Abstract
PB1-F2 is a viral protein that is encoded by the PB1 gene of influenza A virus by alternative translation. It varies in length and sequence context among different strains. The present study examines the functions of PB1-F2 proteins derived from various human and avian viruses. While H1N1 PB1-F2 was found to target mitochondria and enhance apoptosis, H5N1 PB1-F2, surprisingly, did not localize specifically to mitochondria and displayed no ability to enhance apoptosis. Introducing Leu into positions 69 (Q69L) and 75 (H75L) in the C terminus of H5N1 PB1-F2 drove 40.7% of the protein to localize to mitochondria compared with the level of mitochondrial localization of wild-type H5N1 PB1-F2, suggesting that a Leu-rich sequence in the C terminus is important for targeting of mitochondria. However, H5N1 PB1-F2 contributes to viral RNP activity, which is responsible for viral RNA replication. Lastly, although the swine-origin influenza virus (S-OIV) contained a truncated form of PB1-F2 (12 amino acids [aa]), potential mutation in the future may enable it to contain a full-length product. Therefore, the functions of this putative S-OIV PB1-F2 (87 aa) were also investigated. Although this PB1-F2 from the mutated S-OIV shares only 54% amino acid sequence identity with that of seasonal H1N1 virus, it also increased viral RNP activity. The plaque size and growth curve of the viruses with and without S-OIV PB1-F2 differed greatly. The PB1-F2 protein has various lengths, amino acid sequences, cellular localizations, and functions in different strains, which result in strain-specific pathogenicity. Such genetic and functional diversities make it flexible and adaptable in maintaining the optimal replication efficiency and virulence for various strains of influenza A virus.
Influenza A viruses contain eight negative-stranded RNA segments that encode 11 known viral proteins. The 11th viral protein was originally found in a search for unknown peptides during influenza A virus infection recognized by CD8+ T cells. It was termed PB1-F2 and is the second protein that is alternatively translated by the same PB1 gene (8). PB1-F2 can be encoded in a large number of influenza A viruses that are isolated from various hosts, including human and avian hosts. The size of PB1-F2 ranges from 57 to 101 amino acids (aa) (41). While strain PR8 (H1N1) contains a PB1-F2 with a length of 87 aa, PB1-F2 is terminated at amino acid position 57 in most human H1N1 viruses and is thus a truncated form compared with the length in PR8. Human H3N2 and most avian influenza A viruses encode a full-length PB1-F2 protein, which is at least 87 aa (7). Many cellular functions of the PB1-F2 protein, and especially the protein of the PR8 strain, have been reported (11, 25). For example, PR8 PB1-F2 localizes to mitochondria in infected and transfected cells (8, 15, 38, 39), suggesting that PB1-F2 enhances influenza A virus-mediated apoptosis in human monocytes (8). The phosphorylation of the PR8 PB1-F2 protein has been suggested to be one of the crucial causes of the promotion of apoptosis (30).
The rates of synonymous and nonsynonymous substitutions in the PB1-F2 gene are higher than those in the PB1 gene (7, 20, 21, 37, 42). Recent work has shown that both PR8 PB1-F2 and H5N1 PB1-F2 are important regulators of influenza A virus virulence (1). Additionally, the expression of the 1918 influenza A virus (H1N1) PB1-F2 increases the incidence of secondary bacterial pneumonia (10, 28). However, PB1-F2 is not essential for viral replication because the knockout of PB1-F2 in strain PR8 has no effect on the viral titer (40), suggesting that PB1-F2 may have cellular functions other than those that were originally thought (29).
PB1-F2 was translated from the same RNA segment as the PB1 protein, whose function is strongly related to virus RNP activity, which is responsible for RNA chain elongation and which exhibits RNA-dependent RNA polymerase activity (2, 5) and endonuclease activity (9, 16, 26). Previous research has already proved that the knockout of PR8 PB1-F2 reduced virus RNP activity, revealing that PR8 PB1-F2 contributes to virus RNP activity (27), even though PB1-F2 has no effect on the virus growth rate (40). In the present study, not only PR8 PB1-F2 but also H5N1 PB1-F2 and putative full-length swine-origin influenza A virus (S-OIV) PB1-F2 contributed to virus RNP activity. However, PR8 PB1-F2 and H5N1 PB1-F2 exhibit different biological behaviors, including different levels of expression, cellular localizations, and apoptosis enhancements. The molecular determinants of the different localizations were also addressed. The function of the putative PB1-F2 derived from S-OIV was also studied. The investigation described here reveals that PB1-F2 proteins derived from various viral strains exhibited distinct functions, possibly contributing to the variation in the virulence of influenza A viruses.
MATERIALS AND METHODS
Sequence analysis.
All sequences were aligned and analyzed using BioEdit software (version 7.0.5.2) and GeneDoc software (version 2.7.000). The amphipathic helical structure was predicted using GCG Lite software.
Cell lines, antibodies, and reagents.
Ching-Ho Wang, Department of Veterinary Medicine, National Taiwan University, Taipei, Taiwan, kindly provided the viral RNA (vRNA) of A/chicken/Taiwan/01/2000 (H6N1). The two plasmids expressing PB1 of A/Hong Kong/156/1997 (H5N1) and A/Netherlands/219/2003 (H7N7) were generous gifts from Ron A. M. Fouchier, National Influenza Center and Department of Virology, Erasmus Medical Center, Rotterdam, Netherlands. MDCK, 293T, and HeLa cells were obtained from ATCC and were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, San Diego, CA) with 10% fetal bovine serum (Gibco). U937 cells were obtained from ATCC and were maintained in RPMI 1640 medium (Gibco). Green fluorescent protein (GFP) mouse monoclonal antibody (B2) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and FLAG-M2 monoclonal antibody for FLAG epitopes was obtained from Sigma (St. Louis, MO). Influenza A virus nucleoprotein (NP) mouse monoclonal antibody was obtained from Abcam (Cambridge, United Kingdom). Anti-mouse IgG horseradish peroxidase (HRP)-linked species-specific whole antibody was obtained from GE Healthcare (Buckinghamshire, United Kingdom). Anti-goat IgG-HRP antibody was purchased from Santa Cruz Biotechnology. Influenza A virus PB1 was obtained from Santa Cruz Biotechnology. Fluorescein isothiocyanate (FITC)-conjugated antimouse secondary antibody, the MitoTracker Red probe, and Hoechst 33258 dye were obtained from Molecular Probes (Eugene, OR).
RNA extraction from clinical isolates and RT-PCR.
Clinical specimens containing A/Taiwan/3355/1997 (H1N1), A/Taiwan/1184/1999 (H1N1), A/Taiwan/3351/1997 (H3N2), and A/Taiwan/1748/1997 (H3N2) viruses were collected from the Clinical Virology Laboratory, Chang Gung Memorial Hospital Contracted Laboratory of the Center for Disease Control and Prevention, Taiwan. The isolates were passed in MDCK cells, and the supernatant was used to extract viral RNA with a viral RNA extraction miniprep system kit (Virogene, Sunnyvale, CA). Viral RNA was amplified into double-stranded DNA with a segment-specific primer using a Reverse-It one-step kit (ABgene, United Kingdom). The reverse transcription (RT)-PCR program for all clinical isolates was as follows: 50°C for 30 min and 94°C for 2 min, followed by 40 cycles of 94°C for 0.5 min, 50°C for 0.5 min, and 72°C for 1.5 min and a final elongation step of 72°C for 7 min. The final product was stored at 4°C.
Plasmid constructions and mutagenesis.
The coding sequences of the PB1-F2 variants were amplified from the virus RNA genome, which was extracted from clinical isolates using subtype-specific primers. Each PB1-F2 fragment was inserted into the pFLAG-CMV2 vector and flanked by EcoRI and HindIII. Single or double Leu mutations at positions 69 and 75 were introduced into plasmid pFLAG-H5N1. Three mutations were introduced to produce a PR8 PB1-F2-knockout PB1 gene and an H5N1 PB1-F2-knockout PB1 gene in polymerase I-polymerase II transcription (Pol I-Pol II) system plasmids (18, 19). Two mutations (A129C and A267G) were introduced into the wild-type S-OIV PB1 gene, which was cloned into Pol I-Pol II system plasmids to generate full-length S-OIV PB1-F2. All mutant constructs in this work were produced using a QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX), according to the manufacturer's instructions. Mutations were made only to affect the length or amino acid sequence of PB1-F2. No alteration of an amino acid or the length of the PB1 protein occurred, ensuring that the effects of mutations on polymerase activity originate from the PB1-F2 protein and not the PB1 protein. Each mutant construct was verified by full-length sequencing before functional assays of the mutants were performed. vRNAs were extracted from recombinant viruses and sequenced again before examination of the growth curves and apoptosis levels.
Western blot analysis.
Total cell lysate was collected from transfected 293T cells, and total protein was extracted by treatment with CA630 lysis buffer (150 mM NaCl, 1% CA630 detergent, 50 mM Tris base [pH 8.0]) on ice for 30 min. Total cell proteins were analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences, Germany). The PVDF membranes were blocked with Tris-buffered saline with 0.1% Tween 20 that contained 5% nonfat dry milk and probed with the indicated antibodies. Antibodies against the FLAG epitope (1:1,000), GFP (1:5,000), actin (1:5,000), influenza A virus NP (1:500), and influenza A virus PB1 (1:200) were used. After the membrane was washed, it was incubated with HRP-conjugated antimouse antibody or HRP-conjugated antigoat antibody (diluted 1:5,000), as appropriate. HRP was detected using a Western Lightning chemiluminescence kit (Amersham Pharmacia, Freiburg, Germany), following the manufacturer's protocol.
RT-PCR for detecting levels of transcripts of FLAG-tagged PB1-F2 constructs.
293T cells were transfected with 1 μg FLAG-tagged PB1-F2 expression plasmids, which were extracted using a QIAprep spin miniprep kit (Qiagen, Valencia, CA). Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used as a transfection agent. At 24 h posttransfection, 293T cells were collected to extract total cellular RNA using a PureLink Micro-to-Midi total RNA purification system (Invitrogen). Total cellular RNA was reverse transcribed using high-capacity cDNA reverse transcription kits (Applied Biosystems, Foster City, CA) and amplified with Pfu Turbo DNA polymerase (Strategene, Cedar Creek, TX).
Immunofluoresence and confocal microscopy.
HeLa cells were transfected with 2 μg FLAG-tagged PB1-F2 expression plasmids using Lipofectamine 2000. At 24 h posttransfection, the cells were stained with MitoTracker Red (100 nM) and Hoechst 33258 (100 nM) at room temperature for 15 min, fixed with 4% formaldehyde in phosphate-buffered saline for 15 min at room temperature, and then permeabilized with 0.5% Triton X-100 for 15 min at room temperature. Cells were probed with FLAG-M2 monoclonal antibody for 2 h at room temperature and labeled with secondary FITC-conjugated antimouse secondary antibody for 1 h at room temperature. Confocal microscopy was performed with a Zeiss LSM510 Meta microscope. Ten fields (magnification, ×630) were randomly selected to calculate the percentage of the cells that expressed the pFLAG-H5N1-Q69L/H75L expression plasmids.
Generation of recombinant viruses by reverse genetics.
The A/Puerto Rico/8/1934 and other recombinant viruses used in this study were generated using an eight-plasmid reverse genetics system (18, 19). The plasmids were kindly provided by Robert G. Webster, Department of Infectious Diseases, St. Jude Children's Research Hospital, Memphis, TN. To generate recombinant viruses, eight plasmids were cotransfected into 293T cells and the cells were supplemented with a minimal essential medium (Opti-MEM) using Lipofectamine 2000. At 18 h posttransfection, the transfection medium was replaced by DMEM without fetal bovine serum. At 32 h posttransfection, the supernatant was collected and amplified in MDCK cells. The virus titer was measured by plaque assay on MDCK cells.
Plaque assay.
Confluent MDCK cells in six-well plates were washed with phosphate-buffered saline, and serial dilutions of the virus were adsorbed onto cells for 1 h at 37°C. Unadsorbed virus was removed by washing the cells with phosphate-buffered saline, and the cells were overlaid with 3 ml of overlay Dulbecco's modified Eagle's medium that had been supplemented with 3% agarose. After incubation for 48 h at 37°C, the cells were fixed with 10% formalin for 1 h. Following formalin removal, the cells were stained with crystal violet and the plaques were visualized. The visible plaques were counted, and the concentrations of virus (in PFU/ml) were determined. The numbers and sizes of plaques were determined from at least three independent experiments.
Apoptosis assay with U937 cells.
U937 cells were infected with recombinant virus at a multiplicity of infection (MOI) of 2 at 37°C in 5% CO2. At 12 h postinfection, infected U937 cells were washed with phosphate-buffered saline on ice, and the apoptosis level was determined using a TACS annexin V-FITC kit (R&D Systems, Minneapolis, MN), according to the manufacturer's instructions.
Recombinant virus growth curve on U937 cells.
U937 cells were infected with recombinant virus at an MOI of 0.01 for 1 h at 37°C. Unadsorbed virus was removed by centrifuging and washing the cells with phosphate-buffered saline, and the U937 cells were added to fresh RPMI 1640 medium without fetal bovine serum. Supernatant was collected at different time points, and the virus titer was determined by plaque assay on MDCK cells.
CAT ELISA.
RNP activity was measured using a chloramphenicol acetyltransferase (CAT) enzyme-linked immunosorbent assay (ELISA; Roche, Penzberg, Germany). First, 293T cells were cotransfected with 1 μg Pol I-Po lII system expression plasmids and 1 μg pPol I-CAT-RT plasmid. At 48 h posttransfection, cell lysate was extracted using the lysis buffer that was provided with the CAT ELISA kit. After the protein had been quantified, each sample was diluted to 5 μg/μl and then serially diluted 500-fold using lysis buffer. The sample was then assayed to determine the CAT level, according to the manufacturer's instructions.
Infection of mice.
Recombinant viruses were serially diluted 10-fold in phosphate-buffered saline and intranasally inoculated into 6-week-old female BALB/c mice. Each group included six mice that were anesthetized by intraperitoneal injection of 0.12 ml of 0.3 mg tiletamine chlorhydrate and 0.3 mg zolazepam chlorhydrate (Zoletil; Virbac, Carros, France) and inoculated intranasally with 50 μl recombinant viruses. The infected mice were weighed and observed daily for illness or death. Mice that were found to be moribund were considered to have died on that day. The lethal doses for 50% of the mice (MLD50) were calculated by the method of Reed and Muench and are presented in Table 1.
TABLE 1.
MLD50 of recombinant viruses in BABL/c mice
| Virus straina | Cellular (%) localization of PB1-F2 | PB1-F2 subtype/length (aa) | MLD50 (PFU) |
|---|---|---|---|
| PR8 | Mitochondria | H1N1/87 | 125.6 |
| PH | Whole cell | H5N1/90 | 16 |
| PHdelF2b | 49 | ||
| PHQ69LH75Lc | Mitochondria (40.7) and whole cell (59.3) | H5N1/90 | 19 |
Recombinant viruses were generated by reverse genetics.
H5N1 PB1-F2 was deleted.
H5N1 PB1-F2 contains double Leu mutations at positions 69 and 75.
RESULTS
Sequence diversity of PB1-F2 in various influenza A virus subtypes.
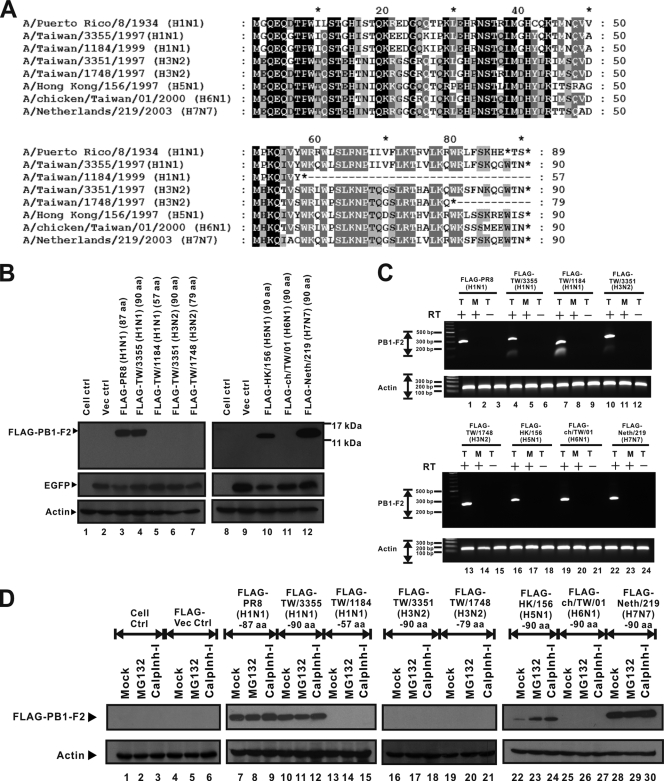
Eight PB1-F2 variants derived from A/Puerto Rico/8/1934 (PR8) (H1N1), A/Taiwan/3355/1997 (H1N1), A/Taiwan/1184/1999 (H1N1), A/Taiwan/3351/1997 (H3N2), A/Taiwan/1748/1997 (H3N2), A/Hong Kong/156/1997 (H5N1), A/chicken/Taiwan/01/2000 (H6N1), and A/Netherlands/219/2003 (H7N7) were selected for use in the present work. The Kozak PB1-F2-coding sequences (24) of the eight strains under investigation have the same nucleotide at the −3 and +4 positions, indicating that all of these PB1-F2 variants have the same translation potential as that of PR8 PB1-F2. Figure 1A presents the amino acid sequences of the translated PB1-F2 proteins, showing various product lengths, from 57 to 90 aa; the completely conserved residues are shown in black, and many of these are close to the N terminus. On the basis of the length of PR8 PB1-F2, the PB1-F2 proteins of five viruses, A/Taiwan/3355/1997 (H1N1), A/Taiwan/3351/1997 (H3N2), A/Hong Kong/156/1997 (H5N1), A/chicken/Taiwan/01/2000 (H6N1), and A/Netherlands/219/2003 (H7N7), were defined to be full length, while the proteins of A/Taiwan/1184/1999 (H1N1) and A/Taiwan/1748/1997 (H3N2) were defined to be truncated. While the PB1-F2 proteins of the three H1N1 strains share high levels of amino acid sequence similarity of over 80%, those of strain PR8 and two H3N2 strains had a lower level of similarity of 60%, as did the PR8 PB1-F2 protein with the other three PB1-F2 proteins from the avian strains of H5N1, H6N1, and H7N7. Most of the sequence variation was at the C terminus of PB1-F2, and the sequences had variable lengths. Other than the PB1-F2 protein of A/Taiwan/3351/1997 (H3N2), which contains only four hydrophobic amino acids, the remaining five PB1-F2 variants have at least five hydrophobic amino acids. In terms of the basic residue count, PR8 PB1-F2 contains the highest at five, while the other PB1-F2 variants contain four or fewer. The residue variability in the amphipathic helical structure of PB1-F2 suggests that distinct PB1-F2 variants may function differently in mitochondrial localization, as observed in PR8.
FIG. 1.
Sequence differentials and various expression levels of eight PB1-F2 variants. (A) Amino acid sequences of PB1-F2 variants that represent all influenza A virus subtypes available in this work. Sequence alignment and analysis were performed using BioEdit (version 7.0.5.2) and GeneDoc (version 2.7.000) software. Black shading, 100% identity; dark gray shading, 80% identity; pale gray shading, 60% identity. Sequence variations are caused by differences in protein lengths and amino acid sequences, especially in the C-terminal region. However, the amino acid sequence in the N-terminal region is more conserved than that in the C-terminal region. (B) FLAG-tagged PB1-F2 variant expression plasmids and the pEGFP-N2 vector were cotransfected into 293T cells. The EGFP expression level represents transfection efficiency, and the actin expression level represents the loading control level. Western blotting shows that expression of the FLAG-PR8 (H1N1) (87 aa) (lane 3), FLAG-TW/3355 (H1N1) (90 aa) (lane 4), FLAG-HK/156 (H5N1) (90 aa) (lane 10), and FLAG-Neth/219 (H7N7) (90 aa) (lane 12) PB1-F2 proteins was detectable. PB1-F2 protein expression by the other four PB1-F2 variants, from FLAG-TW/1184 (H1N1) (57 aa) (lane 5), FLAG-TW/3351 (H3N2) (90 aa) (lane 6), FLAG-TW/1748 (H3N2) (79 aa) (lane 7), and FLAG-ch/TW/01 (H6N1) (90 aa) (lane 11), was not detectable by Western blotting. ctrl, control. (C) FLAG-tagged PB1-F2 variant expression plasmids were transfected into 293T cells. At 24 h posttransfection, RNAs were extracted from transfected 293T cells. T, RNAs extracted from transfected cells; M, RNAs prepared from mock-transfected cells; +, RNAs that were transcribed by reverse transcriptase; −, RNAs that were not treated with reverse transcriptase before PCR. The level of expression of actin gene RNA is presented as the level for the loading control. Each transcript of the FLAG-tagged PB1-F2 expression plasmids was detectable by RT-PCR. (D) FLAG-tagged PB1-F2 expression plasmids were transfected into 293T cells. At 8 h posttransfection, MG132 and calpain inhibitor I (calplnh-I) were added to transfected 293T cells. At 16 h after treatment, total cell lysate was extracted and assayed by Western blotting. As shown in the Western blot, the proteins of FLAG-TW/1184 (H1N1) (57 aa) (lanes 13 to 15), FLAG-TW/3351 (H3N2) (90 aa) (lanes 16 to 18), FLAG-TW/1748 (H3N2) (79 aa) (lanes 19 to 21), and FLAG-ch/TW/01 (H6N1) (90 aa) (lanes 25 to 27) could not be rescued by proteasome inhibitors. However, the amount of PB1-F2 protein of FLAG-HK/156 (H5N1) (90 aa) (lanes 22 to 24) increased upon treatment with proteasome inhibitors.
Various levels of protein expression by PB1-F2 variants in 293T cells.
Following the observation of different compositions of hydrophobic and basic amino acids at the C termini of these PB1-F2 variants, we further examined how they may exhibit different cellular localizations. Since sequence diversity may result in different efficiencies of epitope recognition by anti-PB1-F2 antibody, FLAG-tagged PB1-F2 was generated and transfected into 293T cells. The detection of FLAG-tagged PB1-F2 by Western blotting indicated that the discrepancy in sequence diversity at the potential epitope recognition site had been eliminated. In addition, the enhanced GFP (EGFP) vector was cotransfected with FLAG-tagged PB1-F2 as a control to determine the transfection efficiency. The results demonstrated that the expression of FLAG-tagged PB1-F2 proteins derived from PR8 (H1N1) (Fig. 1B, lane 3, FLAG-PR8), A/Taiwan/3355/1997 (H1N1) (Fig. 1B, lane 4, FLAG-TW/3355), A/Hong Kong/156/1997 (H5N1) (Fig. 1B, lane 10, FLAG-HK/156), and A/Netherlands/219/2003 (H7N7) (Fig. 1B, lane 12, FLAG-Neth/219) was detected by Western blotting with anti-FLAG antibody. The PB1-F2 protein derived from A/Taiwan/1184/1999 (H1N1), which was 57 aa, was not detected (Fig. 1B, lane 5, FLAG-TW/1184). Three PB1-F2 proteins, those of A/Taiwan/3351/1997 (H3N2) (Fig. 1B, lane 6, FLAG-TW/3351), A/Taiwan/1748/1997 (H3N2) (Fig. 1B, lane 7, FLAG-TW/1748), and A/chicken/Taiwan/01/2000 (H6N1) (Fig. 1B, lane 11, FLAG-ch/TW/01), were also not detected. Understanding that any defects at the transcription level may have been responsible for the different PB1-F2 protein expression levels, shown in Fig. 1B, total cellular RNAs were also extracted from transfected or mock-transfected cell lysates and analyzed by RT-PCR. In addition to the four strains whose PB1-F2 proteins were already detected, as shown in Fig. 1B, the RNAs of FLAG-TW/1184 (H1N1), FLAG-TW/3351 (H3N2), FLAG-TW/1748 (H3N2), and FLAG-ch/TW/01 (H6N1) (whose protein levels were undetectable) were clearly observed (Fig. 1C, lanes 7, 10, 13, and 19). Therefore, not all FLAG-tagged PB1-F2 proteins derived from different strains could be detected, although all of their RNAs were transcribed in the transfected cells.
To investigate whether proteasome degradation may have caused the different levels of expression of FLAG-tagged PB1-F2 protein, MG132 or calpain inhibitor I was added to the transfected 293T cells at 8 h posttransfection. As shown in lanes 22 to 24 of Fig. 1D, in the presence of proteasome inhibitors, the expression of FLAG-tagged PB1-F2 protein derived from H5N1 was significantly rescued. The levels of expression of FLAG-tagged PB1-F2 derived from PR8 (Fig. 1D, lanes 7 to 9), TW/3355 (H1N1) (Fig. 1D, lanes 10 to 12), and Neth/219 (H7N7) (Fig. 1D, lanes 28 to 30) showed no clear increase after the treatment with these inhibitors. FLAG-TW/1184 (H1N1) (Fig. 1D, lanes 13 to 15), FLAG-TW/3351 (H3N2) (Fig. 1D, lanes 16 to 18), FLAG-TW/1748 (H3N2) (Fig. 1D, lanes 19 to 21), and FLAG-ch/TW/01 (H6N1) (Fig. 1D, lanes 25 to 27), on the other hand, were not rescued upon addition of proteasome inhibitors. The results in Fig. 1B to D suggest that not every FLAG-tagged PB1-F2 protein derived from the different strains can be expressed and that the proteins of H3N2 and H6N1 cannot be rescued by proteasome inhibitors.
Cellular localization of PB1-F2 variants in HeLa cells.
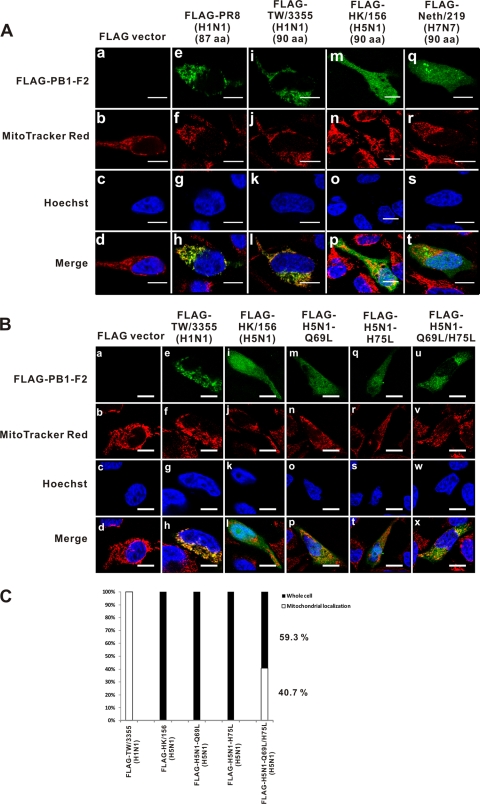
The cellular localization of four PB1-F2 variants whose expression was detected by Western blotting was further examined. FLAG-tagged PB1-F2 expression plasmids were transfected into HeLa cells, and at 24 h posttransfection, transfected HeLa cells were stained with MitoTracker Red and Hoechst 33258. After they were stained, HeLa cells were probed with anti-FLAG antibody and labeled with FITC-conjugated antimouse secondary antibody. The cellular localization of the PB1-F2 variants was determined by confocal microscopy. The FLAG-TW/3355 (H1N1) PB1-F2 has a cellular distribution pattern similar to that of the PR8 PB1-F2, which localized to the mitochondria (Fig. 2 A, panels i to l and panels e to h, respectively). Surprisingly, the FLAG-HK/156 (H5N1) (Fig. 2A, panels m to p) and FLAG-Neth/219 (H7N7) (Fig. 2A, panels q to t) proteins did not specifically localize to the mitochondria. Instead, they were distributed throughout the cell, including the cytoplasm and the nucleus. Additionally, the lack of protein expression by the PB1-F2 variants in transfected HeLa cells was confirmed by immunofluorescence using confocal microscopy.
FIG. 2.
FLAG-tagged PB1-F2 proteins derived from H5N1 and H7N7 viruses exhibit different cellular localization than the protein from strain PR8. (A) FLAG-tagged expression plasmids were transfected into HeLa cells. At 24 h posttransfection, transfected HeLa cells were analyzed by immunofluorescence assay and stained with MitoTracker Red (100 nM) and Hoechst 33258 (100 nM). Bars, 10 μm. The FLAG-TW/3355 (H1N1) (90 aa) (i to l) protein has a cellular localization similar to that of the protein from strain PR8 (e to h). The PB1-F2 proteins derived from FLAG-HK/156 (H5N1) (90 aa) (m to p) and FLAG-Neth/219 (H7N7) (90 aa) (q to t) were expressed in the whole cell, including the cytoplasm and nucleus. (B) At 24 h posttransfection, HeLa cells were analyzed by immunofluorescence assay and stained with MitoTracker Red (100 nM) and Hoechst 33258 (100 nM). Bars, 10 μm. A single Leu mutation at either position 69 (m to p) or position 75 (q to t) results in the same cellular localization of PB1-F2 as that of wild-type H5N1 PB1-F2 (i to l); however, double Leu mutations (u to x) altered cellular localization from the whole cell to the mitochondria. (C) FLAG-tagged wild-type or H5N1 PB1-F2 expression plasmids with Leu mutations were transfected into HeLa cells. The percent cellular localization was determined by randomly selecting 10 files (magnification, ×630) and calculating the results for 100 transfected positive cells. Double Leu mutations drive 40.7% of the mutant H5N1 PB1-F2 proteins to localize to the mitochondria.
Leu is an important residue at amino acids 69 to 87 in PR8 PB1-F2, and this region is responsible for mitochondrial localization (15). There are four or five Leu residues in the C terminus of the PB1-F2 protein derived from PR8 or TW/3355 (H1N1). The Leu-rich domain was considered to be responsible for mitochondrial targeting by interacting with the hydrophobic binding groove of the TOM 20 import receptor on the outer membrane of mitochondria (31). While only three Leu residues occur between positions 65 and 90, at positions 72, 77, and 82 of the H5N1 PB1-F2 and positions 72, 75, and 77 of the H7N7 PB1-F2 (Fig. 1A), whether reducing the number of Leu residues would have changed the localization pattern is of interest. Therefore, we generated mutant H5N1 PB1-F2 with a Q69L, H75L, or Q69L/H75L mutation and noticed that only mutant H5N1 PB1-F2 with double Leu mutations at both position 69 and position 75 could change the cellular localization (Fig. 2B, panels u to x). Figure 2C shows 40.7% mitochondrial localization, determined from calculations for 100 randomly chosen transfected positive cells, indicating that the Leu residues at positions 69 and 75 are important to drive the localization of H5N1 PB1-F2 to mitochondria.
Alteration of cellular localization of H5N1 PB1-F2 does not visibly affect virulence in mouse model.
The present study also attempts to determine whether the cellular localization of H5N1 PB1-F2 is related to its higher level of pathogenicity by generating and inoculating the recombinant viruses with or without H5N1 PB1-F2 into BALB/c mice; the MLD50 values were determined as well (Table 1). Virus strain PR8 was used as the experimental control, and the MLD50 value of PR8 exceeds that of PH virus (PR8 with its PB1 replaced by the H5N1 PB1 gene). The MLD50 of PH virus was 16 PFU, while that of PHdelF2 virus (PH with the H5N1 PB1-F2 gene deletion) was 49 PFU. However, the PH virus and the PHQ69H75L virus (PH virus with double Leu mutations in the H5N1 PB1-F2 protein) exhibited similar levels of virulence in mice (MLD50 values, 16 PFU and 19 PFU, respectively). The experimental results suggest that changing the cellular localization by the incorporation of double Leu mutations did not significantly affect viral virulence in mice.
Induction of apoptosis by H5N1 PB1-F2 in U937 cells.
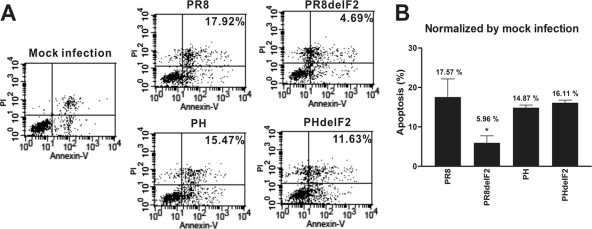
Apoptosis enhancement is one of the documented cellular functions of PR8 PB1-F2, and this phenomenon is cell type specific (8, 39). To evaluate the apoptosis-enhancing activity of H5N1 PB1-F2, each of the recombinant viruses was generated with or without the H5N1 PB1-F2. Human monocytic U937 cells were challenged by each of the wild-type PR8 recombinant virus, PR8delF2 virus (PR8 with the PR8 PB1-F2 gene deletion), PH virus (PR8 with its PB1 replaced by the H5N1 PB1 gene), or PHdelF2 virus (PH with the H5N1 PB1-F2 gene deletion). Flow cytometry was applied for the apoptosis analysis, and the level of apoptosis induction of PR8delF2 virus was significantly (3-fold) lower than that of wild-type PR8 virus (Fig. 3A and B). However, the levels of apoptosis induction were found to be comparable when U937 cells were challenged by recombinant viruses PH and PHdelF2 (14.87% and 16.11%, respectively; Fig. 3B) after three independent experiments, suggesting that H5N1 PB1-F2 does not contribute to the apoptosis enhancement.
FIG. 3.
H5N1 PB1-F2 has no effect on apoptosis enhancement. (A) U937 was challenged with strains PR8, PR8delF2, PH, and PHdelF2 at an MOI of 2. At 12 h postinfection, infected U937 cells were assayed by determining the amount of annexin V-FITC. Infected U937 cells undergo apoptosis, as determined by comparison of infected cells with mock-infected cells. However, the apoptosis level for PH does not differ significantly from that for PHdelF2. (B) The percentages were normalized according to the results for mock infection. Mean values and standard errors from three independent experiments are shown. *, P < 0.05, Student's two-tailed unpaired t test.
Growth curve of H5N1 PB1-F2-deficient virus in U937 cells.
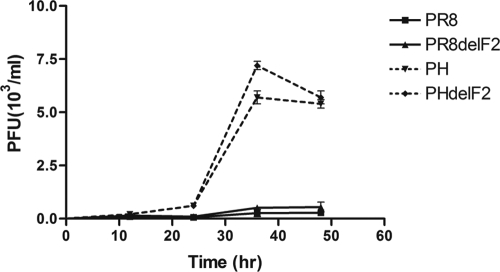
Following the observation that the recombinant virus with PR8 PB1-F2 induced a significantly higher apoptosis level than that induced by recombinant virus without PR8 PB1-F2 and that this effect was immune cell specific, we evaluated virus growth. To determine whether the curves for wild-type and PB1-F2-knockout recombinant virus growth in immune cells would differ, the curves for PR8, PR8delF2, PH, and PHdelF2 virus growth in U937 cells were further examined. Whereas no difference between the growth curves of the recombinant viruses with or without PB1-F2 existed at early and later time points postinfection (Fig. 4), a significant increase in virus titer was observed at a late stage postinfection, and the titers of both the PH and PHdelF2 viruses peaked at 36 h. The results in Fig. 4 suggested that the PB1 gene segment derived from H5N1 enhanced the growth rate of PR8 virus. The existence of PB1-F2, however, did not seem to alter viral growth in U937 cells.
FIG. 4.
Neither PR8 PB1-F2 nor H5N1 PB1-F2 can alter virus growth in U937 cells. U937 cells were infected with PR8, PR8delF2, PH, or PHdelF2 virus at an MOI of 0.01. At 12, 24, 36, and 48 h postinfection, the supernatants were harvested and the virus titers were determined by plaque assay on MDCK cells. Each point represents the mean value and standard error from two independent experiments.
H5N1 PB1-F2 enhanced viral polymerase activity.
The RNP complex of influenza A virus is known to be responsible for viral genome replication in the nucleus of the host cell. PR8 PB1-F2 has recently been reported to promote RNP activity (27). To determine whether the H5N1 PB1-F2 protein, which has been shown to be expressed in the cytoplasm and nucleus, can increase the opportunity to promote virus RNP activity, plasmids expressing the Pol I-Pol II system were cotransfected with PR8 PB2, PR8 PA, PR8 NP, and H5N1 PB1, with or without H5N1 PB1-F2, as well as wild-type PR8, with or without its PB1-F2, together with a plasmid that enabled the expression of a pseudoviral reporter RNA (plasmid pPolI-CAT-RT) into 293T cells. Mutant PR8 PB1 (without PB1-F2) had weaker RNP activity than wild-type PR8 PB1 (Fig. 5A), as expected, and the knockout of H5N1 PB1-F2 reduced the mutant H5N1 RNP activity by nearly 35%, as shown in Fig. 5B, suggesting that the H5N1 PB1-F2 protein can also enhance viral RNP activity, as observed in PR8.
FIG. 5.
Lack of H5N1 PB1-F2 reduces polymerase activity in transfected cells. 293T cells were cotransfected with Pol I-Pol II system expression plasmids with PR8 PB2, PR8 PA, PR8 NP, and PR8 PB1 with or without PR8 PB1-F2 (A) or H5N1 PB1 with or without H5N1 PB1-F2 (B), together with a plasmid that enabled the expression of a pseudoviral reporter RNA (plasmid pPolI-CAT-RT). Vec ctrl, vector control, in which 293T cells were cotransfected with PR8 PB2, PR8 PA, PR8 NP, and pPolI-CAT-RT plasmids without PR8 PB1. At 48 h posttransfection, total cell lysate was extracted with lysis buffer and was assayed using a CAT ELISA. The optical density at 405 nm for PR8 PB1delF2 (A, top panel) or H5N1 PB1delF2 (B, top panel) was normalized according to that for PR8 PB1 (A, top panel) or H5N1 PB1 (B, top panel), which was considered 100%. Mean values and standard errors from three independent experiments are shown. **, P < 0.001, Student's two-tailed unpaired t test. (A, lower panel) Expression of PR8 PB1 and PR8 NP in 293T transfected cells, monitored in Western blots with polyclonal anti-PB1 and monoclonal anti-NP antibody; (B, lower panel) expression of H5N1 PB1 and PR8 NP in 293T transfected cells, monitored in Western blots with polyclonal anti-PB1 and monoclonal anti-NP antibodies.
Cellular function of putative PB1-F2 derived from the 2009 S-OIV.
Given the observation that the H5N1 PB1-F2 protein exhibits functions different from those of the PR8 PB1-F2 protein, it became of interest to investigate the possible role of S-OIV PB1-F2. Although the 2009 S-OIV (H1N1) contains only a truncated form of PB1-F2 (12 aa), potential mutation may in the future enable it to contain a full-length and functional product. In order to generate an S-OIV PB1 gene capable of coding for a putative full-length PB1-F2, two single mutations were introduced into the S-OIV PB1 gene at nucleotide positions A129C and A267G to bypass two stop codons. Sequencing of the full length of the S-OIV PB1 gene was conducted, and analysis of the two point mutations at positions 129 (A to C) and 267 (A to G) was performed, indicating that the coding sequence of the PB1 protein does not change. Table 2 shows the sequence identity between the resulting S-OIV PB1-F2 and the six full-length PB1-F2 proteins that were studied earlier. It has been reported that the PB1 gene of the 2009 S-OIV was derived from human H3N2 influenza A virus (23), which is reflected by detection of the highest level of sequence identity (78%) between this putative S-OIV PB1-F2 and the PB1-F2 protein derived from A/Taiwan/3351/1997 (H3N2). On the other hand, this putative S-OIV PB1-F2 has the lowest level of sequence identity with PB1-F2 derived from A/Taiwan/3355/1997 (H1N1) (54%), even though they are of the same H1N1 subtype.
TABLE 2.
Sequence identity between putative S-OIV PB1-F2 and six other full-length PB1-F2 variants
| Virus strain | Subtype | PB1-F2 length (aa) | % sequence identitya |
|---|---|---|---|
| A/Puerto Rico/8/33 | H1N1 | 87 | 56 |
| A/Taiwan/3355/1997 | H1N1 | 90 | 54 |
| A/Taiwan/3351/1997 | H3N2 | 90 | 78 |
| A/Hong Kong/156/1997 | H5N1 | 90 | 64 |
| A/chicken/Taiwan/01/2000 | H6N1 | 90 | 74 |
| A/Netherlands/219/2003 | H7N7 | 90 | 65 |
Sequence identity was determined by comparison of the sequence with the sequences of six full-length PB1-F2 proteins, which were investigated earlier in the present study, and the S-OIV putative PB1-F2 (87 aa). The value of the sequence identity was calculated by BioEdit (version 7.0.5.2) software.
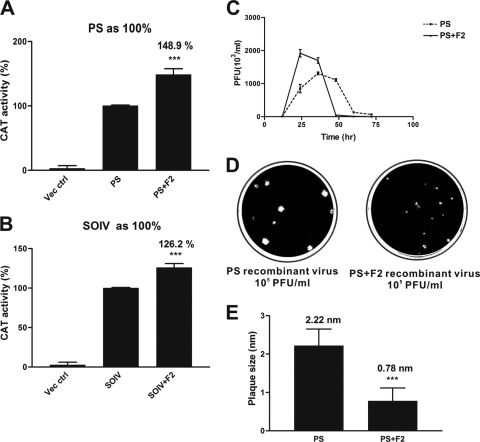
Virus growth and RNP activity were investigated as described for the earlier experiments that yielded Fig. 4 and 5. Figure 6 A (obtained using PR8 PB2, PA, and NP) and Fig. 6B (obtained using S-OIV PB2, PA, and NP) show that the S-OIV PB1 gene with the putative PB1-F2 protein increased virus RNP activity by up to 48.9% (Fig. 6A) and 26.2% (Fig. 6B), respectively, compared with that of the wild-type S-OIV PB1 gene without PB1-F2. U937 cells were challenged with PS (PR8 virus with its PB1 gene replaced by S-OIV PB1) and PS-F2 (PS virus that contains the putative full-length S-OIV PB1-F2) recombinant viruses. The growth curve for the PS-F2 virus reached a maximum at 24 h postinfection, followed by an abrupt drop in virus titer (Fig. 6C). On the other hand, the growth curve for the PS virus peaked at a lower titer and at a later time of 36 h postinfection, after which the titer gradually declined. Interestingly, the plaque size of the PS virus differs significantly from that of the PS-F2 virus (P < 0.005), with the plaque of the former being more than twice as large as that of the latter (Fig. 6D and E).
FIG. 6.
S-OIV full-length PB1-F2 affects virus growth and virus RNP activity and reduces the sizes of the plaques. 293T cells were cotransfected with Pol I-Pol II system expression plasmids with PR8 PB2, PR8 PA, or PR8 NP (A) or S-OIV PB2, S-OIV PA, or S-OIV NP (B) and S-OIV PB1 with or without S-OIV full-length PB1-F2, together with plasmid pPolI-CAT-RT. Vec ctrl, vector control, in which 293T cells were cotransfected with PB2, PA, NP, and pPolI-CAT-RT plasmids without S-OIV PB1. The optical densities at 405 nm for the PS-F2 (A) and S-IOV-F2 (B) proteins were normalized to those for the PS (A) and S-OIV (B) proteins, respectively. The optical density values of the PS and S-OIV proteins were set equal to 100%. (A and B) ***, P < 0.001, Student's two-tailed unpaired t test. (C) U937 cells were infected with PS or PS-F2 virus at an MOI of 0.01. At 12, 24, 36, 48, 60, and 72 h postinfection, the supernatants were harvested and the virus titers were determined by plaque assay on MDCK cells. Each point represents the mean and standard error from two independent experiments. (D) MDCK cells were infected with dilutions of recombinant influenza viruses PS (left panel) and PS-F2 (right panel). At 1 h postinfection, the cells were overlaid with 3% agarose and incubated at 37°C in 5% CO2. At 48 h postinfection, the cells were fixed with 10% formalin for 1 h. Following formalin removal, the cells were stained with 0.5% crystal violet and the plaques were visualized. (E) MDCK cells were grown on a six-well plate and infected with serial dilutions of recombinant influenza viruses PS (left bar) and PS-F2 (right bar). At 1 h postinfection, the cells were overlaid with 3% agarose and incubated on 37°C in 5% CO2. At 48 h postinfection, the cells were fixed with 10% formalin for 1 h and stained with 0.5% crystal violet; the sizes of the plaques were calculated. The mean plaque diameter was calculated using 100 plaques. Mean values and standard errors from two independent experiments are shown. ***, P < 0.001, Student's two-tailed unpaired t test.
DISCUSSION
Whereas the early human influenza A viruses, such as the 1918 Spanish flu and WSN strains, contained an intact PB1-F2 with a length of at least 87 aa, the truncated form of PB1-F2 emerged after 1956, when one mutation caused the early termination of PB1-F2 at 57 residues (7). The early termination of PB1-F2 was found in the recent 2009 S-OIV, which has been reported to have a 12-aa-long PB1-F2. The PB1-F2 protein uses the same RNA segment as the PB1 protein and overlaps with the PB1 protein in a different reading frame. Although the regularly translated PB1 genes are highly similar among all subtypes, the amino acid sequence of PB1-F2 in various subtypes is more diverse than that of PB1. Most avian influenza A virus PB1 proteins encode intact PB1-F2 proteins, with the exceptions being the H5N2, H6N6, H9N2, and H13N2 subtypes, which contain truncated PB1-F2 proteins (42). The truncated PB1-F2 is considered to reduce the ability for mitochondrial localization and lack the function for apoptotic induction because it lacks the putative mitochondrial targeting signal (MTS) at the C terminus of PB1-F2 (15, 32). In order to know whether these PB1-F2 variants derived from different subtypes have distinct cellular functions, eight influenza A virus strains of five subtypes were used for a series of comprehensive investigations. These eight strains contained diverse PB1-F2 proteins in terms of their lengths as well as their amino acid compositions (Fig. 1A). All of these PB1-F2 variants contain a perfect Kozak sequence for efficient translation into a PB1-F2 product and similar amphipathic alpha-helical wheel structures for the formation of a putative mitochondrial localization signal, but only the TW/3355 (H1N1) PB1-F2 and the PR8 PB1-F2 displayed mitochondrial localization. Two PB1-F2 variants, the H5N1 and H7N7 PB1-F2 variants, localized in whole cells. The different cellular localizations of the PB1-F2 proteins exhibited by both H5N1 and H7N7 viruses may reveal that these PB1-F2 proteins have cellular functions different from those of PR8 PB1-F2. In fact, unlike PR8 PB1-F2, H5N1 PB1-F2 has no apoptosis-enhancing capacity, perhaps because of its cellular localization. It has been reported that phosphorylation of PR8 PB1-F2 is critical to the promotion of apoptosis in monocytes; Thr-27 and Ser-35 are the phosphorylation sites of PR8 PB1-F2 (30). Although the H5N1 PB1-F2 has the same residues as the PR8 PB1-F2 at positions 27 and 35, H5N1 PB1-F2 cannot promote apoptosis, according to the results of the present work. The H5N1 PB1-F2 does not contribute to apoptosis, possibly because of the absence of a specific MTS. The Leu residues of the mitochondrial targeting signal are thought to be a key factor for the mitochondrial localization of PR8 PB1-F2 (15); however, H5N1 PB1-F2 contains fewer Leu residues than PR8 PB1-F2 and TW/3355 (H1N1) PB1-F2 in the corresponding region, which may explain the lack of a specific mitochondrial localization. To determine whether the numbers and positions where Leu resides affect the cellular localization of H5N1 PB1-F2, single and double Leu mutations at positions 69 and 75 in H5N1 PB1-F2 were generated. Only mutant H5N1 PB1-F2 that contains double Leu mutations changes its cellular localization. A single Leu amino acid mutation probably does not affect the folding and structure of PB1-F2, but double Leu mutations do. Moreover, other residues close to positions 69 and 75 may also contribute to cellular localization. When proteins, except for those with hydrophobic amino acids (such as Leu), are imported to the mitochondria, basic amino acids also play an important role in the interaction with the TOM complex on the lipid membrane of the mitochondria (3, 4, 31).
In this study, different PB1-F2 variants had different expression levels. Codon usage related to the gene expression level, GC content, and amino acid conservation (6, 12, 22) may be one of the factors affecting the expression of PB1-F2. An analysis of the relative adaptiveness of codon usage (33) revealed one unpreferred synonymous codon, CTA, at position 37 in H5N1 PB1-F2. However, no unpreferred synonymous codon was found in the PB1-F2 derived from the PR8, A/Taiwan/3355/1997 (H1N1), and A/Netherlands/219/2003 (H7N7) strains. The proteasome inhibitors used in this study did not affect the rescue of the expression ability of FLAG-tagged H3N2 PB1-F2 and FLAG-tagged H6N1 PB1-F2 (Fig. 1D), suggesting that the lack of PB1-F2 expression by these two subtypes may be not due to protein degradation by the proteasome. According to Fig. 1B, Western blot analysis cannot detect several PB1-F2 variants, despite the fact that the amount of loaded protein reached 100 μg per sample. We believe that these PB1-F2 proteins are insoluble. Therefore, 8 M urea was added to CA630 lysis buffer (150 mM NaCl, 1% CA630, 50 mM Tris base [pH 8.0]), radioimmunoprecipitation assay lysis buffer (150 mM NaCl, 1% CA630, 50 mM Tris base [pH 8.0], 0.5% sodium deoxycholate, 0.1% SDS), and sample loading buffer (4× Tris-HCl-SDS [pH 6.8], 30% glycerol, 10% SDS, 0.6 M dithiothreitol, 0.012% bromphenol blue, and double-distilled H2O to make the total volume 10 ml). However, only FLAG-PR8 is detected after addition of the 8 M urea, while the other four PB1-F2 variants, i.e., those of FLAG-TW/1184 (H1N1), FLAG-TW/3351 (H3N2), FLAG-TW/1748 (H3N2), and FLAG-ch/TW/01 (H6N1), still cannot be detected (data not shown). In addition, an attempt was made to increase the rate of detection of these PB1-F2 variants and determine whether the rapid degradation of the proteins contributes to the inability to detect the PB1-F2 proteins of several strains through radiolabeling of the transfected cells by 35S for 24 h at 37°C, following immunoprecipitation to concentrate the radiolabeled cell lysate using a FLAG-tagged protein immunoprecipitation kit (Sigma). Radioactivity was detected by film exposure for 48 h, and the strength of the 35S signal represented the level of synthesis of PB1-F2 variants. While the 35S signal was detected in the FLAG-PR8 and FLAG-TW/1184 transfected cell lysates, this strategy did not detect the FLAG-TW/3351 (H3N2), FLAG-TW/1748 (H3N2), and FLAG-ch/TW/01 (H6N1) proteins (data not shown). Besides using immunoprecipitation and radiolabeling, a linker sequence that includes three continuous Ala-Pro pairs was added between the FLAG epitope and the PB1-F2 sequence (data not shown). This strategy was used to increase the flexibility of the FLAG-tagged proteins. Interestingly, FLAG-tagged TW/1184 and FLAG-tagged H3N2 PB1-F2 could be detected after a linker sequence was added; however, the H6N1 PB1-F2 remained undetected.
An outbreak caused by an H1N1 influenza A virus occurred in early 2009 in North America. It became a human pandemic in the following months (23). This new H1N1 virus has been found to originate from recent triple-reassortant swine viruses. Unlike seasonal influenza A viruses, which contain a PB1-F2 with a length of either 57 aa or 90 aa (7, 41, 42), this new H1N1 virus has the potential to code for only a 12-aa peptide (13, 35, 36). Considering the possibility that it may mutate into full-length PB1-F2 in the future, point mutations were artificially introduced into the two stop codons of the S-OIV PB1-F2 to make it capable of translating into an 87-aa product. This human-made PB1-F2 was capable of enhancing viral RNP activity, consistent with our observation that a recombinant virus that contains a full-length PB1-F2 has a higher replication rate than wild-type H1N1 virus with only a 12-aa-long PB1-F2. However, the yield of the recombinant virus with a full-length PB1-F2 reached a higher peak value within only 24 h, even though the time usually observed to be required to reach a peak is later (36 h) in PR8-infected U937 cells (Fig. 6C). Following this quick peak, the titer of recombinant virus (with a full-length S-OIV PB1-F2) in U937 cells dropped dramatically at 48 h postinfection, possibly due to the production of too many viruses, causing rapid cell death. This phenomenon may explain why PB1-F2 was absent from this new H1N1 virus: since PB1-F2 may enhance viral replication through promoting viral RNP activity, the unusually high speed of viral replication causes the host cells to die too fast to enable the infected cells to continue harboring daughter viruses. This speculation was drawn from a recent study (17), in which PB1-F2 derived from S-OIV only slightly affected virus pathogenicity in two mouse models.
It has been reported that the 2009 pandemic H1N1 virus acquired its PB1 gene from human H3N2 virus in about 1998 (14, 34). However, only 78% amino acid identity was found between the PB1-F2 protein of the artificially architected pandemic H1N1 strains and the PB1-F2 proteins of the currently circulating seasonal human H3N2 viruses. This significant genetic disparity (22%) found in the 2009 H1N1 PB1-F2 may have been caused by adaptive mutations that accumulated during the evolutionary course of the PB1 gene of the ancestral H1N1 swine viruses prior to the emergence of the new H1N1 human virus. These adaptive mutations are used to maintain the functional compatibility of the S-OIV PB1 gene with other RNP gene segments (PB2, PA, and NP) of various genetic origins, where PB2 and PA of the new H1N1 virus are known to be derived from North American avian H1N1 virus and NP is known to be derived from the classical swine H1N1 virus (14).
Different PB1-F2 variants from various virus strains have been hypothesized to exhibit variable cellular functions. The present investigation analyzed eight PB1-F2 proteins that were derived from different hosts (human, avian) and different virus subtypes (H1N1, H3N2, H5N1, H6N1, and H7N7). Not only sequence diversity but also cellular localization and functions vary among the PB1-F2 variants. Although H5N1 PB1-F2 is considered a regulator of influenza A virus virulence, H5N1 PB1-F2 was expressed in whole transfected cells. It did not significantly influence the promotion of apoptosis and did not affect the growth of the virus in human monocytes. A mutant H5N1 PB1-F2 which contains double Leu mutations at positions 69 and 75 exhibited a cellular localization different from that of wild-type H5N1 PB1-F2 but was not associated with a different pathogenicity in mice. However, the H5N1 PB1-F2 contributed to increased viral polymerase activity in 293T cells.
Rescue of putative full-length S-OIV PB1-F2 has a significant influence on promotion of virus RNP activity, which was also found in PR8 PB1-F2 and H5N1 PB1-F2. However, full-length S-OIV PB1-F2 enables virus to reach the peak titer in human monocytes at an early time and produces a smaller plaque than the wild-type recombinant virus. In conclusion, PB1-F2 proteins have various lengths, amino acid sequences, cellular localizations, and functions and display strain-specific pathogenicity. Such genetic and functional diversity makes it flexible and adaptable in maintaining the optimal replication efficiency and virulence for various strains of influenza A virus.
Acknowledgments
We thank the National Science Council of Taiwan, Republic of China (grant NSC-98-2321-B-182-005), and Chang Gung Memorial Hospital (grant CMRPD160283) for financially supporting this research.
Ron A. M. Fouchier, National Influenza Center and Department of Virology, Erasmus Medical Center, kindly provided the Pol I-Pol II system expression plasmids with A/Hong Kong/156/1997 (H5N1) PB1 and A/Netherlands/219/2003 (H7N7) PB1. Ted Knoy is appreciated for his editorial assistance.
Footnotes
Published ahead of print on 21 July 2010.
REFERENCES
- 1.Basler, C. F., and P. V. Aguilar. 2008. Progress in identifying virulence determinants of the 1918 H1N1 and the Southeast Asian H5N1 influenza A viruses. Antiviral Res. 79:166-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas, S. K., and D. P. Nayak. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohnert, M., N. Pfanner, and M. van der Laan. 2007. A dynamic machinery for import of mitochondrial precursor proteins. FEBS Lett. 581:2802-2810. [DOI] [PubMed] [Google Scholar]
- 4.Bolender, N., A. Sickmann, R. Wagner, C. Meisinger, and N. Pfanner. 2008. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braam, J., I. Ulmanen, and R. M. Krug. 1983. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell 34:609-618. [DOI] [PubMed] [Google Scholar]
- 6.Carlini, D. B., and W. Stephan. 2003. In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics 163:239-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, G. W., C. C. Yang, K. C. Tsao, C. G. Huang, L. A. Lee, W. Z. Yang, Y. L. Huang, T. Y. Lin, and S. R. Shih. 2004. Influenza A virus PB1-F2 gene in recent Taiwanese isolates. Emerg. Infect. Dis. 10:630-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 9.Cianci, C., L. Tiley, and M. Krystal. 1995. Differential activation of the influenza virus polymerase via template RNA binding. J. Virol. 69:3995-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, J. R. 2007. The PB1-F2 protein of influenza A virus: increasing pathogenicity by disrupting alveolar macrophages. Virol. J. 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conenello, G. M., and P. Palese. 2007. Influenza A virus PB1-F2: a small protein with a big punch. Cell Host Microbe 2:207-209. [DOI] [PubMed] [Google Scholar]
- 12.Ermolaeva, M. D. 2001. Synonymous codon usage in bacteria. Curr. Issues Mol. Biol. 3:91-97. [PubMed] [Google Scholar]
- 13.Fraser, C., C. A. Donnelly, S. Cauchemez, W. P. Hanage, M. D. Van Kerkhove, T. D. Hollingsworth, J. Griffin, R. F. Baggaley, H. E. Jenkins, E. J. Lyons, T. Jombart, W. R. Hinsley, N. C. Grassly, F. Balloux, A. C. Ghani, N. M. Ferguson, A. Rambaut, O. G. Pybus, H. Lopez-Gatell, C. M. Alpuche-Aranda, I. B. Chapela, E. P. Zavala, D. M. Guevara, F. Checchi, E. Garcia, S. Hugonnet, and C. Roth. 2009. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324:1557-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St. George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs, J. S., D. Malide, F. Hornung, J. R. Bennink, and J. W. Yewdell. 2003. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J. Virol. 77:7214-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagen, M., T. D. Chung, J. A. Butcher, and M. Krystal. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol. 68:1509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hai, R., M. Schmolke, Z. T. Varga, B. Manicassamy, T. T. Wang, J. A. Belser, M. B. Pearce, A. Garcia-Sastre, T. M. Tumpey, and P. Palese. 2010. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J. Virol. 84:4442-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann, E., and R. G. Webster. 2000. Unidirectional RNA polymerase I-polymerase II transcription system for the generation of influenza A virus from eight plasmids. J. Gen. Virol. 81:2843-2847. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, E. C., D. J. Lipman, D. Zamarin, and J. W. Yewdell. 2006. Comment on “Large-scale sequence analysis of avian influenza isolates.” Science 313:1573. [DOI] [PubMed] [Google Scholar]
- 21.Hsu, C. N., and C. H. Wang. 2006. Sequence comparison between two quasi strains of H6N1 with different pathogenicity from a single parental isolate. J. Microbiol. Immunol. Infect. 39:292-296. [PubMed] [Google Scholar]
- 22.Ikemura, T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151:389-409. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, Y., K. Shinya, M. Kiso, T. Watanabe, Y. Sakoda, M. Hatta, Y. Muramoto, D. Tamura, Y. Sakai-Tagawa, T. Noda, S. Sakabe, M. Imai, Y. Hatta, S. Watanabe, C. Li, S. Yamada, K. Fujii, S. Murakami, H. Imai, S. Kakugawa, M. Ito, R. Takano, K. Iwatsuki-Horimoto, M. Shimojima, T. Horimoto, H. Goto, K. Takahashi, A. Makino, H. Ishigaki, M. Nakayama, M. Okamatsu, K. Takahashi, D. Warshauer, P. A. Shult, R. Saito, H. Suzuki, Y. Furuta, M. Yamashita, K. Mitamura, K. Nakano, M. Nakamura, R. Brockman-Schneider, H. Mitamura, M. Yamazaki, N. Sugaya, M. Suresh, M. Ozawa, G. Neumann, J. Gern, H. Kida, K. Ogasawara, and Y. Kawaoka. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867-19870. [PubMed] [Google Scholar]
- 25.Lamb, R. A., and M. Takeda. 2001. Death by influenza virus protein. Nat. Med. 7:1286-1288. [DOI] [PubMed] [Google Scholar]
- 26.Li, M. L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazur, I., D. Anhlan, D. Mitzner, L. Wixler, U. Schubert, and S. Ludwig. 2008. The proapoptotic influenza A virus protein PB1-F2 regulates viral polymerase activity by interaction with the PB1 protein. Cell. Microbiol. 10:1140-1152. [DOI] [PubMed] [Google Scholar]
- 28.McAuley, J. L., F. Hornung, K. L. Boyd, A. M. Smith, R. McKeon, J. Bennink, J. W. Yewdell, and J. A. McCullers. 2007. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAuley, J. L., K. Zhang, and J. A. McCullers. 2010. The effects of influenza A virus PB1-F2 protein on polymerase activity are strain specific and do not impact pathogenesis. J. Virol. 84:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitzner, D., S. E. Dudek, N. Studtrucker, D. Anhlan, I. Mazur, J. Wissing, L. Jansch, L. Wixler, K. Bruns, A. Sharma, V. Wray, P. Henklein, S. Ludwig, and U. Schubert. 2009. Phosphorylation of the influenza A virus protein PB1-F2 by PKC is crucial for apoptosis promoting functions in monocytes. Cell. Microbiol. 11:1502-1516. [DOI] [PubMed] [Google Scholar]
- 31.Pfanner, N. 2000. Protein sorting: recognizing mitochondrial presequences. Curr. Biol. 10:R412-R415. [DOI] [PubMed] [Google Scholar]
- 32.Ramakrishnan, M. A., M. R. Gramer, S. M. Goyal, and S. Sreevatsan. 2009. A Serine12Stop mutation in PB1-F2 of the 2009 pandemic (H1N1) influenza A: a possible reason for its enhanced transmission and pathogenicity to humans. J. Vet. Sci. 10:349-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp, P. M., and W. H. Li. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15:1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinde, V., C. B. Bridges, T. M. Uyeki, B. Shu, A. Balish, X. Xu, S. Lindstrom, L. V. Gubareva, V. Deyde, R. J. Garten, M. Harris, S. Gerber, S. Vagasky, F. Smith, N. Pascoe, K. Martin, D. Dufficy, K. Ritger, C. Conover, P. Quinlisk, A. Klimov, J. S. Bresee, and L. Finelli. 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N. Engl. J. Med. 360:2616-2625. [DOI] [PubMed] [Google Scholar]
- 35.Smith, G. J., J. Bahl, D. Vijaykrishna, J. Zhang, L. L. Poon, H. Chen, R. G. Webster, J. S. Peiris, and Y. Guan. 2009. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 106:11709-11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, G. J., D. Vijaykrishna, J. Bahl, S. J. Lycett, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, S. Bhatt, J. S. Peiris, Y. Guan, and A. Rambaut. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122-1125. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki, Y. 2006. Natural selection on the influenza virus genome. Mol. Biol. Evol. 23:1902-1911. [DOI] [PubMed] [Google Scholar]
- 38.Yamada, H., R. Chounan, Y. Higashi, N. Kurihara, and H. Kido. 2004. Mitochondrial targeting sequence of the influenza A virus PB1-F2 protein and its function in mitochondria. FEBS Lett. 578:331-336. [DOI] [PubMed] [Google Scholar]
- 39.Zamarin, D., A. Garcia-Sastre, X. Xiao, R. Wang, and P. Palese. 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 1:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zamarin, D., M. B. Ortigoza, and P. Palese. 2006. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J. Virol. 80:7976-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zell, R., A. Krumbholz, A. Eitner, R. Krieg, K. J. Halbhuber, and P. Wutzler. 2007. Prevalence of PB1-F2 of influenza A viruses. J. Gen. Virol. 88:536-546. [DOI] [PubMed] [Google Scholar]
- 42.Zell, R., A. Krumbholz, and P. Wutzler. 2006. Influenza A virus PB1-F2 gene. Emerg. Infect. Dis. 12:1607-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]