Abstract
Infections with human parvoviruses B19 and recently discovered human bocaviruses (HBoVs) are widespread, while PARV4 infections are transmitted parenterally and prevalent specifically in injecting drug users and hemophiliacs. To investigate the exposure and circulation of parvoviruses related to B19 virus, PARV4, and HBoV in nonhuman primates, plasma samples collected from 73 Cameroonian wild-caught chimpanzees and gorillas and 91 Old World monkey (OWM) species were screened for antibodies to recombinant B19 virus, PARV4, and HBoV VP2 antigens by enzyme-linked immunosorbent assay (ELISA). Moderate to high frequencies of seroreactivity to PARV4 (63% and 18% in chimpanzees and gorillas, respectively), HBoV (73% and 36%), and B19 virus (8% and 27%) were recorded for apes, while OWMs were uniformly negative (for PARV4 and B19 virus) or infrequently reactive (3% for HBoV). For genetic characterization, plasma samples and 54 fecal samples from chimpanzees and gorillas collected from Cameroonian forest floors were screened by PCR with primers conserved within Erythrovirus, Bocavirus, and PARV4 genera. Two plasma samples (chimpanzee and baboon) were positive for PARV4, while four fecal samples were positive for HBoV-like viruses. The chimpanzee PARV4 variant showed 18% and 15% nucleotide sequence divergence in NS and VP1/2, respectively, from human variants (9% and 7% amino acid, respectively), while the baboon variant was substantially more divergent, mirroring host phylogeny. Ape HBoV variants showed complex sequence relationships with human viruses, comprising separate divergent homologues of HBoV1 and the recombinant HBoV3 species in chimpanzees and a novel recombinant species in gorillas. This study provides the first evidence for widespread circulation of parvoviruses in primates and enables future investigations of their epidemiology, host specificity, and (co)evolutionary histories.
Autonomous parvoviruses known to infect humans comprise parvovirus B19 (18) and the recently discovered PARV4 (22) and human bocavirus (HBoV) (3). Members of the family Parvoviridae are genetically and biologically diverse and are classified into several genera or groups, showing marked differences in host range, pathology, and tissue/cellular tropisms (18). Human parvovirus B19, a member of the Erythrovirus genus, is transmitted primarily by the respiratory route but causes systemic infections. Erythroid progenitor cells are specifically targeted through expression of globoside P antigen, which acts as the B19 virus receptor for entry (5). In common with infections by most parvoviruses, B19 virus infections are acute; a period of intense viremia is followed by seroconversion for antibody to B19 virus and lifelong immunity from reinfection (29). Despite the clearance of viremia and seroconversion for antibody, lifelong persistence of viral DNA in tissues has been shown to occur (12, 20, 26, 28, 43, 58). Three genotypes of B19 virus have been described, differing in nucleotide sequence by approximately 13 to 14% (7, 21, 41, 53); genotypes 1 and 2 have been found in Europe, the United States, and other Western countries, while genotype 3 is restricted to sub-Saharan Africa and South America (7, 47, 49). B19 virus widely circulates in human populations worldwide; in Western countries, several studies have documented increasing frequencies of B19 virus seropositivity with age, rising to approximately 60 to 70% by adulthood (15, 39, 48, 61).
Another human parvovirus, PARV4, shows markedly different epidemiology and transmission routes. It was originally detected in plasma from an individual with an “acute infection syndrome” resembling that of primary human immunodeficiency virus (HIV) infection (22). While this clinical presentation has not been observed again, infection with PARV4 is known to be widespread specifically in individuals with a history of parenteral exposure (injecting drug users [IDUs], hemophiliacs, polytransfused individuals), with a strikingly higher incidence in those infected with HIV-1 (13, 14, 30, 35, 54). These observations suggest that PARV4 is primarily transmitted though parenteral routes in Western countries (54, 56). In common with infection with the better-characterized human parvovirus B19, infection with PARV4 is associated with a period of acute viremia, followed by seroconversion for antibody and long-term persistence of viral DNA sequences in lymphoid and other tissue (33, 37, 52). Circulating variants of PARV4 have been classified into three distinct genotypes exhibiting approximately 8% nucleotide sequence divergence from each other. Genotypes 1 and 2 circulate in Western countries, while genotype 3 has to date been recorded only in sub-Saharan Africa (45, 55).
The third human parvovirus, HBoV (3), shows a number of epidemiological and clinical attributes different from those of both B19 virus and PARV4. HBoV was originally found in the respiratory tract of young children and has been the subject of intense investigation as a potential cause of human respiratory disease (reviewed in references 1, 51, and 62). Although it is frequently detected by PCR in the nasopharynx of viremic individuals with primary infections with lower respiratory tract disease, other coinfecting respiratory viruses are frequently detected (19). HBoV additionally shows long-term, low-level carriage in the respiratory tract after primary infection, which further complicates PCR-based etiological studies (2, 38) and warrants the use of other diagnostic strategies, such as serology (30, 32, 59). In contrast to the rather minimal genetic diversity of B19 virus and PARV4 genotypes, bocaviruses infecting humans are now known to comprise three to four major genetic variants (termed types or species 1 to 4) (23, 24). HBoV1 and HBoV2 show 22%, 33%, and 20% amino acid sequence divergence from each other in the encoded viral nonstructural (NS), NP-1, and structural VP1/VP2 proteins, respectively, the latter potentially leading to antigenic diversity and some loss of antigenic cross-reactivity. A third type/species of HBoV is a chimeric form with a nonstructural gene region (NS, NP1) most similar to HBoV1, a recombination breakpoint in the intergenic region between NP1 and VP1, and structural genes related to those of HBoV2 (4, 23). Current data suggest that only HBoV1 is capable of infecting the respiratory tract; most published large-scale screening studies have failed to detect HBoV2 (or HBoV3) in respiratory samples (10, 11, 60), while all three types/species are detectable in fecal samples, indicating the existence of an alternative or additional site of virus replication (23). Despite extensive inquiry, the exact role of HBoV1 in respiratory disease remains unclear, as is the proposed etiological role of HBoV2 (and possibly HBoV3) in gastroenteritis (4, 11, 23, 50). Very recently, a fourth species/type, HBoV4, has been detected in fecal samples; genetically it also shows evidence for past recombination, with NS and NP1 region sequences grouping with HBoV2, while VP1/VP2 is more closely related to HBoV3 (23).
We have little understanding of the past epidemiology, evolution, and origins of human parvoviruses. For both B19 virus and PARV4, evidence has been obtained for a temporal succession of genotypes over time (37, 43); in Europe, B19 virus genotype 1 largely replaced type 2 in the 1960 and 1970s (43), while current data indicate that a similar replacement of PARV4 genotypes occurred within the last 20 years (37). The highly restricted sequence diversity of currently circulating variants of PARV4 and B19 virus and of HBoV1 variants supports the hypothesis of a relatively recent emergence and spread of these viruses in human populations (36, 42, 64).
The existence and evolution of parvoviruses on a much longer time scale is suggested by the observations that members of the Erythrovirus and Parvovirus genera both contain viruses that are highly host species specific and that the molecular phylogenies of both genera are largely congruent with those of their hosts (34). This has led to the hypothesis of long-term coevolution of parvoviruses with their host over the 90 million years of mammalian evolution and perhaps beyond. Among erythroviruses, simian homologues of B19 virus have been found in cynomolgus monkeys (44) and rhesus and pig-tailed macaques (16) and more genetically distant viruses have been characterized in chipmunks and cows (9, 63). Divergent homologues of PARV4 in pigs and cows have been described (31), while the bovine and canine parvoviruses distantly related to HBoV are the originally described members of the Bocavirus genus. However, the process of virus-host codivergence is known to be punctuated by occasional cross-species transmissions, including the well-documented spread of feline parvovirus to dogs (46). Based on serological evidence, the possible transmission of simian erythroviruses to animal handlers has been proposed (6).
To gain further insights into the origins and evolution of human parvoviruses, we have performed large-scale serological and PCR-based screening of nonhuman primates (chimpanzees and gorillas) and of several species of Old World monkeys (OWMs) for evidence of infection with parvoviruses that are antigenically related to the human B19, PARV4, and HBoV viruses. By PCR, we have sought to genetically characterize homologues of the three autonomous human parvoviruses in apes and Old World monkey species and to analyze their evolutionary relationship to human and other mammalian homologues of these viruses.
MATERIALS AND METHODS
Samples.
Plasma samples were collected from 62 chimpanzees (Pan troglodytes troglodytes), 11 gorillas (Gorilla gorilla), and a range of Old World monkey species: Cercocebus agilis (n = 7), C. torquatus (n = 2), C. cephus (n = 3), C. erythrotis (n = 4), C. preussi (n = 4), C. mona (n = 9), C. nictitans (n = 3), C. pogonias (n = 1), C. tantalus (n = 3), Erythrocebus patas (n = 3), Lophocebus albigena (n = 5), Mandrillus leucophaeus (n = 20), M. sphinx (n = 9), and Papio anubis (n = 20). Sample shipment to the United Kingdom from Cameroon was performed in compliance with Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) regulations and Cameroon and United Kingdom import/export regulations. Samples were collected from captive animals in three wildlife sanctuaries. Animals were primarily wild born and brought to the sanctuaries after confiscation by authorities or abandonment by owners. Blood samples were collected during routine health examinations, quarantine, or recaptures by sanctuary staff after escapes. Whole blood was collected via venipuncture and transferred to an EDTA Vacutainer. Plasma was separated by centrifugation and frozen at −80°C until testing. Fecal samples were obtained from 27 chimpanzees and 27 gorillas from the Cameroon (40); species identification was performed by mitochondrial DNA, microsatellite, and sex marker analyses as described previously (25).
Screening by PCR.
Prior to nucleic acid extraction, fecal samples of approximately 50 to 300 mg were supplemented with 50 μl of sterile PBS and briefly mixed by vortexing. Samples were then cleared by centrifugation at 6,000 rpm in a benchtop microcentrifuge for 1 min to pellet bacteria and other solids. DNA extractions were performed (from either cleared fecal supernatants or 50 μl of plasma) using the AllPrep DNA/RNA minikit (Qiagen) according to the manufacturer's instructions with DNA eluted in a final volume of 50 μl.
Samples were screened by nested PCR using sets of primers conserved within each parvovirus genus (see Table S1 at http://www.virus-evolution.org/Downloads/JVI01304-10/). For detection of PARV4, primers matched all described sequences of PARV4, porcine hokovirus (PHoV), and bovine hokovirus (BHoV) sequences. Erythrovirus primers were conserved between B19 and macaque erythroviruses. Bocavirus primers matched NS or VP1/2 coding regions of all currently published types/species of human bocavirus. First-round reactions were performed using the outer primer sets listed above, 5 μl of extracted DNA as a template, and GoTaq reagents (Promega) according to the manufacturer's instructions. Second-round reactions were performed with inner primer sets and 1 μl of first-round product as a template. Cycling conditions for both rounds were as follows: 30 cycles of 18 s at 94°C, 30 s at 50°C, and 90 s at 72°C and a final extension of 6 min at 72°C.
Direct sequencing of PCR products and sequence analysis.
Positive second-round PCR amplicons were sequenced in both directions using the inner sense and inner antisense primers used in the second round of amplification. Sequencing was carried out using BigDye Terminator v3.1 (Applied Biosystems) according to the manufacturer's instructions. Sequences were read at the Gene Pool facility (University of Edinburgh) and analyzed using Simmonic Sequence Editor v1.9 software.
Serology assays.
B19 virus screening was performed by commercially available enzyme-linked immunosorbent assay (ELISA) (Biotrin, Dublin, Ireland) according to the manufacturer's instructions. Wells were coated with recombinant B19 virus VP2 protein, and antibody binding was detected by peroxidase-labeled rabbit antihuman IgG followed by substrate (2,2 azinobis 3-ethylbenthiazoline-6-sulfonate) incubation. Samples were scored as positive according to the manufacturer's criteria based on calibrator control cutoff values.
PARV4 IgG detection was performed for each sample in replicate by indirect ELISA using baculovirus-expressed VP2 and control antigens as previously described (54). Due to a high background reactivity observed for some samples, an additional stipulation that positive samples must demonstrate a VP2 reactivity to a control reactivity optical density ratio of greater than 1.3 was used and any potentially positive samples falling below this cutoff were excluded from analysis. By this criterion, two samples from chimpanzees were omitted from the PARV4 seroprevalence totals.
HBoV1 VP2 recombinant proteins were expressed as previously described (59). For HBoV2, VP2 sequences (GenBank accession no. ACJ38942 and ACR15792) were synthesized and cloned in pUC57 by GenScript and transferred to the baculovirus vector pAcSG2 (Becton Dickinson Biosciences, Franklin Lakes, NJ) by standard cloning practices. Expression and purification of the proteins were performed as previously described (32, 59). ELISA for HBoV antibody was performed in parallel with wells coated with HBoV1 or HBoV2 antigens; reactivity to one or both antigens greater than two standard deviations above the negative-control mean was scored as positive.
Nucleotide sequence accession numbers.
All sequences have been submitted to GenBank and have been assigned the GenBank numbers HQ113143 to HQ113151.
RESULTS
Seroprevalence for B19 virus, PARV4, and HBoV antibodies.
Serological testing of primates used antibody assays previously developed for human samples. To determine whether anti-human IgG conjugates were reactive with ape and OWM IgG, direct ELISA of dilution series of representative plasma samples from each species along with human plasma controls was performed. Equivalent reactivity of the conjugates used in B19 virus, PARV4, and HBoV serological assays for IgG of human, ape, and OWMs was observed (see Fig. S1 at http://www.virus-evolution.org/Downloads/JVI01304-10/), indicating the applicability of the serology assays for primate screening purposes.
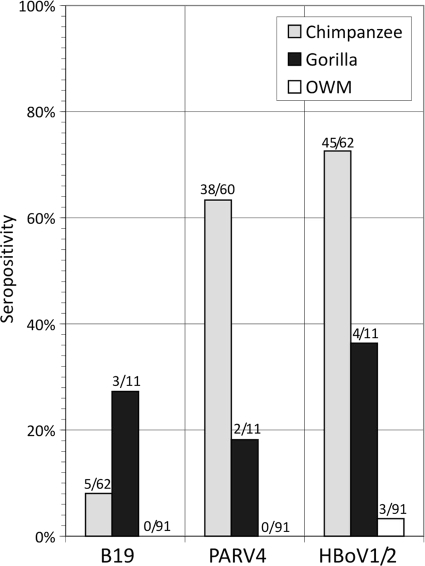
Plasma samples collected from 62 chimpanzees, 11 gorillas, and 91 Old World monkey species were screened for antibody to B19 virus, PARV4, and HBoV1/HBoV2 (Fig. 1). High frequencies of serological reactivity to PARV4, B19 virus, and HBoV were observed for chimpanzees (63%, 73%, and 8%, respectively), lower frequencies were observed for gorillas (18%, 36%, and 27%, respectively), and reactivity was absent in OWMs. In the last group, single samples from a drill (M. leucophaeus), a mona monkey (C. mona), and a Preuss's monkey (C. preussi) were positive for HBoV antibodies.
FIG. 1.
Frequency of serological reactivity of chimpanzee, gorilla, and OWP plasma samples in B19 virus, PARV4, and HBoV antibody ELISAs (number positive/number tested shown above each bar).
Genetic characterization of nonhuman parvoviruses.
The high frequency of serological reactivity to PARV4 in samples from chimpanzees and gorillas suggested the existence of viruses related to human PARV4 circulating in apes. To genetically characterize these variants, plasma and fecal samples from these species were screened for PARV4 DNA using primers conserved between human variants and the PARV4-like viruses described in pigs and cows (see Table S1 at http://www.virus-evolution.org/Downloads/JVI01304-10/). From these, one plasma sample from a (seropositive) chimpanzee (PT-P48) and one from a (seronegative) baboon (Papio anubis; PA-P68) were detectably viremic. Nested primers generated overlapping fragments of PARV4 spanning the genome from the chimpanzee sample, while the baboon sample was positive only with the screening primers originally used.
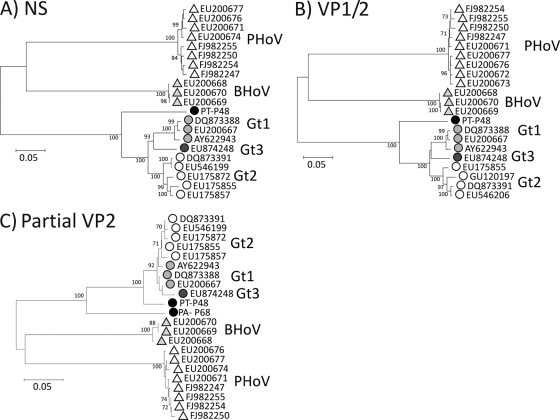
The chimpanzee samples yielded a near-complete genomic sequence of the chimpanzee PARV4 homologue, which was compared to PARV4 variants infecting humans and with the more distantly related porcine and bovine viruses (Fig. 2). In both genes (NS and VP1/VP2), nucleotide sequences from the chimpanzee variant were substantially more similar to those of human viruses (mean 18.2% and 15.2% divergence in NS and VP1/VP2 genes, respectively, to human genotypes 1 to 3) than to porcine and bovine viruses (41.5% and 43.3% and 36.9% and 37.8%, respectively, in the two regions). The chimpanzee variant was, however, substantially more divergent from human viruses than the three genotypes were from each other (mean intertype pairwise distances of 8.7% and 6.6% in NS and VP1/VP2, respectively). All phylogenetic analyses place the chimpanzee strain as a close outgroup to the human genotypes, with strong bootstrap support (Fig. 2). Translated sequences from the VP2 gene of the chimpanzee variant showed 6.9% amino acid sequence divergence from human variants, a level of diversity consistent with the evident cross-reactivity observed by serological screening. Although only a relatively short region could be amplified from the baboon sample, comparison of the VP2 sequences revealed an intermediate phylogenetic position; the baboon strain was more divergent from human sequences than the chimpanzee variant but more similar than the porcine or bovine homologues (Fig. 2C).
FIG. 2.
Phylogenetic analysis of (A) complete NS and (B) VP1/VP2 gene sequences and (C) partial VP2 sequence (positions 3067 to 3310) of the chimpanzee and baboon variants (PT-P48 and PA-P68; solid circles) and available complete genome sequences of PARV4 genotypes (Gt) 1 to 3 and of porcine and bovine homologues (PHoV, BHoV). Different species and genotypes are represented with different symbols and shading. PARV4 sequences showing <0.5% divergence from other sequences were excluded from analysis. The trees were constructed by neighbor joining of pairwise Jukes-Cantor corrected distances between nucleotide sequences; bootstrap values of ≥70% are shown.
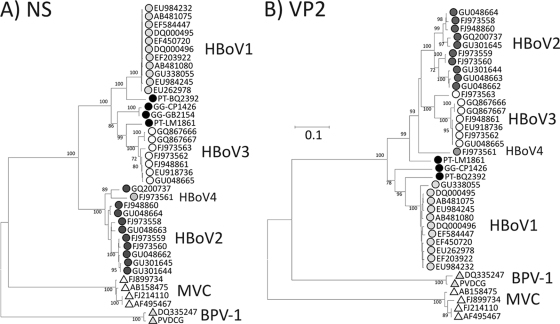
For analysis of HBoV-like viruses in apes and OWMs, plasma and fecal samples were screened for HBoV DNA using primers conserved between HBoV1 and HBoV2. HBoV sequences were detected in fecal samples from two chimpanzees (PT-BQ2392 and PT-LM1861) and two gorillas (GG-CP1426 and GG-GB2154). Sequences could be amplified from the NS gene (positions 1096 to 1581) for all four positive samples, while three were amplifiable in VP2 (positions 4343 to 4828). Comparison of these sequences with available (nonredundant) complete genome sequences of HBoV1 to HBoV4 showed phylogenetic relationships with human-derived viruses that were more complex than those exhibited by the chimpanzee homologues of human PARV4 (Fig. 3). The sequence heterogeneity among chimpanzee and gorilla HBoV variants was considerable, at least matching that observed among human viruses.
FIG. 3.
Phylogenetic analysis of partial NS (positions 1096 to 1581) and VP1/VP2 (positions 4343 to 4828) gene sequences of bocavirus-like sequences amplified from chimpanzee (prefixed PT) and gorilla (GG) positive fecal samples. Sequences were compared with available complete genome sequences of HBoV1-HBoV4 and the more divergent canine (MVC) and bovine (BPV-1) members of the Bocavirus genus. Different species and genotypes are represented with different symbols and shading. HBoV1 and HBoV2 sequences showing <0.5% divergence from other sequences were excluded from analysis. The trees were constructed by neighbor joining of pairwise Jukes-Cantor corrected distances between nucleotide sequences; bootstrap values of ≥70% are shown.
The chimpanzee variant BQ2392 was most similar to HBoV1 in both NS and VP2 regions (9.6% and 13.0% nucleotide sequence divergence, substantially greater than observed within human-derived HBoV1 sequences [0.2% and 1.7%]). The degree of divergence between this chimpanzee variant and HBoV1 was much less than between HBoV types/species (e.g., mean HBoV1-HBoV2 distances were 24.6% and 24.4%, respectively). BQ2392 might therefore be provisionally regarded as the chimpanzee homologue of HBoV1. The other chimpanzee-derived variant (LM1861) was genetically distinct. It also fell within the HBoV1/HBoV3 clade in the NS region and was indeed most similar to HBoV3 (human-derived) sequences (9.0%). However, in the VP2 region, it showed an outgroup relationship to HBoV2, HBoV3, and HBoV4 (15.4% to 16.2% divergence), rather that retaining its clustering with HBoV3. These inconsistent phylogenetic relationships provide evidence for further recombination events in the evolution of HBoV infecting apes.
Finally, the two gorilla-derived viruses were highly similar to each other in the NS region but showed no clear grouping with any single human (or chimpanzee) type species in either genome region. In NS, they were was most similar to HBoV1 and HBoV3 (15.5% and 13.6% divergence, respectively), while in the VP2 region analyzed its closest relative was HBoV1 (although showing 16.2% sequence divergence). Using the criteria that differentiate HBoV1-HBoV3, the gorilla-derived viruses would represent a further, equivalently different bocavirus type or species.
Using B19 virus-specific primers conserved among primate erythroviruses, all samples of plasma and feces from apes and OWMs were PCR negative.
DISCUSSION
This study provides the first evidence for the existence and widespread infection of nonhuman primates with parvoviruses genetically and antigenically related to PARV4, HBoV, and B19 virus. Our initial strategy of screening for anti-parvovirus antibodies was dictated by the acute/resolving nature of infections and absence of persistent viremia or fecal shedding. Population exposure in nonhuman primates was therefore estimated through serological testing for antibodies to each virus rather than screening samples by PCR to detect acute infections.
Serological screening methods.
The effectiveness of serological screening for estimating population or species exposure to a virus is dependent on both the durability of IgG responses after acute infection (i.e., whether the response is lifelong), and the extent of antigenic cross-reactivity between the virus in the target population and the protein used in ELISA. In the current study, we were reassured in both of these aspects.
For B19 virus, it is well established that high-affinity IgG reactivity to nondenatured VP2 antigen (such as that used in the current study) has a lifelong persistence after the initial infection (57). The continuing presence of B19 virus in a wide variety of tissues and minor reactivations that restimulate the immune system potentially contribute to this long-term seropositivity. PARV4 shows evidence for tissue persistence (33, 37, 52) and likely lifelong seropositivity after resolution of acute infections. Although several studies have failed to document an equivalent persistence of HBoV in lymphoid tissue, bone marrow, or skin (27, 37, 43), high frequencies of serological reactivity in human adults have been observed using a VP2-based ELISA similar to that used in the current study (8, 17, 30). These indicate a likely durable immune response and/or the occurrence of multiple infections that restimulate humoral antibody responses (17, 38).
In terms of cross-reactivity of human and primate viruses, the high PARV4 seroprevalence (63%) in chimpanzees provides evidence that extensive serological cross-reactivity with human PARV4 antigen does indeed occur. This supposition is supported by the observation of a relatively low level of amino acid sequence divergence between chimpanzee-derived and human PARV4 VP2 proteins (Table 1). For bocaviruses, sequence divergence of 10% or less between chimpanzee-derived and human viruses similarly predicts antibody cross-reactivity. However, the variant detected in the two positive gorilla samples showed no clear grouping with any of the four human types/species and its greater divergence in the region of VP2 sequenced (16.2%) may reduce cross-reactivity considerably. The 36% seroprevalence detected for gorillas (Fig. 1) may there represent an underestimate of their true rate of exposure.
TABLE 1.
Sequence divergence of ape-derived parvoviruses to human and animal homologues of PARV4 and HBoV
| Ape virus and parvovirus homologue | % Sequence divergence |
|||
|---|---|---|---|---|
| NS region |
VP1/VP2 region |
|||
| Nucleotide | Amino acid | Nucleotide | Amino acid | |
| PT-P48 (chimpanzee) | ||||
| PARV4 Gt 1 | 18.6 | 8.6 | 15.1 | 6.8 |
| Gt 2 | 17.9 | 8.2 | 15.3 | 7.0 |
| Gt 3 | 18.0 | 9.2 | 15.3 | 7.0 |
| BHoV | 41.5 | 42.2 | 36.9 | 34.7 |
| PhoV | 43.3 | 42.8 | 37.8 | 36.4 |
| BQ2392 (chimpanzee) | ||||
| HBoV1 | 9.6 | 3.1 | 13.0 | 4.5 |
| HBoV2 | 25.5 | 19.2 | 25.1 | 21.3 |
| HBoV3 | 17.4 | 5.7 | 25.4 | 21.7 |
| HBoV4 | 26.7 | 20.4 | 24.7 | 21.5 |
| MVC | 47.6 | 53.9 | 57.9 | 70.4 |
| BPV-1 | 50.5 | 58.0 | 51.6 | 66.2 |
| LM1861 (chimpanzee) | ||||
| HBoV1 | 16.3 | 5.6 | 20.2 | 18.6 |
| HBoV2 | 25.0 | 19.5 | 16.0 | 10.2 |
| HBoV3 | 9.0 | 4.0 | 15.4 | 11.0 |
| HBoV4 | 26.3 | 20.4 | 16.2 | 13.3 |
| MVC | 46.9 | 52.0 | 56.5 | 71.6 |
| BPV-1 | 50.1 | 58.0 | 51.8 | 64.6 |
| CP1426/GB2154 (gorilla) | ||||
| HBoV1 | 15.5 | 4.2 | 17.3 | 16.2 |
| HBoV2 | 23.7 | 19.6 | 24.8 | 20.8 |
| HBoV3 | 13.6 | 3.8 | 22.8 | 22.9 |
| HBoV4 | 25.4 | 20.4 | 23.4 | 22.8 |
| MVC | 46.7 | 51.4 | 57.7 | 71.5 |
| BPV-1 | 50.8 | 57.3 | 49.7 | 66.6 |
The B19 virus serology data contain some interpretation difficulties, since our PCR-based screening of plasma and fecal samples failed to detect ape or OWM homologues of B19 virus that would have allowed us to characterize them genetically. To date, the only nonhuman primate B19 viruses in the Erythrovirus genus have been obtained from macaques (16, 44). These show approximately 34% sequence divergence in VP2 from human B19 virus, a level of divergence that likely precludes cross-reactivity in the serology assay for OWM species investigated in the current study. Indeed, the uniformly negative (PARV4, B19 virus) or low frequency of (3% for HBoV) reactivity observed on screening OWMs in the current study most likely arose because of serological non-cross-reactivity rather than an absence of parvovirus homologues in these species. For PARV4, this hypothesis is strengthened by the finding by PCR of a virus variant in a baboon that was seronegative for anti-PARV4 antibodies. For future studies, screening of tissues rather than plasma may provide a better opportunity to detect and genetically characterize these viruses, since B19 virus and PARV4 can be found in the tissues of nonviremic humans (37, 43).
Population exposure of nonhuman primates to parvoviruses.
Notwithstanding the caveats discussed in the previous section concerning serological cross-reactivity, our seroprevalence results for apes represent a minimum estimate of population exposure to homologues of B19 virus, PARV4, and HBoV in wild ape populations. Based on these data, gorilla and chimpanzee homologues of all three human parvoviruses circulate extensively among wild ape populations, with particularly high rates of exposure to PARV4- and HBoV-like viruses in chimpanzees. While the seroprevalence for HBoV matches that of human populations (8, 17), the observation of 63% seroprevalence for PARV4 in chimpanzees appears at odds with that observed for human populations studied so far (54). As described in the introduction, PARV4 has a very specific risk group association with parenteral exposure in Western countries, with high rates of exposure recorded only in IDUs and those exposed to plasma products, such as hemophiliacs (13, 14, 35, 54). However, there is increasing evidence for a different epidemiology and possible alternate transmission routes for human PARV4 in sub-Saharan Africa. A very recent study has described high viremia frequencies in young children in Ghana without a history of parenteral exposure (45), while PARV4 infection has been detected in HIV-infected African men from Central/West Africa without an obvious past parenteral exposure (33, 55). We have additionally recently performed a large-scale serological screen for PARV4 antibodies of over 900 hepatitis C virus (HCV)- and HIV-uninfected blood donor/adult population samples collected from several countries in sub-Saharan Africa. Seroprevalences of PARV4 in HCV- and HIV-uninfected groups were 37% in Burkina Faso, 35% in the Cameroon, 25% in the Democratic Republic of Congo, and 8% in South Africa. This findings contrast markedly with a zero seroprevalence (0 of 360 samples) in French blood donors and United Kingdom low-risk controls (54a), and among 115 low-risk controls in Finland (30). The transmission route(s) responsible for such high exposure in Africa is currently undetermined, although it is most unlikely to be parenteral (as it is in Western countries) and is additionally not associated with HIV infection in Central Africa. If there is an environmental source of infection, this may also underlie the high rate of seropositivity in nonhuman primates. Longitudinal studies documenting when PARV4 infections occur in both humans and apes are required to better characterize the sources and risk factors for this virus infection. Given the seroprevalance of PARV4 in humans and nonhuman primates in sub-Sarahan Africa, it seems reasonable to posit that PARV4 infections observed in IDUs most likely originated from this region. However, the timescale and route of PARV4 emergence in Western countries is not yet known.
Genetic diversity and evolution of parvoviruses.
The newly characterized PARV4- and HBoV-like viruses recovered from nonhuman primates showed a consistently closer evolutionary relationship to human viruses than to homologues infecting pigs and cows (PARV4) or to the host species of viruses that formed the original Bocavirus genus (dogs, cows). It has been suggested that the parvoviruses have coevolved and codiverged with their mammalian hosts, implying that the common ancestor of this group of viruses is at least 90 million years old (34). For example, the relationships among human B19 virus and erythroviruses from three macaque species corresponded to relationships among their hosts as revealed by mitochondrial sequences (34). Although we have not been able to genetically characterize ape homologues of B19 virus, the PARV4 and HBoV-like viruses found in apes in the current study are substantially more genetically similar to human variants than macaque B19-like viruses are to human B19 virus. This lends further, although indirect, support to the coevolution hypothesis.
On the other hand, specific instances of cross-species transmission have been identified in the evolution of parvoviruses, most obviously the recent transmission of feline parvovirus to dogs (46). There are additionally murine and hamster parvoviruses showing mosaicism between genome regions (34), comparable to the chimeric nature of HBoV3 and HBoV4 (23). Phylogenetic relationships of the ape-derived HBoV-like viruses were complex, with further examples of inconsistent phylogenetic relationships between NS and VP2 regions indicative of recombination events in the evolutionary history of ape bocaviruses. Further genetic characterization and a larger sample set will be required to accurately document the evolutionary process underlying the current diversity of HBoV variants detected in apes. One intriguing interpretation of the data obtained in the current study is that chimpanzee and gorilla-associated viruses represent separate, divergent ape homologues for HBoV1, HBoV2, and the recombinant HBoV3; the phylogeny of these outliers indeed recapitulates sequence relationships between chimpanzee and human PARV4 variants. This would logically place the evolution of bocaviruses, including the recombination event that created HBoV3, before ape speciation 5 million years ago if we follow the coevolution model. However, we acknowledge that much more extensive characterization of parvovirus homologues in OWMs and those in the New World is required as additional data points to fully substantiate the hypothesis of parvovirus/primate coevolution in the three parvovirus genera we have analyzed.
Acknowledgments
We thank the Cameroon Ministry of Scientific Research and Innovation and the Ministry of Forestry and Wildlife, which provided the necessary permits for this work. We thank the U.S. Embassy in Cameroon for their support. Additional thanks to the management and staff of Limbe Wildlife Centre, Mfou National Park, and Ape Action Africa. We thank the staff from PRESICA for logistical support in Cameroon and the Cameroonian Ministries of Health, Environment and Forestry, and Research for permission to collect fecal samples in Cameroon. We thank Lea Hedman, Department of Virology, Haartman Institute, University of Helsinki, for help with serology assay development.
N.D.W. is supported by the NIH Director's Pioneer Award (DP1-OD000370), google.org, the Skoll Foundation, the Henry M. Jackson Foundation for the Advancement of Military Medicine, and the Global Emerging Infections Surveillance and Response System (GEIS)—a Division of the United States Armed Forces Health Surveillance Center. Additional support was provided to GVFI by the United States Agency for International Development (USAID) Emerging Pandemic Threats Program, PREDICT project, under the terms of cooperative agreement number GHN-A-OO-09-00010-00. The fecal sample collection was supported by grants from National Institutes of Health (R01 AI50529), the Agence National de Recherches sur le SIDA, France (ANRS 12125 and ANRS 12182), and the Institut de Recherche pour le Développement (IRD). Development of HBoV serology assays was supported by the Helsinki University Central Hospital Research and Education, and Research and Development Funds, the Sigrid Jusélius Foundation, the Medical Society of Finland (FLS), and the Academy of Finland (project 1122539). Development of ELISA for PARV4 antibody was supported by a grant from Baxter (Los Angeles, CA). O.G.P. is supported by the Royal Society.
Footnotes
Published ahead of print on 28 July 2010.
REFERENCES
- 1.Allander, T. 2008. Human bocavirus. J. Clin. Virol. 41:29-33. [DOI] [PubMed] [Google Scholar]
- 2.Allander, T., T. Jartti, S. Gupta, H. G. Niesters, P. Lehtinen, R. Osterback, T. Vuorinen, M. Waris, A. Bjerkner, A. Tiveljung-Lindell, B. G. van den Hoogen, T. Hyypia, and O. Ruuskanen. 2007. Human bocavirus and acute wheezing in children. Clin. Infect. Dis. 44:904-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur, J. L., G. D. Higgins, G. P. Davidson, R. C. Givney, and R. M. Ratcliff. 2009. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 5:e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, K. E., S. M. Anderson, and N. S. Young. 1993. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262:114-117. [DOI] [PubMed] [Google Scholar]
- 6.Brown, K. E., Z. Liu, G. Gallinella, S. Wong, I. P. Mills, and M. G. O'Sullivan. 2004. Simian parvovirus infection: a potential zoonosis. J. Infect. Dis. 190:1900-1907. [DOI] [PubMed] [Google Scholar]
- 7.Candotti, D., N. Etiz, A. Parsyan, and J. P. Allain. 2004. Identification and characterization of persistent human erythrovirus infection in blood donor samples. J. Virol. 78:12169-12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cecchini, S., A. Negrete, T. Virag, B. S. Graham, J. I. Cohen, and R. M. Kotin. 2009. Evidence of prior exposure to human bocavirus as determined by a retrospective serological study of 404 serum samples from adults in the United States. Clin. Vaccine Immunol. 16:597-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, K. C., B. C. Shull, E. A. Moses, M. Lederman, E. R. Stout, and R. C. Bates. 1986. Complete nucleotide sequence and genome organization of bovine parvovirus. J. Virol. 60:1085-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chieochansin, T., A. Kapoor, E. Delwart, Y. Poovorawan, and P. Simmonds. 2009. Absence of detectable replication of human bocavirus species 2 in the respiratory tract. Emerg. Infect. Dis. 15:1503-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow, B. D., Z. Ou, and F. P. Esper. 2010. Newly recognized bocaviruses (HBoV, HBoV2) in children and adults with gastrointestinal illness in the United States. J. Clin. Virol. 47:143-147. [DOI] [PubMed] [Google Scholar]
- 12.Eis-Hubinger, A. M., U. Reber, T. Abdul-Nour, U. Glatzel, H. Lauschke, and U. Putz. 2001. Evidence for persistence of parvovirus B19 DNA in livers of adults. J. Med. Virol. 65:395-401. [DOI] [PubMed] [Google Scholar]
- 13.Fryer, J. F., A. Kapoor, P. D. Minor, E. Delwart, and S. A. Baylis. 2006. Novel parvovirus and related variant in human plasma. Emerg. Infect. Dis. 12:151-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fryer, J. F., S. B. Lucas, D. Padley, and S. A. Baylis. 2007. Parvoviruses PARV4/5 in hepatitis C virus-infected patient. Emerg. Infect. Dis. 13:175-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, N. L., T. W. Gyorkos, C. Beliveau, E. Rahme, C. Muecke, and J. C. Soto. 2005. Seroprevalence of parvovirus B19 infection in daycare educators. Epidemiol. Infect. 133:299-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, S. W., I. Malkovska, M. G. O'Sullivan, and K. E. Brown. 2000. Rhesus and pig-tailed macaque parvoviruses: identification of two new members of the erythrovirus genus in monkeys. Virology 269:105-112. [DOI] [PubMed] [Google Scholar]
- 17.Hedman, L., M. Soderlund-Venermo, T. Jartti, O. Ruuskanen, and K. Hedman. 2010. Dating of human bocavirus infection with protein-denaturing IgG-avidity assays—secondary immune activations are ubiquitous in immunocompetent adults. J. Clin. Virol. 48:44-48. [DOI] [PubMed] [Google Scholar]
- 18.Heegaard, E. D., and K. E. Brown. 2002. Human parvovirus B19. Clin. Microbiol. Rev. 15:485-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hindiyeh, M. Y., N. Keller, M. Mandelboim, D. Ram, J. Rubinov, L. Regev, V. Levy, S. Orzitzer, H. Shaharabani, R. Azar, E. Mendelson, and Z. Grossman. 2008. High rate of human bocavirus and adenovirus coinfection in hospitalized Israeli children. J. Clin. Microbiol. 46:334-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hokynar, K., J. Brunstein, M. Soderlund-Venermo, O. Kiviluoto, E. K. Partio, Y. Konttinen, and K. Hedman. 2000. Integrity and full coding sequence of B19 virus DNA persisting in human synovial tissue. J. Gen. Virol. 81:1017-1025. [DOI] [PubMed] [Google Scholar]
- 21.Hokynar, K., M. Soderlund-Venermo, M. Pesonen, A. Ranki, O. Kiviluoto, E. K. Partio, and K. Hedman. 2002. A new parvovirus genotype persistent in human skin. Virology 302:224-228. [DOI] [PubMed] [Google Scholar]
- 22.Jones, M. S., A. Kapoor, V. V. Lukashov, P. Simmonds, F. Hecht, and E. Delwart. 2005. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 79:8230-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapoor, A., P. Simmonds, E. Slikas, L. Li, L. Bodhidatta, O. Sethabutr, H. Triki, O. Bahri, B. S. Oderinde, M. M. Baba, D. N. Bukbuk, J. Besser, J. Bartkus, and E. Delwart. 2010. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 201:1633-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapoor, A., E. Slikas, P. Simmonds, T. Chieochansin, A. Naeem, S. Shaukat, M. M. Alam, S. Sharif, M. Angez, S. Zaidi, and E. Delwart. 2009. A newly identified bocavirus species in human stool. J. Infect. Dis. 199:196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keele, B. F., F. Van Heuverswyn, Y. Li, E. Bailes, J. Takehisa, M. L. Santiago, F. Bibollet-Ruche, Y. Chen, L. V. Wain, F. Liegeois, S. Loul, E. M. Ngole, Y. Bienvenue, E. Delaporte, J. F. Brookfield, P. M. Sharp, G. M. Shaw, M. Peeters, and B. H. Hahn. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr, J. R., J. P. Cartron, M. D. Curran, J. E. Moore, J. R. Elliott, and R. A. Mollan. 1995. A study of the role of parvovirus B19 in rheumatoid arthritis. Br. J. Rheumatol. 34:809-813. [DOI] [PubMed] [Google Scholar]
- 27.Kuethe, F., J. Lindner, K. Matschke, J. J. Wenzel, P. Norja, K. Ploetze, S. Schaal, V. Kamvissi, S. R. Bornstein, U. Schwanebeck, and S. Modrow. 2009. Prevalence of parvovirus B19 and human bocavirus DNA in the heart of patients with no evidence of dilated cardiomyopathy or myocarditis. Clin. Infect. Dis. 49:1660-1666. [DOI] [PubMed] [Google Scholar]
- 28.Kuhl, U., M. Pauschinger, M. Noutsias, B. Seeberg, T. Bock, D. Lassner, W. Poller, R. Kandolf, and H. P. Schultheiss. 2005. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation 111:887-893. [DOI] [PubMed] [Google Scholar]
- 29.Kurtzman, G. J., B. J. Cohen, A. M. Field, R. Oseas, R. M. Blaese, and N. S. Young. 1989. Immune response to B19 parvovirus and an antibody defect in persistent viral infection. J. Clin. Invest. 84:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahtinen, A., P. Kivela, L. Hedman, A. Kumar, A. Kantele, M. Lappalainen, K. Liitsola, M. Ristola, E. Delwart, C. Sharp, P. Simmonds, M. Soderlund-Venermo, and K. Hedman. Human parvovirus PARV4—comprehensive serodiagnostics and identification of primary infections. Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 31.Lau, S. K., P. C. Woo, H. Tse, C. T. Fu, W. K. Au, X. C. Chen, H. W. Tsoi, T. H. Tsang, J. S. Chan, D. N. Tsang, K. S. Li, C. W. Tse, T. K. Ng, O. T. Tsang, B. J. Zheng, S. Tam, K. H. Chan, B. Zhou, and K. Y. Yuen. 2008. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J. Gen. Virol. 89:1840-1848. [DOI] [PubMed] [Google Scholar]
- 32.Lindner, J., L. Karalar, S. Zehentmeier, A. Plentz, H. Pfister, W. Struff, M. Kertai, H. Segerer, and S. Modrow. 2008. Humoral immune response against human bocavirus VP2 virus-like particles. Viral Immunol. 21:443-449. [DOI] [PubMed] [Google Scholar]
- 33.Longhi, E., G. Bestetti, V. Acquaviva, A. Foschi, R. Piolini, L. Meroni, C. Magni, S. Antinori, C. Parravicini, and M. Corbellino. 2007. Human parvovirus 4 in the bone marrow of Italian patients with AIDS. AIDS 21:1481-1483. [DOI] [PubMed] [Google Scholar]
- 34.Lukashov, V. V., and J. Goudsmit. 2001. Evolutionary relationships among parvoviruses: virus-host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses. J. Virol. 75:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lurcharchaiwong, W., T. Chieochansin, S. Payungporn, A. Theamboonlers, and Y. Poovorawan. 2008. Parvovirus 4 (PARV4) in serum of intravenous drug users and blood donors. Infection 36:488-491. [DOI] [PubMed] [Google Scholar]
- 36.Manning, A., V. Russell, K. L. G. H. Eastick, N. Hallam, K. E. Templeton, and P. Simmonds. 2006. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J. Infect. Dis. 194:1283-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manning, A., S. J. Willey, J. E. Bell, and P. Simmonds. 2007. Comparison of tissue distribution, persistence, and molecular epidemiology of parvovirus B19 and novel human parvoviruses PARV4 and human bocavirus. J. Infect. Dis. 195:1345-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin, E. T., M. P. Fairchok, J. Kuypers, A. Magaret, D. M. Zerr, A. Wald, and J. A. Englund. 2010. Frequent and prolonged shedding of bocavirus in young children attending daycare. J. Infect. Dis. 201:1625-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mossong, J., N. Hens, V. Friederichs, I. Davidkin, M. Broman, B. Litwinska, J. Siennicka, A. Trzcinska, P. V. Damme, P. Beutels, A. Vyse, Z. Shkedy, M. Aerts, M. Massari, and G. Gabutti. 2008. Parvovirus B19 infection in five European countries: seroepidemiology, force of infection and maternal risk of infection. Epidemiol. Infect. 136:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neel, C., L. Etienne, Y. Li, J. Takehisa, R. S. Rudicell, I. N. Bass, J. Moudindo, A. Mebenga, A. Esteban, F. Van Heuverswyn, F. Liegeois, P. J. Kranzusch, P. D. Walsh, C. M. Sanz, D. B. Morgan, J. B. Ndjango, J. C. Plantier, S. Locatelli, M. K. Gonder, F. H. Leendertz, C. Boesch, A. Todd, E. Delaporte, E. Mpoudi-Ngole, B. H. Hahn, and M. Peeters. 2010. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J. Virol. 84:1464-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen, Q. T., S. Wong, E. D. Heegaard, and K. E. Brown. 2002. Identification and characterization of a second novel human erythrovirus variant, A6. Virology 301:374-380. [DOI] [PubMed] [Google Scholar]
- 42.Norja, P., A. M. Eis-Hubinger, M. Soderlund-Venermo, K. Hedman, and P. Simmonds. 2008. Rapid sequence change and geographical spread of human parvovirus B19; comparison of B19 evolution in acute and persistent infections. J. Virol. 82:6427-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norja, P., K. Hokynar, L. M. Aaltonen, R. Chen, A. Ranki, E. K. Partio, O. Kiviluoto, I. Davidkin, T. Leivo, A. M. Eis-Hubinger, B. Schneider, H. P. Fischer, R. Tolba, O. Vapalahti, A. Vaheri, M. Soderlund-Venermo, and K. Hedman. 2006. Bioportfolio: lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc. Natl. Acad. Sci. U. S. A. 103:7450-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Sullivan, M. G., D. C. Anderson, J. D. Fikes, F. T. Bain, C. S. Carlson, S. W. Green, N. S. Young, and K. E. Brown. 1994. Identification of a novel simian parvovirus in cynomolgus monkeys with severe anemia. A paradigm of human B19 parvovirus infection. J. Clin. Invest. 93:1571-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panning, M., R. Kobbe, S. Vollbach, J. F. Drexler, S. Adjei, O. Adjei, C. Drosten, J. May, and A. M. Eis-Hubinger. 2010. Novel human parvovirus 4 genotype 3 in infants, Ghana. Emerg. Infect. Dis. 16:1143-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parrish, C. R. 1999. Host range relationships and the evolution of canine parvovirus. Vet. Microbiol. 69:29-40. [DOI] [PubMed] [Google Scholar]
- 47.Parsyan, A., C. Szmaragd, J. P. Allain, and D. Candotti. 2007. Identification and genetic diversity of two human parvovirus B19 genotype 3 subtypes. J. Gen. Virol. 88:428-431. [DOI] [PubMed] [Google Scholar]
- 48.Rohrer, C., B. Gartner, A. Sauerbrei, S. Bohm, B. Hottentrager, U. Raab, W. Thierfelder, P. Wutzler, and S. Modrow. 2008. Seroprevalence of parvovirus B19 in the German population. Epidemiol. Infect. 136:1564-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanabani, S., W. K. Neto, J. Pereira, and E. C. Sabino. 2006. Sequence variability of human erythroviruses present in bone marrow of Brazilian patients with various parvovirus B19-related hematological symptoms. J. Clin. Microbiol. 44:604-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos, N., T. C. Peret, C. D. Humphrey, M. C. Albuquerque, R. C. Silva, F. J. Benati, X. Lu, and D. D. Erdman. 2010. Human bocavirus species 2 and 3 in Brazil. J. Clin. Virol. 48:127-130. [DOI] [PubMed] [Google Scholar]
- 51.Schildgen, O., A. Muller, T. Allander, I. M. Mackay, S. Volz, B. Kupfer, and A. Simon. 2008. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin. Microbiol. Rev. 21:291-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider, B., J. F. Fryer, U. Reber, H. P. Fischer, R. H. Tolba, S. A. Baylis, and A. M. Eis-Hubinger. 2008. Persistence of novel human parvovirus PARV4 in liver tissue of adults. J. Med. Virol. 80:345-351. [DOI] [PubMed] [Google Scholar]
- 53.Servant, A., S. Laperche, F. Lallemand, V. Marinho, M. G. De Saint, J. F. Meritet, and A. Garbarg-Chenon. 2002. Genetic diversity within human erythroviruses: identification of three genotypes. J. Virol. 76:9124-9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharp, C. P., A. Lail, S. Donfield, R. Simmons, C. Leen, P. Klenerman, E. Delwart, E. D. Gomperts, and P. Simmonds. 2009. High frequencies of exposure to the novel human parvovirus, PARV4 in haemophiliacs and injecting drug users detected by a serological assay for PARV4 antibodies. J. Infect. Dis. 200:1119-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54a.Sharp, C. P., M. Vermeulen, Y. Nébie, C. F. Djoko, M. LeBreton, U. Tamoufe, A. W. Rimoin, P. K. Kayembe, J. K. Carr, A. Servant-Delmas, S. Laperche, A. Harrison, O. Pybus, E. Delwart, N. D. Wolfe, A. Saville, J. Lefrère, and P. Simmonds. Widespread exposure to PARV4 infection in sub-Saharan Africa identified by serological screening for IgG antibodies to viral capsid protein. Emerg. Infect. Dis., in press.
- 55.Simmonds, P., J. Douglas, G. Bestetti, E. Longhi, S. Antinori, C. Parravicini, and M. Corbellino. 2008. A third genotype of the human parvovirus PARV4 in sub-Saharan Africa. J. Gen. Virol. 89:2299-2302. [DOI] [PubMed] [Google Scholar]
- 56.Simmonds, P., A. Manning, R. Kenneil, F. W. Carnie, and J. E. Bell. 2007. Parenteral transmission of the novel human parvovirus, PARV4. Emerg. Infect. Dis. 13:1386-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soderlund, M., C. S. Brown, W. J. Spaan, L. Hedman, and K. Hedman. 1995. Epitope type-specific IgG responses to capsid proteins VP1 and VP2 of human parvovirus B19. J. Infect. Dis. 172:1431-1436. [DOI] [PubMed] [Google Scholar]
- 58.Soderlund, M., R. von Essen, J. Haapasaari, U. Kiistala, O. Kiviluoto, and K. Hedman. 1997. Persistence of parvovirus B19 DNA in synovial membranes of young patients with and without chronic arthropathy. Lancet 349:1063-1065. [DOI] [PubMed] [Google Scholar]
- 59.Soderlund-Venermo, M., A. Lahtinen, T. Jartti, L. Hedman, K. Kemppainen, P. Lehtinen, T. Allander, O. Ruuskanen, and K. Hedman. 2009. Clinical assessment and improved diagnosis of bocavirus-induced wheezing in children, Finland. Emerg. Infect. Dis. 15:1423-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song, J. R., Y. Jin, Z. P. Xie, H. C. Gao, N. G. Xiao, W. X. Chen, Z. Q. Xu, K. L. Yan, Y. Zhao, Y. D. Hou, and Z. J. Duan. 2010. Novel human bocavirus in children with acute respiratory tract infection. Emerg. Infect. Dis. 16:324-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vyse, A. J., N. J. Andrews, L. M. Hesketh, and R. Pebody. 2007. The burden of parvovirus B19 infection in women of childbearing age in England and Wales. Epidemiol. Infect. 135:1354-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams, J. V. 2010. Deja vu all over again: Koch's postulates and virology in the 21st century. J. Infect. Dis. 201:1611-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoo, B. C., D. H. Lee, S. M. Park, J. W. Park, C. Y. Kim, H. S. Lee, J. S. Seo, K. J. Park, and W. S. Ryu. 1999. A novel parvovirus isolated from Manchurian chipmunks. Virology 253:250-258. [DOI] [PubMed] [Google Scholar]
- 64.Zehender, G., C. De Maddalena, M. Canuti, A. Zappa, A. Amendola, A. Lai, M. Galli, and E. Tanzi. 2010. Rapid molecular evolution of human bocavirus revealed by Bayesian coalescent inference. Infect. Genet. Evol. 10:215-220. [DOI] [PubMed] [Google Scholar]