Abstract
The lyssavirus matrix (M) protein induces apoptosis. The regions of the M protein that are essential for triggering cell death pathways are not yet clearly defined. We therefore compared the M proteins from two viruses that have contrasting characteristics in terms of cellular apoptosis: a genotype 3 lyssavirus, Mokola virus (MOK), and a genotype 1 rabies virus isolated from a dog from Thailand (THA). We identified a 20-amino-acid fragment (corresponding to positions 67 to 86) that retained the cell death activities of the full-length M protein from MOK via both the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and inhibition of cytochrome c oxidase (CcO) activity. We found that the amino acids at positions 77 and 81 have an essential role in triggering these two cell death pathways. Directed mutagenesis demonstrated that the amino acid at position 77 affects CcO activity, whereas the amino acid at position 81 affects TRAIL-dependent apoptosis. Mutations in the full-length M protein that compromised induction of either of these two pathways resulted in delayed apoptosis compared with the time to apoptosis for the nonmutated control.
In most viral infections, the death of infected cells is a key event in the host-pathogen interaction (5, 7, 14, 34, 44). This event is often induced by the immune system recognizing particular signals and may limit the spread of infection (9, 23, 40, 43). Therefore, the ability to avoid detection by either the innate or the acquired immune system can contribute to viral pathogenicity (12). Many pathogenic viruses cause undetectable cellular changes during viral production (13, 19, 39). Others, such as animal RNA viruses, including alphaviruses and vesicular stomatitis virus (VSV), reproduce rapidly and escape possible interference from the apoptotic response, thereby facilitating virion release (18, 26, 28).
In the case of lyssavirus infection, the integrity of the host cell is maintained, enabling rapid spread of the virus to the central nervous system (CNS) and the subsequent development of rabies (27). The matrix (M) protein is a small multifunctional protein of 202 amino acids which is essential for virus maturation and budding. Through regulating the expression of viral and host proteins, it also has key roles in viral morphogenesis and modulating replication and transcription of the viral genome. The structure of the M protein from the Lagos bat virus (M-LAG), a genotype 2 lyssavirus, has recently been resolved and is similar to the structures of M proteins from other rhabdoviruses, such as VSV (2). Different lyssavirus proteins, including glycoprotein (35, 36, 41), phosphoprotein (20), and M protein (15, 22), have been reported to play a role in induction of cell death. We have previously shown that the M protein activates caspase 8 and induces apoptosis via the tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) (22). Lyssavirus M protein also causes mitochondrial defects by compromising the respiratory chain through cytochrome c (cyt-c) oxidase (CcO) inhibition (15).
In the present study, we compared the M protein from Mokola virus (M-MOK; a low-pathogenicity genotype 3 lyssavirus) with the M protein from a rabies virus isolated from a dog from Thailand (M-THA; a genotype 1 lyssavirus) (3). We defined a small fragment of 20 amino acids (aa) located between residues 67 and 86 of the M protein that was able to induce TRAIL-mediated cell death as well as inhibit CcO activity. We also characterized the roles of 2 aa at positions 77 and 81 on a solvent-exposed face of the M protein (2) involved in TRAIL-mediated cell death (15). The residues at these two positions appear to contribute to two different cell death pathways.
MATERIALS AND METHODS
Antibodies.
Anti-enhanced green fluorescent protein (anti-EGFP) monoclonal antibody (clone JL-8) and CcO4 antibodies were purchased from Clontech Laboratories. Monoclonal anti-β-actin (clone AC-74) antibody was purchased from Sigma.
Cells and viruses.
Human carcinoma epithelial (HeLa) cells were cultured in Dulbecco's minimal essential medium supplemented with 0.2% glutamic acid from Gibco and 10% heat-inactivated fetal bovine serum from Eurobio. Two lyssaviruses, genotype 1 THA and genotype 3 MOK, were used to generate clones coding for the M gene, as described previously (15, 22).
Recombinant plasmid construction and site-directed mutagenesis.
The M-MOK and M-THA fragments were amplified by reverse transcription-PCR performed with viral RNA. The sequences of the sense and antisense primers designed to amplify wild-type M-MOK and M-THA and the MOK deletion mutants are shown in Table 1. PCR products were inserted into pEGFP-C1 plasmids (Clontech), as described previously (15). Point mutations in wild-type M-MOK and M-THA were made using a QuikChange II site-directed mutagenesis kit (Stratagene) (Table 1). The M1-7 MOK mutants were made by hybridization of two synthesized complementary oligonucleotides containing the desired mutations and the restriction sites used for the cloning experiments (Table 1).
TABLE 1.
Primers used for mutant constructionsa
| Construct | Name | Primer sequence (5′ → 3′)b |
|---|---|---|
| Total M | ||
| M-MOK | MMOKeGFP-S | F: GAGGAGATCTATGAATTTCCTCAAGAAAATG |
| MMOKeGFP-AS | R: TGGGAAGCTTGCTACTCTAATAAAAGTGAGGTGTT | |
| M-THA | MTHAeGFP-S | F: GAGGAGATCTATGAACTTTCTACGCAAAATC |
| MTHAeGFP-AS | R: TGGGAAGCTTGCTATTCTAGGAGCAGGGAAGAGTC | |
| M1-2 truncated fragments | ||
| M1-5 | M1-2MOKS | F: GAGGAGATCTGGCAAGGCCAGTGTGAGAAAC |
| M1-5MOKAS | R: TGGGAAGCTTGCTAGTTCCCAGAGTAAACAT | |
| M1-6 | M1-6MOKS | F: GAGGAGATCTTACTCCTTCAAGATACTC |
| M1MOKAS | R: TGGGAAGCTTGCTAATGCCCTCCGGGACTG | |
| M1-7 | M1-7MOKS | F: GAGGAGATCTTACTCCTTCAAGATACTCAGGCATATTTTGGAGTCGTTTGATAATGTTTACTCTGGGAACTAGCAAGCTTCCCA |
| M1-8MOKAS | R: TGGGAAGCTTGCTAGTTCCCAGAGTAAACATTATCAAACGACTCCAAAATATGCCTGAGTATCTTGAAGGAGTAAGATCTCCTC | |
| M1-8 | M1-8MOKS | F: GAGGAGATCTTACTCCTTCAAGATACTCAGGCATATTTTGAAGTCGTTTTAGCAAGCTTCCCA |
| M1-8MOKAS | R: TGGGAAGCTTGCTAAAACGACTTCAAAATATGCCTGAGTATCTTGAAGGAGTAAGATCTCCTC | |
| M1-9 | M1-9MOKS | F: GAGGAGATCTTTTGATAATGTTTACTCTGGGTAGCAAGCTTCCCA |
| M1-9MOKAS | R: TGGGAAGCTTGCTACCCAGAGTAAACATTATCAAAAGATCTCCTC | |
| Mutants of M M1-7, M1-8, and M1-9 with point mutations | ||
| M1-7 EN | M1-7ENS | F: GAGGAGATCTTACTCCTTCAAGATACTCAGGCATATTTTGGAGTCGTTTGATAATGTTTACTCTGGGAACTAGCAAGCTTCCCA |
| M1-7ENAS | R: TGGGAAGCTTGCTAGTTCCCAGAGTAAACATTATCAAACGACTCCAAAATATGCCTGAGTATCTTGAAGGAGTAAGATCTCCTC | |
| M1-7 KE | M1-7KES | F: GAGGAGATCTTACTCCTTCAAGATACTCAGGCATATTTTGAAGTCGTTTGATGAGGTTTACTCTGGGAACTAGCAAGCTTCCCA |
| M1-7KEAS | R: TGGGAAGCTTGCTAGTTCCCAGAGTAAACCTCATCAAACGACTTCAAAATATGCCTGAGTATCTTGAAGGAGTAAGATCTCCTC | |
| M1-7 RE | M1-7RES | F: GAGGAGATCTTACTCCTTCAAGATACTCAGGCATATTTTGAGATCGTTTGATGAGGTTTACTCTGGGAACTAGCAAGCTTCCCA |
| M1-7REAS | R: TGGGAAGCTTGCTAGTTCCCAGAGTAAACCTCATCAAACGATCTCAAAATATGCCTGAGTATCTTGAAGGAGTAAGATCTCCTC | |
| M1-8E | M1-8ES | F: GAGGAGATCTTACTCCTTCAAGATACTCAGGCATATTTTGGAGTCGTTTTAGCAAGCTTCCCA |
| M1-8EAS | R: TGGGAAGCTTGCTAAAACGACTCCAAAATATGCCTGAGTATCTTGAAGGAGTAAGATCTCCTC | |
| M1-8R | M1-8RS | F: GAGGAGATCTTACTCCTTCAAGATACTCAGGCATATTTTGAGATCGTTTTAGCAAGCTTCCCA |
| M1-8RAS | R: TGGGAAGCTTGCTAAAACGATCTCAAAATATGCCTGAGTATCTTGAAGGAGTAAGATCTCCTC | |
| M1-8 P74 | M1-8P74S | F: GAGGAGATCTTACTCCTTCAAGATACTCAGGCCGATTTTGAAGTCGTTTTAGCAAGCTTCCCA |
| M1-8PAS | R: TGGGAAGCTTGCTAAAACGACTTCAAAATCGGCCTGAGTATCTTGAAGGAGTAAGATCTCCT | |
| M1-9E | M1-9ES | F: GAGGAGATCTTTTGATGAGGTTTACTCTGGGTAGCAAGCTTCCCA |
| M1-9EAS | R: TGGGAAGCTTGCTACCCAGAGTAAACCTCATCAAAAGATCTCCTC | |
| M-MOK RE | M-MOKR77E81S | F: CTCAGGCATATTTTGCGGTCGTTTGATGAGGTTTACTCTGGG |
| M-MOKR77E81AS | R: CCCAGAGTAAACCTCATCAAACGACCGCAAAATATGCCTGAG | |
| M-THA KN | M-THAK77N81S | F: GGATCCTGCGGCACATTCTAAAGTCATTCGATAATATATATTCTGG |
| M-THAK77N81AS | R: CCAGAATATATATTATCGAATGACTTTAGAATGTGCCGCAGGATCC |
All constructs were cloned in the pEGFP-C1 vector.
The sequences of the different primers were directed from 5′ to 3′. F, forward; R, reverse; boldface, the stop codon was added for the end of the protein expression; underlining, restriction sites for cloning.
Cotransfection and cellular localization of M-MOK deletion mutants.
Transfections were performed with the Lipofectamine 2000 reagent (Invitrogen), in accordance with the manufacturer's instructions. HeLa cells were transfected with plasmids carrying the M-protein deletion mutants or the M1-7 mutants and pDsRed2-Mito (Clontech) coding for the mitochondrial marker CcO7 (15). After the appropriate incubation times, transfected cells were analyzed using a Zeiss Axioplan (version 2.2) fluorescence microscope equipped with a Zeiss ApoTome system, as described previously (15).
Subcellular fractionation and Western blot (WB) analysis.
The ApoAlert cell fractionation kit (Clonetech) and radioimmunoprecipitation assay buffer (Santa Cruz) were used for protein extraction and subcellular fractionation, as described previously (15). Protein concentration was determined using a bicinchoninic acid protein assay reagent (Pierce). Protein samples were resolved on 10% NuPAGE gels and transferred to nitrocellulose membranes using an iBlot gel transfer stack and iBlot transfer system (Invitrogen).
Quantification of CcO activity.
CcO activity was measured using a CcO assay kit (Sigma), as described previously (15).
TRAIL neutralizing assays.
HeLa cells were transfected with the various constructs and incubated with either mouse anti-human TRAIL neutralizing monoclonal antibody (clone N2B2) or recombinant human decoy receptors, TRAIL-R1-Fc and TRAIL-R2-Fc (Alexis Corporation). Apoptosis was determined at 24 and 48 h posttransfection by terminal deoxynucleotidyltransferase dUTP nick end labeling (TUNEL) assay using an ApoDetect kit (Q-biogen), in accordance with the manufacturer's instructions and with minor modifications to the protocol, as described previously (15).
Statistical analyses.
The Mann-Whitney test (nonparametric and unpaired) in Prism (version 3.03) software was used. Statistical significance was defined as a P value of <0.05.
RESULTS
Subfragments of the M-MOK sequence: amino acids 67 to 86 retain all apoptotic characteristics of the total protein.
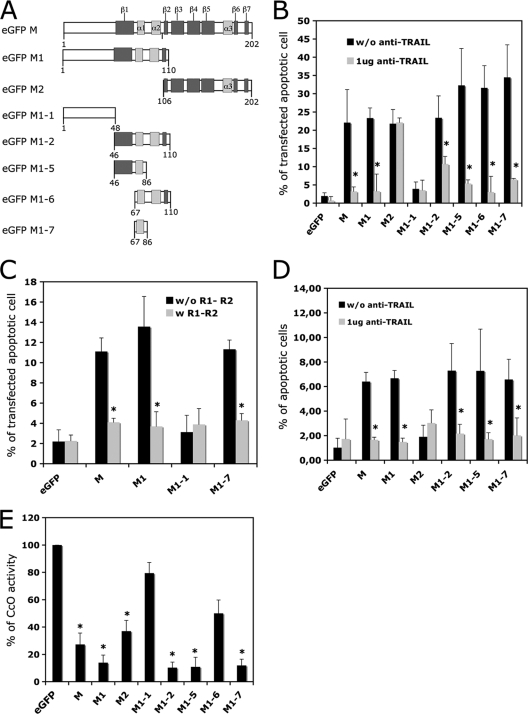
A series of truncated mutants was produced from the M-MOK sequence: M1 (aa 1 to 110), M2 (aa 106 to 202), M1-1 (aa 1 to 48), M1-2 (aa 46 to 110), M1-5 (aa 46 to 86), M1-6 (aa 67 to 110), and M1-7 (aa 67 to 86). All of these mutants respected the secondary structures of M in the related Lagos bat virus (2) (Fig. 1 A). The truncated mutants were tested for their ability to induce apoptosis through the TRAIL-dependent pathway (22) or through inhibition of CcO activity (15).
FIG. 1.
M1-7-induced apoptosis by the extrinsic and intrinsic pathways. (A) Schematic representation of M-MOK mutants with deletions N-terminally fused to an EGFP protein tag. The α helix (light gray) and β sheets (dark gray) were determined by the study of M-LAG (15). HeLa cells were transfected with plasmids encoding EGFP alone (eGFP), full-length M (M), or truncated forms (M1, M2, M1-1, M1-2, M1-5, M1-6, and M1-7). Cells were harvested after 24 h of incubation at 37°C. (B) The percentage of transfected cells undergoing apoptosis was measured by TUNEL assay. Transfections were performed in the presence or absence of a neutralizing anti-TRAIL antibody (1 μg of anti-TRAIL for 104 cells); this antibody inhibits TRAIL-mediated apoptosis. (C) HeLa cells were transfected in the presence of soluble TRAIL receptors R1 and R2. Their effect on apoptosis was measured by TUNEL assay. (D) HeLa cells were transfected with the various mutant constructs and cultured; the supernatants were collected, clarified, and added to freshly prepared cell cultures. The cultures were incubated at 37°C for 24 h in the presence or absence of anti-TRAIL antibody (1 μg of anti-TRAIL for 104 cells). The percentage of cells undergoing apoptosis was measured by TUNEL assay. The results are expressed as the mean value for three independent experiments. Error bars indicate standard deviations of these values. Significant differences before and after treatment with anti-TRAIL antibody or R1 and R2 receptors are indicated by asterisks (P < 0.05). (E) CcO activity was evaluated after 24 h. The reported values are the percentages of CcO activity relative to that of cells expressing EGFP alone normalized to the protein concentration. Mean values for three independent experiments and error bars are shown. Significant differences between cells expressing EGFP and the mutants are indicated by asterisks (P < 0.05).
HeLa cells were transfected with the plasmid constructs in the presence or absence of neutralizing anti-TRAIL or TRAIL R1 and R2 decoy receptors (Fig. 1B and C). On day 1, apoptosis was quantified by TUNEL assay. In accordance with previous findings obtained for the M1 fragment (15), the M1-1 fragment did not induce apoptosis and was used here as a negative control. However, M1-2 and all other mutants derived from it (M1-5, M1-6, and M1-7) induced apoptosis in at least 30% of the transfected cells, as assessed by TUNEL assay. In the presence of neutralizing anti-TRAIL, the extent of apoptosis induced by M, M1, M1-2, M1-5, M1-6, or M1-7 was less than 10% (Fig. 1B). Similarly, the extent of apoptosis in cells expressing M, M1, and M1-7 was significantly (P < 0.05) lower in the presence of TRAIL R1 and R2 decoy receptors (Fig. 1C), consistent with involvement of the extrinsic pathway. We tested the apoptotic effect of clarified supernatants recovered from transfected cells and added to fresh HeLa cells. The apoptotic effect of cells expressing M1-2, M1-5, or M1-7 was significantly lower (P<0.05) in the presence of neutralizing anti-TRAIL antibody (Fig. 1D). This suggests that the supernatants of cells expressing M1-2, M1-5, or M1-7 contain active soluble TRAIL, as already observed for the wild-type M protein (17). In contrast and as we previously reported, the M2 fragment induced TRAIL-independent apoptosis (15). We assayed the total protein extracts from HeLa cells for their CcO activity 24 h after transfection (Fig. 1E). CcO activity was significantly (P < 0.05) lower in cells transfected with M1-2 (aa 46 to 110) or M1-5 (aa 46 to 86) than in cells transfected with M1-6 (aa 67 to 110). However, CcO activity was also significantly lower (P < 0.05) following transfection with M1-7 (aa 67 to 86), which overlaps with both the M1-5 and M1-6 fragments.
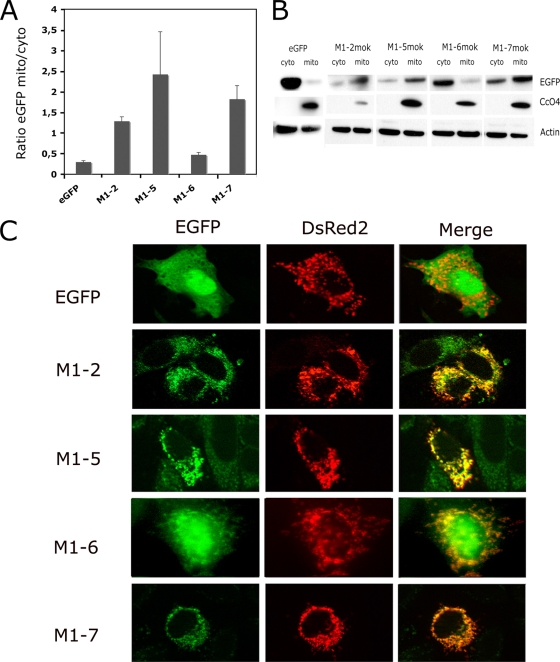
The subcellular localization of M1-5, M1-6, and M1-7 was studied by Western blot analyses of the cytosolic and mitochondrion-enriched fractions (Fig. 2 A and B). A mitochondrial marker, CcO4, and actin were used as controls to normalize the different subcellular fractions. The EGFP signal (indicating protein expression) for M1-5 and M1-7 was greater in the mitochondrion-enriched fraction than in the cytosolic fraction. In contrast, the signal for M1-6 was higher in the cytosolic fraction (Fig. 2B). This finding may explain why M1-6 did not inhibit CcO activity, unlike the M1-5 and M1-7 constructs. The subcellular localization of these fragments was confirmed in HeLa cells transfected with both reporter plasmid DsRed2-Mito and each of the truncated mutants (Fig. 2C). For M1-5 and M1-7, the EGFP and DsRed2-Mito signals were colocalized, whereas a more diffuse signal involving the mitochondria as well as the cytosol and nucleus was found for M1-6 (Fig. 2C). Transfection with a plasmid expressing EGFP alone did not result in any specific subcellular localization of the signal (Fig. 2C). Therefore, the mutant M1-7, which consists of only 20 aa of the total protein, is targeted to the mitochondria and is able to inhibit CcO activity and induce apoptosis via a TRAIL-dependent mechanism.
FIG. 2.
Localization of M-truncated mutants and M1-7 mutants in the transfected cells. (A) Mitochondrion-enriched fractions (mito) and cytosolic fractions (cyto) were analyzed by Western blotting and densitometry. For each mutant, the ratio of the signal in the mitochondrion-enriched fraction to that in the cytosolic fraction was determined, and the mean for two independent experiments is reported. (B) Representative immunoblot with anti-EGFP antibody: different times of exposure to the film are used to show EGFP-tagged constructs. However, CcO4 and β-actin bands are obtained from single exposure times. (C) HeLa cells were cotransfected with plasmid vectors expressing DsRed2-Mito and constructs encoding mutants with deletions (M1-2, M1-5, M1-6, M1-7) or the vector alone (EGFP). The subcellular localization of these constructs was assessed after 24 h with a Zeiss Axioplan 2.2 fluorescence microscope equipped with a Zeiss ApoTome system. EGFP, the EGFP signal, DsRed2 (DsRed2-Mito), the mitochondrial marker; Merge, overlay image composed of the signals for various constructs and the mitochondrial marker.
Positions 77 and 81 in the M1-7 fragment are essential for induction of apoptosis.
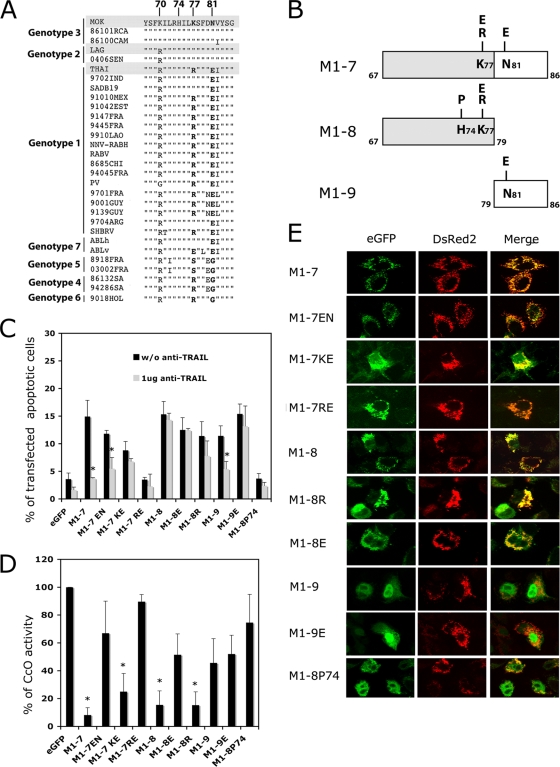
The M1-7 sequences (aa 67 to 86) from THA and MOK were compared with those from other lyssavirus genotypes. A highly conserved region was identified (Fig. 3A). The main differences between these sequences were at positions 70, 77, 81, and 82. Despite the amino acid sequence difference at position 70, M-MOK and M-LAG show similar apoptotic phenotypes (22), suggesting that the type of amino acid at position 70 has little impact on apoptosis. However, different lyssaviruses show conservative substitutions (I, V, or L) at position 82. We therefore investigated positions 77 and 81.
FIG. 3.
Role of positions K77 and N81 and the α1 helix of the M1-7 fragment in apoptosis pathways. (A) Sequence alignment of sequence positions 67 to 86 of lyssavirus matrix protein. The amino acids at positions 77 and 81 are indicated in boldface. (B) Schematic representation of M1-7 mutants with deletions N-terminally fused to an EGFP protein tag. Mutations were introduced into each subfragment at position 77 or 81. M1-8 was also mutated at position 74 (M1-8 P74). (C). HeLa cells were transfected with plasmids expressing EGFP alone (eGFP), M1-7 KN (wild type for MOK sequence) or its mutated forms (M1-7 EN, KE, and RE), or truncated mutants M1-8 K and its E77 (M1-8 E), R77 (M1-8 R), and P74 (M1-P74) mutated forms and M1-9 and its E81 mutated form (M1-9 E). The transfected cells were cultured in the presence or in the absence of a neutralizing anti-TRAIL antibody (1 μg of anti-TRAIL). After 24 h of incubation at 37°C the percentage of transfected cells undergoing apoptosis was measured by TUNEL assay. The results are expressed as mean values for three independent experiments. Error bars indicate standard deviations of these values. Significant differences in the presence and absence of treatment with anti-TRAIL antibody are indicated with asterisks (P < 0.05). (D) CcO activity was assayed after 24 h of culture. Reported results are percentages of activity relative to that of cells expressing EGFP alone (arbitrarily defined to be 100%) and normalized for protein concentration. Values are means for three independent experiments, and standard deviations are shown as error bars. Significant differences between cells expressing EGFP and the mutants are indicated by asterisks (P < 0.05). (E) HeLa cells were cotransfected with plasmid vectors encoding DsRed2-Mito and plasmids encoding mutants M1-7 EN, KE, and RE and mutants M1-8 and M1-9 or their mutants with point mutations (M1-8 E, M1-8 R, and M1-9 E). The subcellular localization of these mutants was assessed after 24 h with a Zeiss Axioplan 2.2 fluorescence microscope equipped with a Zeiss ApoTome system. eGFP, EGFP signal, DsRed2 (DsRed2-Mito), mitochondrial marker; Merge, overlay image between the various constructs and the mitochondrial pattern of the cell.
We constructed mutants of M1-7 MOK by hybridization of synthetic oligonucleotides. We replaced the K at position 77 (K77) and/or the N at position 81 (N81) (the wild-type sequence of MOK) with R or E (the acidic amino acid present in the Australian bat lyssavirus sequence) at position 77 (R77 or E77) and/or E at position 81 (E81) (Fig. 3A and B). With these substitutions, we obtained three different mutants, named M1-7 EN, M1-7 KE, and M1-7 RE (corresponding to the amino acids present in the THA sequence). We tested the effects of these variants on cell death and CcO activity. Except for M1-7 RE, all of the M1-7 mutants induced apoptosis (>8% of transfected cells were TUNEL assay positive at 24 h posttransfection) (Fig. 3C). We then tested the effects of TRAIL antagonists on reducing cell death. Anti-TRAIL antibody did not significantly influence the apoptotic effect of M1-7 KE but did, however, reduce the apoptotic effects of wild-type M1-7 KN and mutant M1-7 EN (P < 0.05) (Fig. 3C; Table 2). Therefore, N81 seemed to be important for induction of apoptosis through a TRAIL-mediated pathway.
TABLE 2.
Effects of mutations at positions 77 and 81 of M1-7 and its truncated fragments
| Strain | Phenotype observeda |
||
|---|---|---|---|
| Apoptotic effect | TRAIL-mediated pathway | CcO inhibition | |
| M1-7 KN | + | + | + |
| M1-7 KE | + | − | + |
| M1-7 EN | + | + | − |
| M1-7 RE | − | − | − |
| M1-8 K | + | − | + |
| M1-8 E | + | − | − |
| M1-8 R | + | − | + |
| M1-9 N | + | + | − |
| M1-9 E | + | − | − |
| M1-8 P74 | − | − | − |
The various phenotypes (apoptotic effect, TRAIL-mediated pathway, and CcO inhibition) observed in HeLa cells 24 h after transfection with each construct are reported as present (+) or absent (−).
We then analyzed the inhibition of CcO activity in cells transfected with plasmids expressing different mutants. In contrast, compared with the CcO activity of cells transfected with M1-7 EN and M1-7 RE, transfection of cells with M1-7 KN (wild type) and M1-7 KE resulted in a significant decrease (P < 0.05) in CcO activity (Fig. 3D). Therefore, K77 is important for inhibition of CcO activity (Fig. 3D; Table 1). The M1-7 RE variant (with R at position 77 and E at position 81, as in the Thai sequence) did not trigger TRAIL-mediated apoptosis or inhibit CcO activity (Fig. 3C and D).
The subcellular localization of these different M1-7 mutants was then studied in HeLa cells cotransfected with the DsRed2-Mito reporter plasmid. All mutants except M1-7 KE exhibited a strict colocalization of the EGFP and DsRed2-Mito signals; M1-7 KE also showed a diffuse EGFP signal outside the mitochondria (Fig. 3E).
Role of the α1 helix in the truncated mutants of M1-7.
To determine the proapoptotic regions in the M1-7 fragment, we generated two new truncated mutants, M1-8 (aa 67 to 79) and M1-9 (aa 79 to 86) (Fig. 3B). We also created mutants of M1-8 and M1-9 with different point mutations, similar to those analyzed for M1-7 (Fig. 3B). First, for M1-8, we changed K77 in R to obtain the THA sequence. Second, the basic amino acid at position 77 was replaced by an acidic one (E77) (Fig. 3A and B). In M1-9, we changed N81 in E to obtain the THA sequence. Thus, three constructs were made: M1-8 R, M1-8 E, and M1-9 E. These mutants were tested for their ability to induce apoptosis via a TRAIL-dependent pathway (Fig. 3C) or via inhibition of CcO activity (Fig. 3D). Both the M1-8 and M1-9 mutants retained the ability to induce rates of apoptosis similar to that for M1-7 (10% to 15% of transfected cells). However, apoptosis in cells transfected with different M1-8 constructs switch to a TRAIL-independent mechanism which did not occur with M1-7. In contrast, transfection with M1-9 still strongly induces TRAIL-mediated apoptosis, though this effect is lost after substitution of N81 for E81 (Fig. 3C).
With respect to inhibition of CcO activity, this action was retained only in the M1-8 and M1-8 R variants and was compromised in M1-8 E, M1-9, and M1-9 E (Fig. 3D). This provides further confirmation that the N-terminal part of M1-7 has an important role in inhibition of CcO activity (Table 2). We found that only fragments containing aa 67 to 79 (M1-8, M1-8 E, and M1-8 R) gave an EGFP signal which colocalized with the DsRed2-Mito signal (Fig. 3E). Despite having a reduced effect on CcO activity, the M1-8 E fragment is still able to target the protein to mitochondria. Furthermore, M1-9 and M1-9 E showed diffuse EGFP signals with no specific localization (Fig. 3E). To disturb the structure of the α1 helix in M1-8, we replaced the histidine at position 74 with a proline (M1-8 P74) (Fig. 3B). This P74 variant did not sustain any more induction of apoptosis and inhibition of CcO activity, even though it retained the wild-type K at position 77 (Fig. 3C and D). However, disruption of the α-helix structure clearly modified the targeting, which was no longer mitochondrial but, rather, nuclear (Fig. 3E). These results give support to the notion that the α-helix structure in the M protein is essential for mitochondrial targeting.
Modification of positions 77 and 81 in the full-length matrix protein.
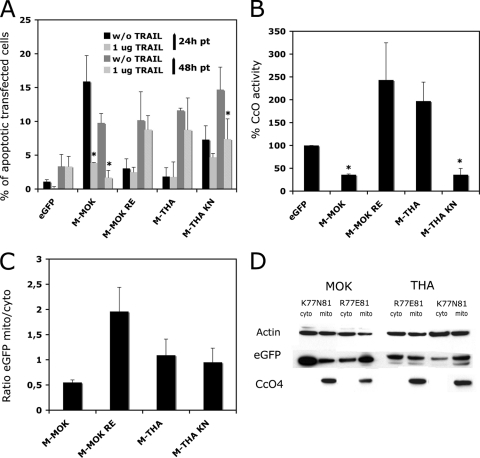
In the final part of this investigation, we tested for apoptotic effects 24 h and 48 h after transfection with full-length M-MOK and M-THA and with the M-MOK RE (representing M-MOK with the amino acids of THA at positions 77 and 81) and M-THA KN (representing M-THA with the amino acids of MOK at positions 77 and 81) mutants (Fig. 4 A). Transfection with M-MOK caused a marked induction of apoptosis as soon as 24 h (∼15% of TUNEL assay-positive cells), whereas transfection with M-MOK RE had less of an effect (<3%). Following transfection with the THA variants, M-THA showed no apoptotic effect (<2%) at 24 h, whereas M-THA KN induced a level of apoptosis of over 7% which increased to 15% at 48 h (Fig. 4A; Table 3). The presence of anti-TRAIL significantly inhibited M-MOK-induced apoptosis at 24 h and 48 h (P < 0.05) and M-THA KN-induced apoptosis at 48 h (Fig. 4A; Table 3).
FIG. 4.
Pivotal role of positions 77 and 81 of M protein on the induction of apopotosis by the intrinsic and extrinsic pathways. M-MOK KN was mutated by site-directed mutagenesis to M-MOK RE, and M-THA RE was mutated to M-THA KN. HeLa cells were transfected with the constructs encoding these mutants and with the vector alone. (A) The transfected cells were cultured in the presence or absence of a neutralizing anti-TRAIL antibody (1 μg of anti-TRAIL) for 24 and 48 h, and the percentage of transfected cells undergoing apoptosis was measured by TUNEL assay. (B) The CcO activity was measured after 24 h of culture. CcO activity is reported as a percentage of that in cells expressing eGFP alone and normalized to the protein concentration. Values are means for three independent experiments, and error bars indicate the standard deviations. Significant differences between cells expressing EGFP and the mutants are indicated with asterisks (P < 0.05). (C) Cytosol and mitochondrion-enriched fractions were tested for the presence of the mutants by Western blotting and densitometry, and the results reported are the ratio of the EGFP signal in the mitochondrion-enriched fraction to that in the cytosol fraction. (D) A representative immunoblot of fractions of cells expressing M-MOK and M-THA mutants probed with anti-EGFP antibody is shown. Different times of exposure to the film were used to reveal EGFP-tagged constructs. However, CcO-4 and β-actin were obtained from single exposure times.
TABLE 3.
Effects of mutations at positions 77 and 81 of M-MOK and M-THAa
| Strain | Phenotype observed |
|||
|---|---|---|---|---|
| Apoptotic effect |
TRAIL-mediated pathwayb | CcO inhibitionc | ||
| 24 h p.t. | 48 h p.t. | |||
| M-MOK (KN)d | + | + | + | + |
| M-MOK RE | − | + | − | − |
| M-THA (RE)d | − | + | − | − |
| M-THA KN | +/− | + | + | + |
The various phenotypes (apoptotic effect, TRAIL-mediated pathway, and CcO inhibition) observed in HeLa cells 24 h and 48 h posttransfection (p.t.) with each construct are reported as present (+) or absent (−).
Data for strains M-MOK (KN), M-MOK RE, and M-MOK (RE) are from 24 h after transfection; data for M-THA KN are from 48h after transfection.
Data for CcO activity are from 24 h after transfection.
Wild type.
We then analyzed CcO activity. After 24 h, the level of CcO activity was significantly lower for M-MOK and M-THA KN (P < 0.05) than for M-MOK RE and M-THA, for which the CcO activity was not significantly different from the control value, despite a slight tendency to be higher (Fig. 4B; Table 3). We determined the subcellular localization of all the M mutants: HeLa cells were transfected with the various constructs, and after 24 h cytosolic and mitochondrion-enriched cell fractions were tested for EGFP expression by Western blotting (Fig. 4C and D). All constructs except M-MOK RE had similar localizations, including mitochondrial targeting, whereas M-MOK RE appeared to be even more localized to the mitochondria than the others. This suggests that the failure of M-MOK RE to inhibit CcO activity was not due to inappropriate localization but to the modification of positions 77 and 81.
DISCUSSION
Apoptosis is inversely correlated with the pathogenicity of lyssaviruses (32, 42). Studies of neuronal cell cultures show that some lyssavirus genotypes, in particular, LAG and MOK, induce cell death, unlike the classical rabies virus of genotype 1 (THA), which does not kill host cells (15, 22). The lyssavirus glycoprotein has an important role in the induction of apoptosis (10, 32, 36). However, the viral M protein is also able to induce apoptosis in a manner independent of that for other viral proteins via a TRAIL and caspase 8-dependent pathway (22). A comparison of M proteins from different lyssaviruses indicated that the M protein from MOK, a genotype 3 virus with low virulence, induces cell death by dysregulating the respiratory chain (15). In cells expressing M-MOK, CcO activity was significantly inhibited and caspase 9 was activated. A segment of M-MOK (aa 46 to 110) also retains the CcO-inhibiting and apoptosis-inducing activities through the TRAIL-dependent mechanism of the intact molecule (15). The aim of the present study was to describe the apoptotic functions of M-MOK in more detail and to identify the domains of the protein involved in these functions. Using a strategy of deletion and site-directed mutagenesis, we identified a domain of 20 residues (aa 67 to 86) in M-MOK that retained the capacity to induce TRAIL-mediated cell death and inhibit CcO activity.
Our mutation analyses show that the K at position 77 in M-MOK is essential for inhibition of CcO activity: M-THA mutants presenting K77 inhibited CcO activity and all the mutations of K77 except a K-to-R conserved-charge mutation in a small subfragment spanning aa 67 to 79 restored the CcO activity. Similarly, the N at position 81 in M-MOK is necessary to induce apoptosis via the TRAIL signaling pathway. Supernatants of cells transfected with constructs expressing apoptotic activity induced TRAIL-mediated cell death in nontransfected cell cultures; the factor responsible was therefore presumably soluble and secreted into the supernatant.
TRAIL is a type II membrane cytokine with a releasable extracellular component that is capable of receptor recognition and apoptosis induction (24, 45). Although death receptors are constitutively expressed in most cell types, expression of their ligands is tightly regulated. Several viruses cause sensitivity to TRAIL or its expression and release from infected cells (6, 17, 29). Sensitivity to TRAIL is influenced by regulation of TRAIL receptor expression by the p53 tumor suppressor protein (16). The induction of type I interferon by viral infection increases the level of expression of TRAIL in various cell types (11, 38), and this can result in cell death from infiltrating immune effector cells (4, 37). However, the mechanism involved during lyssavirus infection does not seem to be related to the overexpression of TRAIL (22; unpublished results). We report on the sensitization of cells to TRAIL after expression of the M1-7 construct, resulting in cell suicide and fratricide. Neutralizing antibodies prevented cell death in this system, indicating that the increased apoptosis is most probably related to a higher level of transduction of the signal generated by constitutive TRAIL. It would be of interest to investigate whether the low TRAIL sensitivity of THA-infected cells is also due to antiapoptotic factors, such as cellular Fas-associated death domain-like interleukin-1beta-converting enzyme inhibitory protein (cFLIP) (31).
Functional signal motifs within pluripotent matrix proteins can be overlapping or situated in close proximity to each other. The active residues at positions 77 and 81 in M-MOK participate in both induction of TRAIL-mediated apoptosis and inhibition of CcO activity. These residues are located at the surface (Fig. 5) of the recently resolved crystal structure of a lyssavirus M protein and are spatially independent of other known functional motifs (2). The effects of these residues on apoptosis and CcO activity could be due to adjacent overlapping motifs or to interaction with points of cross talk between the two signaling pathways, such as B-cell lymphoma 2 (Bcl-2) family members or inhibitor of apoptosis (IAP) proteins (1, 25). The signaling pathways are not always mutually exclusive, and several death receptors activate both proapoptotic and survival pathways.
FIG. 5.
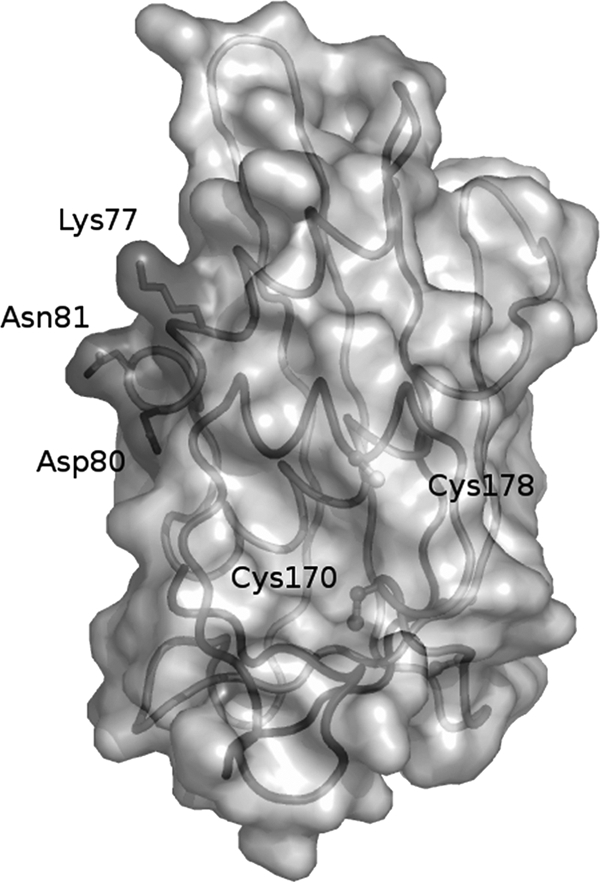
Representation of the structure of the LAG matrix protein from residues 48 to 202 after a crystallography study (2). Positions K77 and N81 are at the outside face of the protein.
Some viral disease mechanisms involve apoptosis in the CNS (33). However, the rabies virus avoids early apoptosis (21, 22). Mutation analyses of M-MOK and M-THA indicate that mutations at positions 77 or 81 modify the activity of the molecule (Fig. 4A). However, exchanging the amino acids at positions 77 and 81 of M-THA with those present in M-MOK did not completely confer the apoptotic activity of M-MOK on M-THA. Therefore, other as yet unidentified motifs within the protein are presumably involved.
Alignments of the M-protein sequences from the seven described lyssavirus genotypes showed that most strains of genotype 1 have R and E at positions 77 and 81, respectively; strains of genotypes 2 and 3 have K and N at these respective positions (Fig. 3A) (3, 22). Therefore, residues R77 and E81 in genotype 1 may correlate with delayed apoptosis and, subsequently, with higher pathogenicity. Indeed, most isolates from genotypes 2 and 3 have lower pathogenicity (30) than isolates from genotype 1 (3, 22). These correlations are consistent with the M protein being important in the modulation of the host response to infection and especially the delayed innate immune response after infection with genotype 1 virus. This delay may be essential for viral infection and colonization of the CNS. The cell death induction process described here would benefit the host by limiting viral dissemination. The sequential induction of TRAIL-mediated apoptosis, particularly the delayed induction observed in the case of THA, is intriguing (15, 22). It remains to be explored whether this delay favors dissemination of THA into the central nervous system, leading to a pathogenesis more severe than that of MOK. This is striking when it is considered that THA is a highly pathogenic dog rabies virus isolated in Asia, where there is a high incidence of rabies, and that MOK, a wild isolate from Africa, is less pathogenic in humans (only two cases in humans have so far been described in the literature) (8).
Acknowledgments
We are grateful to E. Perret and S. Shorte at the Plate-forme d'Imagerie Dynamique (PFID) at the Institut Pasteur for invaluable experimental help, discussion, and advice on data processing. We thank J. M. Grimes for his help with Fig. 5.
J.E. was supported by an ANRS grant. A.G. was supported by a predoctoral scholarship from Total.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Ashkenazi, A., and R. S. Herbst. 2008. To kill a tumor cell: the potential of proapoptotic receptor agonists. J. Clin. Invest. 118:1979-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assenberg, R., O. Delmas, S. C. Graham, A. Verma, N. Berrow, D. I. Stuart, R. J. Owens, H. Bourhy, and J. M. Grimes. 2008. Expression, purification and crystallization of a lyssavirus matrix (M) protein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64:258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badrane, H., C. Bahloul, P. Perrin, and N. Tordo. 2001. Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J. Virol. 75:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baloul, L., and M. Lafon. 2003. Apoptosis and rabies virus neuroinvasion. Biochimie 85:777-788. [DOI] [PubMed] [Google Scholar]
- 5.Benedict, C. A., P. S. Norris, and C. F. Ware. 2002. To kill or be killed: viral evasion of apoptosis. Nat. Immunol. 3:1013-1018. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, P., S. M. Meintzer, S. Gibson, C. Widmann, T. P. Garrington, G. L. Johnson, and K. L. Tyler. 2000. Reovirus-induced apoptosis is mediated by TRAIL. J. Virol. 74:8135-8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clem, R. J., and L. K. Miller. 1993. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J. Virol. 67:3730-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delmas, O., E. C. Holmes, C. Talbi, F. Larrous, L. Dacheux, C. Bouchier, and H. Bourhy. 2008. Genomic diversity and evolution of the lyssaviruses. PLoS One 3:e2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, H., and G. McFadden. 1999. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 7:160-165. [DOI] [PubMed] [Google Scholar]
- 10.Faber, M., R. Pulmanausahakul, S. S. Hodawadekar, S. Spitsin, J. P. McGettigan, M. J. Schnell, and B. Dietzschold. 2002. Overexpression of the rabies virus glycoprotein results in enhancement of apoptosis and antiviral immune response. J. Virol. 76:3374-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanger, N. A., C. R. Maliszewski, K. Schooley, and T. S. Griffith. 1999. Human dendritic cells mediate cellular apoptosis via tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J. Exp. Med. 190:1155-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay, B. B., and G. McFadden. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124:767-782. [DOI] [PubMed] [Google Scholar]
- 13.Gallei, A., S. Blome, S. Gilgenbach, N. Tautz, V. Moennig, and P. Becher. 2008. Cytopathogenicity of classical swine fever virus correlates with attenuation in the natural host. J. Virol. 82:9717-9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galluzzi, L., C. Brenner, E. Morselli, Z. Touat, and G. Kroemer. 2008. Viral control of mitochondrial apoptosis. PLoS Pathog. 4:e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gholami, A., R. Kassis, E. Real, O. Delmas, S. Guadagnini, F. Larrous, D. Obach, M. C. Prevost, Y. Jacob, and H. Bourhy. 2008. Mitochondrial dysfunction in lyssavirus-induced apoptosis. J. Virol. 82:4774-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, B., P. Yue, G. L. Clayman, and S. Y. Sun. 2001. Evidence that the death receptor DR4 is a DNA damage-inducible, p53-regulated gene. J. Cell. Physiol. 188:98-105. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, M., C. Spiropoulou, and P. E. Rollin. 2007. Ebola virus infection of human PBMCs causes massive death of macrophages, CD4 and CD8 T cell sub-populations in vitro. Virology 364:45-54. [DOI] [PubMed] [Google Scholar]
- 18.Hinshaw, V. S., C. W. Olsen, N. Dybdahl-Sissoko, and D. Evans. 1994. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J. Virol. 68:3667-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irie, T., N. Nagata, T. Igarashi, I. Okamoto, and T. Sakaguchi. 2010. Conserved charged amino acids within Sendai virus C protein play multiple roles in the evasion of innate immune responses. PLoS One 5:e10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, N., G. W. Moseley, D. Blondel, K. Shimizu, C. L. Rowe, Y. Ito, T. Masatani, K. Nakagawa, D. A. Jans, and M. Sugiyama. 2010. Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J. Virol. 84:6699-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, A. C., J. P. Rossiter, and M. Lafon. 2006. Expression of Toll-like receptor 3 in the human cerebellar cortex in rabies, herpes simplex encephalitis, and other neurological diseases. J. Neurovirol. 12:229-234. [DOI] [PubMed] [Google Scholar]
- 22.Kassis, R., F. Larrous, J. Estaquier, and H. Bourhy. 2004. Lyssavirus matrix protein induces apoptosis by a TRAIL-dependent mechanism involving caspase-8 activation. J. Virol. 78:6543-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawai, T., and S. Akira. 2007. Antiviral signaling through pattern recognition receptors. J. Biochem. 141:137-145. [DOI] [PubMed] [Google Scholar]
- 24.Kim, M. H., T. R. Billiar, and D. W. Seol. 2004. The secretable form of trimeric TRAIL, a potent inducer of apoptosis. Biochem. Biophys. Res. Commun. 321:930-935. [DOI] [PubMed] [Google Scholar]
- 25.Kominsky, D. J., R. J. Bickel, and K. L. Tyler. 2002. Reovirus-induced apoptosis requires both death receptor- and mitochondrial-mediated caspase-dependent pathways of cell death. Cell Death Differ. 9:926-933. [DOI] [PubMed] [Google Scholar]
- 26.Koyama, A. H. 1995. Induction of apoptotic DNA fragmentation by the infection of vesicular stomatitis virus. Virus Res. 37:285-290. [DOI] [PubMed] [Google Scholar]
- 27.Lafon, M. 2004. Subversive neuroinvasive strategy of rabies virus. Arch. Virol. Suppl., p. 149-159. [DOI] [PubMed]
- 28.Levine, B., Q. Huang, J. T. Isaacs, J. C. Reed, D. E. Griffin, and J. M. Hardwick. 1993. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature 361:739-742. [DOI] [PubMed] [Google Scholar]
- 29.Liang, X., J. Du, Y. Liu, M. Cui, C. Ma, L. Han, Z. Qu, Z. Zhang, Z. Sun, L. Zhang, Y. H. Chen, and W. Sun. 2007. The hepatitis B virus protein MHBs (t) sensitizes hepatoma cells to TRAIL-induced apoptosis through ERK2. Apoptosis 12:1827-1836. [DOI] [PubMed] [Google Scholar]
- 30.Markotter, W., I. V. Kuzmin, C. E. Rupprecht, and L. H. Nel. 2009. Lagos bat virus virulence in mice inoculated by the peripheral route. Epidemiol. Infect. 137:1155-1162. [DOI] [PubMed] [Google Scholar]
- 31.Morales, J. C., M. J. Ruiz-Magana, and C. Ruiz-Ruiz. 2007. Regulation of the resistance to TRAIL-induced apoptosis in human primary T lymphocytes: role of NF-kappaB inhibition. Mol. Immunol. 44:2587-2597. [DOI] [PubMed] [Google Scholar]
- 32.Morimoto, K., D. C. Hooper, S. Spitsin, H. Koprowski, and B. Dietzschold. 1999. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J. Virol. 73:510-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberhaus, S. M., R. L. Smith, G. H. Clayton, T. S. Dermody, and K. L. Tyler. 1997. Reovirus infection and tissue injury in the mouse central nervous system are associated with apoptosis. J. Virol. 71:2100-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilder, S., J. Logan, and T. Shenk. 1984. Deletion of the gene encoding the adenovirus 5 early region 1b 21,000-molecular-weight polypeptide leads to degradation of viral and host cell DNA. J. Virol. 52:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prehaud, C., S. Lay, B. Dietzschold, and M. Lafon. 2003. Glycoprotein of nonpathogenic rabies viruses is a key determinant of human cell apoptosis. J. Virol. 77:10537-10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prehaud, C., N. Wolff, E. Terrien, M. Lafage, F. Megret, N. Babault, F. Cordier, G. S. Tan, E. Maitrepierre, P. Menager, D. Chopy, S. Hoos, P. England, M. Delepierre, M. J. Schnell, H. Buc, and M. Lafon. 2010. Attenuation of rabies virulence: takeover by the cytoplasmic domain of its envelope protein. Sci. Signal 3:ra5. [DOI] [PubMed] [Google Scholar]
- 37.Raftery, M. J., C. K. Behrens, A. Muller, P. H. Krammer, H. Walczak, and G. Schonrich. 1999. Herpes simplex virus type 1 infection of activated cytotoxic T cells: induction of fratricide as a mechanism of viral immune evasion. J. Exp. Med. 190:1103-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sedger, L. M., D. M. Shows, R. A. Blanton, J. J. Peschon, R. G. Goodwin, D. Cosman, and S. R. Wiley. 1999. IFN-gamma mediates a novel antiviral activity through dynamic modulation of TRAIL and TRAIL receptor expression. J. Immunol. 163:920-926. [PubMed] [Google Scholar]
- 39.Smirnova, N. P., H. Bielefeldt-Ohmann, H. Van Campen, K. J. Austin, H. Han, D. L. Montgomery, M. L. Shoemaker, A. L. van Olphen, and T. R. Hansen. 2008. Acute non-cytopathic bovine viral diarrhea virus infection induces pronounced type I interferon response in pregnant cows and fetuses. Virus Res. 132:49-58. [DOI] [PubMed] [Google Scholar]
- 40.Sumbayev, V. V., and I. M. Yasinska. 2006. Role of MAP kinase-dependent apoptotic pathway in innate immune responses and viral infection. Scand. J. Immunol. 63:391-400. [DOI] [PubMed] [Google Scholar]
- 41.Thoulouze, M.-I., M. Lafage, A. Montano-Hirose Juan, and M. Lafon. 1997. Rabies virus infects mouse and human lymphocytes and induces apoptosis. J. Virol. 71:7372-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoulouze, M. I., M. Lafage, V. J. Yuste, G. Kroemer, S. A. Susin, N. Israel, and M. Lafon. 2003. Apoptosis inversely correlates with rabies virus neurotropism. Ann. N. Y. Acad. Sci. 1010:598-603. [DOI] [PubMed] [Google Scholar]
- 43.Uematsu, S., and S. Akira. 2006. Toll-like receptors and innate immunity. J. Mol. Med. 84:712-725. [DOI] [PubMed] [Google Scholar]
- 44.White, E., S. H. Blose, and B. W. Stillman. 1984. Nuclear envelope localization of an adenovirus tumor antigen maintains the integrity of cellular DNA. Mol. Cell. Biol. 4:2865-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiley, S. R., K. Schooley, P. J. Smolak, W. S. Din, C. P. Huang, J. K. Nicholl, G. R. Sutherland, T. D. Smith, C. Rauch, C. A. Smith, et al. 1995. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3:673-682. [DOI] [PubMed] [Google Scholar]