Abstract
Natural evolution in primate lentiviral reverse transcriptase (RT) appears to have been constrained by the necessity to maintain function within an asymmetric protein composed of two identical primary amino acid sequences (66 kDa), of which one is cleaved (51 kDa). In this study, a detailed phylogenetic analysis now segregates groups O and M into clusters based on a cysteine or tyrosine residue located at position 181 of RT and linked to other signature residues. Divergent evolution of two group O (C181 or Y181) and the main (Y181 only) HIV-1 lineages did not appreciably impact RT activity or function. Group O RT structural models, based on group M subtype B RT crystal structures, revealed that most evolutionarily linked amino acids appear on a surface-exposed region of one subunit while in a noncatalytic RT pocket of the other subunit. This pocket binds nonnucleoside RT inhibitors (NNRTI); therefore, NNRTI sensitivity was used to probe enzyme differences in these group O and M lineages. In contrast to observations showing acquired drug resistance associated with fitness loss, the C181Y mutation in the C181 group O lineage resulted in a loss of intrinsic NNRTI resistance and was accompanied by fitness loss. Other mutations linked to the NNRTI-resistant C181 lineage also resulted in altered NNRTI sensitivity and a net fitness cost. Based on RT asymmetry and conservation of the intricate reverse transcription process, millions of years of divergent primate lentivirus evolution may be constrained to discrete mutations that appear primarily in the nonfunctional, solvent-accessible NNRTI binding pocket.
Human immunodeficiency viruses (HIV) are classified into two types, HIV-1 and -2. HIV-1 is further divided into groups M (main), O (outlier), and N (non-M, non-O). Among the HIV-1 groups, group M is the most dominant in the world and consists of 9 subtypes (A to D, F to H, J, and K) and 43 circulating recombinant forms (CRFs [http://www.hiv.lanl.gov/content/index]). In contrast, HIV-2 has eight groups (A through H) with only groups A and B establishing human-to-human transmission chains. Geographical distribution of HIV-2, unlike type 1, is limited primarily to West Africa and India (11, 41). The origin of HIV-1 and -2 has been linked to central African chimpanzees (Pan troglodytes troglodytes) and West African monkeys (sooty mangabeys), respectively, which are both infected with simian immunodeficiency viruses (SIV) (18). Sequences similar to those of HIV-1 group O have been isolated from fecal material of gorillas living in the wild, an unlikely source of this HIV-SIV lineage. Although isolated from gorillas, these group O-like strains may still have originated in chimpanzees (40, 45). Recently, a similar group O-like virus was isolated from a Cameroonian HIV-infected patient living in France (30). This strain has been tentatively termed HIV-1 group P, based on its distinct clustering with the HIV-1 group O and the group O-like sequences from gorillas (30).
HIV-1 group O was first described for individuals of West Central African origin in the 1990s (15, 44) but has also been identified in Europe (8, 25), the United States (35), and several African countries (29). Aside from the Cameroon/Gabon epidemic, founder events, including the earliest-known group O infection, that of a Norwegian sailor during the early 1960s, have not established extensive epidemics in other geographical regions (17). The origin of the group O radiation has been dated to the 1920s (1890 to 1940), a timing which is similar to that of the estimated human introduction to HIV-1 group M, 1931 (1915 to 1941) (19, 22). HIV-2, in contrast, is believed to have emerged later in the 20th century, with the most recent common ancestors (MRCA) for HIV-2 groups A and B dated to 1940 ± 16 years and 1945 ± 14 years, respectively (23). It has been difficult to subdivide group O strains into clades, in part due to high genetic diversity and the availability of relatively few sequences for phylogenetic analyses. Three studies have attempted to classify these viruses into subtypes: Quinones-Mateu et al. (32) suggested two subtypes, A-O and non-A-O; Roques et al. (37) proposed subtypes O:A, O:B, and O:C; and Yamaguchi et al. (46) classified group Os into clusters I to V.
Group O viruses are genetically distinct from group M, with nucleotide sequence differences between M and O being 24 to 32% in gag, 33 to 37% in pol, and 39 to 49% in env genes. These differences translate to approximately 33.5% amino acid diversity in the reverse transcriptase (RT) coding region of pol (12, 32). Group O strains also carry natural polymorphisms such as A98G, V179E, and Y181C in the RT (12, 32, 42), thus rendering them resistant to nonnucleoside reverse transcriptase inhibitors (NNRTI). Group O viruses are, however, sensitive to protease inhibitors (PI) and nucleoside reverse transcriptase inhibitors (NRTI) as well as to newly described fusion and integrase inhibitors (7, 12, 31). The “resistance” to NNRTI in group O viruses is intrinsic and arose during the divergent evolution of HIV-1 groups M and O viruses in the simian/human immunodeficiency virus lineage of lentiviruses (19, 22, 39). Intrinsic resistance in this lineage suggests a flexibility/accommodation of the NNRTI binding pocket of RT to genetic change(s). Evidence for such flexibility may be best described by the relatively low fitness cost of NNRTI-resistant mutations K103N and Y181C compared to the higher fitness cost of most other drug-resistant mutations in HIV-1 group M subtype B isolates (10).
In this study, the evolutionary history of HIV-1/SIV lineages was compared to phenotypic characteristics (fitness and drug susceptibility) of HIV-1 group M and O clones/mutants. We observed that the majority of SIVs (including HIV-2) found in some Old World primates could be classified by an isoleucine/valine/phenylalanine (I/V/F) at position 181 in HIV-1 RT, a site related to NNRTI drug resistance. A second cluster had a tyrosine or cysteine at position 181 and included all HIV-1 isolates (groups M, N, O, and P) as well as SIV sequences from gorillas (SIVgor), chimpanzees (SIVcpz), mandrills (SIVmnd), red-capped mangabeys (SIVrcm), and l'Hoest monkeys (SIVlst). A detailed phylogenetic analysis of 43 group O and related SIVgor sequences showed two distinct genetic clusters in pol, which were also maintained in gag, int/vif, and env. These two clusters, regardless of gene, were most tightly linked to the presence of cysteine (C) versus tyrosine (Y) at position 181 of RT, hence the new terms C181- and Y181-like group O subtypes. As described herein, this divergent group O evolution does not appear to alter the secondary structures or RT activity compared to that of HIV-1 group M subtype B RT (i.e., the HXB2 RT) (28).
To further explore possible phenotypic differences of these C181- and Y181-like subtypes of group O, we carried out site-directed mutagenesis using the only available group O infectious molecular clone (pCMO2.41) (42) to generate the combinations of C181, Y181, and various other linked substitutions (A98, V179, N103, R103, S103) of the two lineages. Mutant viruses related to the group O C181 and Y181 lineages as well as group M lineage (subtype B) were tested for their susceptibility to NNRTI. All the wild-type (WT) and mutant viruses were also used in pairwise competitions/dual infections to evaluate relative fitness. Our results reveal that amino acid substitutions in the NNRTI-resistant group O clone, which resulted in an NNRTI-sensitive virus, were also associated with a cost in replicative fitness. In contrast, mutations in group M viruses conferring NNRTI resistance resulted in fitness losses (10). These findings suggest that different primate lentiviral lineages evolved with neutral alterations in a solvent-accessible pocket in RT. Consequentially, modulation in NNRTI susceptibility provides an excellent tool to model this high degree of divergent evolution.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
U87 human glioma cells expressing CD4 and either CCR5 or CXCR4 were obtained through the AIDS Research and Reference Reagent Program. These cells were maintained in Dulbecco modified Eagle medium (Mediatech, Inc., Herndon, VA) supplemented with 15% fetal bovine serum (FBS; Mediatech, Inc.), penicillin (100 U/ml), streptomycin (100 μg/ml), puromycin (1 μg/ml) and G418 (300 μg/ml). 293 T cells were grown in Dulbecco modified Eagle medium containing 10% FBS and penicillin-streptomycin.
HIV-1 group O proviral plasmid pCMO2.41, a replication-competent clone (42) was used to generate 13 different mutants. Single mutants included G98A, K103N/R/S, E179V, and C181I/V/Y. Double mutants were generated in the Y181 backbone with a G98A, K103N/R/S, or E179V mutation (data not shown). Mutant plasmids were generated using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The pNL4-3 plasmids carrying either a K103N or Y181C mutation were kindly provided by Lisa Demeter and Carrie Dykes (Rochester, NY) (1). Infectious chimeric viruses were produced by transfecting 293T cells using Effectene transfection reagent (Qiagen, Valencia, CA). Supernatant-containing virus was collected 2 days posttransfection, clarified by centrifugation at 2,500 rpm for 10 min, and purified through a 0.45-μm-pore-size filter (Millipore, Billerica, MA). Viruses were subsequently passaged through U87-CD4/CCR5 or U87-CD4/CXCR4 cells to remove debris produced in transfection. Sequence analysis was performed with virus stocks to confirm that reversion or new mutations did not emerge during propagations. Virus titers were calculated by using a limited-dilution 50% tissue culture infective dose (TCID50) method (Reed-Muench endpoint titration) with U87-CD4/CCR5 or U87-CD4/CXCR4 cells as described previously (24). Virus production was measured with a reverse transcription assay utilizing radiolabeled dTTP ([α-32P]TTP) as described previously (43). YU2, a CCR5-using laboratory-adapted HIV-1 group M subtype B strain, was used as a control in the drug susceptibility assay.
Drugs and drug sensitivity assays.
Nevirapine (NVP), efavirenz (EFV), delavirdine (DLV), TMC-125 (etravirine, ETV), and lamivudine (3TC) were obtained from the AIDS Research and Reference Reagent Program. Stocks were initially diluted in dimethyl sulfoxide (DMSO) and then into RPMI medium as working concentrations. All drugs were validated against reference strain NL4-3. Sensitivity to inhibitors of HIV-1 replication was determined by infection of U87-CD4/CCR5 or U87-CD4/CXCR4 cells for R5- and X4-utilizing HIV-1 viruses, respectively. Cells were added to 96-well plates at a density of 1 × 104 cells/well and allowed to adhere overnight. Cells were treated with NVP (1 pM to 10 μM), EFV (10 fM to 1 μM), DLV (10 fM to 1 μM), ETV (10 fM to 1 μM), or 3TC (1 pM to 10 μM) for 1 h prior to infection. Cells were then exposed to virus at a multiplicity of infection (MOI) of approximately 0.01 infectious units/ml for 24 h, after which cells were washed with phosphate-buffered saline (PBS) and fresh medium containing drug was added. Supernatant aliquots were taken at time intervals from days 3 to 10, and virus production was quantified by a radioactive reverse transcriptase assay in bulk. Fifty percent inhibitory concentration (IC50) curves were constructed for supernatant samples taken at the time of peak virus production. IC50s were determined directly from the curves as previously described (24).
Growth competition assays and PCR.
Full pairwise dual-infection/competition assays were performed with 14 group O viruses (13 site-directed mutant-derived viruses and the wild-type CMO2.41). NL4-3-derived viruses (WT, K103N, and Y181C) were used as controls and competed against each other as previously described (5). Competitions were performed in 48-well plates using 1 × 105 cells by adding the two viruses at equal MOIs of 0.0001. A monoinfection representing each of the viruses in the competition was included at the same MOI. Virus production was monitored by RT assays using cell-free supernatants. At peak RT activity (day 9), cells and supernatants were harvested and stored at −80°C. DNA was extracted from harvested cells using a Qiagen DNA extraction kit. Nested PCR was performed for pol to cover the region containing residues 103 and 181 of RT using the following previously described primers: first round, RTS-1gpO and RTA-9 gp O; second round, RTS-2 gp O and RTA-8 gpO for the group O samples (2). The primers for the NL4-3 controls, also previously described, were as follows: first round, RTS-1 and RTA-9; second round, RTS2 and RTA-8 (21). PCR products were run on a gel to confirm these products and then used for ligation detection reactions (LDR) as outlined below.
Oligonucleotide design and LDR.
Oligonucleotides for LDR were matched to anneal upstream and downstream to an HIV-1 isolate-specific polymorphism at 60 to 61°C under the appropriate conditions. Downstream interrogator oligonucleotides (with bead associations) and upstream reporter capture oligonucleotides (with 5′-phosphate and 3′-biotin modifications) were used (data not shown). Reverse transcriptase codon 98 sequences GGGG and AGCG, 103 sequences KAAG, NAAC, SAGC, and RAGG, and 181 sequences VGTY, IATY, YTAY, and CTGY were used to discriminate and quantify, by LDR, the level of one and the other strains in a dual infection.
Ligase discrimination reactions used 5 units of Ampligase DNA ligase (Epicentre Biotechnologies, Madison, WI) (as defined in reference 6), 7.5 nM concentrations of each oligonucleotide, and 10 ng template DNA in a buffer containing 15 mM Tris-HCl (pH 8.3), 0.06% Triton X-100, 1 mM dithiothreitol, 40 mM KCl, 7.5 mM MgCl2, 0.3 mM NAD+ sodium salt, and 0.08% PEG 6000 in a final volume of 12 μl. Reactions were subjected to 300 cycles at 95°C for 10 s and 37°C for 40 s. Detections for HIV-1 reverse transcriptase codon 103 were performed by subjecting reactions to 300 cycles of 95°C for 10 s and 66°C for 40 s.
Luminex detection of ligated oligonucleotides for estimation of replicative fitness.
Approximately 500 of the appropriate xTAG beads (Luminex Corporation, Austin, TX) were added to each LDR in 1× TMAC buffer (2.5 M tetramethyl ammonium chloride [TMAC], 0.1 M Tris-HCl [pH 8.0], 3 mM EDTA, and 0.1% SDS) in a volume of 60 μl. Reactions were subjected to 95°C for 5 min followed by 37°C for 45 min. One hundred nanograms of streptavidin-phycoerythrin conjugate (SAPE; Invitrogen S-866) was then added to each reaction in a solution of 1× TMAC buffer in a volume of 6 μl. Reactions were subjected to 37°C for 45 min. Seventy-five beads per region were measured with a Luminex 200 instrument at the highest RP1 photomultiplier tube setting (typically 700 to 750 V). The bead counts obtained from the Luminex instrument were used to determine fitness. The cutoff/background values were set as 2 standard deviations from the mean result for replicate negative controls. Estimation of viral fitness was performed as previously described (5). Briefly, the final ratio of the two viruses produced from each dual infection was determined by comparing the virus production in the competition to the virus production in the monoinfection. The production of each group O virus in a dual infection (f0) was divided by the initial proportion of the inocula (i0) to determine the relative fitness (W = f0/i0). The fitness difference (Wd) is the ratio of the relative fitness values of each group O virus in the competition (Wd = wM/wL) (5).
Fitness relationships and rank order of group O NNRTI mutants.
Integer programming is often used to solve various types of ranking problems; this method was applied using the Lingo optimization system in order to represent the fitness relationships between the strains. Viruses were ranked based on the binary outcomes of the pairwise competition events, with the correspondence of the ranking with the competition results maximized; under the constraint of transitivity in the ranking, e.g., a given virus must be fitter than all of the viruses ranked below it. The resulting “closest match” transitive ranking and the fitness relationships between each strain were then represented in figures using Visio (version 2007; Microsoft Corporation).
Sequence and phylogenetic analyses.
Genotypic relationships between the RT coding region of HIV and SIV and within the group O RT sequences were determined by examining the available nucleotide sequences of at least 719 bp of the RT coding region retrieved from the Los Alamos National Laboratory HIV database. Three gorilla SIV sequences (CP684.1, CP2149.287, and CP2135.1), five HIV-2 sequences, the HIV-1 subtype B reference sequence (HXB2), and two group O reference sequences (Ant70 and MVP5180) were included in the RT lentiviral analyses. Forty-three RT sequences were used to construct the group O trees. For each analysis, sequences were aligned and used for the construction of phylogenetic trees by neighbor-joining and Bayesian methods, using MEGA and MrBayes (version 3.0; http://mrbayes.csit.fsu.edu/) programs, respectively. As a preliminary analysis, the neighbor-joining method in MEGA was used to construct a phylogenetic tree from the nucleotide alignment using the Kimura-2-parameter model with default parameters. The tree was bootstrapped with 1,025 repetitions. A phylogenetic tree was constructed next using MrBayes, with SIVcpz_US_Marilyn defined as the out group. The general time reversible substitution model was used, with a gamma-shaped rate of variation and a proportion of invariable sites. The data set was partitioned by codons, with model parameters unlinked across partitions. The model parameters were kept at default values. The number of generations was 300,000, with burning of 1,250. Convergence was achieved with 0.023316 as the average standard deviation of split frequencies.
For analysis of the relationship between the ancient Norwegian group O sequence spanning the 3′ integrase to the 5′ vif (817 bp), HIV-1 group O nucleotide sequences spanning the length of the ancient sequence were retrieved from the Los Alamos HIV National Laboratory database (http://hiv-web.lanl.gov). Group O-like gorilla sequences, HXB2, and SIVcpz_Marilyn were aligned with group O sequences, and a maximum-likelihood phylogenetic tree was constructed using PhyML (version 3.0). The HKY substitution model was used with all parameters estimated from the data. A BIONJ tree was used as the starting tree, and nonparametric bootstrapping with 100 repetitions was performed in order to estimate branch support. TreeRate (version beta; http://www.hiv.lanl.gov) with single time point optimization was used to root the tree.
Structural analysis.
To assess the effect of mutations on the overall structure of HIV-1 RT, we generated mutant models by sequentially changing the amino acid residues followed by minimization protocols as described below. Two models of HIV-1 reverse transcriptase were generated using the crystal structure of nevirapine (NVP)-bound RT (Protein Data Bank [PDB] file 1VRT) (36). These group O models contain C181 and Y181 backgrounds. The Y181 background contained R28, R103, V142, N174, and I178. The C181 background contained K28, K103, I142, D174, and L178. This structure contains the Y181 background residues. To generate the mutant model, the amino acid residues were mutated, and a local minimization was carried out by using the “annealing” application of the SYBYL molecular modeling package. The Tripos force field and AMBER parameters (these are parts of SYBYL) were used to local minimization. After the generation of all mutants, the entire structures were minimized until the energy gradient difference between two subsequent steps of minimization became less than 0.1. The structures of the WT and mutant HIV-1 RT were superimposed using the Cα coordinates of the β strands in the palm subdomain.
RESULTS
Phylogenetic analysis of RT sequence and clustering among SIV and HIV.
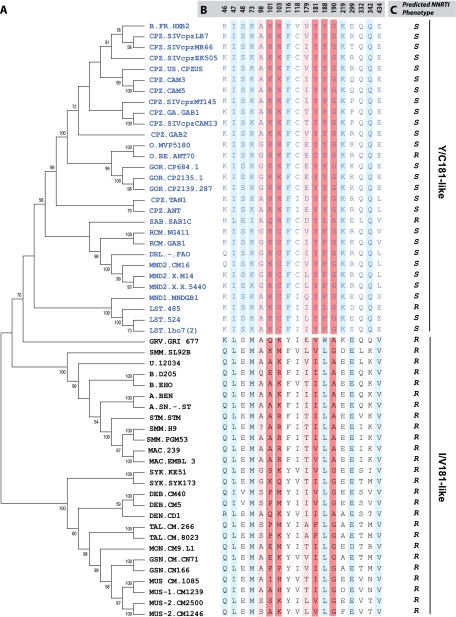
SIV from nonhuman primates are the source of HIV, as indicated by the high degree of viral sequence similarities among humans infected with HIV and primates carrying SIV. pol is the most conserved gene of lentiviruses and is useful in tracing the evolutionary path of these viruses. The entire RT sequences from different SIV and all HIV lineages were aligned and used to construct phylogenetic trees by the neighbor-joining and Bayesian methods. HIV-1, SIVcpz, SIVrcm, SIVsab, SIVmnd, and SIVlst clustered together, while HIV-2, SIVsmm, SIVmac, SIVsyk, SIVdeb, SIVtal, SIVmon, SIVgsn, and SIVmus formed a second cluster (Fig. 1A), confirming earlier reports (9). To determine if these two clusters (HIV-1 and HIV-2) had distinguishing amino acids residues, we aligned the amino acid sequences using the VESPA program (20). Eighteen RT positions (46, 47, 48, 73, 98, 101, 103, 116, 118, 179, 181, 188, 190, 219, 299, 332, 342, and 434) were selected based on at least 70% conservation with one or the other cluster (Fig. 1B). The highest conservation between clusters mapped to amino acid residue 181 in RT and, as a consequence, lineages in primate lentiviruses were classified as Y/C181-like (i.e., the cluster containing HIV-1) or I/V181-like (i.e., the cluster containing HIV-2) (Fig. 1B). In the I/V181-like cluster, only the SIV RT sequences from the talapoin monkey (SIVtal) were exceptions, with phenylalanine at position 181 (Fig. 1A and B). Interestingly, SIVtal also contained an alanine at position 179, which is unique to this SIV lineage and suggests a possible coevolution with F181 (Fig. 1A and B).
FIG. 1.
Phylogenetic analyses of primate lentivirus evolution in the reverse transcriptase gene. Phylogenetic analyses were performed with the RT of HIV and SIV strains (A). Sequences obtained from the HIV Los Alamos database were aligned to construct trees using neighbor-joining and Bayesian methods as described in Materials and Methods. The tree was bootstrapped with 1,025 repetitions. VESPA was used to identify conserved signature amino acid residues among SIV and HIV RT (B), leading to the identification of two distinct clusters, (Y/C181), which include all HIV-1 strains and a set of SIVs (SIVcpz, SIVmnd, SIVlst, SIVrcm), and (I/V181), which consists of HIV-2 and all monkey-derived SIVs. The numbers at the top of the alignment represent the RT amino acid positions. Amino acids known to confer high- and low-level resistance to NNRTIs are indicated in dark red and light red, respectively. Conserved positions linked to the Y/C181 or I/V181 lineages are indicated in sky blue. Linkage was performed using VESPA to identify conserved amino acid residues, i.e., those unique to the consensus sequence of each cluster. (C) Predicted NNRTI phenotypes are shown as R (resistant) or S (susceptible). Abbreviations for primates are as follows: CPZ, chimpanzees; HUM, humans; GOR, gorilla; RCM, red-capped mangabey; SAB, sabeus; DRL, drill; MND, mandrill; LST, l'Hoest monkey; GRV, grivet; SMM, sooty mangabey; STM, stump-tailed macaques; MAC, rhesus macaques; SYK, Sykes monkey; DEB, De Brazza monkey; TAL, talapoin; MON, mona monkey; GSN, greater spot-nosed monkey; MUS, mustached monkey.
In these two primate lentivirus clusters (Y181 and C181), there are five positions that appear to diverge in the lineages, that are found in the solvent-accessible RT pocket, and that are also associated with primary resistance to NNRTI, namely, 101, 103, 181, 188, and 190 (Fig. 1B). Almost all the SIV strains from monkeys in the Y/C-181 cluster possess a phenylalanine or tyrosine residue at position 188 and a L188 in the I/V-181-like group (>95% among each group) (Fig. 1B and C). Positions 101, 103, and 190 show evidence of linked evolution but were less conserved among the two groups. It is important to note that even though the majority of directed evolution in primate lentiviruses occurred in this solvent-accessible pocket of RT, possible NNRTI resistance would be an artifact, since none of these viruses were ever exposed to these drugs.
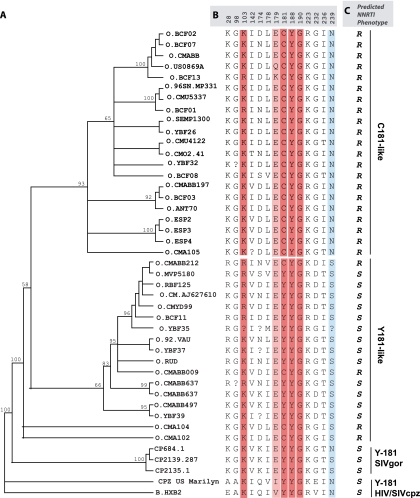
Earlier reports by Descamps et al. (12) and Quinones-Mateu et al. (32) showed that intrinsically NNRTI-resistant group O strains formed a single cluster in a phylogenetic tree (12, 32). With the expansion of group O sequences in the HIV database, we now aligned 38 group O RT sequences and constructed a phylogenetic tree using MrBayes (version 3.0, http://mrbayes.csit.fsu.edu/). Two distinct group O clusterings were observed, namely, for C181 and Y181 strains (Fig. 2A). The cluster containing C181 group O sequences showed a more distinct branching, with a higher bootstrap value (93%) than that for the Y181 cluster (bootstrap value of 66%) (Fig. 2A). Few C181-like isolates, namely, 96CMABB009, CMABB212, CMA104, and CMA102, were found to be in the Y181 cluster of RT (Fig. 2A). Isolate 96CMABB009 had been described as unclassified in RT (37). CMABB497, CMA102, and CMA104 are recombinants with their apparent Y181-like sequences (Fig. 2A and B) clustering with the C181 group O lineage in the trees constructed from 3′ integrase to 5′ vif gene (int/vif) sequences (Fig. 3).
FIG. 2.
Phylogenetic analyses of human immunodeficiency virus type 1 group O evolution in the reverse transcriptase gene. (A) Neighbor joining phylogenetic analysis was performed with the first 750 nucleotides from 38 HIV-1 group O and 3 SIVgor group O-like sequences obtained from the Los Alamos HIV database (see Materials and Methods and the legend to Fig. 1). (B) Signature amino acid residues were identified using VESPA and are grouped as C181-like or Y181-like sequences. CMO2.41, the clone used in this study, is in the C181-like cluster. The majority of group O sequences are C181-like, and the minority bears Y181 and cluster more closely with HIV-1 group M (HXB2). The amino acid positions are indicated at the top of the sequence. Question marks represent positions with no clear amino acid, as outlined in the Los Alamos database. Amino acids known to confer high- and low-level resistance to NNRTIs are indicated in dark red and light red, respectively. Completely conserved positions (>99%) are shown in sky blue. (C) Predicted NNRTI phenotypes are indicated as R (resistant) or S (susceptible).
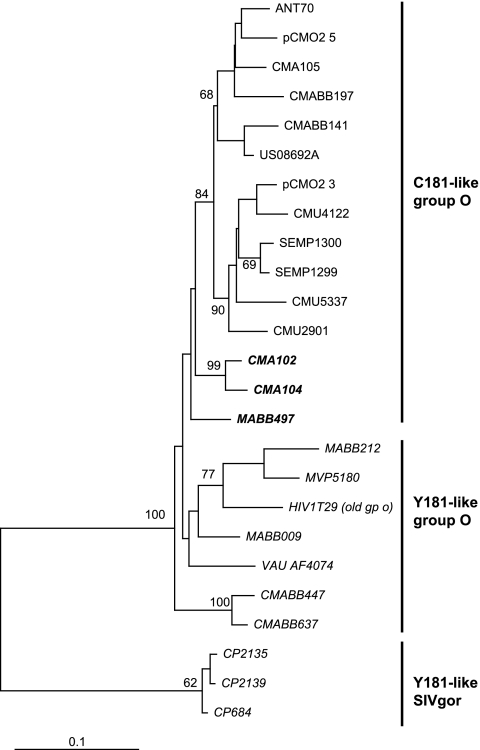
FIG. 3.
Linking the ancient group O strain from a 1960 infection to the Y181 group O lineage. An 817-bp fragment (integrase to vif) of group O variants was aligned with the only sequenced region of the Norwegian HIV-1 group O infection dated to 1960 (HIV1T29). A maximum-likelihood phylogenetic tree was constructed using PhyML, as described in more detail in Materials and Methods. The tree was rooted with HXB2 and SIVcpz_US_Marilyn as the out group.
The most ancient group O sequence (HIV1T29) described to date was obtained from a Norwegian patient infected in the 1960s. A previous neighbor-joining phylogenetic tree analysis of a 432-bp pol sequence from this patient found that it clustered with the non-A-O viruses (Y181-like group O strains in this study) (32). Phylogenetic trees using MrBayes confirmed the inclusion of this ancient int/vif group O sequence with an int/vif cluster of group O isolates whose RT sequences also formed the monophyletic Y181 cluster (Fig. 3). These findings suggest that the Y181 lineage had been circulating in the human population of central Africa before the 1960s and that the first cross-species transmission of SIVgor is at least consistent with the previous molecular clock analyses for an introduction around 1890 to 1940 (22). Interestingly, a simian progenitor of the larger C181 lineage of group O has not been identified (40).
Molecular modeling of HIV-1 group O C181-like and Y181-like RT.
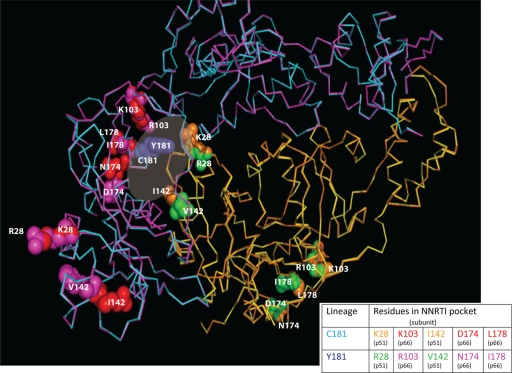
To determine possible structural differences between the C181 and Y181-like group O RTs, we first generated consensus sequences of C181-like and Y181-like RT strains using the panel of 38 group O sequences described above. Using VESPA, five signature group O RT residues were linked to either C181 or Y181 and could differentiate these two clusters in group O. These were K28, K103, I142, D174, and L178 for C181-like and R28, R103, V142, N174, and I178 for Y181-like strains (Fig. 2B). The mutant models of the Y181 and C181 lineages of HIV-1 group O RTs (as described in Materials and Methods) were then superimposed using Cα-coordinated residues of β strands 7, 9, and 10 from the palm subdomain using SYBYL (Tripos Associates, St. Louis, MO) and the X-ray RT crystal structure (36) as the template. Models included unliganded and NVP-bound RT (PDB file 1VRT) (Fig. 4). No significant changes were observed in the overall RT structure of the two lineages (181C and 181Y), with the root mean square deviation (rmsd) of the Cα backbone below 0.5Å for the entire RT structures. However, several striking observations from the modeling are noted. First, the five signature residues in both the 181C and 181Y lineages clustered near the dimer interface and NNRTI binding pocket as part of the p66 or p51 subunits (Fig. 4). Second, the signature residues that are part of the pocket/interface in p66 (i.e., 103, 174, and 178) or p51 (28 and 142) (Fig. 4) are found on the surface-exposed regions of RT. Fine structural differences were observed at these interface surface contacts between these two lineages of RTs (181Y versus 181C). For example, a hydrophobic compensation was necessary due to the V142 in the p51 subunit of the 181Y lineage. Signature residues of the C181 and Y181 lineages appeared necessary to maintain a functional RT structure through both subunits in our models. Interestingly, if these linked mutations of both lineages were placed into the opposing RT subunits (p66 and p51), the models with the p66 of the C181 lineage in a heterodimer with the p51 of the Y181 lineage were unstable.
FIG. 4.
Superimposition of the group O RT structural models for the C181 and Y181 lineages. A superimposition of C181 and Y181 group O RT models was generated by sequentially changing the amino acid residues of a crystal structure of nevirapine (NVP)-bound group M subtype B RT and minimized as described in Materials and Methods. Positions of the amino acid residues of interest in the NNRTI binding pocket in the two structures are indicated as follows. p66 subunits of C181 and Y181 RTs are shown as the Ca backbone in cyan and magenta, respectively, and the p51 subunits are colored yellow and dark orange, respectively. The linked residues for each lineage are rendered as space-filled spheres (red for the C181 and magenta for the Y181 lineages in p66; green for the C181 and orange for the Y181 lineages in p51). The five signature group O RT residues in the p51 and p66 subunits which are linked to C181 or Y181 are labeled in the structure and also shown in a table below the structure.
Translating differential group O evolution to phenotype-replicative fitness, RT structure, and NNRTI sensitivity.
To explore differential group O evolution and the retention of RT activity/function, the CMO2.41 group O molecular clone was mutated to toggle RT amino acids that were found in the C181 and Y181 lineages and that were associated with intrinsic NNRTI resistance in group O. These CMO2.41 virus mutants were then used in drug susceptibility analyses and in pairwise dual-virus competitions to determine how group O has evolved through a fitness landscape. Sensitivity to NNRTIs provides a general probe for group O RT structure, considering that the drugs nevirapine (NVP), delavirdine (DLV), etravirine (ETV), and efavirenz (EFV) bind at slightly different sites in the NNRTI pocket and are more or less dependent on key residues in Y181 and C181 lineages for this binding. By comparing sensitivities to these drugs with the respective structural models of RT as well as impacts on replicative fitness, we can determine how group O evolved as two separate lineages which are both fully functional, distinct from subtype B, and intrinsically resistant to NNRTIs.
In these analyses, the level of resistance differed based on the type of NNRTI (NVP, EFV, DLV, or ETV) and genetic backbone of the group O molecular clone. The CMO2.41 clone belongs to the C181 lineage and carried two other “wild-type” residues known to confer some minimal NNRTI resistance in HIV-1 group M subtype B isolates (Fig. 2A to C) (42). The A98G mutation confers some resistance to NVP, primarily, while the V179E mutation causes minimal resistance to NVP, DLV, and EFV in group M subtype B HIV-1 isolates. Site 103 is also commonly mutated during NNRTI treatment with group M, but natural 103 variation exists with the Y181 group O lineage. R103 occurs frequently, as indicated by the presence of this sequence in BCF01 and BCF13 of the C181 cluster (Fig. 2A and B). R103 has not been observed for HIV-1 group M subtype B but occurs in about 1% of non-subtype B strains from untreated patients.
To determine the role of these residues in NNRTI resistance and replicative fitness, we generated site-directed mutants in either a C181 or mutated Y181 backbone of CMO2.41. Since most primate lentivirus lineages appear linked to the 181 position in RT, we also generated C181I and C181V mutants, i.e., residues found in most SIVs and HIV-2 strains (Fig. 1B and data not shown). Site-directed mutations were introduced into the pCMO2.41 plasmid, and the infectious titers of the resulting viruses were used for drug susceptibility and fitness studies (data not shown).
Fitness analyses.
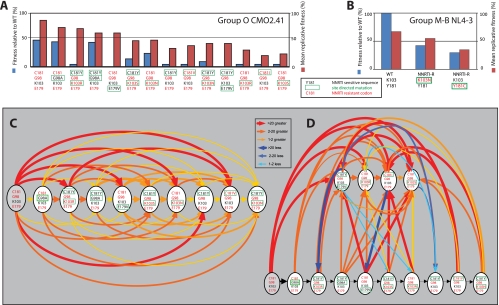
Fitness analyses were first performed as previously described (5) and involved 169 dual infections of U87.CD4.CCR5 cells, for which each HIV-1 group O CMO2.41 virus (13 mutants and the wild-type virus) was competed against each other in a full pairwise analysis. The wild-type HIV-1 group O CMO2.41 was the most fit virus, whereas each mutation (away from the “wild type”) resulted in a decrease in fitness (Fig. 5A). As described below, mutations that changed wild-type CMO2.41 sequences from intrinsically NNRTI resistant to more NNRTI sensitive (e.g., E179V, G98A, and C181Y) resulted in decreases in fitness (Fig. 5A). To understand the effects of various mutations on fitness, we ranked the viruses based on the binary outcomes of pairwise competitions. In these analyses, a given virus must be fitter than those ranked below it. Overall, a transitive relationship in replicative fitness was observed for most of the 13 HIV-1 CMO2.41-derived viruses used in this 169-competition pairwise fitness matrix (Fig. 5C and D). For example, wild-type CMO2.41 (the proverbial “A” virus) was more fit than CMO2.41G98A (the proverbial “B” virus) in direct competition (i.e., A > B), whereas CMO2.41G98A out-competed CMO2.41V179E (the proverbial “C” virus) (Fig. 5A) (i.e., B > C). To complete this transitive relationship (A > B, B > C; thus, A > C), wild-type CMO2.41 did indeed outgrow CMO2.41V179E in direct competitions (Fig. 5C) (A > C). In a comparison of all 169 competitions, a linear rank order was observed for all 13 viruses (Fig. 5C and D, red arrows), but only 6 of the 169 competitions involving CMO2.41V179E/C181Y, CMO2.41K103R, CMO2.41C181I, and CMO2.41K103S did not follow a transitive relationship (blue arrows).
FIG. 5.
Fitness of HIV-1 group O and M WT and derivative drug-resistant mutants. Pairwise competitions were performed among 14 different group O viruses and also the NL4-3-derived viruses. The production of each virus in a dual infection (f0) was divided by the initial proportion of the inocula (i0) to determine the relative fitness (W = f0/i0). (A) Mean relative fitness of group O mutants relative to the wild type (blue bars) and total relative fitness (red bars). Mean relative fitness was calculated from direct competition against only the wild-type group O CMO2.41 clone, whereas mean relative fitness was derived from the average for 13 competitions performed against a specific strain. Each virus is identified by amino acid sequence at positions 181, 98, 103, and 179 but contains CMO.2.41 sequence for the remainder of the genome. Amino acids in red text represent an NNRTI-resistant codon, and black text represents an NNRTI-sensitive codon. The amino acids boxed by a green line represent the change introduced by site-directed mutagenesis. (B) Mean relative fitness of group M (NL4-3) mutants (Y181 and C181) relative to the WT and the total relative fitness are represented as blue and red bars, respectively. (C and D) Fitness rank order of HIV-1 group O NNRTI mutants is illustrated by a transitive relationship. Ten of the 14 mutants formed a transitive chain (shown in panel C), while four others formed a separate chain (D); intermediate relationships are also shown. The red, orange, and yellow arrows represent transitive relationships and point from the more fit to the less fit viruses. As indicated in the key, the “heat map” and the thickness of the arrow lines indicate the relative ability of one virus to outcompete another. The yellow, orange, and red represent transitive competitions, and the “cooler” blue arrows (light to dark blue) represent fallout competitions of this transitive chain in the pairwise competition. Six (blue arrows) of 169 competitions were responsible for the inability to form transitive chains by four viruses. Thick, thicker, and thickest arrows represent total relative fitness values of between 1 and 2, 2 and 20, and 21 and 100, respectively.
Two previous reports had demonstrated the fitness rank order of the group M subtype B laboratory strain NL4-3 and the subtype B clone, 89ES061, using direct dual-virus competitions (10, 16). A fitness order of WT > K103N > Y181C was described by Collins et al. (10), whereas Iglesias-Ussel et al. (16) showed a rank order of Y181C > WT > K103N. Resistance to NNRTIs, RTIs, protease inhibitors, and integrase inhibitors in HIV-1 has always resulted in a fitness cost (10, 13, 26, 27). Use of WT NL4-3 or an NL4-3 harboring Y181C and K103N in competitive fitness assays confirmed the results of Collins et al. (10), in that the WT was more fit than the K103N variant which, in turn, out-competed the subtype B NL4-3 mutant, Y181C (Fig. 5B). As shown in Fig. 5A, the NNRTI-resistant, wild-type group O strain (CMO2.41) was of higher replicative fitness than any of its NNRTI-sensitive derivatives harboring the C181Y, C181I, C181V, G98A, or E179V mutation. Although increased replicative fitness is not associated with the emergence of HIV-1 drug resistance, group O HIV-1 strains attained natural resistance to NNRTI through a divergent evolutionary pathway from group M HIV-1 and definitely not in relation to NNRTI treatment/selection (discussed below). Thus, there was a prediction of decreased fitness due to mutations from the consensus, NNRTI-resistant group O C181 lineage, even though many of these mutations would result in an NNRTI-sensitive virus. For example, the C181Y mutation in CMO2.41 renders the virus sensitive to most NNRTIs but is still associated with a fitness loss (Fig. 5A and 6B to E). The N/R/S103 mutations are not common in the C181 lineage, and all confer EFV resistance in Y181 lineage. When introduced as K103N, R, or S into WT CMO2.41 (EFV sensitive), this mutant CMO2.41 gained resistance to EFV but lost fitness compared to that of the WT (Fig. 5A).
FIG. 6.
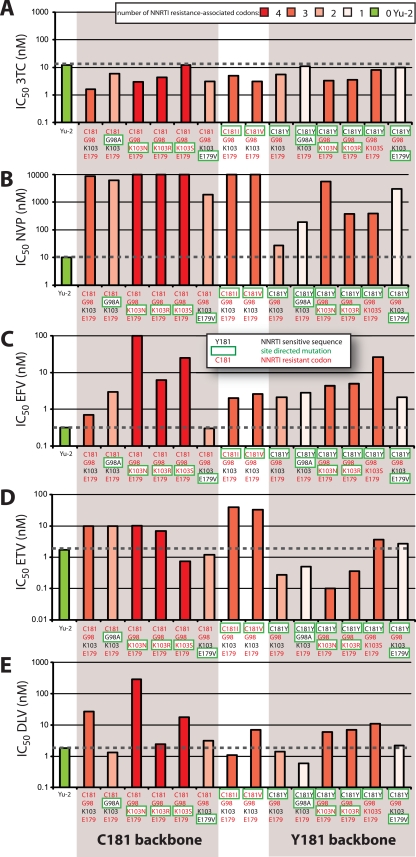
Sensitivity of site-directed mutants of group O CMO2.41 virus to nonnucleoside RT inhibitors. Group O CMO2.41 virus and its derivative mutants as well as the group M subtype B Yu2 were tested for sensitivity to lamivudine (3TC) as an NRTI control and four NNRTIs. U87-CD4/CCR5 cells were incubated with 10-fold dilutions of (A) lamivudin (3TC), (B) nevirapine (NVP), (C) efavirenz (EFV), (D) etravirine (ETV), and (E) delaviride (DLV) and exposed to virus for 24 h before washing. The supernatants were collected at 4, 6, and 9 days postinfection. Drug susceptibility curves (data not shown) were plotted to determine IC50s using the Probit program. The two group O clusters (C181 and Y181 backbones) are shaded. The bars in all panels are shown in different shades of red based on the number of NNRTI resistance-associated codons harbored in each. The Yu2 control is in green and does not harbor NNRTI-resistant codons. Each virus is identified by amino acid sequence at positions 181, 98, 103, and 179 but contains CMO.2.41 sequence for the remainder of the genome. Amino acids in red text represent an NNRTI-resistant codon, and black text represents an NNRTI-sensitive codon. The amino acids boxed by a green line represent the change introduced by site-directed mutagenesis.
Drug susceptibility and relation to RT structure.
Drug susceptibility assays were performed with U87.CD4 CCR5 cells using four NNRTIs (NVP, EFV, DLV, and ETV) and one NRTI (3TC) as a control. As expected, all group O variants and the YU2 control displayed similar susceptibilities to 3TC inhibition, with IC50s ranging from 1.6 to 12 nM (Fig. 6A). In contrast, the subtype B YU-2 strain was highly sensitive to inhibition by all NNRTIs, whereas CMO2.41 and most of the derived mutants showed some degree of resistance to NNRTIs (Fig. 6B to E). With “wild-type” CMO2.41, a C181 lineage, the highest level of resistance was observed to NVP (>1,000-fold over that of Yu-2) compared to EFV, DLV, or ETV (∼100-fold) (Fig. 6B to E).
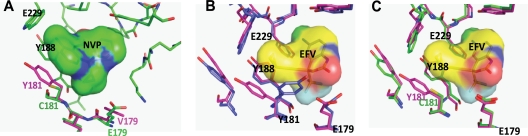
Sensitivity to NVP inhibition was only modestly “restored” (to group M subtype B HIV-1 sensitivity levels) with the G98A and E179V mutations in group O CMO2.41. A98 and V179 are considered wild-type sequences in group M subtype B HIV-1 isolates (Fig. 6B). As residue 179 is at interacting distance with NVP at the NNRTI binding pocket, the mutation of glutamine to hydrophobic valine would improve hydrophobic interactions with NVP (∼5-fold decrease in IC50) (Fig. 7A, magenta). Minimal reversion in NVP resistance with G98A and V179E likely relates to the strong exclusion of NVP in the group O NNRTI binding pocket (Fig. 7A). The impact of these G98A and E179V mutations also appears relatively benign on RT structure/function, considering that these mutations had the least fitness cost in a group O C181 background (Fig. 5A). Mutation E179V might increase susceptibility to EFV (∼4-fold decrease in IC50), based on hydrophobic interactions between valine and the trifluoromethyl group of EFV (Fig. 6C and Fig. 7B).
FIG. 7.
RT models of NVP and EVF bound by various group O variants in the NNRTI binding pocket. Models of group O RT bound to NVP or EFV were generated by sequentially changing the amino acid residues of a crystal structure of a NVP- or EFV-bound subtype B RT, as described in Materials and Methods. (A). The Y181-like group O RT bearing V179 (magenta) was superimposed on the WT CMO2.41 (C181 and E179 [green]). Only the NVP bound to WT CMO2.41 is shown. WT CMO2.41 has an impact on the hydrophobic interactions with NVP at the NNRTI binding pocket, as described in Results. (B) EFV bound to the Y181, E179, and K103 mutant RT (blue) superposed on the Y181, E179, and N103 mutant (magenta). The conformational change caused by mutating residue 181 is important for NVP and not EFV binding. (C) Superposition of Y181 CMO2.41 RT EFV (magenta) onto CMO2.41 WT (C181, green) HIV RT. In panels B and C, only the EVF bound to the Y181-like RT is shown.
Subtype B RT crystal structures with NNRTI describe overlapping binding sites for NVP, DLV, and ETV with contact to the Y181 residue, whereas EFV binds in the NNRTI binding pocket with more dependency on the 103 residue. In subtype B, EFV resistance is commonly conferred by a K103N or, less frequently, a K103S mutation. K103 has been found in all wild-type group M isolates and most C181-like group O isolates, whereas R103 forms a subcluster in the Y181-like group O tree (Fig. 2A and B). In contrast, the 103 site is unlinked to the 181 position in the I/V181-like primate lentivirus lineage, with a toggling at 103 between amino acids K, M, R, Q, P, and H but not asparagine or serine (Fig. 1A and B). Mutating CMO2.41 to harbor K103R in the C181 backbone resulted in EFV resistance but did not appreciably affect resistance to NVP, possibly because of the high level of resistance resulting from the C181 mutation. The K103R mutant actually slightly sensitized the virus to ETV or DLV compared to results with the wild-type CMO2.41 (Fig. 6D and E), whereas the K103R/C181Y double mutant had a minimal effect on ETV and DLV sensitivity but weak NVP resistance (Fig. 6D and E). This result again is consistent with the general control of ETV and with DLV sensitivity by residue 181 as opposed to residue 103 in the NNRTI binding pocket. A K103R mutation occurs only in the C181-like lineage and is one of the least stable mutations in the modeling studies on RT of this group O C181 lineage. Thus, it is not surprising that K103R in CMO2.41 resulted in a demonstrable decrease in replicative fitness (derived from the pairwise competition studies) (Fig. 5A). In contrast, a lysine or arginine is common in the group O Y181 lineage (Fig. 2A), which again supports the observation that the fitness of the mutant CMO2.41 carrying a R103 and Y181 is similar to that of the wild-type CMO2.41 with K103 and C181 (Fig. 5A). It appears that the C181Y can compensate for mutations the R103 in group O.
Mutations K103N/S are not found in the lentivirus lineage in the absence of direct NNRTI treatment/selection. These two mutations confer among the highest costs in replicative fitness for the CMO2.41 backbone. As described earlier, K103N/S confers resistance to EFV in subtype B. K103N/S as well as K103R confer a higher level of resistance to EFV in the CMO2.41 virus (C181-like lineage) than in the CMO2.41C181Y virus (Y181-like lineage) (Fig. 6C). Low-level EFV resistance was observed with the K103S/C181Y mutations (Fig. 6C). Previous studies reported that most group O HIV-1 isolates retain sensitivity to EFV, suggesting that this drug could be employed in highly active antiretroviral treatment (HAART). However, unlike the case with most group M isolates, resistance to EFV might involve a unique pathway in group O isolates due to a high fitness cost of K103 mutations in the C181-like lineage.
The C181Y mutation in CMO2.41 resulted in a virus highly susceptible to NVP, DLV, and ETV but with a modest increase in resistance to EFV (Fig. 6B to E). Modeling of NVP in the binding pocket of group O RT showed that CMO2.41 with C181 is relatively resistant to NVP because of minimal interactions between the short cysteine and the NVP (Fig. 7A, green). Interestingly, C181Y RT gains sensitivity to NVP (∼20 nM compared to 10,000 nM IC50 of WT) because substitution to tyrosine restores contacts between the 181 aromatic ring and NVP (Fig. 7A). As expected, V181 and I181 mutants also presented a high level of resistance to NVP and ETV (Fig. 6B and E) and had no significant effect on susceptibility to DLV or EFV (Fig. 6C and D). In both the group O and group M subtype B RT models, mutations at codon 181 (C181Y/I/V) have minimal effects on EFV resistance due to weak interactions between residue 181 and the cyclopropyl ring of EFV compared to interactions with the larger rings of NVP (Fig. 7B and C). Although the C181I and C181V mutations had mixed and relatively minor effects on NNRTI sensitivity, these mutations did significantly reduce replicative fitness (Fig. 5A). This reduction in fitness could also explain the absence of valine and isoleucine in the HIV-1/SIV lineages (Y/C181-like) compared to those of HIV-2/SIVmac and other SIVs, which cluster with HIV-2 (I/V181-like). In the HIV-2/SIVmac lineage, for example, I181 or V181 could be stabilized to linked L47, E48, M73, A101, I118, D188, L332, K342, and/or E434 residues not found in either of the Y/C181-like lineages of primate lentiviruses (Fig. 1).
DISCUSSION
Recent studies of nonhuman primates have clearly traced the origin of HIV-1 to P. troglodytes troglodytes (18). The central African chimpanzee P.troglodytes troglodytes and its eastern African counterpart P. troglodytes schweinfurthii are the only two chimpanzee species infected with SIVcpz. The other two chimpanzee species, P. troglodytes verus and P. troglodytes vellerosus, both found in West Africa, are not infected with SIVcpz, suggesting a separation of chimpanzees in eastern and central Africa prior to this primate lentivirus transmission. Following SIV transmission and the introduction of HIV-1 groups (M, N, and O) into humans, the subtypes have evolved partially through founder effects, leading to HIV-1 genetic forms showing different geographical distributions (33, 34). However, this human spread was disproportionate, as indicated by the limited group O pandemic, despite a similar estimated date of human introduction (22, 23). Although the group O-like strains were isolated in gorillas (45), it is more likely that these viruses were transmitted from chimpanzees (40).
Bailes et al. (4) have reported the hybrid origin of SIVcpz, the source of HIV-1. In the amino terminus of pol, SIVcpz appears to be a recombinant originating from mostly monkey-derived SIVs, namely, SIVgsn or SIVrcm. Our results confirmed the hypothesis that SIVrcm within the RT region of pol clustered with SIVcpz strains. Our findings also suggest that SIVlst, SIVman, and SIVsab carried RT sequences which are similar to those of HIV-1 groups M, N, O, and P and SIVcpz strains but more distant from the other cluster, containing HIV-2 as well as SIVmac, SIVsmm, SIVstm, SIVsyk, SIVdeb, SIVtal, SIVgsn, and SIVmus. A tyrosine or cysteine versus an isoleucine or valine at position 181 of RT is the most defined amino acid between these two respective clusters of primate lentiviruses. Another five RT positions are linked to these Y/C181- and I/V181-like groups. Interestingly, the recently described, ancient endogenous lentivirus RT from a gray mouse lemur also carried a tyrosine at position 181 (14). As described below, the cluster harboring HIV-1 groups M, O, and N and SIVcpz can be further subdivided into two phylogenetic lineages with a set of signature RT sequences linked to either Y181 or C181 in RT. Recombination and diversification of immunodeficiency viruses in different host species leaves open the possibility that nonhuman primates may carry SIV strains with the C181-like and Y181-like group O lineages but that are still undiscovered. Alternatively, the C181-like and Y181-like lineage may have evolved in humans, but this hypothesis is not supported by the low prevalence of HIV-1 group O even at its peak. In Cameroon, Gabon, and Equatorial Guinea, the number currently infected with group O is less than 20,000 and never reached a prevalence of beyond 0.6% (3). Nonetheless, an initial HIV-1 group O classification by several groups (32, 37, 46) suggested that group O strains most likely arose from a single cross-species transmission.
Several research studies have attempted to accommodate group O genetic diversity based on gag, pol, and/or env phylogenetic trees and, as a consequence, defined two (A-O and non-A-O) (32), three (O:A, O:B, and O:C) (37), and five (I, II, III, IV, and V) (46) different subtypes. Lack of a clear structure in the group O phylogenetic tree could be explained by an early diversification in humans or even prior to its human introduction. Although clusters/subtypes have been defined by nucleotide sequences, previous attempts to identify amino acid residues that correlate with the phylogenetic structure in group M or group O sequences have failed (37, 46). This may be due in part to a focus on more diverse gene regions, such as gp41, C2-V3 in env, and even CA in gag. In our study, we have now defined group O clusters as well as most primate immunodeficiency virus lineages based on signature residues at position 181 in RT. With the exception of some HIV-1 group O strains (defined as C181-like or O:A or Ia/Ib/IV clusters) (32, 37, 46), the Y181 residue is found in a large collection of phylogenetically linked SIV sequences from SIVcpz, SIVgor, SIVrcm, SIVdrl, SIVmnd, and SIVlst as well as HIV-1 group M and some O strains (see above). It is important to note that K28, I142, L178, G232, I236, and N239 dominate in the C181 lineage and that these amino acids are extremely rare in the Y181 lineage of group O. The tight linkage to signature sequences in group O RT lineages are rarely found among the HIV-1 group M subtypes within RT or any other coding sequence/gene.
The impact of divergent group O evolution on phenotype can be difficult to interpret. Previous studies indicate that recombinant group O RT is virtually indistinguishable from group M subtype B RT based on in vitro polymerase or RNase H kinetics using both RNA and DNA templates (28). This conservation in RT function is observed despite a 33% amino acid diversity between group O and M RTs. Upon a close characterization of the HIV-1 evolutionary history in RT, the only distinguishing structural feature between HIV-1 groups in RT found appears to be the alterations in a solvent-accessible, noncatalytic pocket of RT formed by both the p66 and p51 subunits. RT crystal structures of group M subtype B and models of both Y181- and C181-like group O RTs reveal conserved positioning of catalytic residues (e.g., D110, Y183, D185, and D186 in the polymerase active site), even with rather large movements in this noncatalytic, solvent-accessible RT pocket. Of course, this pocket is also the target of many anti-HIV-1 drugs, namely, the class of nonnucleoside RT inhibitors, hence the term NNRTI binding pocket. Surprisingly, the drug resistance mutations (e.g., Y181C) selected during NNRTI therapy (Y181C, A98G, K103N, V179E) are at the same amino acid sites as those that segregate lineages of group O as well as group O from group M and other primate lentivirus species. As a consequence, sensitivity to NNRTIs can be used as a means to explore and probe the impact of divergent evolution in the RT of group O lineages and of HIV-1 group M. In conjunction with NNRTI susceptibility, replicative fitness establishes a baseline for RT function in relation to a replicating virus.
A molecular clone of group O representing the C181 lineage was mutated to harbor signature group O sequences that were linked to C181 or Y181 and associated with NNRTI resistance. In general, any mutation moving the group O clone from the C181 to the Y181 lineage (or HIV-1 group M) resulted in a loss of replicative fitness. However, many of these mutations resulted in loss of NNRTI resistance. In general, a loss of acquired resistance in a pathogen would be associated with a gain in fitness. However, in this case, resistance was intrinsic and the result of divergent RT evolution under pressure to maintain catalytic activity and function. In other words, Wright's fitness landscape would predict that any fitness loss derived from the evolution of the unique C181 lineage would be eventually regained given time. Interestingly, this C181 lineage either converged or never diverged from the Y181 lineage in RT function while undergoing significant structural rearrangements in the NNRTI binding pocket of RT. As described below, evolution of lentiviral RT is unique in that all genetic change must be accommodated by two subunits of identical sequence but with differential structure in an asymmetric dimer.
In summary, a clear pattern appears to have emerged during the evolution of HIV-1 and other primate lentiviral RTs. Aside from nearly identical enzymatic activity, all primate lentivirus utilize tRNALys,3 for initiation of the minus-strand DNA and process the same 3′ and central polypurine tracts for initiation of the plus-strand DNA. This conservation in function is remarkable considering the >30% amino acid diversity between divergent, primate lentiviral RT sequences (e.g., HIV-1 groups O and M). Nonetheless, this genetic diversity can be mapped to specific signature sequences that toggle through discrete amino acid residues between different SIVs, HIV types, and HIV-1 groups. Furthermore, this divergent RT evolution appears to have taken a path of least functional consequence for enzyme activity by introducing amino acid changes in the solvent-accessible NNRTI pocket of one RT subunit in conjunction with change in surface-exposed regions of the opposite subunit. This constrained evolution is likely related to the fact that HIV-1 RT is the only asymmetric dimer in the virus. In fact, an asymmetric protein homodimer or heterodimer (with the same primary amino acid sequence in both subunits) appears to be a relatively rare phenomenon in nature. Given the relatively rapid HIV-1 mutation rate, this constrained evolution with a single gene (forming an asymmetric heterodimer) may be unique and may also be difficult to study for any other organism or system.
Even with characterization of this constrained evolution, a key phenotypic tool to analyze evolutionary change is quite rare for most biological systems. NNRTI drugs provide an effective tool to probe the structural consequences of this natural RT evolution. Again, it must be stressed that each branch point on the primate lentiviral tree may harbor fully functional RT. However, the evolutionary history can be partly reconstructed by mutating key residues in this solvent-accessible RT pocket. These studies indicate that any “swapping” of signature RT sequences within the HIV-1 group O lineages resulted in fitness loss. Thus, any change in the solvent-accessible RT pocket, no matter how subtle, is likely accommodated by compensatory mutations (i.e., the associated signature sequences).
Acknowledgments
Research at Case Western Reserve University was supported by the National Institute of Allergy and Infectious Diseases, NIH (grant AI49170).
All virus work was performed in the biosafety level 2 and 3 facilities of the Case/UH Center for AIDS Research (AI25879).
The authors have no conflicting financial interests.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Archer, R. H., C. Dykes, P. Gerondelis, A. Lloyd, P. Fay, R. C. Reichman, R. A. Bambara, and L. M. Demeter. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arien, K. K., A. Abraha, M. E. Quinones-Mateu, L. Kestens, G. Vanham, and E. J. Arts. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayouba, A., P. Mauclere, P. M. Martin, P. Cunin, J. Mfoupouendoun, B. Njinku, S. Souquieres, and F. Simon. 2001. HIV-1 group O infection in Cameroon, 1986 to 1998. Emerg. Infect. Dis. 7:466-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 5.Ball, S. C., A. Abraha, K. R. Collins, A. J. Marozsan, H. Baird, M. E. Quinones-Mateu, A. Penn-Nicholson, M. Murray, N. Richard, M. Lobritz, P. A. Zimmerman, T. Kawamura, A. Blauvelt, and E. J. Arts. 2003. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 77:1021-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barany, F. 1991. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl. Acad. Sci. U. S. A. 88:189-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briz, V., C. Garrido, E. Poveda, J. Morello, P. Barreiro, C. de Mendoza, and V. Soriano. 2009. Raltegravir and etravirine are active against HIV type 1 group O. AIDS Res. Hum. Retroviruses 25:225-227. [DOI] [PubMed] [Google Scholar]
- 8.Charneau, P., A. M. Borman, C. Quillent, D. Guetard, S. Chamaret, J. Cohen, G. Remy, L. Montagnier, and F. Clavel. 1994. Isolation and envelope sequence of a highly divergent HIV-1 isolate: definition of a new HIV-1 group. Virology 205:247-253. [DOI] [PubMed] [Google Scholar]
- 9.Clewley, J. P., J. C. Lewis, D. W. Brown, and E. L. Gadsby. 1998. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J. Virol. 72:10305-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, J. A., M. G. Thompson, E. Paintsil, M. Ricketts, J. Gedzior, and L. Alexander. 2004. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J. Virol. 78:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damond, F., M. Worobey, P. Campa, I. Farfara, G. Colin, S. Matheron, F. Brun-Vezinet, D. L. Robertson, and F. Simon. 2004. Identification of a highly divergent HIV type 2 and proposal for a change in HIV type 2 classification. AIDS Res. Hum. Retroviruses 20:666-672. [DOI] [PubMed] [Google Scholar]
- 12.Descamps, D., G. Collin, F. Letourneur, C. Apetrei, F. Damond, I. Loussert-Ajaka, F. Simon, S. Saragosti, and F. Brun-Vezinet. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J. Virol. 71:8893-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dykes, C., J. Wang, X. Jin, V. Planelles, D. S. An, A. Tallo, Y. Huang, H. Wu, and L. M. Demeter. 2006. Evaluation of a multiple-cycle, recombinant virus, growth competition assay that uses flow cytometry to measure replication efficiency of human immunodeficiency virus type 1 in cell culture. J. Clin. Microbiol. 44:1930-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gifford, R. J., A. Katzourakis, M. Tristem, O. G. Pybus, M. Winters, and R. W. Shafer. 2008. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U. S. A. 105:20362-20367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurtler, L. G., P. H. Hauser, J. Eberle, B. A. Von, S. Knapp, L. Zekeng, J. M. Tsague, and L. Kaptue. 1994. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J. Virol. 68:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iglesias-Ussel, M. D., C. Casado, E. Yuste, I. Olivares, and C. Lopez-Galindez. 2002. In vitro analysis of human immunodeficiency virus type 1 resistance to nevirapine and fitness determination of resistant variants. J. Gen. Virol. 83:93-101. [DOI] [PubMed] [Google Scholar]
- 17.Jonassen, T. O., K. Stene-Johansen, E. S. Berg, O. Hungnes, C. F. Lindboe, S. S. Froland, and B. Grinde. 1997. Sequence analysis of HIV-1 group O from Norwegian patients infected in the 1960s. Virology 231:43-47. [DOI] [PubMed] [Google Scholar]
- 18.Keele, B. F., H. F. Van, Y. Li, E. Bailes, J. Takehisa, M. L. Santiago, F. Bibollet-Ruche, Y. Chen, L. V. Wain, F. Liegeois, S. Loul, E. M. Ngole, Y. Bienvenue, E. Delaporte, J. F. Brookfield, P. M. Sharp, G. M. Shaw, M. Peeters, and B. H. Hahn. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 20.Korber, B., and G. Myers. 1992. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res. Hum. Retroviruses 8:1549-1560. [DOI] [PubMed] [Google Scholar]
- 21.Lalonde, M. S., R. M. Troyer, A. R. Syed, S. Bulime, K. Demers, F. Bajunirwe, and E. J. Arts. 2007. Sensitive oligonucleotide ligation assay for low-level detection of nevirapine resistance mutations in human immunodeficiency virus type 1 quasispecies. J. Clin. Microbiol. 45:2604-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemey, P., O. G. Pybus, A. Rambaut, A. J. Drummond, D. L. Robertson, P. Roques, M. Worobey, and A. M. Vandamme. 2004. The molecular population genetics of HIV-1 group O. Genetics 167:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemey, P., O. G. Pybus, B. Wang, N. K. Saksena, M. Salemi, and A. M. Vandamme. 2003. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. U. S. A. 100:6588-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobritz, M. A., A. J. Marozsan, R. M. Troyer, and E. J. Arts. 2007. Natural variation in the V3 crown of human immunodeficiency virus type 1 affects replicative fitness and entry inhibitor sensitivity. J. Virol. 81:8258-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loussert-Ajaka, I., M. L. Chaix, B. Korber, F. Letourneur, E. Gomas, E. Allen, T. D. Ly, F. Brun-Vezinet, F. Simon, and S. Saragosti. 1995. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J. Virol. 69:5640-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, J., P. Sista, F. Giguel, M. Greenberg, and D. R. Kuritzkes. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20). J. Virol. 78:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menendez-Arias, L., A. Abraha, M. E. Quinones-Mateu, A. Mas, M. J. Camarasa, and E. J. Arts. 2001. Functional characterization of chimeric reverse transcriptases with polypeptide subunits of highly divergent HIV-1 group M and O strains. J. Biol. Chem. 276:27470-27479. [DOI] [PubMed] [Google Scholar]
- 29.Peeters, M., A. Gueye, S. Mboup, F. Bibollet-Ruche, E. Ekaza, C. Mulanga, R. Ouedrago, R. Gandji, P. Mpele, G. Dibanga, B. Koumare, M. Saidou, E. Esu-Williams, J. P. Lombart, W. Badombena, N. Luo, M. Vanden Haesevelde, and E. Delaporte. 1997. Geographical distribution of HIV-1 group O viruses in Africa. AIDS 11:493-498. [DOI] [PubMed] [Google Scholar]
- 30.Plantier, J. C., M. Leoz, J. E. Dickerson, F. De Oliveira, F. Cordonnier, V. Lemee, F. Damond, D. L. Robertson, and F. Simon. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871-872. [DOI] [PubMed] [Google Scholar]
- 31.Poveda, E., P. Barreiro, B. Rodes, and V. Soriano. 2005. Enfuvirtide is active against HIV type 1 group O. AIDS Res. Hum. Retroviruses 21:583-585. [DOI] [PubMed] [Google Scholar]
- 32.Quinones-Mateu, M. E., J. L. Albright, A. Mas, V. Soriano, and E. J. Arts. 1998. Analysis of pol gene heterogeneity, viral quasispecies, and drug resistance in individuals infected with group O strains of human immunodeficiency virus type 1. J. Virol. 72:9002-9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rambaut, A., D. Posada, K. A. Crandall, and E. C. Holmes. 2004. The causes and consequences of HIV evolution. Nat. Rev. Genet. 5:52-61. [DOI] [PubMed] [Google Scholar]
- 34.Rambaut, A., D. L. Robertson, O. G. Pybus, M. Peeters, and E. C. Holmes. 2001. Human immunodeficiency virus: phylogeny and the origin of HIV-1. Nature 410:1047-1048. [DOI] [PubMed] [Google Scholar]
- 35.Rayfield, M. A., P. Sullivan, C. I. Bandea, L. Britvan, R. A. Otten, C. P. Pau, D. Pieniazek, S. Subbarao, P. Simon, C. A. Schable, A. C. Wright, J. Ward, and G. Schochetman. 1996. HIV-1 group O virus identified for the first time in the United States. Emerg. Infect. Dis. 2:209-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren, J., R. Esnouf, E. Garman, D. Somers, C. Ross, I. Kirby, J. Keeling, G. Darby, Y. Jones, D. Stuart, and. 1995. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat. Struct. Biol. 2:293-302. [DOI] [PubMed] [Google Scholar]
- 37.Roques, P., D. L. Robertson, S. Souquiere, F. Damond, A. Ayouba, I. Farfara, C. Depienne, E. Nerrienet, D. Dormont, F. Brun-Vezinet, F. Simon, and P. Mauclere. 2002. Phylogenetic analysis of 49 newly derived HIV-1 group O strains: high viral diversity but no group M-like subtype structure. Virology 302:259-273. [DOI] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Sharp, P. M., E. Bailes, F. Gao, B. E. Beer, V. M. Hirsch, and B. H. Hahn. 2000. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem. Soc. Trans. 28:275-282. [DOI] [PubMed] [Google Scholar]
- 40.Takehisa, J., M. H. Kraus, A. Ayouba, E. Bailes, F. Van Heuverswyn, J. M. Decker, Y. Li, R. S. Rudicell, G. H. Learn, C. Neel, E. M. Ngole, G. M. Shaw, M. Peeters, P. M. Sharp, and B. H. Hahn. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tebit, D. M., I. Nankya, E. J. Arts, and Y. Gao. 2007. HIV diversity, recombination and disease progression: how does fitness “fit” into the puzzle? AIDS Rev. 9:75-87. [PubMed] [Google Scholar]
- 42.Tebit, D. M., L. Zekeng, L. Kaptue, L. Gurtler, O. T. Fackler, O. T. Keppler, O. Herchenroder, and H. G. Krausslich. 2004. Construction and characterization of an HIV-1 group O infectious molecular clone and analysis of vpr- and nef-negative derivatives. Virology 326:329-339. [DOI] [PubMed] [Google Scholar]
- 43.Torre, V. S., A. J. Marozsan, J. L. Albright, K. R. Collins, O. Hartley, R. E. Offord, M. E. Quinones-Mateu, and E. J. Arts. 2000. Variable sensitivity of CCR5-tropic human immunodeficiency virus type 1 isolates to inhibition by RANTES analogs. J. Virol. 74:4868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanden Haesevelde, M., J. L. Decourt, R. J. De Leys, B. Vanderborght, G. van der Groen, H. H. van, and E. Saman. 1994. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J. Virol. 68:1586-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Heuverswyn, F., Y. Li, C. Neel, E. Bailes, B. F. Keele, W. Liu, S. Loul, C. Butel, F. Liegeois, Y. Bienvenue, E. M. Ngolle, P. M. Sharp, G. M. Shaw, E. Delaporte, B. H. Hahn, and M. Peeters. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. [DOI] [PubMed] [Google Scholar]
- 46.Yamaguchi, J., A. S. Vallari, P. Swanson, P. Bodelle, L. Kaptue, C. Ngansop, L. Zekeng, L. G. Gurtler, S. G. Devare, and C. A. Brennan. 2002. Evaluation of HIV type 1 group O isolates: identification of five phylogenetic clusters. AIDS Res. Hum. Retroviruses 18:269-282. [DOI] [PubMed] [Google Scholar]