Abstract
RNA silencing suppressors (RSSs) are well studied for plant viruses but are not well defined to date for animal viruses. Here, we have identified an RSS from a medically important positive-sense mammalian virus, Severe acute respiratory syndrome coronavirus. The viral 7a accessory protein suppressed both transgene and virus-induced gene silencing by reducing the levels of small interfering RNA (siRNA). The suppression of silencing was analyzed by two independent assays, and the middle region (amino acids [aa] 32 to 89) of 7a was responsible for suppression. Finally, the RNA suppression property and the enhancement of heterologous replicon activity by the 7a protein were confirmed for animal cell lines.
RNA silencing processes restrict viral proliferation and disease manifestation (4, 8, 15). To overcome this antiviral response, viruses have evolved to encode RNA silencing suppressors (RSSs) (46). For plant viruses, RSSs were first described as pathogenicity factors that contribute to high levels of virus accumulation and disease (2, 23). Though the majority of RSS proteins have been identified in plant viruses, only a few are reported to occur in animal viruses. Some of the best-studied mammalian virus RSSs are NS1 of influenza A virus (7, 29), VP35 of Ebola virus, E3L of vaccinia virus, Tat of HIV-1, NSs of La Crosse virus, C of hepatitis C virus, and Tas of primate foamy virus 1 (19, 38, 42). Thus, screening methods need to be developed for identification of novel RSSs in animal viruses.
Severe acute respiratory syndrome coronavirus (SARS-CoV) is one of the highly infectious positive-stranded RNA viruses that spread worldwide and killed hundreds of people in early 2003 (9). Since SARS-CoV generates abundant double-stranded RNA (dsRNA) as replicative and transcriptive intermediates (24), this virus is likely to induce and become a target of gene silencing. Although animal viruses may experience gene silencing onslaught differently from plant viruses (5), we examined whether the SARS-CoV could encode an RSS protein(s) like other positive-strand RNA viruses (30, 31, 45). SARS-CoV contains 8 unique open reading frames (ORFs) that are predicted to encode proteins contributing toward viral replication and pathogenesis (26, 27, 43, 47). SARS-CoV leads to a gradual increase of viral load in the first 10 days postinfection (dpi), and no type I interferon response is observed, despite the onset of viral replication and viral protein synthesis (12, 26). Though early activation of innate immune pathways in SARS-CoV patients is possible, the role of RSSs in immune evasion and interferon agonist activity cannot be ruled out (10, 34).
Since RSSs act in a cross-kingdom fashion, we subjected the viral ORFs to a plant-based RSS screening test (22). These assays have an edge over the mammalian ones, as there are no overlapping immune and interferon-responsive pathways (6, 13, 41). The 7a accessory protein, along with another ORF (3a), was targeted as a candidate RSS protein. 7a induces apoptosis in a caspase-dependent manner (44), blocks cell cycle progression at G0/G1 phase, induces cell cycle arrest (48), inhibits cellular protein synthesis but not transcription and nucleoplasmic transport, and activates p38MAPK but not c-Jun and N-terminal protein kinases (25). For the RSS assay, two known viral RSSs, namely, Mungbean yellow mosaic India virus (MYMIV) AC2 and Flock house virus (FHV) B2, were included as positive controls (40).
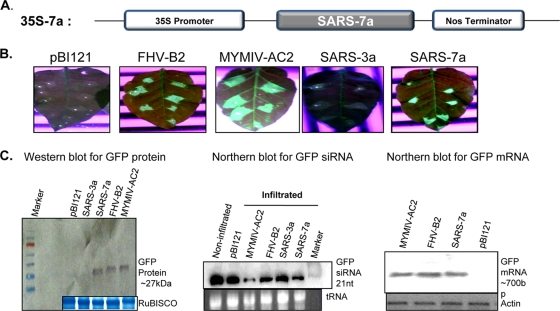
The reversal-of-silencing assay and the replication-based spot assay (22) were applied to screen the SARS-CoV ORFs. All necessary ORFs (7a, 3a, AC2, and B2) were cloned into the plant binary vector pBI121 (Fig. 1 A). The reversal-of-silencing assay was carried out with green fluorescent protein (GFP)-silenced transgenic Nicotiana xanthi lines by agroinfiltration-mediated transient ectopic expression of ORFs in leaf tissues. Infiltrated leaves were analyzed for reversal of GFP activity at 8 days postinfiltration. Only the 35S 7a-infiltrated zones (and not 3a) showed GFP fluorescence like the two positive controls, namely, FHV B2 and MYMIV AC2 (Fig. 1B). Interestingly, the ectopic expression of 7a led to reduction of biogenesis of small interfering RNA (siRNA) (17, 18, 49), as evident from the small RNA Northern blot (Fig. 2 C, middle). In fact, similar reduction has also been reported for many RSSs, viz., the reovirus sigma 3, hepatitis C virus core, and FHV B2 proteins (11, 32, 40). Reduction in siRNA level was associated with corresponding increases in GFP-mRNA and -protein levels (Fig. 1C).
FIG. 1.
Expression construct and reversal-of-silencing assay for the determination of suppressor activity by the SARS-CoV 7a ORF. (A) SARS-CoV 7a ORF under 35S promoter of the binary vector pBI121. (B) GFP fluorescence pictures taken under a UV lamp at 8 dpi of agroinfiltrated leaves. Each of the leaves was labeled at the top for the construct with which it was infiltrated. Empty vector was used as a mock control, while MYMIV AC2 and FHV B2 were positive controls in the assay. The tested SARS-CoV 7a protein showed positive reversal of silencing, while SARS virus 3a has been presented as negative for RSS activity. (C) Molecular analysis of reversal of silencing. Western blot analysis for GFP protein and Northern blot analysis for GFP-mRNA and -siRNA are presented. Fifty micrograms of RNA was extracted at 8 dpi from leaf patches infiltrated with Agrobacterium cultures harboring the corresponding plasmids as indicated. The bottom gels show the respective loading controls (Coomassie brilliant blue-stained RuBISCO band for the protein blot, actin-probed band for the Northern mRNA blot, and ethidium bromide [EtBr]-stained tRNA for the siRNA blot).
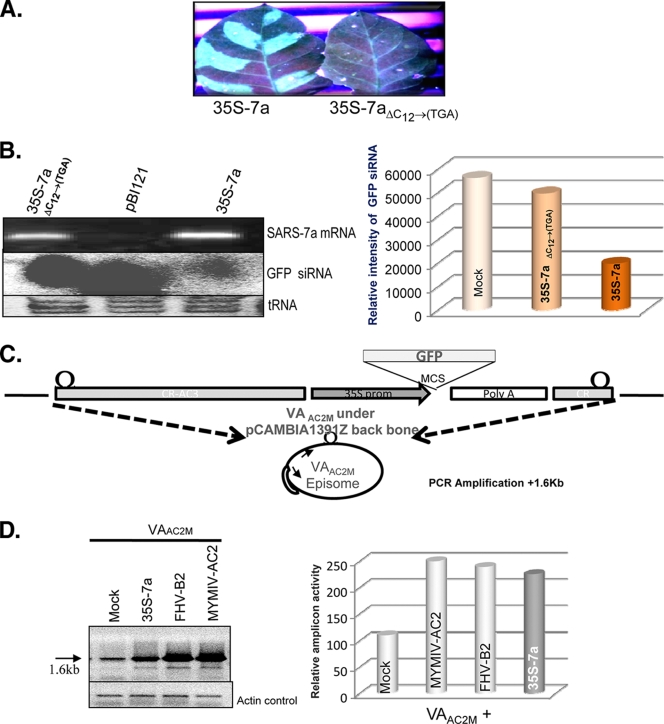
FIG. 2.
The SARS-CoV 7a protein, not the RNA, is responsible for the RNA silencing suppression activity. (A) Visualization under UV light of leaf tissue from GFP-silenced plants agroinfiltrated with 35S 7a and 35-7aΔC12→(TGA) after 8 dpi. (B) The top part of the left panel shows the reverse transcription-PCR (RT-PCR) result for SARS-CoV 7a-specific primers, where 7a mRNA has been generated both in wild-type and 7aΔC12→(TGA)-agroinfiltrated leaves. The middle portion of the panel shows the siRNA level corresponding to the reporter gene GFP upon agroinfiltration with empty vector pBI121, 35S 7a, and 35S 7aΔC12→(TGA). The lower portion shows the tRNA loading control on 15% PAGE gel stained with EtBr. The right panel shows a density graph of the siRNA blot normalized with the tRNA control. (C) Schematic representation of the viral amplicon-based RNA silencing suppressor assay. The VAAC2M vector is shown where the viral amplicon with a reporter GFP gene was cloned into the backbone of the plant binary vector pCAMBIA 1391Z. The VAAC2M vector in planta replication generates an amplicon of 2.4 kb. The bottom panel shows a diagrammatic representation of the PCR strategy used for detection of amplicon levels. (D) PCR-based analysis for determining the enhancement of replication in the presence of AC2, FHV B2, and 35S 7a upon coinfiltration with VAAC2M in the leaves of Nicotiana tabacum cv. Petit Havana at 14 days postinfiltration. The left panel shows an agarose gel stained with EtBr, showing a 1.6-kb band as the PCR product of the viral amplicon, with the respective actin control in the bottom gel. The right panel shows the density graph of the PCR product normalized with the actin control.
The viral RNAs also show RSS activity (3). Adenovirus-associated (VA) RNAs I and II serve as the competitive substrate squelching the Dicer enzyme (3), which in turn affects the normal biogenesis of siRNA. To determine whether the 7a protein or the 7a RNA possesses the RSS activity, we generated a site-specific deletion mutation in the 7a gene to introduce a premature stop codon after the N-terminal 3rd amino acid and the downstream frameshift mutation. This mutant was designated 35S 7aΔC12→TGA. This mutant mRNA was similar to wild-type (wt) 35S 7a (Fig. 2B, left panel, top row) but failed to show reversal of silencing (Fig. 2A and 3 A). In the 35S 7aΔC12→TGA-infiltrated patches, no GFP fluorescence (Fig. 2A) or reduction in siRNA levels was observed (Fig. 2B, left panel, middle row, and right panel). Therefore, the 7a protein, not the RNA, was essential for suppression of RNA silencing.
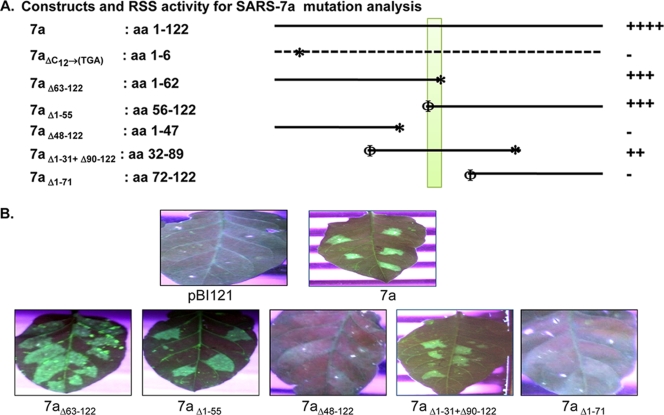
FIG. 3.
Deletion constructs and their corresponding RSS activities. (A) Diagrammatic representation of the various SARS-CoV 7a mutations used in this study. The numbers at the ends of the deletions refer to amino acids. The stop codon (*) and the start codon (Φ) introduced during cloning are indicated. The dotted line indicates the transcript that failed to translate due to a premature stop codon. The yellow rectangle shows the 6-amino-acid mapped domain responsible for RSS activity. Relative RSS activity exhibited by each 7a construct (wt, point mutation, and deletions) are shown on the extreme right, where ++++ represents the highest level of RSS activity, +++ a medium level, ++ a low level, and − the lowest level. (B) Results from the reversal-of-silencing assay, with various deletions of 7a performed to map the suppressor domain in the protein.
An alternative method for observing the RSS activity involved conducting functional complementation of an established RSS (37). Thus, using MYMIV-based viral amplicon mutated for the RSS AC2 (Fig. 2A) (22, 39) VAAC2M, we analyzed the 7a RSS activity. 35S 7a, upon coinfiltration with VAAC2M in a wild-type tobacco (Nicotiana tabacum cv. Petit Havana) plant, was able to rescue the amplicon accumulation (Fig. 2C) in trans to approximately the same extent as the control RSSs, i.e., MYMIV AC2 and FHV B2 (Fig. 2D). The amplicon accumulation was generally diminished by host RNA silencing activity and was quantitated by measuring the intensity of the 1.6-kb PCR band when the PCR was carried out for 26 cycles using the divergent primers (the locations of which are marked at the bottom of Fig. 2C) (35, 39). Thus, 35S 7a was able to suppress the virus-induced gene silencing activated against the viral amplicon during in planta replication as well.
SARS-CoV 7a encodes a 122-amino-acid protein, and we wanted to decipher the region important for its RSS activity. Our deletion mutation analysis showed that both the N-terminal (SARS-CoV 7aΔ63-122) and the C-terminal (SARS-CoV 7aΔ1-55) halves were capable of independently showing RSS activity (Fig. 3A and B). The contribution of two halves of a protein for the same biochemical activity has been reported for many proteins earlier (16, 28). Moreover, on further deletion, the peptide fragment containing amino acids [aa] 32 to 89 (35S 7aΔ1−31+Δ90−122) (Fig. 3B) has also been found to be somewhat essential for the RSS activity. This region primarily constitutes the compact immunoglobulin-like β-sandwich fold (20, 23) of 7a. However, further studies need to be initiated to establish this structure-function correlation. Further, the deletion mutants, namely, 35S 7aΔ48-122 and 35S 7aΔ1-71, failed to show any suppression activity (Fig. 3A and B), indicating that the signal peptide (N-terminal 15 aa), transmembrane domain (aa 96 to 117), and cytoplasmic tail (aa 117 to 122) have no role in RSS activity. This observation points out the importance of the peptide (aa 56 to 62) in RSS activity (Fig. 3).
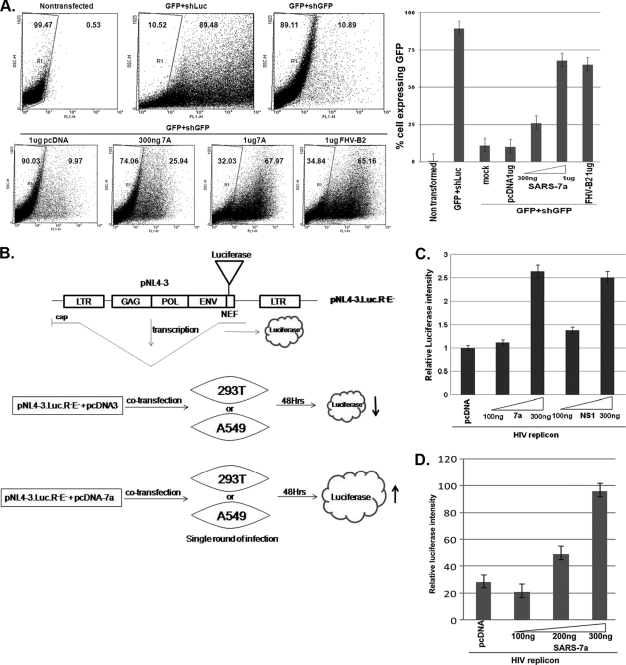
Finally, we confirmed the RSS activity of the 7a protein in animal cell lines. A bicistronic gene vector (RNAi-Ready-pSIREN-RetroQ-ZsGreen, abbreviated Retro-Q; Clonetech, CA) that encodes the GFP under cytomegalovirus (CMV) and the short hairpin RNA (shRNA) under the U6 promoter was used for the reporter assay. In this vector, shRNA for luciferase (an unrelated shRNA control) was cloned for control experiments. The transfection of the Retro-Q vector in HEK293T cells resulted in almost 90% cells showing green fluorescence while the control cells showed no fluorescence, as expected. The expression of shRNA against GFP (nucleotide [nt] positions 973 to 994) resulted in reduction of GFP-expressing cells by 79%. This reporter plasmid DNA with shRNA-GFP (0.5 μg in 1 ml) was cotransfected either with an increasing concentration of the 7a plasmid or with the control RSS, viz., FHV B2 (Fig. 4 A). Cotransfection dramatically restored the number of GFP-expressing cells in a 7a dose-dependent fashion (Fig. 4A, both right and left panels). The same was also true when 1 μg of FHV B2-expressing plasmid was used.
FIG. 4.
RNAi suppression activity of 7a in the mammalian cell line. (A) The left panel represents the FACS analysis of the mammalian cell line HEK293T transfected with 0.5 μg of RNAi-ReadypSIREN-RetroQ-ZsGreen-Retroviral vector (Clonetech) coexpressing GFP along with shRNA against GFP and viral ORFs (7a and FHV B2). All the ORFs were cloned downstream of the human cytomegalovirus (CMV) promoter in the vector pCDNA. The GFP expression of the cells was analyzed at 3 days posttransfection. The control GFP expression was monitored by replacing short hairpin GFP (shGFP) with short hairpin luciferase (shLuc), where 89%of the cells showed GFP fluorescence. The shGFP induced 79% reduction in GFP-expressing cells. The restoration of GFP silencing was monitored in the presence of increasing amounts (300 ng and 1 μg) of 7a as well as the positive control FHV B2. Each transfection experiment was carried out in three replicates. The right panel shows the bar graph representation of the FACS result, with the percentage of cells expressing GFP represented on the y axis and different transfection combinations represented on the x axis. (B) Schematics of the structural features of HIV-1 replicon DNA (pNL4-3.LucR-E-), showing the insertion of the firefly luciferase gene in the Nef-encoding region. Upon transfection into mammalian cells, the luciferase gene is expressed from a spliced transcript. The extent of luciferase is increased when the cells (293T or A549 cells) are cotransfected with the 7a construct, compared to the level for the control DNA (pCDNA). (C) Increase in HIV replicon activity obtained with the 7a protein. HEK293T cells were transfected with 100 ng of the HIV-1 clone pNL-LucR-E- and increasing concentrations of 7a (and the positive-control NS1 gene from influenza virus), along with 10 ng of the pTK-RL plasmid, using Lipofectamine reagent. At 48 h posttransfection, cell lysates were prepared in accordance with standard protocols and the luciferase values were measured using a dual luciferase kit from Promega. The relative luciferase amounts were monitored for the various transfection combinations and are represented in the bar diagram. All the values were normalized to those for Renilla luciferase. (D) Analysis of 7a activity in the SARS-CoV-permissive human lung epithelial cell line A549 by a replicon enhancement assay. One hundred nanograms of HIV-1 clone pNL-LucR-E was transfected with increasing concentrations of 7a by use of Lipofectamine reagent. At 48 h posttransfection, the cell lysate was prepared and luciferase values were measured using a dual luciferase kit from Promega as described for panel C.
We also examined whether 7a could increase the animal viral replicon activity, similar to those seen in Fig. 2D. Hence, we used an HIV-1 reporter gene-based replicon assay system employing the full-length HIV-1 genome-containing luciferase reporter gene (pNL-LucR-E; obtained from NIH AIDS Research & Reference Reagent, MD) (21) (Fig. 4B). The firefly luciferase gene was inserted into the pNL4-3 (infectious HIV-1 DNA) Nef gene to yield pNL4-3.LucR-E- (pNL-Luc). The extent of luciferase reporter gene activity was directly proportional to the extent of HIV-1 replication. HEK293T/A549 cells were cotransfected with a fixed amount of pNL-Luc (0.1 μg in 1 ml for 1 × 105 cells) by use of Lipofectamine along with an increasing concentration of SARS-CoV 7a-encoding plasmid DNA for 48 h. The cells were lysed in 100 μl of lysis buffer, and the reporter activity was measured with a luminometer. A dose-dependent increase in luciferase activity, i.e., HIV replicon activity, was observed (Fig. 4C). Similar enhancement was also observed with the NS1 protein of influenza A virus, a known RSS. A strikingly similar result, i.e., 7a-mediated enhancement of HIV replicon activity, was observed when the SARS-CoV-permissive A549 cells were used for transfection experiments (Fig. 4D). A control dual reporter gene plasmid (pTK-RL) was used to normalize transfection efficiency in the entire transfection-based assay. The results shown in Fig. 4C and D are representative of three independent experiments. A similar type of enhancement of the HIV replicon by the RSS activity of HIV Tat (19), rice hoja blanca virus NS3 protein (37), and influenza virus NS1 protein (14) has been reported earlier. These results suggest that the 7a protein of SARS-CoV might help the viral genome replicate efficiently in vivo, defying the host RNA interference (RNAi)-mediated degradative action toward the replicating viral genome. With the availability of the SARS-CoV replicon, this hypothesis could be put to test and thus the 7a protein could become an important drug target.
Collectively, the above-mentioned experiments establish that SARS-CoV 7a is a potent suppressor of siRNA activity in mammalian cells, including the SARS-CoV-permissive lung carcinoma cells. Its localization in the endoplasmic reticulum (36), where viral dsRNA replicative and transcriptive intermediates also exist (24), also suggests its potential role in protecting the virus from the host RNA silencing mechanism during infection. Moreover, the previously reported loss of virus accumulation upon inactivation of ORF 7 (1) can now be ascribed to the RSS activity of 7a. Thus, the 7a protein of highly pathogenic SARS-CoV has probably evolved to counter the host RNA silencing mechanism, in addition to showing various other biological activities (43, 44).
Acknowledgments
We thank Nurul Islam, Rakhi Agarwal, Subhra Mukhopadyay, Bhavna Varshney, and A. S. Banu for their helpful input and Ranjit DasGupta of the University of Wisconsin, Madison, WI, for providing the FHV RNA II genome.
The financial assistance of CSIR (India) to S.K. is duly acknowledged.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Akerstrom, S., A. Mirazimi, and Y. J. Tan. 2007. Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S. Antiviral Res. 73:219-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marethe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. U. S. A. 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, M. G., P. C. Haasnoot, N. Xu, S. Berenjian, B. Berkhout, and G. Akusjärvi. 2005. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 79:9556-9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baulcombe, D. C. 2004. RNA silencing in plants. Nature 431:356-363. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B., and K. T. Jeang. 2007. RISCy business: microRNAs, pathogenesis, and viruses. J. Biol. Chem. 282:26641-26645. [DOI] [PubMed] [Google Scholar]
- 6.Bridge, A. J., S. Pebernard, A. Ducraux, A. L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. [DOI] [PubMed] [Google Scholar]
- 7.Bucher, E., H. Hemmes, P. D. Haan, R. Goldbach, and M. Prins. 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 85:983-991. [DOI] [PubMed] [Google Scholar]
- 8.Carrington, J. C., and V. Ambros. 2003. Role of microRNAs in plant and animal development. Science 301:336-338. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2003. Revised U.S. surveillance case definition for severe acute respiratory syndrome (SARS) and update on SARS cases—United States and worldwide, December 2003. MMWR Morb. Mortal. Wkly. Rep. 52:1202-1206. [PubMed] [Google Scholar]
- 10.Chang, Y. J., C. Y. Liu, B. L. Chiang, Y. C. Chao, and C. C. Chen. 2004. Induction of IL-8 release in lung cells via activator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: identification of two functional regions. J. Immunol. 173:7602-7614. [DOI] [PubMed] [Google Scholar]
- 11.Chen, W., Z. Zhang, J. Chen, J. Zhang, J. Zhang, Y. Wu, Y. Huang, X. Cai, and A. Huang. 2008. HCV core protein interacts with Dicer to antagonize RNA silencing. Virus Res. 133:250-258. [DOI] [PubMed] [Google Scholar]
- 12.Cheung, C. Y., L. L. Poon, I. H. Ng, W. Luk, S. F. Sia, M. H. Wu, K. H. Chan, K. Y. Yuen, S. Gordon, Y. Guan, and J. S. Peiris. 2005. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 79:7819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cronin, S. J., N. T. Nehme, S. Limmer, S. Liegeois, J. A. Pospisilik, D. Schramek, A. Leibbrandt, M. Simoes Rde, S. Gruber, U. Puc, I. Ebersberger, T. Zoranovic, G. G. Neely, A. von Haeseler, D. Ferrandon, and J. M. Penninger. 2009. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325:340-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries, W., J. Haasnoot, R. Fouchier, P. de Haan, and B. Berkhout. 2009. Differential RNA silencing suppression activity of NS1 proteins from different influenza A strains. J. Gen. Virol. 90:1916-1922. [DOI] [PubMed] [Google Scholar]
- 15.Ding, S. W., H. Li, R. Lu, F. Li, and W. X. Li. 2004. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 102:109-115. [DOI] [PubMed] [Google Scholar]
- 16.Donelan, N. R., B. Dauber, X. Wang, C. F. Basler, T. Wolff, and A. García-Sastre. 2004. The N- and C-terminal domains of the NS1 protein of influenza B virus can independently inhibit IRF-3 and beta interferon promoter activation. J. Virol. 78:11574-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in C. elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 19.Haasnoot, J., W. de Vries, E. J. Geutjes, M. Prins, P. de Haan, and B. Berkhout. 2007. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 3:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanel, K., T. Stangler, M. Stoldt, and D. Willbold. 2006. Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin like fold and suggests a binding activity to integrin I domains. J. Biomed. Sci. 13:281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgon, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karjee, S., M. N. Islam, and S. K. Mukherjee. 2008. Screening and identification of virus-encoded RNA silencing suppressors. Methods Mol. Biol. 442:187-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 24.Knoops, K., M. Kikkert, S. H. E. van den Worm, S. J. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PloS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae and their replication, p. 1163-1185. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 27.Lau, Y. L., and J. S. Peiris. 2005. Pathogenesis of severe acute respiratory syndrome. Curr. Opin. Immunol. 17:404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leliveld, S. R., R. T. Dame, J. L. Rohn, M. H. M. Noteborn, and J. P. Abrahams. 2004. Apoptin's functional N- and C-termini independently bind DNA. FEBS Lett. 557:155-158. [DOI] [PubMed] [Google Scholar]
- 29.Li, W. X, H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. García-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. U. S. A. 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 31.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 32.Lichner, Z., D. Silhavy, and J. Burgyan. 2003. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol. 84:975-980. [DOI] [PubMed] [Google Scholar]
- 33.Nelson, C. A., A. Pekosz, C. A. Lee, M. S. Diamond, and D. H. Fremont. 2005. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure 13:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng, L. F., M. L. Hibberd, E. E. Ooi, K. F. Tang, S. Y. Neo, J. Tan, K. R. Murthy, V. B. Vega, J. M. Chia, E. T. Liu, and E. C. Ren. 2004. A human in vitro model system for investigating genome-wide host responses to SARS coronavirus infection. BMC Infect. Dis. 4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey, P., N. R. Choudhury, and S. K. Mukherjee. 2009. A geminiviral amplicon (VA) derived from Tomato leaf curl virus (ToLCV) can replicate in a wide variety of plant species and also acts as a VIGS vector. Virol. J. 6:152-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pekosz, A., S. R. Schaecher, M. S. Diamond, D. H. Fremont, A. C. Sims, and R. S. Baric. 2006. Structure, expression, and intracellular localization of the SARS-CoV accessory proteins 7a and 7b. Adv. Exp. Med. Biol. 581:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnettler, E., W. de Vries, H. Hemmes, J. Haasnoot, R. Kormelink, R. Goldbach, and B. Berkhout. 2009. The NS3 protein of rice hoja blanca virus complements the RNAi suppressor function of HIV-1 Tat. EMBO Rep. 10:258-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schutz, S., and P. Sarnow. 2006. Interaction of viruses with the mammalian RNA interference pathway. Virology 344:151-157. [DOI] [PubMed] [Google Scholar]
- 39.Singh, D. K., M. N. Islam, N. R. Choudhury, S. Karjee, and S. K. Mukherjee. 2007. The 32 kDa subunit of replication protein A (RPA) participates in the DNA replication of Mung bean yellow mosaic India virus (MYMIV) by interacting with the viral Rep protein. Nucleic Acids Res. 35:755-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh, G., S. Popli, Y. Hari, P. Malhotra, S. Mukherjee, and R. K. Bhatnagar. 2009. Suppression of RNA silencing by Flock house virus B2 protein is mediated through its interaction with the PAZ domain of Dicer. FASEB J. 23:1845-1857. [DOI] [PubMed] [Google Scholar]
- 41.Sledz, C., M. Holko, M. de Veer, R. Silverman, and B. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 42.Soldan, S. S., M. L. Plassmeyer, M. K. Matukonis, and F. Gonzalez-Scarano. 2005. La Crosse virus nonstructural protein NSs counteracts the effects of short interfering RNA. J. Virol. 79:234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surjit, M., and S. K. Lal. 2010. The nucleocapsid protein of the SARS coronavirus: structure, function and therapeutic potential, p. 129-151. In S. K. Lal (ed.), Molecular biology of the SARS-coronavirus, 4th ed. Springer, Germany.
- 44.Tan, Y. J., B. C. Fielding, P. Y. Goh, S. Shen, T. H. P. Tan, S. G. Lim, and W. Hong. 2004. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-Dependent pathway. J. Virol. 78:14043-14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants—defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 46.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206-220. [DOI] [PubMed] [Google Scholar]
- 47.Yamate, M., M. Yamashita, T. Goto, S. Tsuji, Y. G. Li, J. Warachit, M. Yunoki, and K. Ikuta. 2005. Establishment of Vero E6 cell clones persistently infected with severe acute respiratory syndrome coronavirus. Microbes Infect. 7:1530-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan, X., J. Wu, Y. Shan, Z. Yao, B. Dong, B. Chen, Z. Zhao, S. Wang, J. Chen, and Y. Cong. 2006. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology 346:74-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]