to the fidelity of HIV-1 replication have also been developed. Using an HIV-1 vector containing a lacZα reporter gene, Mansky and Temin reported a mutation rate of 3.4 × 10−5 to 4 × 10−5 mutations/bp/cycle (42, 46), some 10- to 20-fold lower than the rates reported for purified HIV-1 RT in vitro (9, 39, 62, 68) but within 3-fold of the retroviral average of 1.5 × 10−5 mutations/bp/cycle (46, 47, 54-56). To date, most of the measurements of HIV-1 fidelity in cell culture have been based solely on the frequency of phenotypic inactivation of LacZα; there has been relatively little information about the nature of the underlying mutations (15, 43-45). Thus, little is known about the specificity of the errors made in vivo (46).
We used the inactivation of a lacZα reporter gene to identify mutants generated in a single cycle of HIV-1 replication in cultured cells; however, we also determined the nature, position, and frequency of the underlying mutations. Our HIV-1 vector was designed so that circular forms of the HIV-1 DNA can replicate as plasmids in E. coli. More than 4 × 104 independent replicated copies of the genome of an HIV-1 vector can be recovered from a single plate of infected HOS cells, a nearly 20-fold improvement (42, 46, 47, 55, 56). Individual colonies were screened for the presence of functional LacZα, and mutant HIV lacZα DNAs were sequenced. We characterized the mutations that arose in both the forward and reverse orientations of the lacZα reporter gene. The orientation of lacZα affected the types and positions of mutations but not the rate, suggesting that the nature of the mutations depends on the strand present in viral RNA.
MATERIALS AND METHODS
Plasmid construction.
Two different vector-based systems were used to generate virus stocks for the analysis of HIV-1 fidelity. A NotI-MluI shuttle cassette, derived from pHIV-SH (52), was modified for use in both vector systems. To eliminate redundancy and to shorten the cassette, both the Pol II promoter and the lacO sequence were deleted, and a 477-bp sequence containing lacZα derived from Litmus-29 (New England Biolabs, Ipswich, MA) was inserted in place of lacO. The final cassette contained a zeocin resistance gene with an upstream EM-7 promoter (EM-zeor), a lacZα sequence (lacZα), and a ColE1 origin of replication (oriE). Two versions of the shuttle cassette were created, with the lacZα sequence in opposite orientations: the forward (lacZα-F, LZF) and reverse (lacZα-R, LZR) orientations (Fig. 1A).
FIG. 1.
Vector-based strategies used to measure HIV-1 replication fidelity. (A) Shuttle cassette. EM-zeor, EM-7 promoter zeocin resistance gene; lacZα-F and lacZα-R, α-complementing peptides of E. coli β-galactosidase in the forward and reverse orientations, respectively; ColE1 (oriE), E. coli bacterial origin of replication. Shuttle cassettes carrying lacZα as a mutational target in both orientations were cloned into the indicated vectors (see below), creating the substrates for transcription by RNA Pol II, RNA packaging, reverse transcription, and circularization of unintegrated viral lacZα DNA. (B and C) The 2-vector and 4-vector systems. The plasmids were transfected into 293T cells to produce pseudotyped HIV-1 viruses which were used to infect HOS cells. In panel B, on the left is shown the viral RNA-encoding vector plasmid for the 2-vector system. Two shaded boxes separated by an FspI site represent the HIV-1 long terminal repeats (LTRs). Retroviral genes are indicated in rectangular boxes as gag, pol, and env. On the right is shown the VSV-G expression plasmid, which contains a CMV promoter. In the 4-vector system, shown in panel C, the viral RNA-encoding vector plasmid does not contain the gag and pol genes, which are expressed from a second plasmid that contains a CMV promoter. The Rev protein is also expressed from a separate plasmid, but from a Rous sarcoma virus (RSV) promoter. In both the 2-vector and 4-vector systems, the viral vector plasmid has a nonviral sequence between the 3′ LTR and the 5′ LTR; this insert is not present in replicated viral DNAs. Both of the viral vector systems are limited to a single round of replication by an HIV-1 Env deletion. An integrase inactivating mutation, D116N, was used to increase the amount of replicated circular viral lacZα DNA present in HOS cells infected with these vectors. WRE, woodchuck regulatory element.
The 2-vector system (Fig. 1B) included either pNLZeoIN-R-E-.LZF or pNLZeoIN-R-E-.LZR and pCMV-VSV-G (Clontech Laboratories, Mountain View, CA) to pseudotype virions with vesicular stomatitis virus G (VSV-G) envelope glycoprotein. The replication-defective HIV-1 vector used to produce viral RNA, pNLZeoIN-R-E-.LZF or pNLZeoIN-R-E-.LZR, contained deletions in vpr (R-) and env (E-) and was derived from pHIV1-SH (52) by replacement of the NotI-MluI shuttle cassette in the nef reading frame. To facilitate recovery of unintegrated circular viral DNAs, an active site mutation (D116N) was also introduced into integrase (IN-) by using a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA).
The 4-vector system included pMDL-SH.IN-, pRSV-REV, pCMV-VSV-G, and either pSICO-LZF or pSICO-LZR. The pMDL-SH.IN- vector, which encodes HIV-1 Gag and Pol and contains a Rev-response element (RRE), was derived from pMDLg/pRRE by replacing the gag and pol genes with the equivalent sequences from pHIV1-SH (52), followed by the introduction of an active site mutation (D116N) in integrase (IN-), as described above. Both the pRSV-REV and pMDLg/pRRE lentiviral vectors were obtained from Didier Trono (EPFL SV-DO, Lausanne, Switzerland) (21) through Addgene, Inc. (Cambridge, MA). The vector used to produce viral RNA, pSICO-LZF or pSICO-LZR, was derived from pSICO-XBX (74) by introduction of our shuttle cassette as a NotI-MluI fragment and contained a chimeric 5′ LTR with a cytomegalovirus (CMV) promoter in place of U3 and an HIV Psi packaging/RRE RNA export signal.
Cells, transfection, and infection.
HEK (human embryonic kidney) 293T and HOS (human osteosarcoma) cells were maintained in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 5% (vol/vol) fetal bovine serum, 5% (vol/vol) newborn calf serum, 100 μg/ml penicillin G, and 100 μg/ml streptomycin (Quality Biological, Gaithersburg, MD). Recombinant virus stocks were generated by calcium phosphate-mediated cotransfection of 293T cells seeded at 9 × 105 cells in 100-mm culture plates by using either the 2-vector- or the 4-vector-based system: (i) 2-vector system-derived viral particles were produced by cotransfection with 15 μg pNLZeoIN-R-E.LZF (or pNLZeoIN-R-E.LZR) and 4 μg pCMV-VSV-G, and (ii) 4-vector system-derived viral particles were produced by cotransfection with 10 μg pMDL-SH.IN-, 5 μg pRSV-REV, 15 μg pSICO-LZF (or pSICO-LZR), and 4 μg pCMV-VSV-G per plate. Virus-containing culture supernatants were harvested 48 h posttransfection, clarified by filtration through a 50-ml Steriflip 0.22-μm-pore-size filter unit (Millipore, Billerica, MA), and stored at −80°C until use. The amount of recombinant virus was determined using HIV-1 p24 antigen enzyme-linked immunosorbent assay kits, obtained from PerkinElmer (Boston, MA). On the day of infection, virus stocks were thawed and treated with 100 U DNase I (Invitrogen, Carlsbad, CA) plus 5 mM MgCl2 for 1 h at 37°C to remove any vector DNA carried over from the transfection. Recombinant virus equivalent to 500 ng of p24 was then added to 150-mm culture plates along with 8 μg/ml polybrene (Sigma-Aldrich, St. Louis, MO), followed by the addition of 2 × 106 HOS cells per plate.
Recovery of unintegrated HIV lacZα DNAs.
Circularized, unintegrated HIV lacZα DNAs were recovered from infected HOS cells 48 h postinfection by use of a modified Hirt extraction procedure. Briefly, trypsinized HOS cells were resuspended in 125 μl of P1 buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 100 μg/ml RNase A). Cells were lysed for 5 min at 25°C with 125 μl P2 buffer (200 mM NaOH, 1% SDS), gently mixed with 175 μl P3 buffer (3 M potassium acetate [pH 5.5]), and placed on ice for 5 min. Lysates were then centrifuged (16,000 × g, 20 min, 4°C) to separate soluble and insoluble fractions, and the supernatants were clarified by a second centrifugation step. The supernatants were extracted with phenol-chloroform-isoamyl alcohol (25:24:1) (pH 8.1) and chloroform-isoamyl alcohol (24:1) (pH 8.1) and then adjusted to a final concentration of 0.3 M sodium acetate. Viral DNA was recovered by ethanol precipitation and centrifugation (16,000 × g, 60 min, 4°C, washed with 70% ethanol), and the pellets were resuspended in 90 μl 10 mM Tris-Cl (pH 8.0). Any residual vector DNA was digested with 30 U DpnI for 2 h at 37°C, and the replicated viral DNA was recovered by phenol-chloroform extraction and ethanol precipitation with 1 vol 5 M ammonium acetate.
The presence of intact and mutant lacZα in the recovered viral DNA was determined by cloning the DNA directly into an E. coli strain that contained the ω-complementing segment of β-galactosidase. In brief, the viral DNAs were resuspended in 6 μl of double-distilled water (ddH2O), and 1 μl was electroporated into 45 μl of ElectroMax E. coli DH10B bacterial cells (Invitrogen, Carlsbad, CA) by using a BTX Electro cell manipulator 600 (Biotechnologies and Experimental Research, Inc., San Diego, CA). Recipient E. coli cells were subjected to a single 7.5-ms pulse (field strength, 1.5 kV/cm; capacitance, 25 μF; resistance, 186 Ω) by using a 1-mm-gap E-Shot Standard electroporation cuvette (Invitrogen, Carlsbad, CA) at room temperature. To prevent clonal expansion, transformed E. coli was allowed to recover for only 10 min prior to plating onto 15-cm-diameter low-salt Luria-Bertani plates containing 100 mM IPTG (isopropyl-β-d-thiogalactopyranoside), 50 μg/ml zeocin, and 250 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (InVivoGen, San Diego, CA). The colonies were allowed to grow for 24 h at 37°C, and the β-galactosidase activity was monitored by the color of the colonies: white to light blue (inactive/reduced activity) or blue (fully active) was scored using an ECount electronic colony counter (Heathrow Scientific LLC, Vernon Hills, IL). Individual mutant colonies were inoculated into 96-well blocks containing 1.5 ml low-salt Luria-Bertani medium containing 50 μg/ml zeocin and cultured for 48 to 72 h at 37°C. Plasmid DNA was extracted using a Biorobot 3000 instrument (Qiagen, Valencia, CA), and the lacZα coding region was sequenced (Macrogen USA Inc., Rockville, MD). Restriction enzyme digestion of plasmids recovered from 192 clones showed that less than 1% of the recovered plasmids were derived from vector DNA carried over from transfected 293T cells. Transformation of vector plasmid pSICO-LZF into DH10B yielded only a small fraction of white to light-blue colonies (14 out of 206,400), suggesting that the transformation and growth of the E. coli cells made an insignificant contribution to the mutations obtained in lacZα.
Classification of mutations and compilation of lacZα mutation spectra.
Sequenced viral DNA products encoding the entire full-length LacZα fusion protein (477 nucleotides [nt]) were compiled by alignment and separated from aberrant deleted products by using Sequencher v4.1.4 software (Gene Codes Corporation, Ann Arbor, MI). Pairwise blast analysis of each recovered lacZα sequence and the reference wild-type sequence (174 nt) was conducted using the command line BLAST program blastall (http://www.ncbi.nlm.nih.gov/staff/tao/URLAPI/blastall/) installed on a Linux computer. A Perl script was written to parse the alignment and sort the lacZα sequences into separate classes according to the types of mutations detected: class 1, single nucleotide substitutions; class 2, single nucleotide frameshifts (+1/−1); class 3, multiple nucleotide substitutions, including spaced singlets and doublets (cutoff of two consecutive bases); and class 4, indel (insertion/deletion) mutations, including deletions, deletions with insertions, duplications, and multiples of single nucleotide substitutions plus frameshifts (cutoff of 3 or more consecutive bases). Mutation spectra were compiled from the sequences in each mutation class to summarize the total number, types, and positions of errors detected within the 174-nt lacZα target sequence. Frameshifts and transition/transversion substitutions were tabulated for each type of mutation.
To validate the nature of class 3 multiple nucleotide substitutions, 414 nearly complete HIV-1 subtype B genomes (1 per patient) were retrieved from the Los Alamos HIV database (http://www.hiv.lanl.gov/) and compiled by alignment. Using B.FR.1983.HXB2-LAI-IIIB-BRU as a reference, a Perl script was written to assess the fraction of multiple A-to-G substitutions compared to multiple C-to-T substitutions within each sequence.
Determination of mutation frequencies and rates.
In the vectors we used, LacZα was expressed as part of a fusion protein derived from a 477-nt region of Litmus-29 (New England Biolabs, Ipswich, MA). This 477-nt region was comprised of a regulatory region (108 nt: CAP site, promoter, lac operator, and ribosome binding site), the first 5 codons of lacZα (15 nt), a multiple cloning site/polylinker region (180 nt), and 58 codons of lacZα (174 nt). Although the actual ATG start site was upstream of the polylinker, the total length of the mutational lacZα target sequence was defined as the 174 nt including the first glycine GGA codon after the polylinker to the first TAA termination codon. This 174-nt region is consistent with the lacZα targets previously used to measure HIV-1 RT fidelity (4, 9, 46). Because the polylinker region of the LacZα fusion protein makes no functional contribution to LacZα activity, mutations detected in this region (typically termination codons and indels) were excluded from analysis of the overall mutation rate. The mutation frequency was determined by dividing the number of mutational events by the total number of colonies screened. The mutation rate was determined by dividing the mutation frequency by the length of the lacZα target (174 nt). Because some of the mutations are silent, this strategy necessarily underestimates the actual mutation rate. A small number of mutations which did not alter the protein sequence (typically third base codon positions) was detected; these were invariably found with another mutation that did alter the protein sequence.
Comparative analysis of lacZα-F and lacZα-R mutation spectra.
Data were analyzed by log linear categorical analysis, contingency table analysis, and related methods. The number of mutations in the 174-nt sequence of lacZα was detailed in 2 × 174-cell tables for each pairwise comparison of lacZα mutational profiles. These tables were further collapsed, to include only those sites that contained nonzero counts (incidences) of mutations, to 2 × n tables in which n was typically less than 174. A likelihood-ratio chi-square (LR chi-square) test was performed to ascertain whether counts across each pair of profiles compared were homogeneous. This provided a conservative test to determine whether a statistically significant but perhaps spurious event(s) might be present. Positions where there were frequent mutations (hot spots) were thus identified by a “search and discovery” strategy, in which contingency tables were subsequently decomposed into orthogonal components to permit a series of independent LR chi-square tests. Prospective hot spots were then subjected to global Fisher's exact tests to determine statistical significance. Compounding type I error rates were controlled through use of the Bonferroni correction. To that end, minimal criterion probability levels of at least 0.01 were obtained. Statistical significance of hot spot differences between lacZα-F and lacZα-R mutation spectra is reported at levels of significance of 0.01, 0.001, and 0.0001.
RNA secondary-structure predictions.
RNA secondary-structure predictions were made using mfold v3.2 software (http://www.bioinfo.rpi.edu/applications/mfold), based on algorithms and methodologies originally published by Michael Zuker (Institute for Biomedical Computing, Washington University, St. Louis, MO) (78). Conditions were fixed by the program at 1 M NaCl and 37°C.
RESULTS
Vector-based measurement of HIV-1 replication fidelity.
To accurately assess the nature and position of errors made during viral replication, we compared two vector expression systems designed to facilitate the recovery of large numbers of replicated HIV-1 vector DNAs carrying a copy of a lacZα reporter that can be expressed in E. coli (Fig. 1B and C). Both vectors contain the same shuttle cassettes (Fig. 1A) comprised of the mutational target lacZα, the zeocin resistance gene (zeor) for selection, and a bacterial origin of replication (oriE). A pair of vectors with the forward (lacZα-F, LZF) or reverse (lacZα-R, LZR) orientation of the lacZα gene were prepared for each expression system to generate virus particles with complementary strands of lacZα in their viral RNA. The 2-vector system includes the HIV-1 vector pNLZeoIN-R-E-.LZF/R (Fig. 1B), containing the lacZα shuttle cassette in the nef region, and a VSV-G envelope expression plasmid. The reporter cassette is in different contexts in the 2 systems, and we wanted to ask whether this affected the nature of the mutations or the error rate. In the 4-vector system, the HIV-1 proteins are expressed from two different plasmids (pMDL-SH.IN- and pRSV-REV), and the lacZα RNA-expressing vector (pSICO-LZF/R) is smaller than the corresponding pNLZeoIN-R-E-.LZF/R HIV-1 vector. The smaller genome may have the advantage that it could permit RT mutants to make more complete copies of the vector DNA (Fig. 1C). Both vector systems carry out a single cycle of HIV-1 replication. The 2-vector system contains an Env deletion and is Vif+, while the 4-vector system does not express either Env or Vif. HIV-1 viral stocks pseudotyped with VSV-G were produced in APOBEC3G-negative 293T human kidney cells (26, 58). An inactivating mutation in integrase (D116N) was used to increase the amount of unintegrated circular viral DNA, facilitating recovery from infected HOS cells. The circular viral DNAs were electroporated into E. coli DH10B cells and scored for mutations in lacZα by screening for white or light-blue bacterial colonies on X-Gal plates.
Recovery and analysis of phenotypic mutant HIV lacZα DNAs.
HOS cells were transduced with virus from either vector system at approximately 0.2 pg p24/cell. This typically yielded a total of approximately 4 × 104 E. coli colonies from a single 15-cm plate of infected HOS cells. The 4-vector system required about twice as many viral particles as the 2-vector system to produce approximately the same number of E. coli colonies (data not shown). The frequencies of LacZα inactivation in the forward and reverse orientations were first determined by counting the number of white and light-blue colonies. Of the 172,906 LZF bacterial colonies recovered from HOS cells infected with 4-vector system-derived virus, 776 (0.45%) were white or light blue (Table 1). A similar frequency (0.44%) was seen with lacZα in the reverse (LZR) orientation. The frequency of LacZα inactivation was slightly higher for the 2-vector system (Table 1): the forward orientation of lacZα (LZF) was inactivated at a rate of 0.50% (537 out of 106,940), while the reverse orientation of lacZα (LZR) was inactivated at a rate of 0.77% (450 out of 58,672). Although the average levels of LacZα phenotypic inactivation were similar for both vector systems, only about one-half of the mutant lacZα DNAs could be used to determine the mutation rate in the forward or reverse lacZα orientation. As described in Materials and Methods, LacZα was expressed as part of a fusion protein derived from a 477-nt region of Litmus-29. The C-terminal part of this fusion protein is the LacZα peptide (174 nt), while the N-terminal portion is derived from regulatory (105-nt) and polylinker (195-nt) regions and makes no functional contribution to the activity of LacZα. Although the entire 477-nt sequence of the inactive lacZα DNAs was aligned to the reference sequence, only the errors in the 174-nt target region were used in the fidelity analysis. Based on the number of mutations in the 174-nt lacZα target region, the frequencies of lacZα mutations were similar for the forward lacZα orientation (0.25% versus 0.22%; P = 0.086) and the reverse lacZα orientation (0.22% versus 0.18%; P = 0.12) for both the 4-vector and 2-vector systems. These results indicated that either vector system can be used to measure HIV-1 replication fidelity and show that the context of the reporter cassette is not a critical factor.
TABLE 1.
Effect of the vector on LacZα phenotypic inactivation frequencies
| Vector system | lacZα orientation | Recoverya |
Analysisb |
Final interpretationc |
|||
|---|---|---|---|---|---|---|---|
| No. of mutant colonies/total no. of recovered colonies | Frequency (%) of lacZα inactivation | No. of aligned lacZα DNAs/no. of mutant colonies | Relative fraction (%) | No. of mutant lacZα DNAs/total no. of recovered colonies | Frequency (%) of lacZα inactivation | ||
| 4-Vector | LZF | 776/172,906 | 0.45 | 432/776 | 56 | 432/172,906 | 0.25 |
| LZR | 687/155,904 | 0.44 | 340/687 | 49 | 340/155,904 | 0.22 | |
| 2-Vector | LZF | 537/106,940 | 0.50 | 232/537 | 43 | 232/106,940 | 0.22 |
| LZR | 450/58,672 | 0.77 | 107/450 | 24 | 107/58,672 | 0.18 | |
The number of mutant colonies (white or light blue) was divided by the total number of colonies recovered × 100 to determine the frequency of LacZα phenotypic inactivation.
The number of recovered lacZα DNAs that aligned with the reference sequence (477 nt) was divided by the number of mutant colonies. The number of lacZα DNAs with a mutation(s) in the lacZα target sequence (174 nt) was divided by the number of recovered lacZα DNAs that aligned with the reference sequence (477 nt).
The number of lacZα DNAs with a scorable mutation(s) in the lacZα target sequence (174 nt) was divided by the corrected total number of recovered colonies × 100 to determine the actual frequency of lacZα mutations.
Effect of lacZα orientation on the types of in vivo mutations.
Although the integrated HIV-1 genome is double-stranded DNA, only one strand is copied into viral RNA by the host DNA-dependent RNA Pol II. This also means that RT uses different templates to synthesize the two strands of the viral DNA, the minus strand from RNA and the plus strand from DNA. Our assay does not discriminate between errors made by RNA Pol II and those made by RT. Because the mutations were recovered after one round of viral DNA synthesis by transforming a multicopy plasmid into E. coli, the assay may preferentially detect mutations generated during minus-strand DNA synthesis compared to those made during plus-strand DNA synthesis (for a more complete explanation, see Text S1 in the supplemental material). Experiments with a spleen necrosis virus (SNV) vector (in which mutations were scored directly in cultured eukaryotic cells) showed that approximately two-thirds of the observed mutations were generated either by Pol II or during minus-strand DNA synthesis (32, 57).
We compared all of the mutations that arose in the forward and reverse orientations of lacZα (see Fig. 2 to 5 and Tables 2 and 3). To simplify the comparison, the data presented in the main text were all derived from the 4-vector system. Similar results were obtained using the 2-vector system (see Tables S1 and S2 and Fig. S1 to S4 in the supplemental material). As described in Materials and Methods, the lacZα DNA mutations were divided into classes according to the types of mutations detected. Single nucleotide substitution errors (class 1) were the most frequent (75 to 83% of the total) (Table 2). Single nucleotide frameshifts (class 2) accounted for 5 to 6% of mutations. We found multiple nucleotide substitutions (class 3) in another 9 to 16% of mutant lacZα DNA sequences, which, if they were counted independently, would have represented 34 to 46% of the total mutations detected. The final type of mutations detected (class 4) were indel mutations (insertions, deletions, and deletions with insertions), comprising 3 to 4% of lacZα mutations.
FIG. 2.
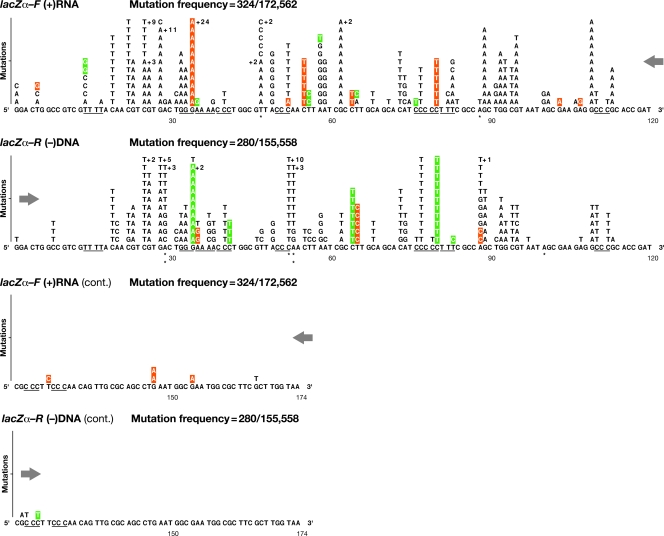
Class 1 mutations: single nucleotide substitutions detected in lacZα (4-vector system). The numbers, types, and locations of the independent substitution errors are shown for both the forward lacZα orientation (plus-strand nucleotide sequence) and the reverse lacZα orientation (negative-strand nucleotide sequence). Opposing directional arrows indicate the actual sequence context and direction of minus-strand DNA synthesis during reverse transcription. The total length of the lacZα target sequence was defined as 174 nt, representing codons 6 to 63 from GGA to the first TAA termination codon. Single nucleotide substitution errors are shown as letters above the original wild-type template sequence, limited to 11 per position, with additional errors indicated by +n. Runs of 3 or more identical nucleotides are underlined. Misalignment/slippage of the primer or template strand that could result in a substitution error is highlighted in orange or green, respectively. Mutational hot spots for which there are significant differences in the forward and reverse lacZα orientations are indicated by asterisks: *, P < 0.01; **, P < 0.001; ***, P < 0.0001 (see Materials and Methods). The mutation frequency is the number of lacZα DNAs with a mutation(s) divided by the total number of adjusted recovered clones.
TABLE 2.
Orientation of lacZα does not affect HIV-1 mutation rate (4-vector system)
| lacZα orientation and mutation type(s) | Overall totald |
Class 1 |
Class 2 |
Class 3 |
Class 4 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation frequencya | Mutation rateb | Mutation frequency | Mutation rate | Mutation fractionc (%) | Mutation frequency | Mutation rate | Mutation fraction (%) | Mutation frequency | Mutation rate | Mutation fraction (%) | Mutation frequency | Mutation rate | Mutation fraction (%) | |
| LZF | ||||||||||||||
| All − class 3 | 361/172,562 | 1.2 × 10−5 | 324/172,562 | 1.1 × 10−5 | 90 | 26/172,562 | 8.6 × 10−7 | 7 | 11/172,562 | 3.6 × 10−7 | 3 | |||
| All + class 3 dep. | 432/172,562 | 1.4 × 10−5 | 75 | 6 | 71/172,562 | 2.4 × 10−6 | 16 | 3 | ||||||
| A-to-G substitution | 64/172,562 | 2.1 × 10−6 | 90 | |||||||||||
| Others | 7/172,562 | 2.3 × 10−7 | 10 | |||||||||||
| All + class 3 indep. | 669/172,562 | 2.2 × 10−5 | 48 | 4 | 308/172,562 | 1.0 × 10−5 | 46 | 2 | ||||||
| A-to-G substitution | 289/172,562 | 9.6 × 10−6 | 94 | |||||||||||
| Others | 19/172,562 | 6.3 × 10−7 | 6 | |||||||||||
| LZR | ||||||||||||||
| All − class 3 | 309/155,558 | 1.1 × 10−5 | 280/155,558 | 1.0 × 10−5 | 91 | 16/155,558 | 5.9 × 10−7 | 5 | 13/155,558 | 4.8 × 10−7 | 4 | |||
| All + class 3 dep. | 340/155,558 | 1.3 × 10−5 | 83 | 5 | 31/155,558 | 1.1 × 10−6 | 9 | 4 | ||||||
| A-to-G substitution | 25/155,558 | 9.2 × 10−7 | 81 | |||||||||||
| Others | 6/155,558 | 2.2 × 10−7 | 19 | |||||||||||
| All + class 3 indep. | 471/155,558 | 1.7 × 10−5 | 60 | 3 | 162/155,558 | 6.0 × 10−6 | 34 | 3 | ||||||
| A-to-G substitution | 150/155,558 | 5.5 × 10−6 | 93 | |||||||||||
| Others | 12/155,558 | 4.4 × 10−7 | 7 | |||||||||||
The number of mutations was divided by the total number of adjusted recovered clones (adjusted to exclude the number of inactive clones that did not align with the reference sequence or that contained mutations outside the 174-nt lacZα target). Mutations are represented as dependent (related) or independent (separate) errors.
The mutation rate is the mutation frequency divided by the size of the lacZα target sequence (174 nt), expressed as mutations/bp/cycle.
The mutation fraction is the number of mutations in a particular class, expressed as a percentage.
The overall total mutation frequencies and rates were calculated as the sum of the frequencies and rates from the individual classes of mutations (classes 1 to 4). Three approaches were used to determine overall mutation rates of HIV-1, as follows. All − class 3, all of the mutations except class 3 were included because the class 3 multiple mutations might not have been made by host RNA Pol II or RT; class 4 indels were counted as dependent errors (single mutational events). All + class 3 dep., all of the mutations, including the class 3 multiple errors, were counted; class 3 and class 4 mutations were counted as dependent errors (single mutational events). All + class 3 indep., all of the mutations were included; class 3 multiple errors were counted as independent mutations and class 4 as dependent mutations.
TABLE 3.
Nucleotide substitution and frameshift errors in lacZα (4-vector system)a
The numbers and types of mutations in the forward and reverse orientations of lacZα (shown in the plus-strand nucleotide sequence) are given in the upper and lower panels, respectively, for class 1, single nucleotide substitutions (A); class 2, single nucleotide frameshifts (B); and class 3, multiple nucleotide substitutions (C). The original wild-type plus-strand template nucleotides are given on the left side of each panel, with the corresponding substitutions along the top. Transition and transversion substitutions are highlighted in yellow and green, respectively. Single nucleotide addition [(+)1 f/s] and deletion [(−)1 f/s] frameshift mutations are highlighted along the bottom in pink and blue, respectively. The relative percentage of each type of error was determined by dividing the number of each type of mutation by the total number of mutations detected. Total lacZα, total number of mutant lacZα DNA sequences scored.
We first compared the frequencies of the specific types of errors generated in the forward and reverse lacZα orientations (Table 3A to C). To facilitate a comparison of the errors, the inverse complements of the reverse lacZα sequence data are presented in Fig. 2 to 5. For each class of mutation, the positions of mutational hot spots were compiled, both for a particular lacZα orientation (intragenic) and between the lacZα-F and lacZα-R orientations (intergenic), based on the relative frequency of occurrence and the sample size. Successive runs of 3 or more nucleotides are underlined in the figures (10 in total), and dinucleotide runs (29 in total) are indicated.
Effect of lacZα orientation on the mutations arising in vivo. (i) Class 1: single nucleotide substitutions.
A total of 324 and 280 independent single nucleotide substitutions were detected in the forward and reverse lacZα orientations, respectively. These mutations occurred at 61 and 56 positions of the 174-nt target, respectively (Fig. 2). Although there was more than one type of substitution at some locations, the overall numbers of different substitutions were similar in both orientations (LZF, 82; LZR, 81; P = 0.36). Nearly all of the single nucleotide substitutions (99%) resulted in a change in an amino acid codon within the 174-nt lacZα open reading frame. Those substitutions which resulted in silent changes (<1%) were invariably linked to a substitution in the polylinker region (LZF, 3 of 324 at 2 sites; LZR, 1 of 280 at 1 site). Based on these data, there are at least 71 positions at which single nucleotide mutations can be detected and scored within the 174-nt lacZα target (Table 4).
TABLE 4.
Positions in lacZα where mutations were detected (4-vector system)
| lacZα orientation | Categorya | No. of nucleotide template positions |
|||
|---|---|---|---|---|---|
| Class 1 | Class 2 (independent) | Class 3 | Class 4 (dependent) | ||
| LZF | Detected | 61 | 9 | 44 | 174 |
| Scorable | 59 | 9 | 30 (14) | 174 | |
| LZR | Detected | 56 | 8 | 29 | 174 |
| Scorable | 55 | 8 | 19 (8) | 174 | |
| LZF and LZR | Identical | 43 | 5 | 3 | 174 |
| Mutatable | 71 | 12 | 46 | 174 | |
Detected, the number of independent nucleotide template positions in lacZα where mutations were detected; scorable, the number of positions where a mutation caused a change in an amino acid (the number of unique sites is shown in parentheses); identical, the number of “scorable” positions in lacZα that were identical in both orientations; mutatable, the minimal number of “scorable” positions in lacZα, as determined from the combined data for lacZα-F and lacZα-R.
Transition mutations comprised 76 to 85% of the single nucleotide substitutions (LZF, 274 of 324; LZR, 212 of 280). The frequencies of transition and transversion substitutions differed considerably in the two lacZα orientations (P = 0.0074). Consistent with genomic analyses of HIV-1 from infected patients (38, 71, 72), we detected twice as many G-to-A as C-to-T substitutions (Table 3A). The remaining 15 to 24% of the single nucleotide substitutions were transversions (LZF, 50 of 324; LZR, 68 of 280), primarily C to A and T to A. Although the target sequence contained nearly equal proportions of nucleotides (A, 36; C, 56; G, 45; T, 37), there were slightly more overall substitutions at C and G sites. Of the 61 positions in lacZα-F where mutations were detected, there were approximately twice as many C and G sites as A and T sites (A, 10; C, 19; G, 20; T, 12). Similar results were seen at the 56 positions where mutations were detected in the reverse lacZα orientation in the plus-strand nucleotide sequence (A, 7; C, 16; G, 20; T, 13).
For both lacZα orientations, there was a strong preference for mutations at specific sites (Fig. 2). The majority of single nucleotide substitutions were located within a 100-nt region of lacZα, from positions 14 to 114 (LZF, 313 of 324; LZR, 273 of 280). This is consistent with the minimum length of lacZα required for complementation (residues 3 to 41) (37). More than half of the sites within this segment were hot spots at which there were 3 or more of the same mutation (LZF, 28 of 61; LZR, 32 of 56). Only a small fraction (16 to 20%) of single nucleotide substitutions were detected near the ends of homopolymeric nucleotide runs (that is, either in the last nucleotide of a run or in the nucleotide immediately adjacent to a run) (LZF, 64 of 324; LZR, 44 of 280); the frequencies were similar for both lacZα orientations (P = 0.20). Of the mutations that occurred near a run, most occurred at or near the end of runs of 3 or more nucleotides (LZF, 46 of 324; LZR, 30 of 280) in the direction of minus-strand DNA synthesis. In most cases, the mutated nucleotide could be paired with the preceding adjacent template nucleotide in the lacZα-F orientation and the succeeding adjacent template nucleotide in the lacZα-R orientation (P < 0.001), suggesting that the mutations could have arisen by a slippage mechanism (see Discussion). Although the overall patterns and compilations of hot spots for the two complementary lacZα orientations appeared similar, there were clear intergenic differences in the hot spot positions. There were 15 positions with at least a 2-fold difference in the number of substituted nucleotides: 7 sites were favored in lacZα-F, and 8 sites were favored in lacZα-R. Statistically significant differences occurred at 5 of the 15 total sites: 3 positions in lacZα-F and 2 positions in lacZα-R (see the legend for Fig. 2 for the corresponding P values). These data show that the single substitution errors, and thus the majority of mutations, are affected by which strand of lacZα is expressed as viral RNA.
(ii) Class 2: single nucleotide frameshifts.
A total of 26 and 16 independent single nucleotide frameshift mutations were detected in the forward and reverse lacZα orientations, respectively. The frequencies of insertion and deletion frameshifts were surprisingly similar in the two lacZα orientations (P = 0.45). The majority were single nucleotide insertions (LZF, 22 of 26; LZR, 12 of 16) in which the inserted nucleotide could have base paired with the preceding adjacent template nucleotide in the direction of minus-strand DNA synthesis (Table 3B and Fig. 3). While single nucleotide frameshifts appeared to have occurred at 9 lacZα-F and 8 lacZα-R nucleotide template positions (Table 4), a large fraction (88 to 94%) were detected in 5 nucleotide runs (LZF, 21 of 26; LZR, 13 of 16). It is not possible to be certain where in a run a frameshift mutation occurs; for convenience, they are shown at the end of the run. The relative positions of frameshifts in either lacZα orientation revealed strong intragenic site preferences for runs of A's and T's compared to C's and G's. This result is similar to a previous report based on an in vitro assay with HIV-1 RT (11). Only one run in lacZα-F had a significantly different number of frameshift mutations compared to lacZα-R (P < 0.001).
FIG. 3.
Class 2 mutations: single nucleotide frameshifts detected in lacZα (4-vector system). Single nucleotide frameshift errors are indicated as triangles above the wild-type sequence: single nucleotide deletions (−1) are shown as upright triangles and single nucleotide additions (+1) as inverted triangles. Runs of 3 or more identical nucleotides are underlined. If a frameshift occurred within a run, its exact position within the run cannot be determined, and the deletions and additions are marked over the last nucleotide in the run in the direction of minus-strand DNA synthesis. Frameshift errors in nucleotide runs that could have arisen by a misalignment/slippage of the primer or template strand are highlighted in orange or green, respectively. Mutational hot spots for which there are significant differences in the forward and reverse lacZα orientations are indicated by asterisks: *, P < 0.01; **, P < 0.001; ***, P < 0.0001 (see Materials and Methods). The mutation frequency is the number of lacZα DNAs with a mutation(s) divided by the total number of adjusted recovered clones.
(iii) Class 3: multiple nucleotide substitutions.
Despite the relatively low overall mutation frequency, we recovered numerous lacZα DNAs containing two or more substitution mutations (71 lacZα-F and 31 lacZα-R DNAs). A total of 308 and 162 multiple nucleotide substitutions were detected in the forward and reverse lacZα orientations, at 44 and 30 nucleotide template positions, respectively (Fig. 4). Transitions accounted for nearly all multiple nucleotide substitutions in both lacZα orientations (LZF, 300 of 308; LZR, 157 of 162), a major fraction of which (94%) were A to G (LZF, 289 of 308; LZR, 150 of 162), (Table 3C). In contrast to class 1 single nucleotide substitutions, a significant proportion (32 to 52%) of the detected class 3 multiple nucleotide substitutions were silent changes (LZF, 100 of 308 at 14 sites; LZR, 84 of 162 at 10 sites). Nearly half of the remaining scorable class 3 substitutions occurred at the same positions as all forms of class 1 substitutions, leaving only 14 lacZα-F and 8 lacZα-R sites unique to class 3 (Table 4).
FIG. 4.
Class 3 mutations: multiple nucleotide substitutions detected in lacZα (4-vector system). Multiple mutations are shown as letters above the original wild-type template sequence, limited to 11 per position, with additional errors indicated by +n. Runs of 3 or more identical nucleotides are underlined. Mutational hot spots for which there are significant differences in the forward and reverse lacZα orientations are indicated by asterisks: *, P < 0.01; **, P < 0.001; ***, P < 0.0001 (see Materials and Methods). The mutation frequency is the number of lacZα DNAs with a mutation(s) divided by the total number of adjusted recovered clones.
Indirect evidence suggests that the multiple A-to-G mutations were caused by a host enzyme, ADAR (see Discussion). There were up to 10 A-to-G mutations in the lacZα DNA sequences (average, 6; median, 3); there were mutations in similar proportions of template adenosine residues in the two lacZα orientations (LZF, 28 of 36; LZR, 20 of 37). The positions of these class 3 A-to-G mutations had minimal overlap with the sites of class 1 A-to-G substitutions (LZF, 6 of 28 sites; LZR, 5 of 20 sites). Multiple A-to-G substitutions found in experiments with lacZα in both orientations revealed a strong intragenic preference for template adenosines preceded 5′ by an adenosine or a uridine in the viral RNA (LZF, 18 of the 36 A nucleotides were preceded by an A or a C; LZR, 16 of the 37 A nucleotides were preceded by an A or a C) and for template adenosines predicted to reside within RNA secondary structures (LZF, 20 sites; LZR, 17 sites [data not shown]). Because the mutations occurred in adenosines in the viral RNA strand, the positions at which these substitutions occurred were strongly affected by the orientation of lacZα. Although there were too few A-to-G substitutions at some positions, statistically significant differences in the two orientations were found at 22 of the 48 total positions (LZF, 12 of 28; LZR, 10 of 20) (see the legend for Fig. 4 for the corresponding P values). Taken together, these findings indicate that the sequence and/or structural context of the mutational target in the viral RNA determines the specificity of the multiple A-to-G mutations.
A minor fraction (6%) of multiple nucleotide substitutions (LZF, 19 of 308; LZR, 12 of 162) in the class 3 lacZα DNAs (LZF, 7 of 71; LZR, 6 of 31) involved a variety of multiple transition and transversion mutations (Table 3C). Indirect evidence suggests that some of these mutations could have been caused by an error-prone polymerase and/or a host enzyme, APOBEC3G (see Discussion). In contrast to multiple A-to-G substitutions, in this subclass of multiple nucleotide substitutions there were only 2 changes in the lacZα DNA, and there was considerable overlap with the sites of the class 1 single nucleotide substitutions (LZF, 9 of 16 sites; LZR, 6 of 9 sites). About half of these multiple nucleotide substitutions were G-to-A transitions (LZF, 8 of 19; LZR, 6 of 12), and an examination of their positions in either lacZα orientation revealed an intragenic site preference for template guanosines proceeded 3′ by a guanosine (LZF, 3 sites; LZR, 4 sites).
(iv) Class 4: insertions, deletions, and deletions with insertions (indels).
Finally, we recovered a small number of lacZα DNAs containing insertions or deletions (small or large) or deletions with insertions (11 lacZα-F and 13 lacZα-R DNAs). All of the indel mutations altered the reading frame of lacZα (Fig. 5). Because these mutations did not localize to any specific nucleotide positions, the entire 174-nt length of lacZα was considered scorable for class 4 mutants (Table 4).
FIG. 5.
Class 4 mutations: insertions, deletions, and/or deletions with insertions (indels) detected in lacZα (4-vector system). Indel mutations (deletions, deletions with insertions, and duplications) are shown as red lines extending over the positions of the mutations, with each lacZα DNA sequence numbered accordingly. The lines approximate the sizes of the indels, with the actual number of nucleotides inserted or deleted preceded by an inverted or upright triangle, respectively. Additional substitution errors within indels are shown as letters above the original wild-type template sequence. Runs of 3 or more identical nucleotides are underlined. The mutation frequency is the number of lacZα DNAs with a mutation(s) divided by the total number of adjusted recovered clones.
The frequencies of indels were quite similar in the two lacZα orientations (P = 0.54), with deletions substantially outnumbering insertions (LZF, 10 of 11; LZR, 8 of 13). The deletions ranged from 4 to 44 nt. The insertion or deletion junction in both large and small indels typically involved a short direct repeat of 1 to 3 nt and/or a preceding run of identical nucleotides in the direction of minus-strand synthesis. A minor fraction of indels involved a deletion of 2 to 3 nt followed by an insertion of a similar length (LZF, 0 of 11; LZR, 2 of 13). Finally, two lacZα-R DNAs were found to contain a single nucleotide substitution and a frameshift separated by 3 to 9 nucleotides.
The orientation of lacZα does not affect the in vivo mutation frequency.
Three separate approaches were used to determine the overall in vivo mutation rate for a single round of HIV-1 replication (Table 2). Since our measurements include only mutations that inactivate lacZα, these determinations underestimate the actual mutation rate (see Discussion). We classified individual errors as either related (dependent) or separate (independent) mutations. Single nucleotide changes (classes 1 and 2) and indels (class 4) were considered single mutational events. However, the multiple substitutions in class 3 were treated as either dependent or independent events, because it was unclear whether they were caused by a host enzyme that modifies the nucleic acid or by an error-prone polymerase (see Discussion). If the class 3 mutations were excluded, the overall mutation rate in the forward lacZα orientation was 1.2 × 10−5 mutations/bp/cycle. If each class 3 lacZα DNA sequence was counted as a single mutational event, the overall mutation rate was 1.4 × 10−5 mutations/bp/cycle (P = 0.013). Alternatively, if each individual change in the class 3 mutants was counted separately, there were an additional 237 mutations (308 total), giving rise to a 1.8-fold-higher overall mutation rate of 2.2 × 10−5 mutations/bp/cycle (P < 0.0001). Class 3 mutations (as dependent or independent events) appeared to modestly increase the overall mutation rate, and similar increases were seen in both the forward and reverse lacZα orientations. If the multiple A-to-G substitutions of class 3 were introduced by a host enzyme (ADAR), then their relative contribution to the overall forward lacZα mutation rate would be 2.1 × 10−6 mutations/bp/cycle (see Discussion). Whether the few remaining class 3 multiple nucleotide substitutions were dependent events caused by a host enzyme (for example, APOBEC3G) or independent events caused by an error-prone polymerase, their relative contribution would not have appreciably affected the final overall lacZα-F mutation rate of 1.4 × 10−5 mutations/bp/cycle. Finally, we compared the overall total mutation rates for the forward and reverse orientations of lacZα and found no significant difference between 1.4 × 10−5 mutations/bp/cycle (lacZα-F) and 1.3 × 10−5 mutations/bp/cycle (lacZα-R) (P = 0.061). These results suggest that while the specific mutations are affected by which strand of lacZα is expressed in viral RNA, the overall mutation rate is not.
DISCUSSION
Nature and position of mutational hot spots in vivo.
Many of the analyses of mutations generated in vitro by RT have used a mutational target similar to lacZα-F. There have been relatively few cell-free in vitro studies using an RNA template with the lacZα-F orientation (8, 30) and no studies using either an RNA or a DNA template equivalent to the lacZα-R orientation. Although similar lacZα DNA sequences have been used as targets in multiple in vitro studies, the patterns of reported mutations are vastly different. Furthermore, the overall mutation rate reported for RT in vitro is significantly higher than the overall mutation rate in vivo. Our findings extend the available in vivo observations and provide information about the nature, position, and frequency of the mutations. Because the published in vivo data do not provide detailed information about the spectrum of mutations, it has not been possible to consider the mechanisms by which the mutations arose (42, 46). The classes and proportions of mutations we obtained using HIV-1 lacZα were consistent with results from previous reports for HIV-1 (42, 46), SNV (11, 55, 56), and bovine leukemia virus (BLV) (47). Base substitutions are the most common mutations, with transitions predominating over transversions. Although the proportions of single nucleotide frameshifts and indels have been reported to make up 10 to 25% of the total, we observed much lower fractions of 5 to 6% and 3 to 4%, respectively. We also found more multiple mutations (including both G-to-A and A-to-G mutations) than were previously reported.
The mutational hot spots seen in vivo depended upon the strand of lacZα that was expressed in the viral RNA. Although the natures and relative frequencies of the various classes of mutations were similar in both lacZα orientations, the sequence and/or structural context of the template influenced the location of hot spots. Both single (class 1) and multiple (class 3) nucleotide substitutions are orientation dependent. Although the class 1 (single) G-to-A and C-to-T substitutions were broadly distributed, class 3 A-to-G and G-to-A substitutions usually occurred next to specific neighboring nucleotides. In contrast, frameshift (class 2) and indel (class 4) mutations were much less frequent, and most were in the same nucleotide runs in both lacZα orientations. This result differs from in vitro findings, where single nucleotide frameshift deletions were much more frequent (4, 5, 8, 33, 35) and frameshift insertions did not necessarily match adjacent template nucleotides (9, 10). The pattern of mutational hot spots seen in vivo in lacZα-F or lacZα-R did not match any of the published in vitro hot spot data obtained using purified RT to copy lacZα (4, 8-10, 30, 39). Although Mansky and Temin (46) reported relatively few in vivo mutations, making it difficult to unambiguously identify hot spots, their data are more similar to ours than any of the in vitro data are.
Possible underlying mechanism(s) of in vivo mutations.
The in vivo-based system detects the mutations made by both RT and RNA Pol II but does not reveal which enzyme made any individual mutation. The underlying mechanism(s) that contributes to the mutations that arise during HIV-1 replication has not been established. Our data suggest that the majority of in vivo lacZα mutations (80 to 84% of single nucleotide substitutions) arise by nucleotide misincorporation rather than by misalignment or slippage of the template primer strands at the ends of nucleotide runs (LZF, 260 of 324; LZR, 236 of 280). This result matches the findings of a study of SNV mutations (32, 56, 57) but differs from the initial in vitro reports that most of the missense errors made by HIV-1 RT are due to misalignment/slippage (4, 5, 8). The slippage model for missense mutagenesis suggests that polymerization through a homopolymeric sequence may produce a transiently misaligned intermediate with an unpaired extrahelical base in either the template or the primer strand, which could result in two different outcomes. If the alignment error leads to polymerization without realignment, a deletion or an insertion frameshift will arise in the nucleotide run if the unpaired extrahelical base is in the template or primer strand, respectively. If the alignment error is followed by incorporation of a single nucleotide and then realignment, the single nucleotide is often a mismatch with the template, causing a specific missense substitution. Although a slippage mechanism cannot account for the majority of the missense mutations, it can account for the majority of the frameshift mutations.
Although slippage can occur anywhere in a homopolymeric run, whether the template or primer strand is the strand that slips should depend on the nature of the adjacent nucleotide at the ends of the homopolymeric run and on the interactions of the nucleic acid substrate with the polymerase. Interestingly, while 46 to 52% of all class 1 substitutions were detected near the ends of runs (LZF, 167 of 324; LZR, 128 of 280), only 34 to 38% of these mutations matched the sequence context for slippage (LZF, 64 of 167; LZR, 44 of 128). The majority of class 2 frameshift mutations were found in nucleotide runs and matched the sequence context for slippage (LZF, 20 of 26; LZR, 11 of 16). While most frameshifts reported from in vitro assays using purified RT were template-based (deletion) slippages (4, 5, 11, 33), most of the events seen in vivo were primer-based (insertion) slippages. Slippage could explain the generation of some of the indels located in nucleotide runs. However, large deletions occurred near direct repeats, and the mutant sequences suggested that the mutations arose by template switching during reverse transcription. Because the vast majority of the mutations are missense substitutions, these results strongly suggest that most lacZα mutations are due to misincorporation, made either by RT or RNA Pol II or by some combination of the two.
A significant fraction (9 to 16%) of mutant lacZα DNAs contained multiple nucleotide substitutions. Based on the relatively low number of single mutations, this is an unexpected result. The most obvious explanation for multiple nucleotide substitutions is an error-prone polymerase or a mutagenic host editing enzyme. G-to-A hypermutations have been reported in the genomes of a number of retroviruses and can lead to incomplete reverse transcription and/or the production of nonfunctional viral proteins (32, 72, 73). The most common change, 5′-GG to -GA within the viral plus-strand DNA, has been attributed to the deamination of cytidines to uridines in the viral minus-strand DNA by the host enzyme APOBEC3G (A3G) (24, 38, 41). Because the 293T producer cells used to generate HIV-1 in our in vivo-based system have little or no APOBEC3G (26, 58), we found very few multiple G-to-A mutations. Most of the class 3 multiple nucleotide substitutions involved A-to-G transitions (94%). The small residual fraction of the multiple mutations involving G-to-A changes (6%) could have been caused by APOBEC3G. Some or all of the rare multiple transitions and transversions could have been caused by an error-prone RT or RNA polymerase II. However, we suggest that the much more common A-to-G hypermutations were caused by a host adenosine deaminase that acts on RNA (ADAR). ADARs catalyze the deamination of adenosine to inosine in structured or double-stranded regions of RNA, with a sequence preference for 5′-UA or -AA (3, 40, 67). Any A-to-I changes caused by ADAR in the viral RNA would appear as A-to-G mutations in the viral plus-strand DNA. The nature and position of most class 3 substitutions matched the sequence and structural preference of ADARs. ADAR-induced A-to-G and U-to-C hypermutations have been documented in many negative-strand viruses (3, 14). While ADAR-induced mutations have not been directly correlated with an antiviral effect, it has been suggested that the mutations enhance viral persistence (14) or the degradation (64) or nuclear export (76) of hyperedited dsRNA. In contrast, A-to-G hypermutations are not as prevalent in positive-strand RNA viruses or in retroviruses. To date, only a few Rous-associated virus type 1 (RAV-1) (22), avian leukosis virus (ALV) (23), and SNV (32) proviral sequences have been reported to contain A-to-G hypermutations that could have been caused by ADAR. Recent reports have proposed that ADARs may stimulate the release and the infectious potential of HIV-1 either directly, by editing adenosines in the 5′ untranslated region (5′ UTR) (TAR), Rev, and Tat coding regions, or indirectly, by suppressing the proapoptotic dsRNA-dependent kinase activities of protein kinase R (PKR) from acting on translation initiation factor eIF-2α (α subunit of eukaryotic initiation factor) (16, 20, 49). Our survey of 414 HIV-1 full genomic sequences showed that ADAR-like A-to-G hypermutations are rare. Although multiple A-to-G substitutions were detected when the HIV-1 subtype B genomic sequences were compared to a reference sequence (126 of 3,411 adenosines), the intrasequence variation of these changes was not significant compared to that for multiple C-to-T substitutions (69 of 1,772 cytidines) (P = 0.76). This suggests that ADAR-induced A-to-G hypermutations are rare in the HIV-1 genomes isolated from patients. A-to-G hypermutations may lead to the generation of viruses with little or no replication capacity, which could contribute to the rarity of A-to-G hypermutations in HIV-1 genomes in patients.
Determination of an in vivo mutation rate for HIV-1 by using lacZα.
Although it is widely believed that HIV-1 replication is exceptionally error-prone, we show here that the in vivo forward mutation rate of HIV-1, measured by the inactivation of lacZα, is approximately 1.4 × 10−5 mutations/bp/cycle. This rate is approximately 20-fold lower than the in vitro error rate for RT measured using a DNA template (9, 10, 29, 30, 62, 68) and 17-fold lower than the rate measured using an RNA template (29, 30). Our reported mutation rate for HIV-1 in HOS cells is also about 3-fold lower than earlier reported rates in HeLa and CEM cells (42, 46) and is similar to the average rate reported for other retroviruses (1.5 × 10−5 mutations/bp/cycle) (69).
The primary advantage of using the lacZα-based system to study HIV-1 replication fidelity is that the nature of the mutations that arise in vivo can be determined. An accurate mutation rate depends on (i) the number of mutable positions in lacZα, (ii) whether mutations in both DNA strands are scored, and (iii) whether mutants with multiple changes in the lacZα DNA (classes 3 and 4) are counted as having single or multiple mutations. One limitation of using lacZα to determine the in vivo mutation rate of HIV-1 is that silent mutations are not detected. Based on the collection of mutations generated by recombinant RT in vitro, it has been proposed that mutations can be detected in approximately 40% of the nucleotides in the promoter and coding regions of lacZα. The specific numbers of mutable target sites in a 280-nt lacZα have been reported as 113 for substitutions, 150 for frameshifts, and 280 for indels (4, 8, 34, 55, 63). These numbers could not be used to analyze the data from our in vivo assay because only 174 nt of lacZα were the same in our experiments and the published data. However, we have confirmed that single nucleotide missense mutations could be detected at a minimum of 71 out of 174 nt positions (Table 4). This suggests that our measured mutation rate may be 2- to 3-fold lower than the actual overall mutation rate. The mutation rate of HIV-1 was previously determined, using different constraints, to be 3.4 × 10−5 mutations/bp/cycle (46). When the rate was recalculated using the 174-nt target, the mutation rate was 2.4 × 10−5 mutations/bp/cycle, which is similar to our result (for details, see Text S3 and Table S3 in the supplemental material). Based on the published in vivo data and our data, HIV-1 replication is not error-prone relative to that of other retroviruses. This implies that, at least in the context of the replicating virus, HIV-1 RT is not significantly more error-prone than other retroviral RTs.
Supplementary Material
Acknowledgments
This study was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
We are grateful to Dwight Nissley for helpful discussions and Teresa Burdette for editing and help in preparing the manuscript. We also thank Octavio A. Quiñones for helpful assistance in data preparation and statistical analyses and Al Kane for assistance in preparing publication-quality figures and tables.
Footnotes
Published ahead of print on 21 July 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Avidan, O., M. E. Meer, I. Oz, and A. Hizi. 2002. The processivity and fidelity of DNA synthesis exhibited by the reverse transcriptase of bovine leukemia virus. Eur. J. Biochem. 269:859-867. [DOI] [PubMed] [Google Scholar]
- 2.Bakhanashvili, M., and A. Hizi. 1993. Fidelity of DNA synthesis exhibited in vitro by the reverse transcriptase of the lentivirus equine infectious anemia virus. Biochemistry 32:7559-7567. [DOI] [PubMed] [Google Scholar]
- 3.Bass, B. L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71:817-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bebenek, K., J. Abbotts, J. D. Roberts, S. H. Wilson, and T. A. Kunkel. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 264:16948-16956. [PubMed] [Google Scholar]
- 5.Bebenek, K., J. Abbotts, S. H. Wilson, and T. A. Kunkel. 1993. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 268:10324-10334. [PubMed] [Google Scholar]
- 6.Bonhoeffer, S., E. C. Holmes, and M. A. Nowak. 1995. Causes of HIV diversity. Nature 376:125. [DOI] [PubMed] [Google Scholar]
- 7.Bowman, R. R., W.-S. Hu, and V. K. Pathak. 1998. Relative rates of retroviral reverse transcriptase template switching during RNA- and DNA-dependent DNA synthesis. J. Virol. 72:5198-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, J. C., K. Bebenek, and T. A. Kunkel. 1992. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc. Natl. Acad. Sci. U. S. A. 89:6919-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer, P. L., and S. H. Hughes. 2000. Effects of amino acid substitutions at position 115 on the fidelity of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 74:6494-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer, P. L., C. R. Stenbak, D. Hoberman, M. L. Linial, and S. H. Hughes. 2007. In vitro fidelity of the prototype primate foamy virus (PFV) RT compared to HIV-1 RT. Virology 367:253-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns, D. P., and H. M. Temin. 1994. High rates of frameshift mutations within homo-oligomeric runs during a single cycle of retroviral replication. J. Virol. 68:4196-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler, I. F., I. Pandrea, P. A. Marx, and C. Apetrei. 2007. HIV genetic diversity: biological and public health consequences. Curr. HIV Res. 5:23-45. [DOI] [PubMed] [Google Scholar]
- 13.Campbell, T. B., K. Schneider, T. Wrin, C. J. Petropoulos, and E. Connick. 2003. Relationship between in vitro human immunodeficiency virus type 1 replication rate and virus load in plasma. J. Virol. 77:12105-12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cattaneo, R. 1994. Biased (A→I) hypermutation of animal RNA virus genomes. Curr. Opin. Genet. Dev. 4:895-900. [DOI] [PubMed] [Google Scholar]
- 15.Chen, R., M. Yokoyama, H. Sato, C. Reilly, and L. M. Mansky. 2005. Human immunodeficiency virus mutagenesis during antiviral therapy: impact of drug-resistant reverse transcriptase and nucleoside and nonnucleoside reverse transcriptase inhibitors on human immunodeficiency virus type 1 mutation frequencies. J. Virol. 79:12045-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clerzius, G., J. F. Gelinas, A. Daher, M. Bonnet, E. F. Meurs, and A. Gatignol. 2009. ADAR1 interacts with PKR during HIV infection of lymphocytes and contributes to viral replication. J. Virol. 83:10119-10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 18.Coffin, J. M. 1986. Genetic variation in AIDS viruses. Cell 46:1-4. [DOI] [PubMed] [Google Scholar]
- 19.de Mercoyrol, L., Y. Corda, C. Job, and D. Job. 1992. Accuracy of wheat-germ RNA polymerase II. General enzymatic properties and effect of template conformational transition from right-handed B-DNA to left-handed Z-DNA. Eur. J. Biochem. 206:49-58. [DOI] [PubMed] [Google Scholar]
- 20.Doria, M., F. Neri, A. Gallo, M. G. Farace, and A. Michienzi. 2009. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 37:5848-5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, et al. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felder, M. P., D. Laugier, B. Yatsula, P. Dezelee, G. Calothy, and M. Marx. 1994. Functional and biological properties of an avian variant long terminal repeat containing multiple A to G conversions in the U3 sequence. J. Virol. 68:4759-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajjar, A. M., and M. L. Linial. 1995. Modification of retroviral RNA by double-stranded RNA adenosine deaminase. J. Virol. 69:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 25.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 26.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, et al. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 27.Jern, P., R. A. Russell, V. K. Pathak, and J. M. Coffin. 2009. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 5:e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji, J., and L. A. Loeb. 1994. Fidelity of HIV-1 reverse transcriptase copying a hypervariable region of the HIV-1 env gene. Virology 199:323-330. [DOI] [PubMed] [Google Scholar]
- 30.Ji, J. P., and L. A. Loeb. 1992. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31:954-958. [DOI] [PubMed] [Google Scholar]
- 31.Kantor, R., R. W. Shafer, S. Follansbee, J. Taylor, D. Shilane, L. Hurley, et al. 2004. Evolution of resistance to drugs in HIV-1-infected patients failing antiretroviral therapy. AIDS 18:1503-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, T., R. A. Mudry, Jr., C. A. Rexrode, and V. K. Pathak. 1996. Retroviral mutation rates and A-to-G hypermutations during different stages of retroviral replication. J. Virol. 70:7594-7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkel, T. A. 1990. Misalignment-mediated DNA synthesis errors. Biochemistry 29:8003-8011. [DOI] [PubMed] [Google Scholar]
- 34.Kunkel, T. A., and P. S. Alexander. 1986. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J. Biol. Chem. 261:160-166. [PubMed] [Google Scholar]
- 35.Kunkel, T. A., and K. Bebenek. 2000. DNA replication fidelity. Annu. Rev. Biochem. 69:497-529. [DOI] [PubMed] [Google Scholar]
- 36.Kunkel, T. A., K. Bebenek, J. D. Roberts, M. P. Fitzgerald, and D. C. Thomas. 1989. Analysis of fidelity mechanisms with eukaryotic DNA replication and repair proteins. Genome 31:100-103. [DOI] [PubMed] [Google Scholar]
- 37.Langley, K. E., M. R. Villarejo, A. V. Fowler, P. J. Zamenhof, and I. Zabin. 1975. Molecular basis of beta-galactosidase alpha-complementation. Proc. Natl. Acad. Sci. U. S. A. 72:1254-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 39.Lewis, D. A., K. Bebenek, W. A. Beard, S. H. Wilson, and T. A. Kunkel. 1999. Uniquely altered DNA replication fidelity conferred by an amino acid change in the nucleotide binding pocket of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 274:32924-32930. [DOI] [PubMed] [Google Scholar]
- 40.Maas, S., A. Rich, and K. Nishikura. 2003. A-to-I RNA editing: recent news and residual mysteries. J. Biol. Chem. 278:1391-1394. [DOI] [PubMed] [Google Scholar]
- 41.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 42.Mansky, L. M. 1996. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS Res. Hum. Retroviruses 12:307-314. [DOI] [PubMed] [Google Scholar]
- 43.Mansky, L. M., and L. C. Bernard. 2000. 3′-Azido-3′-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1. J. Virol. 74:9532-9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansky, L. M., R. E. Le, S. Benichou, and L. C. Gajary. 2003. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J. Virol. 77:2071-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansky, L. M., D. K. Pearl, and L. C. Gajary. 2002. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutant frequencies. J. Virol. 76:9253-9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mansky, L. M., and H. M. Temin. 1994. Lower mutation rate of bovine leukemia virus relative to that of spleen necrosis virus. J. Virol. 68:494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez, M. A., J. P. Vartanian, and S. Wain-Hobson. 1994. Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc. Natl. Acad. Sci. U. S. A. 91:11787-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nie, Y., G. L. Hammond, and J. H. Yang. 2007. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J. Virol. 81:917-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nowak, M. 1990. HIV mutation rate. Nature 347:522. [DOI] [PubMed] [Google Scholar]
- 51.Nowak, M. A., R. M. Anderson, A. R. McLean, T. F. Wolfs, J. Goudsmit, and R. M. May. 1991. Antigenic diversity thresholds and the development of AIDS. Science 254:963-969. [DOI] [PubMed] [Google Scholar]
- 52.Oh, J., M. J. McWilliams, J. G. Julias, and S. H. Hughes. 2008. Mutations in the U5 region adjacent to the primer binding site affect tRNA cleavage by human immunodeficiency virus type 1 reverse transcriptase in vivo. J. Virol. 82:719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Neil, P. K., G. Sun, H. Yu, Y. Ron, J. P. Dougherty, and B. D. Preston. 2002. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J. Biol. Chem. 277:38053-38061. [DOI] [PubMed] [Google Scholar]
- 54.Parthasarathi, S., A. Varela-Echavarria, Y. Ron, B. D. Preston, and J. P. Dougherty. 1995. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J. Virol. 69:7991-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc. Natl. Acad. Sci. U. S. A. 87:6024-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl. Acad. Sci. U. S. A. 87:6019-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pathak, V. K., and H. M. Temin. 1992. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J. Virol. 66:3093-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piroozmand, A., Y. Yamamoto, B. Khamsri, M. Fujita, T. Uchiyama, and A. Adachi. 2007. Generation and characterization of APOBEC3G-positive 293T cells for HIV-1 Vif study. J. Med. Invest. 54:154-158. [DOI] [PubMed] [Google Scholar]
- 59.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 60.Pulsinelli, G. A., and H. M. Temin. 1994. High rate of mismatch extension during reverse transcription in a single round of retrovirus replication. Proc. Natl. Acad. Sci. U. S. A. 91:9490-9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rezende, L. F., and V. R. Prasad. 2004. Nucleoside-analog resistance mutations in HIV-1 reverse transcriptase and their influence on polymerase fidelity and viral mutation rates. Int. J. Biochem. Cell Biol. 36:1716-1734. [DOI] [PubMed] [Google Scholar]
- 62.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171-1173. [DOI] [PubMed] [Google Scholar]
- 63.Roberts, J. D., B. D. Preston, L. A. Johnston, A. Soni, L. A. Loeb, and T. A. Kunkel. 1989. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol. Cell. Biol. 9:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scadden, A. D., and C. W. Smith. 2001. Specific cleavage of hyper-edited dsRNAs. EMBO J. 20:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah, F. S., K. A. Curr, M. E. Hamburgh, M. Parniak, H. Mitsuya, J. G. Arnez, et al. 2000. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 275:27037-27044. [DOI] [PubMed] [Google Scholar]
- 66.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, et al. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steele, E. J., R. A. Lindley, J. Wen, and G. F. Weiller. 2006. Computational analyses show A-to-G mutations correlate with nascent mRNA hairpins at somatic hypermutation hotspots. DNA Repair (Amst.) 5:1346-1363. [DOI] [PubMed] [Google Scholar]
- 68.Stuke, A. W., O. Ahmad-Omar, K. Hoefer, G. Hunsmann, and K. D. Jentsch. 1997. Mutations in the SIV env and the M13 lacZa gene generated in vitro by reverse transcriptases and DNA polymerases. Arch. Virol. 142:1139-1154. [DOI] [PubMed] [Google Scholar]
- 69.Svarovskaia, E. S., S. R. Cheslock, W. H. Zhang, W. S. Hu, and V. K. Pathak. 2003. Retroviral mutation rates and reverse transcriptase fidelity. Front. Biosci. 8:d117-d134. [DOI] [PubMed] [Google Scholar]
- 70.Taube, R., O. Avidan, M. Bakhanashvili, and A. Hizi. 1998. DNA synthesis exhibited by the reverse transcriptase of mouse mammary tumor virus: processivity and fidelity of misinsertion and mispair extension. Eur. J. Biochem. 258:1032-1039. [DOI] [PubMed] [Google Scholar]
- 71.Turner, D., B. Brenner, D. Mosis, C. Liang, and M. A. Wainberg. 2005. Substitutions in the reverse transcriptase and protease genes of HIV-1 subtype B in untreated individuals and patients treated with antiretroviral drugs. MedGenMed 7:69. [PMC free article] [PubMed] [Google Scholar]
- 72.Vartanian, J. P., M. Henry, and S. Wain-Hobson. 2002. Sustained G→A hypermutation during reverse transcription of an entire human immunodeficiency virus type 1 strain Vau group O genome. J. Gen. Virol. 83:801-805. [DOI] [PubMed] [Google Scholar]
- 73.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G—-A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ventura, A., A. Meissner, C. P. Dillon, M. McManus, P. A. Sharp, P. L. Van, et al. 2004. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. U. S. A. 101:10380-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wabl, M., P. D. Burrows, A. von Gabain, and C. Steinberg. 1985. Hypermutation at the immunoglobulin heavy chain locus in a pre-B-cell line. Proc. Natl. Acad. Sci. U. S. A. 82:479-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, Z., and G. G. Carmichael. 2001. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106:465-475. [DOI] [PubMed] [Google Scholar]
- 77.Zhuang, J., A. E. Jetzt, G. Sun, H. Yu, G. Klarmann, Y. Ron, et al. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J. Virol. 76:11273-11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.