Abstract
So far there has been no report of any clinical or preclinical evidence for chromosomal vector integration following adenovirus (Ad) vector-mediated gene transfer in vivo. We used liver gene transfer with high-capacity Ad vectors in the FAHΔexon5 mouse model to analyze homologous and heterologous recombination events between vector and chromosomal DNA. Intravenous injection of Ad vectors either expressing a fumarylacetoacetate hydrolase (FAH) cDNA or carrying part of the FAH genomic locus resulted in liver nodules of FAH-expressing hepatocytes, demonstrating chromosomal vector integration. Analysis of junctions between vector and chromosomal DNA following heterologous recombination indicated integration of the vector genome through its termini. Heterologous recombination occurred with a median frequency of 6.72 × 10−5 per transduced hepatocyte, while homologous recombination occurred more rarely with a median frequency of 3.88 × 10−7. This study has established quantitative and qualitative data on recombination of adenoviral vector DNA with genomic DNA in vivo, contributing to a risk-benefit assessment of the biosafety of Ad vector-mediated gene transfer.
Recombinant adenovirus (Ad) vectors are under clinical development for different applications, including tumor therapy, vaccination, and gene therapy. Today, the largest number of clinical gene transfer trials has been based on Ad vectors (http://www.wiley.co.uk/genmed/clinical). Several Ad vectors are in phase III clinical trials, and two products have already been approved in China. The occurrence of malignancies due to retroviral integration and oncogene activation in a clinical trial for the treatment of children with SCID-X1 (10) has pointed to the need for a thorough preclinical evaluation of potential genotoxic effects due to chromosomal integration of gene transfer vectors as an important part of the overall risk-benefit analysis. Detailed information on genotoxicity following gene transfer is available for vectors derived from viruses of the Retroviridae and Parvoviridae families (2, 20, 23, 26, 46). Between 60 and 75% of integrations of retrovirus, lentivirus, or adeno-associated virus (AAV)-based vectors take place in or close to genes.
Chromosomal integration of Ad vector DNA following gene transfer in cell culture has been analyzed in only a few studies, and even less is known about Ad vector integration in vivo. Since the life cycle of wild-type adenovirus is extrachromosomal, Ad vectors are perceived to be nonintegrating vectors. However, in earlier studies it was observed that injection of hamsters with wild-type adenovirus type 12 (Ad12) resulted in tumor formation due to chromosomal integration of virus DNA and expression of the E1A/E1B oncoproteins (33). Recent in vitro studies with Ad vectors with E1 deletions have demonstrated the occurrence of vector integration following transduction of transformed cell lines and primary cells, with the frequencies of homologous and heterologous recombination being between 10−3 and 10−6 and between 10−3 and 10−5 per cell, respectively, depending on the conditions used (12, 14, 28, 36, 37, 42, 43). Since clinical gene transfer trials, including prophylactic vaccination of healthy volunteers against infectious diseases, are performed with large amounts of vector (in general, between 1010 and 1013 particles), it is possible that substantial integration of adenoviral vector DNA might also occur in vivo even if integration rates were low. However, so far there has been no attempt to experimentally address the issue of Ad vector integration in vivo. We used the FAHΔexon5 mouse model (8) of tyrosinemia type I (MIM 27670) to analyze potential homologous and heterologous recombination events between Ad vector DNA and chromosomal DNA in vivo. Tyrosinemia type I is caused by the lack of fumarylacetoacetate hydrolase, an enzyme that is involved in the tyrosine degradation pathway and that converts fumarylacetoacetate into fumaric acid and acetoacetic acid in hepatocytes (38). Loss of fumarylacetoacetate hydrolase (FAH) activity in hepatocytes results in the accumulation of toxic and mutagenic metabolites in a cell-autonomous fashion, leading after birth to an acute hepatopathy and later in life to a chronic hepatopathy. Liver damage can be prevented both in humans and in FAH-deficient animals by the administration of 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC), which blocks the tyrosine degradation pathway by inhibiting 4-hydroxphenyl pyruvate dioxygenase, thereby preventing the accumulation of the toxic compounds. The murine FAH gene is located on chromosome 7, contains 14 exons, and spans 20.5 kb.
The autosomal recessive FAHΔexon5 mouse model, in which exon 5 is disrupted by the insertion of a NeoR gene (8), has been a useful system to analyze chromosomal integration of AAV, retrovirus, Sleeping Beauty transposon, and plasmid DNA in hepatocytes (13, 25, 27, 31). Similar to human tyrosinemia type I patients with spontaneous reversions of point mutations (18), FAH-expressing hepatocytes have a strong growth advantage over FAH−/− hepatocytes, and the developing nodules, consisting of FAH-positive [FAH+] hepatocytes, can be easily distinguished in an environment of FAH−/− hepatocytes. Following injection of an FAH-expressing Ad vector with the E1 deletion (30) into FAH−/− mice, the development of FAH+ nodules in the livers of the experimental animals was observed, suggesting potential chromosomal integration of vector DNA. Since transgene expression from vectors with the E1 deletion is transient, in part due to viral toxicity and an immune response directed to viral proteins expressed from the vector, integration events and their characterization were not possible. We reasoned that the use of high-capacity Ad (HC-Ad) vectors (also called “helper-dependent” or “gutless” Ad vectors) (41) not expressing any viral proteins would allow reliable data on Ad vector integration in vivo to be obtained.
MATERIALS AND METHODS
Plasmid construction.
To generate an HC-Ad vector expressing the FAH cDNA from the respiratory syncytial virus (RSV) promoter, plasmid pmFAH4AR1 containing the murine FAH cDNA in pTZ19U was digested with EcoRI and the cDNA was cloned into the HindIII site of pRc/RSV, generating pSLS15. pSLS15 was digested with NruI and PvuII, and the RSV-FAH cDNA-expressing cassette was cloned into the SwaI site of pSTK120 (an HC-Ad vector plasmid carrying a 16-kb human hypoxanthine-guanine phosphoribosyltransferase [HPRT] stuffer [nucleotides {nt} 1799 to 17853 of the HPRT gene] and a 9-kb c346 stuffer [nt 12421 to 21484 of cosmid c346]) to generate the HC-Ad vector plasmid pSLS16.
To generate an HC-Ad vector carrying a genomic fragment of the murine FAH locus, the bacteriophage lambda vector λmFAH6 was digested with NotI and ClaI (beginning at nt 91754097 of the murine FAH gene) and a 12.3-kb genomic fragment of the FAH gene containing the FAH region from introns 1 to 9 was cloned into pBKRSV (Stratagene), generating pSLS9. pSLS13 is an HC-Ad vector shuttle plasmid carrying a 16-kb HPRT stuffer DNA (nt 1799 to 17853 of the human HPRT gene) and was generated by cloning the PmeI fragment from pSTK68 into the PmeI site of pSTK5. pSLS13 was cleaved with EcoRV, and the 12.3-kb genomic DNA fragment derived from pSLS9 was inserted, generating the HC-Ad vector plasmid pSLS14.
Vector production and titration.
The HC-AdSLS16 vector (carrying the FAH cDNA) and the HC-AdSLS14 vector (carrying the FAH genomic DNA fragment from introns 2 to 9) were produced as described previously (32). The physical and infectious titers were determined as described previously (17). The particle/infectious unit (i.u.) ratio of the HC-AdSLS16 vector was 20:1, the particle/i.u. ratio of the HC-AdSLS14 vector was 10:1, and the level of helper virus contamination was less than 1% in both vector preparations.
Mice.
Immunodeficient Rag1−/− FAHΔexon5 mice of strain 129SvJ (8) were used in all experiments. When the animals were kept on NTBC, it was administered in the drinking water at a concentration of 7.6 mg/ml. The animals were monitored daily and were weighed weekly. Care and experiments were performed according to the guidelines of the Department of Animal Care at the Oregon Health Sciences University.
Vector injection and serial transplantation of hepatocytes into recipient livers.
Vectors were administered to the mice via tail vein injection. The serial transplantation of viable hepatocytes into the liver of the recipient mouse via splenic injection was carried out as described previously (29).
Histology.
Histology studies were carried out with liver tissues, as described previously (30).
Calculation of number of vector molecules reaching the liver following injections of different doses of HC-Ad vector in the tail vein.
The HC-AdSLS16 vector was injected via the tail vein into FAHΔexon5 mice at doses ranging from 1 × 1010 to 5 × 108 i.u. per mouse, and the animals were killed 10 days later (to avoid the readout from the vector DNA that is known to be present inside Kupffer cells shortly after gene transfer due to phagocytosis of viral particles). The livers were harvested, the DNA was extracted, and the vector copy number per cell was determined using the slot blot method (17), assuming that a diploid mouse cell has 6 pg of DNA.
Calculation of rate of recombination.
Details of the method to calculate nodule frequency are described in reference 44. Briefly, a mathematical calculation is used to estimate the frequency of clonal events after FAH+ nodules are counted and their sizes are noted in two dimensions. The maximal nodule size is then used to correct for the fact that larger nodules are more likely to be scored in two-dimensional sections, even though they represent only one clonal event. The calculation makes the assumption that nodules are round spheres and that the largest nodules found are representative for the average size of all nodules. Thus, the calculation provides a lower estimate of clonal frequency.
The histology slides containing the liver samples were scanned on a Canon flatbed scanner along with a size standard. The slides were examined for brown FAH+ cells and nodules. Photos were taken using a Zeiss inverted microscope and a CCD camera from Hamamatsu (Open Lab [version 3.03] software). Image-J software was used to determine the size of the liver samples and the cell numbers, as described in reference 44.
The calculations are exemplified for mouse 5 in Table 3, which received an injection of AdSLS14. In the six sections studied, a total of 31 FAH+ nodules were counted. The total surface area scanned was 476.82 mm2, and the total hepatocyte number was 890,222.9. Intravenous injection of 1 × 1010 transducing Ad vector units resulted in 10 Ad vector genomes per cell, so that a total of 8,902,229 Ad vector genomes were present in 890,222.9 cells. The correction factor is based on the largest nodule observed in the injected cohort, being eight in this case. Together, the formula N × 1/H × 1/C, where N is the number of colonies, H the number of vector genomes in a given number of cells, and C is the correction factor, is used to calculate the integration rates. In mouse 5 (see Table 3) the integration rate is 4.35 × 10−7.
Statistical analysis.
The exact confidence intervals for the median integration rates were calculated as described in reference 21.
PCR.
Oligodeoxyribonucleotides were purchased from MWG Biotech. To demonstrate homologous recombination following HC-Ad vector-mediated gene transfer, the DNA extracted from mice receiving HC-AdSLS14 vector injections and serial hepatocyte transplants was analyzed by PCR using the primers and conditions described previously (8).
For isolation of junctions between vector DNA and chromosomal DNA, an inverse PCR protocol followed by nested PCR, adapted for adenoviral termini, was carried out as described previously (36). The following primers were used for the right ITR: RITR1 (5′-CATCACTCCGCCCTAAAACCT-3′), RITR1nest (5′-TAAAACCTACGTCACCCGCC-3′), RITR4Tsp (5′-CCCTCGAGGTCTCGACGGTAT-3′), and RITR4nestTsp (5′-CGACGGTATCGATAAGCTTGA-3′); for the left ITR, LITR1 (5′-GCAACATCACACTTCCGCCAC-3′), LITR1nest (5′-TACTACGTCACCCGCCCCGT-3′), LITR4Tsp (5′-TGACGTTTTTGGTGTGCGCCG-3′), and LITR4nestTsp (5′-GCCGGTGTACACAGGAAGTGA-3′).
Typically, 1/100 of the product of the inverse PCR was used for the nested PCR. The PCRs were carried out using Hot Star Taq polymerase (Qiagen). DNA bands were cut out from agarose gel and cloned into a TOPO TA cloning vector (Invitrogen) for DNA sequencing.
ENSEMBL version.
The ENSEMBL program, release 56 (September 2007), was used in this study.
RESULTS
Frequency of heterologous recombination between vector and chromosomal DNA in vivo.
Since FAHΔexon5 mice have a hepatopathy, which could influence the uptake of vectors in hepatocytes following systemic administration, we first determined the exact number of genomes with the HC-Ad vector reaching the hepatocytes following injection in this mouse model. Rag1−/− FAHΔexon5 mice were injected via the tail vein with 1 × 1010, 2 × 109, and 5 × 108 i.u. of the HC-AdSLS16 vector carrying the murine FAH cDNA under the control of the RSV promoter (Fig. 1 A). Ten days later (when there is no vector DNA left in Kupffer cells), the livers were harvested and the DNA was extracted. The vector load was quantified by slot blot analysis (17) and was found to be 10, 3.2, and 0.45 vector genomes per cell, about two-thirds of which are hepatocytes, following injection of 1 × 1010, 2 × 109, and 5 × 108 infectious vector units, respectively.
FIG. 1.
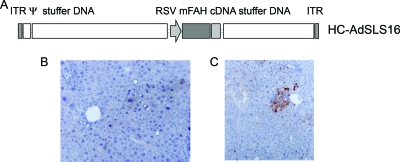
Analysis of random integration of HC-Ad vector DNA into chromosomal DNA in vivo. (A) Map of HC-AdSLS16 vector expressing the murine FAH cDNA from the RSV promoter. ITR, Ad5 inverted terminal repeat; Ψ, Ad5 packaging signal; stuffer DNA, 16-kb stuffer from human HPRT genomic DNA (nt 1799 to 17853 of human HPRT gene); RSV, RSV promoter; mFAH cDNA, murine FAH cDNA; stuffer DNA, 9-kb c346 stuffer (nt 12421 to 21484 of cosmid c346). The HC-AdSLS16 vector was used to analyze random integration events. (B and C) Representative photomicrographs of liver sections stained with hematoxylin-eosin. FAH expression is detected with a polyclonal rabbit antibody to rat FAH.
The HC-AdSLS16 vector was then used to determine the rate of heterologous recombination of vector and chromosomal DNA in vivo. Because the FAH-expressing hepatocytes have a strong selective advantage in the context of a tyrosinemic liver microenvironment, vector treatment following chromosomal integration would give rise to FAH+ nodules, which are easily observable in an FAH-deficient background. However, in case of extensive liver transduction, this would not allow the hepatocytes in which integration occurred to be distinguished from those in which the vector was still in episomal form. Therefore, we tested different vector doses to obtain FAH+ nodules that could be easily detected and counted following immunostaining of liver sections. Since a dose of 5 × 108 i.u. was found to result in nodules that could be easily distinguished and counted following immunostaining of liver sections, this dose was used in subsequent experiments. Following injection, the mice were kept on NTBC for 30 days to allow their recovery. Then, selective pressure for the formation of FAH+ liver nodules in the livers of the mice was induced by depriving the animals of NTBC for 25 days. Thus, NTBC was restored to the drinking water when mice exhibited physical conditions of a reduced health state (loss of body weight, presence of ruffled, unkempt fur). The mice were again taken off NTBC when they had recovered and killed, and the livers were harvested. Four of the mice were rotated on and off NTBC for 9 months, following which they were killed and the livers were harvested for in situ detection of FAH+ cells by immunohistochemical analysis.
The rate of recombination between vector and chromosomal DNA was calculated on the basis of the number of FAH+ nodules in the histology slides (minimum of six slides per mouse); the number of vector genomes in hepatocytes, determined as described above; and the correction factor used in counting FAH+ nodules, as described previously (44). HC-Ad vector DNA integrated into the chromosomal DNA at a median rate of 6.72 × 10−5/i.u./hepatocyte (median, 6.72 × 10−5; 96.9% confidence interval [CI], 3.20 × 10−5 to 1.37 × 10−4) in six mice (Table 1). In other words, about 1 vector genome out of 14,880 hepatocyte-transducing vector particles integrated into the chromosomal DNA.
TABLE 1.
Heterologous recombination of HC-Ad vector DNA with chromosomal DNA in vivoa
| Mouse identifier | No. of hepatocytes | No. of vector particles transducing the hepatocytes | No. of colonies | Correction factor | Integration rate/i.u./hepatocyte |
|---|---|---|---|---|---|
| 28 | 244,573 | 110,058 | 55 | 6.4 | 7.81 × 10−5 |
| 29 | 784,993 | 353,247 | 79 | 6.4 | 3.49 × 10−5 |
| 30 | 211,113 | 95,001 | 41 | 6.4 | 6.74 × 10−5 |
| 58 | 630,406 | 283,683 | 97 | 2.5 | 1.37 × 10−4 |
| 59 | 1,923,542 | 865,594 | 145 | 2.5 | 6.70 × 10−5 |
| 11 | 884,958 | 398,231 | 51 | 4 | 3.20 × 10−5 |
Summary of the results following injection of 5 × 108 i.u. of the HC-AdSLS16 vector expressing the FAH cDNA. Details of the calculation are provided in the Materials and Methods section.
Molecular characterization of integration events following heterologous recombination between vector and chromosomal DNA in vivo.
At 55 days after injection of the HC-AdSLS16 vector, two mice were used for serial transplantation. Part of the liver was removed for in situ detection of FAH+ cells by immunohistochemical analysis (Fig. 1B and C) and measurement of the nodule frequency. The rest of the liver was perfused, hepatocytes were isolated, and 1 × 106 viable cells were transplanted into secondary FAHΔexon5 recipient mice via splenic injection. By performing transplantation experiments, we aimed at obtaining a larger amount of clonal cells that would facilitate identification of junction sequences between chromosomal and viral DNA.
The secondary recipients were cycled on and off NTBC until complete repopulation with FAH+ hepatocytes had occurred. To characterize chromosomal junctions between vector and chromosomal DNA as direct evidence for vector integration, DNA was isolated from the livers of several FAHΔexon5 mice that had been repopulated with hepatocytes isolated from primary animals injected with the HC-AdSLS16 vector, as described above. Since earlier work on HC-Ad vector recombination indicated that the vector genome integrated mainly as an intact molecule through the vector termini (12, 14, 36), as also observed previously with wild-type adenovirus DNA (6), we designed our PCR primers accordingly (36). The junctions between the viral termini and the chromosomal DNA were amplified using inverse PCR (22, 35, 39), and the PCR products were cloned and sequenced. Except in one case, small deletions of between 1 and 78 nucleotides of the Ad vector terminus were detected (Table 2). Two of the seven integrations analyzed had taken place in genes (defined as regions between the transcriptional start site and the termination signal) (Table 2). In five of seven of the junction sites, the presence of microhomologies (3 or more nucleotides) between the vector DNA and chromosomal DNA was noticed.
TABLE 2.
Summary of analysis of the nucleotide sequences of the junction sites between vector and chromosomal DNAa
| Mouse identifier | Ad5 term | Integration in chromosome | Deletion from Ad terminus | Nucleotide sequence of 20 nt at junction between vector and chromosomal DNA | Integration in gene | Comment(s) on integration, names of genes where integration took place |
|---|---|---|---|---|---|---|
| 16.2 | R | Unknown | 31 | 5′-ACATTGGCTTTGAGGACAGC-3′ | Yes | Substitution at position −9; 45S pre-rRNA |
| 511 | L | 5, nt 65264899 | 15 | 5′-AGATACACAAACCTTATTTT-3′ | No | 2 microhomologies of 3 nt |
| 327 | L | 10, nt 36736768 | 1 | 5′-ATAGAGTTATATCATCAATA-3′ | No | 1 microhomology of 3 nt; vector integrated 15,074 nt downstream of the Hdac2 gene |
| S21 | R | 2, nt 89512808 | 10 | 5′-AAGGTATATTTAGTCTCTTT-3′ | No | 1 microhomology of 4 nt; vector integrated 11,435 nt upstream of the Olfr1250 gene |
| S23 | L | 2, nt 181002607 | 64 | 5′-TGGCAAGAGAGACGTGGCGC-3′ | Yes | Gmeb2 |
| S23 | R | 12, nt 113549717 | 16 | 5′-CAAAATAAGGCACACAAACA-3′ | No | 1 microhomology of 3 nt and 1 microhomology of 4 nt |
| S24 | R | 18, nt 60584686 | 78 | 5′-CGTTCCCACGAGGGTGTATA-3′ | N | 2 microhomologies of 3 nt; vector integrated |
| 34,205 kb upstream of the ligp1 gene |
The table lists the integration sites of AdSLS16, the Ad5 terminus which was sequenced during the inverse PCR, the details of the site where the vector DNA integrated, the deletions from the vector terminus, the nucleotide sequences of the junctions between cellular and vector DNA, and additional information. BLAST searches of the NCBI and ENSEMBL databases were performed. By convention, when the same nucleotides were found in the junction that corresponded to both the Ad5 DNA and genomic DNA, they were taken as having originated from the Ad terminus. Ad5 terminus sequences are underlined. R, right Ad5 terminus; L, left Ad5 terminus. The names of the genes in which vector DNA integration had taken place are given in boldface letters.
Homologous recombination between vector and chromosomal DNA in vivo.
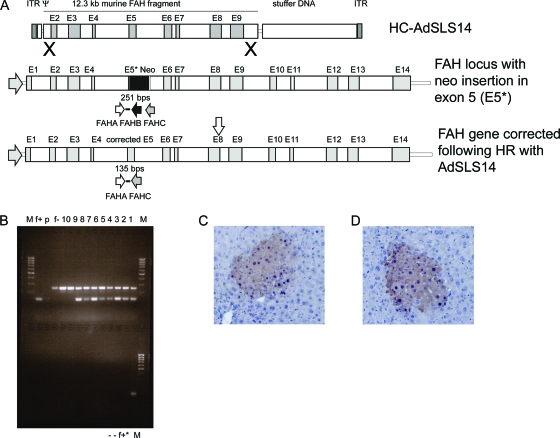
To determine the rate of homologous recombination of the vector DNA with the chromosomal DNA in vivo, we constructed the Ad vector HC-AdSLS14 carrying a genomic 12.3-kb fragment of the murine FAH locus spanning the region between the first and the ninth introns of the FAH gene (Fig. 2A ). FAH+ nodules were expected to arise in the liver only when, upon homologous recombination between the FAH locus and the HC-AdSLS14 vector, the disrupted exon 5 in the mouse chromosome was replaced by the normal allele. Note that random integration of the HC-AdSLS14 vector would not result in FAH expression, since the construct is without a promoter; lacks exon 1, including the ATG codon; and also lacks exons 10 to 14. FAHΔexon5 mice were injected via the tail vein with 1 × 1010 and 2 × 109 i.u. of the HC-AdSLS14 vector. Following injection, the mice were kept on NTBC for 2 weeks. Thereafter, selective pressure for the formation of FAH-positive nodules in the livers was induced by removing NTBC from the diet for 4 weeks. Four mice were killed at this point, and for the others NTBC was restored in the drinking water, until they were killed 2 weeks later. Following immunohistochemical analysis of the livers of these animals, FAH-positive groups of hepatocytes were detected (Fig. 2C and D). The rate of homologous recombination between the vector DNA and the chromosomal DNA was determined as described above (Table 3). Accordingly, HC-Ad vector DNA underwent homologous recombination with the chromosomal DNA at a median rate of 3.89 × 10−7/i.u./hepatocyte (median, 3.89 × 10−7; 92.9% CI, 2.13 × 10−7 to 6.03 × 10−7). In other words, about 1 out of 2.6 × 106 hepatocyte-transducing vector genomes recombined with the chromosomal DNA via homologous recombination.
FIG. 2.
Analysis of homologous recombination of HC-Ad vector DNA with chromosomal DNA in vivo. (A) Schematic of the HC-AdSLS14 vector carrying a 12.3-kb genomic fragment of the murine DNA and the scheme of homologous recombination (X) whereby exon 5 in the FAH locus may be replaced by the targeting vector. ITR, Ad5 inverted terminal repeat; Ψ, Ad5 packaging signal; murine FAH fragment, 12.3-kb murine FAH genomic DNA fragment beginning at nt 91754097 of the murine FAH gene from introns 1 to 9; E2 to E9, wild-type exons 2 to 9, respectively; E5*, mutated exon 5 with NeoR insertion; Neo, NeoR gene; stuffer DNA, 16-kb stuffer from human HPRT genomic DNA (nt 1799 to 17853 of human HPRT gene). The positions of the primers used in the PCR to demonstrate the presence of an uninterrupted exon 5 in the livers are indicated as the horizontal arrows FAHA, FAHB, and FAHC. (B) The 2% agarose gel used for separation of the PCR products; Lanes: −, water controls; f+* and f+, C57/BL6 and 129 mouse controls; respectively; p, pSLS14 plasmid control; f−, FAH−/− mouse negative control; 10, FAH−/− mouse injected with the first-generation Ad vector negative control; 9, transplantation recipient of HC-AdSLS16 vector negative control; 4 to 8, transplantation recipients; 1 to 3, injected animals. (C and D) Representative photomicrographs of liver sections stained with hematoxylin-eosin. FAH expression is detected with a polyclonal rabbit antibody to rat FAH.
TABLE 3.
Homologous recombination of HC-Ad vector DNA with chromosomal DNA in vivoa
| Mouse identifier | No. of AdSLS14 i.u. units | No. of hepatocytes | No. of vector particles transducing the hepatocytes | No. of colonies | Correction factor | Integration rate/i.u./hepatocyte |
|---|---|---|---|---|---|---|
| 20 | 2 × 109 | 803,559 | 2,571,388 | 28 | 6.4 | 1.70 × 10−6 |
| 22 | 2 × 109 | 1,236,768 | 3,957,658 | 13 | 6.4 | 5.13 × 10−7 |
| 23 | 2 × 109 | 1,133,067 | 3,625,816 | 14 | 6.4 | 6.03 × 10−7 |
| 25 | 2 × 109 | 1,253,017 | 4,009,653 | 7 | 6.4 | 2.73 × 10−7 |
| 5 | 1 × 1010 | 890,222.9 | 8,902,229 | 31 | 8 | 4.35 × 10−7 |
| 6 | 1 × 1010 | 1,115,924.6 | 11,159,246 | 19 | 8 | 2.13 × 10−7 |
| 7 | 2 × 109 | 1,369,930 | 4,383,776 | 12 | 8 | 3.42 × 10−7 |
| 8 | 2 × 109 | 1,572,518 | 5,032,058 | 4 | 8 | 9.94 × 10−8 |
Summary of the results following injection of the HC-AdSLS14 vector carrying the FAH genomic DNA fragment from introns 2 to 9. Details of the calculation are provided in the Materials and Methods section.
To demonstrate the correction of the exon 5 disruption in the knockout mice following homologous recombination by the vector DNA, PCR was carried out with the DNA extracted from the livers of mice that had been either injected with the HC-AdSLS14 vector or serially transplanted with hepatocytes from mice injected with the HC-AdSLS14 vector (Fig. 2B and C). The PCR was designed to produce only a 180-bp product from the corrected locus and a 240-bp product from the knockout allele (8). As expected, in addition to the 240-bp fragment, a smaller product was observed in the livers of the injected mice (Fig. 2B), and sequencing demonstrated that FAH exon 5 had been corrected. However, we cannot formally rule out the possibility that the smaller band was amplified from episomal vector DNA or a heterologous integrant, even though it is unlikely that the episome or a random integrant (which could not have a selective advantage in this context) would have survived the rounds of transplantation and amplification of the hepatocytes in the recipient livers, where the FAH+ nodules were clearly visible.
DISCUSSION
Following the occurrence of leukemia in several children with SCID-X1 undergoing ex vivo hematopoietic gene therapy with a retroviral vector (9, 10), the consideration of genotoxic effects due to insertional mutagenesis has become an essential part of any risk-benefit analysis of treatments with integrating vectors. However, although hundreds of individuals have received large doses of adenovirus vectors by either local or systemic injection, only little information on Ad vector integration in vitro is available and no information on vector DNA integration after in vivo gene transfer is available.
Our study, being the first to address this issue in vivo, was designed to allow the analysis of homologous and heterologous recombination events between adenoviral vector DNA and chromosomal DNA, aiming not only to establish quantitative data on in vivo Ad vector integration but also to provide a first characterization of the quality of vector integration.
To detect random integration events due to heterologous recombination between vector and chromosomal DNA, we injected FAH-deficient mice with a dose resulting in an average of 0.5 vector genomes per hepatocyte. This dose is similar to the vector amount we previously used in vitro for transduction of primary human cells and cell lines (36). In vivo we calculated an integration frequency of 6.72 × 10−5/i.u./hepatocyte, meaning approximately 1 integration event in 14,880 vector-transduced hepatocytes. This number is less than the 5.5 × 10−3 to 1.1 × 10−4/i.u./cell (corresponding to 1 integration event for every 200 to 10,000 transduced cells) that we had observed in our in vitro studies (36). Although the FAH model is sensitive for detection of FAH-expressing hepatocytes in an FAH-negative background, it is possible that the actual integration frequency is somewhat higher, since the calculated rate of recombination was dependent on detection of FAH+ nodules and occasional promoter silencing of the transgene expression cassette cannot be excluded. On the other hand, since fumarylacetoacetate and maleylacetoacetate are DNA-damaging agents, their presence in the precorrected cell might promote an increase in the number of recombination events compared to the number in normal hepatocytes.
Interestingly, the FAH model was even suitable to detect rare homologous recombination events between adenoviral vector and chromosomal DNA. Using higher vector doses to increase the sensitivity, we measured a homologous recombination rate of 3.89 × 10−7/ i.u./cell, meaning that 1 out of 2.6 × 106 vector genomes that had been taken up by hepatocytes underwent homologous recombination with one of the knockout alleles. For comparison, in our in vitro study using vector at a multiplicity of infection of (MOI) of 1, homologous recombination occurred at a rate of 2 × 10−5 to 1.6 × 10−6 per i.u./cell (36). Together, the heterologous and homologous recombination frequencies in vivo appeared to be lower than those in vitro, and as expected, the rate of homologous recombination was considerably lower than the rate of heterologous recombination, confirming previous data obtained from in vitro studies (12, 14, 36, 42, 43), although in studies with embryonic stem cells, the percentage of homologous recombination was found to be quite high (28, 37).
The molecular characterization of random vector integration at the sequence level, although it has so far been performed with only a very limited data set, revealed many features that mirrored those previously observed following both wild-type Ad12 DNA integration, resulting in tumor formation in hamsters (4, 6), and Ad5 vector DNA integration in cultured cells (12, 14, 28, 36, 42, 43). Since our in vitro study (36) suggested that Ad vector integration takes place mainly through the vector termini, we used PCR primer pairs designed for amplification of junction sequences between vector and chromosomal DNA. Frequently, small deletions of terminal viral nucleotides were detected (Table 2), and microhomologies between the viral termini and the chromosomal preintegration sites were often observed. In one out of the seven integrations analyzed, a substitution was observed 9 nucleotides downstream of the integration event. The presence of microhomologies between the viral terminus and chromosomal DNA has been observed in recombination studies of both viral vectors (28, 36, 43) and wild-type adenovirus (5, 6). Two out of seven integrations had occurred within genes, and in three of the remaining five integrations, the vector DNA had recombined into a locus between 11 and 34 kb upstream or downstream of a gene. While the overall number of integrations that we analyzed was low, not justifying any conclusion relating to potential integration preferences into genes to be made, we note that in our previous in vitro study (36), half of the integration events had occurred within genes, indicating a preference for integration into gene loci. Preferential integration into genes has been observed with all vector systems tested so far (2, 14, 24, 26, 47), apart from foamy virus-derived vectors (28 to 31% in human, 30% in dog) (1, 40).
Another observation of potentially significant concern which has been documented upon Ad12 DNA integration in vitro (6, 34) and in vivo (15) and also in some instances following Ad vector integration in cultured cells (43) has been chromosomal deletions, sometimes of substantial size, and rearrangements (12, 28, 36). We believe that with current technology, this cannot be easily demonstrated in liver tissue. However, given that all other documented and quality-related features suggest nonhomologous end joining (NHEJ) as the mechanism involved in Ad vector integration, we consider it very likely that such mutations would also occur following gene transfer in vivo.
A liver of an adult human with a weight of 1.2 to 1.5 kg and a hepatocellularity of about 1.07 × 108 cells/g (45) consists of about 1.5 × 1011 hepatocytes. Assuming that the liver of a patient is transduced in such a way that each hepatocyte will receive one vector genome (cell-specific MOI = 1), an integration rate of 6.72 × 10−5/i.u./hepatocyte, as determined in this study, would translate to a total of 1.01 × 107 hepatocytes carrying a chromosomally integrated vector genome. Due to the size of the adenoviral vector genome, we can assume that most of the integration events, even when they are in introns, would result in gene inactivation.
According to published data from the human genome sequencing effort, the human genome has a size of 2.9 × 109 nt and harbors a maximum of 25,000 genes (16), with the mean gene size being 27 kb (19) and the mean size of the protein-coding parts being 1.35 kb. If integration were random, in one liver there would be a total of 94 integrations into every gene and of 4 to 5 integrations into the coding part of every gene. Since Ad vectors have an up to 2-fold preference for integration into genes in vitro, depending on the cell type used (28, 36, 43), we can assume up to 188 integrations into every gene and about 9 integrations in the coding part of every gene. At first sight, these numbers may appear to be very high. However, to be meaningful within a risk analysis, these numbers have to be compared with the spontaneous mutation frequency in liver tissue, i.e., the number of gene mutations in a specific locus at a given time. In transgenic rodents bearing the Escherichia coli lacI transgene, a spontaneous mutation frequency of about 3 × 10−5 has been documented (48). To our knowledge, corresponding quantitative data for hepatocytes from humans are not available. In humans, hematopoietic cells have been the most extensively studied system. The spontaneous mutation frequency can vary, depending on the model system, from >1 × 10−4 to <1 × 10−8 (7). For the HPRT system, the most thoroughly studied system, the overall mean mutation frequency (for inactivating mutations) calculated from different studies has been determined to be about 6 × 10−6 (corresponding to 1 mutant HPRT allele in 1.7 × 105 cells) (7). We now consider the likelihood that the Ad vector will integrate into the HPRT locus and assume that most integrations of the adenoviral vector genome will result in inactivation of the HPRT gene. With a 40-kb size of the HPRT gene and a genome size of 2.9 × 109 bp, the overall probability of integrating into/disrupting the HPRT gene is about 0.9 × 10−9 in case of random integration and 1.8 × 10−9 in case of preferred integration into genes. Therefore, compared to the spontaneous mutation frequency, the likelihood that Ad vector integration would result in the inactivation of genes is relatively low.
Although these findings are reassuring, they does not exclude the possibility of a genotoxic effect through mechanisms other than gene inactivation; for example, (i) a genotoxic effect may occur through inactivation of tumor suppressor genes, in particular, in individuals who are already heterozygous at a tumor suppressor locus. (ii) It may occur through gene activation; for example, to our knowledge it has not been addressed directly whether the well-known E1A enhancer, located at the left terminus of the vector genome, can activate genes in the neighborhood of the integration side, although we believe that this may well occur. (iii) A genotoxic effect may occur through disruption of microRNA genes; this may substantially influence the expression of large numbers and networks of genes (11), and the effects will not be restricted to single transcription units. (iv) It may occur through chromosomal deletions and structural rearrangements; these may have more significant consequences than simple gene inactivation. The frequency of their occurrence has not been determined so far, although in vitro studies (6, 28, 34, 36, 43) suggest that they may well occur. (v) A genotoxic effect may occur through the effects of foreign DNA insertion on remote parts of the genome, resulting in alterations to epigenetic and transcriptional patterns (3).
We acknowledge that, depending on the route of delivery, current Ad vector technology is relatively inefficient in specific gene transfer due to barriers preventing efficient gene transfer into target cells, so that a very small fraction (probably less than 0.1%, in most cases) of the injected vector, whether it is given intramuscularly for vaccination or intravenously for liver gene transfer, will reach target or nontarget cells. We suggest, however, that concurrent with improvements in vector technology resulting in improved gene delivery, genotoxic side effects due to less well studied consequences (see above) should be investigated in greater detail.
Acknowledgments
We thank G. Pfitzer and D. Grosskopf-Kroiher for logistical support during a part of the experiments; D. Escors, F. Kreppel, K. Schwarz, F. Radecke, and R. Geyer for discussions; H. Cox for her expert help in preparing the manuscript; and S. Lindstedt for his generous gift of NTBC.
The experimental animals were maintained according to the rules of Oregon Health Sciences University.
S.L.S. was supported by a travel bursary from the Boehringer Ingelheim Fonds. This work was supported by grants from the NIH (grant HL059314 to S.K. and grant DK18252 to M.G.) and the European Union (NoE-Clinigene) to S.K.
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Beard, B. C., K. A. Keyser, G. D. Trobridge, L. J. Peterson, D. G. Miller, M. Jacobs, R. Kaul, and H. P. Kiem. 2007. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, or foamy virus. Hum. Gene Ther. 18:423-434. [DOI] [PubMed] [Google Scholar]
- 2.Bushman, F., M. Lewinski, A. Ciuffi, S. Barr, J. Leipzig, S. Hannenhalli, and C. Hoffmann. 2005. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3:848-858. [DOI] [PubMed] [Google Scholar]
- 3.Doerfler, W. 2006. De novo methylation, long-term promoter silencing, methylation patterns in the human genome, and consequences of foreign DNA insertion. Curr. Top. Microbiol. Immunol. 301:125-175. [DOI] [PubMed] [Google Scholar]
- 4.Doerfler, W. 1968. The fate of the DNA of adenovirus type 12 in baby hamster kidney cells. Proc. Natl. Acad. Sci. U. S. A. 60:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doerfler, W., H. Burger, J. Ortin, E. Fanning, D. T. Brown, M. Mestphal, U. Winterhoff, B. Weiser, and J. Schick. 1975. Integration of adenovirus DNA into the cellular genome. Cold Spring Harbor Symp. Quant. Biol. 39 (Pt 1):505-521. [DOI] [PubMed] [Google Scholar]
- 6.Doerfler, W., R. Gahlmann, S. Stabel, R. Deuring, U. Lichtenberg, M. Schulz, D. Eick, and R. Leisten. 1984. On the mechanism of recombination between adenoviral and cellular DNAs: the structure of junction sites. Curr. Top. Microbiol. Immunol. 109:193-228. [DOI] [PubMed] [Google Scholar]
- 7.Drake, J. W., B. Charlesworth, D. Charlesworth, and J. F. Crow. 1998. Rates of spontaneous mutation. Genetics 148:1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grompe, M., M. al-Dhalimy, M. Finegold, C. N. Ou, T. Burlingame, N. G. Kennaway, and P. Soriano. 1993. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 7:2298-2307. [DOI] [PubMed] [Google Scholar]
- 9.Hacein-Bey-Abina, S., C. von Kalle, M. Schmidt, F. Le Deist, N. Wulffraat, E. McIntyre, I. Radford, J. L. Villeval, C. C. Fraser, M. Cavazzana-Calvo, and A. Fischer. 2003. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 348:255-256. [DOI] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 11.Hagan, J. P., and C. M. Croce. 2007. MicroRNAs in carcinogenesis. Cytogenet. Genome Res. 118:252-259. [DOI] [PubMed] [Google Scholar]
- 12.Harui, A., S. Suzuki, S. Kochanek, and K. Mitani. 1999. Frequency and stability of chromosomal integration of adenovirus vectors. J. Virol. 73:6141-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Held, P. K., E. C. Olivares, C. P. Aguilar, M. Finegold, M. P. Calos, and M. Grompe. 2005. In vivo correction of murine hereditary tyrosinemia type I by phiC31 integrase-mediated gene delivery. Mol. Ther. 11:399-408. [DOI] [PubMed] [Google Scholar]
- 14.Hillgenberg, M., H. Tonnies, and M. Strauss. 2001. Chromosomal integration pattern of a helper-dependent minimal adenovirus vector with a selectable marker inserted into a 27.4-kilobase genomic stuffer. J. Virol. 75:9896-9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohlweg, U., M. Hosel, A. Dorn, D. Webb, K. Hilger-Eversheim, R. Remus, B. Schmitz, R. Buettner, A. Schramme, L. Corzilius, A. Niemann, and W. Doerfler. 2003. Intraperitoneal dissemination of Ad12-induced undifferentiated neuroectodermal hamster tumors: de novo methylation and transcription patterns of integrated viral and of cellular genes. Virus Res. 98:45-56. [DOI] [PubMed] [Google Scholar]
- 16.International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431:931-945. [DOI] [PubMed] [Google Scholar]
- 17.Kreppel, F., V. Biermann, S. Kochanek, and G. Schiedner. 2002. A DNA-based method to assay total and infectious particle contents and helper virus contamination in high-capacity adenoviral vector preparations. Hum. Gene Ther. 13:1151-1156. [DOI] [PubMed] [Google Scholar]
- 18.Kvittingen, E. A., H. Rootwelt, P. Brandtzaeg, A. Bergan, and R. Berger. 1993. Hereditary tyrosinemia type I. Self-induced correction of the fumarylacetoacetase defect. J. Clin. Invest. 91:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 20.Laufs, S., K. Z. Nagy, F. A. Giordano, A. Hotz-Wagenblatt, W. J. Zeller, and S. Fruehauf. 2004. Insertion of retroviral vectors in NOD/SCID repopulating human peripheral blood progenitor cells occurs preferentially in the vicinity of transcription start regions and in introns. Mol. Ther. 10:874-881. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann, E. L. 2006. Nonparametrics: statistical methods based on ranks. Springer, Berlin, Germany.
- 22.Lund, A. H., M. Duch, and F. S. Pedersen. 1996. Increased cloning efficiency by temperature-cycle ligation. Nucleic Acids Res. 24:800-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, D. G., G. D. Trobridge, L. M. Petek, M. A. Jacobs, R. Kaul, and D. W. Russell. 2005. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J. Virol. 79:11434-11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montini, E., P. K. Held, M. Noll, N. Morcinek, M. Al-Dhalimy, M. Finegold, S. R. Yant, M. A. Kay, and M. Grompe. 2002. In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol. Ther. 6:759-769. [DOI] [PubMed] [Google Scholar]
- 26.Nakai, H., E. Montini, S. Fuess, T. A. Storm, M. Grompe, and M. A. Kay. 2003. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 34:297-302. [DOI] [PubMed] [Google Scholar]
- 27.Nakai, H., X. Wu, S. Fuess, T. A. Storm, D. Munroe, E. Montini, S. M. Burgess, M. Grompe, and M. A. Kay. 2005. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J. Virol. 79:3606-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohbayashi, F., M. A. Balamotis, A. Kishimoto, E. Aizawa, A. Diaz, P. Hasty, F. L. Graham, C. T. Caskey, and K. Mitani. 2005. Correction of chromosomal mutation and random integration in embryonic stem cells with helper-dependent adenoviral vectors. Proc. Natl. Acad. Sci. U. S. A. 102:13628-13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overturf, K., M. Al-Dhalimy, M. Finegold, and M. Grompe. 1999. The repopulation potential of hepatocyte populations differing in size and prior mitotic expansion. Am. J. Pathol. 155:2135-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overturf, K., M. al-Dhalimy, C. N. Ou, M. Finegold, R. Tanguay, A. Lieber, M. Kay, and M. Grompe. 1997. Adenovirus-mediated gene therapy in a mouse model of hereditary tyrosinemia type I. Hum. Gene Ther. 8:513-521. [DOI] [PubMed] [Google Scholar]
- 31.Overturf, K., M. Al-Dhalimy, R. Tanguay, M. Brantly, C. N. Ou, M. Finegold, and M. Grompe. 1996. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat. Genet. 12:266-273. [DOI] [PubMed] [Google Scholar]
- 32.Schiedner, G., F. Kreppel, and S. Kochanek. 2006. Production and quality control of high-capacity adenoviral vectors. In J. E. Celis (ed.), Cell biology: a laboratory handbook. Elsevier Academic Press, Burlington, MA.
- 33.Schirm, S., and W. Doerfler. 1981. Expression of viral DNA in adenovirus type 12-transformed cells, in tumor cells, and in revertants. J. Virol. 39:694-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz, M., and W. Doerfler. 1984. Deletion of cellular DNA at site of viral DNA insertion in the adenovirus type 12-induced mouse tumor CBA-12-1-T. Nucleic Acids Res. 12:4959-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silver, J., and V. Keerikatte. 1989. Novel use of polymerase chain reaction to amplify cellular DNA adjacent to an integrated provirus. J. Virol. 63:1924-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephen, S. L., V. G. Sivanandam, and S. Kochanek. 2008. Homologous and heterologous recombination between adenovirus vector DNA and chromosomal DNA. J. Gene Med. 10:1176-1189. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki, K., K. Mitsui, E. Aizawa, K. Hasegawa, E. Kawase, T. Yamagishi, Y. Shimizu, H. Suemori, N. Nakatsuji, and K. Mitani. 2008. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc. Natl. Acad. Sci. U. S. A. 105:13781-13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanguay, R. M., R. Jorquera, J. Poudrier, and M. St-Louis. 1996. Tyrosine and its catabolites: from disease to cancer. Acta Biochim. Pol. 43:209-216. [PubMed] [Google Scholar]
- 39.Triglia, T., M. G. Peterson, and D. J. Kemp. 1988. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16:8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trobridge, G. D., D. G. Miller, M. A. Jacobs, J. M. Allen, H. P. Kiem, R. Kaul, and D. W. Russell. 2006. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. U. S. A. 103:1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volpers, C., and S. Kochanek. 2004. Adenoviral vectors for gene transfer and therapy. J. Gene Med. 6(Suppl. 1):S164-S171. [DOI] [PubMed] [Google Scholar]
- 42.Wang, H., and A. Lieber. 2006. A helper-dependent capsid-modified adenovirus vector expressing adeno-associated virus rep78 mediates site-specific integration of a 27-kilobase transgene cassette. J. Virol. 80:11699-11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, H., D. M. Shayakhmetov, T. Leege, M. Harkey, Q. Li, T. Papayannopoulou, G. Stamatoyannopolous, and A. Lieber. 2005. A capsid-modified helper-dependent adenovirus vector containing the beta-globin locus control region displays a nonrandom integration pattern and allows stable, erythroid-specific gene expression. J. Virol. 79:10999-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, X., E. Montini, M. Al-Dhalimy, E. Lagasse, M. Finegold, and M. Grompe. 2002. Kinetics of liver repopulation after bone marrow transplantation. Am. J. Pathol. 161:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson, Z. E., A. Rostami-Hodjegan, J. L. Burn, A. Tooley, J. Boyle, S. W. Ellis, and G. T. Tucker. 2003. Inter-individual variability in levels of human microsomal protein and hepatocellularity per gram of liver. Br. J. Clin. Pharmacol. 56:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 47.Yant, S. R., X. Wu, Y. Huang, B. Garrison, S. M. Burgess, and M. A. Kay. 2005. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 25:2085-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, S., B. W. Glickman, and J. G. de Boer. 2001. Spontaneous mutation of the lacI transgene in rodents: absence of species, strain, and insertion-site influence. Environ. Mol. Mutagen. 37:141-146. [DOI] [PubMed] [Google Scholar]