Abstract
The heterotrimeric RNA-dependent RNA polymerase of influenza viruses catalyzes RNA replication and transcription activities in infected cell nuclei. The nucleotide polymerization activity is common to both replication and transcription processes, with an additional cap-snatching function being employed during transcription to steal short 5′-capped RNA primers from host mRNAs. Cap-binding, endonuclease, and polymerase activities have long been studied biochemically, but structural studies on the polymerase and its subunits have been hindered by difficulties in producing sufficient quantities of material. Recently, because of heightened effort and advances in expression and crystallization technologies, a series of high resolution structures of individual domains have been determined. These shed light on intrinsic activities of the polymerase, including cap snatching, subunit association, and nucleocytoplasmic transport, and open up the possibility of structure-guided development of new polymerase inhibitors. Furthermore, the activity of influenza polymerase is highly host- and cell type-specific, being dependent on the identity of a few key amino acid positions in the different subunits, especially in the C-terminal region of PB2. New structures demonstrate the surface exposure of these residues, consistent with ideas that they might modulate interactions with host-specific factors that enhance or restrict activity. Recent proteomic and genome-wide interactome and RNA interference screens have suggested the identities of some of these potential regulators of polymerase function.
Keywords: RNA Viruses, Viral Polymerase, Viral Protein, Virus Replication, Viral Transcription, X-ray Crystallography, Influenza
Introduction
Influenza A viruses are important viral pathogens of humans and animals. In humans, they cause both yearly seasonal influenza epidemics and more extensive global outbreaks termed pandemics. The 1918 pandemic killed 50 million people, and those in 1957 and 1968 also caused serious mortalities. Although of relatively low virulence, the current swine origin H1N1 pandemic has shown that outbreaks can occur suddenly and unexpectedly despite constant worldwide surveillance. Influenza is also a major animal pathogen affecting domestic poultry and pigs, resulting in significant economic impact with, for example, sudden loss of whole flocks from sickness or preventative culling. All influenza A viruses originate from wild waterfowl that are generally asymptomatic during infection. From this origin, they can infect humans, either directly or via a domestic animal intermediate such as poultry or pigs. The infection of humans by avian viruses does not result in a sustainable pathogen because of poor human-to-human transmission; however, there are continual concerns that avian viruses such as the current highly virulent H5N1 avian strain may adapt and become a serious threat.
The influenza A virus is a member of the Orthomyxoviridae family possessing a negative-sense single-stranded RNA genome that is divided into eight viral RNA (vRNA)2 genomic segments that encode 10 major proteins (1). The three largest vRNAs encode the three subunits of the RNA-dependent RNA polymerase: the acidic subunit PA and the two basic subunits PB1 and PB2. There are also two minor pb1 gene products: an N-terminally truncated form of PB1 originating from an alternative start codon (2) and PB1-F2, a short polypeptide expressed from an alternative +1 reading frame that seems to increase the virulence of some strains (3). The medium segments encode the nucleoprotein (NP) that, together with the polymerase subunits and the vRNA, forms the ribonucleoprotein (RNP) and the two viral glycoproteins (4) hemagglutinin (HA) and neuraminidase (NA). HA is the major protein on the viral surface, and it binds cellular sialic acid receptors, leading to virus uptake. Once in the cell, fusion of viral and endosomal membranes occurs, and RNPs are released into the cytoplasm. NA is the enzyme that cleaves these same sugars from the surfaces of cells and new viruses during viral budding. There are two short vRNA segments that each encode two proteins. The first generates matrix protein (M1) that lines the internal surface of the viral lipid membrane and an ion channel (M2) that mediates the uncoating of the viral particle during infection and is the target for the drug amantadine. The second short segment encodes NS1, which is a significant virulence factor involved in evasion of the innate immune system (1), and NS2 (also known as nuclear export protein), which exports viral RNPs from the nucleus into the cytoplasm (5).
The influenza polymerase has no proofreading activity, resulting in a high gene mutation rate of approximately one error per replicated genome (6), so each cell can produce 10,000 new viral mutants to infect neighboring cells. This is crucial to the virus's evolutionary strategy, as continual changes in the glycoprotein sequences, notably HA, lead to evasion of the host antibody response, so-called “antigenic drift,” which underlies the inevitable escape from seasonal vaccines. Likewise, mutations in genes of the M2 and NA proteins have rapidly rendered the antiviral drugs amantadine and oseltamivir inactive against these targets. The polymerase itself is the target for new antiviral drugs (7), including T-705, an inhibitor in current late stage development (8). Experience suggests that, under selective pressure from drug use, the error-prone aspect of the very activity against which the inhibitors are targeted will lead to resistance. The segmented structure of the genome also contributes to the rapid evolution of new influenza viruses through a process called reassortment, where segments from different co-infecting viruses are packaged into new viral particles. The generation of reassortment viruses containing genes from avian and human viruses is thought to arise most commonly in pigs because these animals are susceptible to both viral types due to the presence of avian-like α2,3-linked and human-like α2,6-linked sialic acid cell-surface receptors. Through reassortment, an avian virus may evolve suddenly into a human pathogen by combining polymerase subunits and HA (and also other viral proteins, e.g. NS1) that function efficiently in human cells (9). If the HA protein is immunologically distinct from circulating varieties, “antigenic shift” can occur, resulting in a pandemic strain to which the population has no immunological protection. The current novel pandemic H1N1 strain arose through such a triple-reassortment process, combining avian PB2 and PA polymerase subunits, human PB1, and classic swine HA (10).
The influenza polymerase is therefore essential to the biological processes of (i) virus replication in cells by replicating the vRNA segments and transcribing their genes and (ii) virus evolution through its error-prone RNA replication, producing variants of the viral proteins, including the glycoproteins and the polymerase subunits themselves, which leads to viruses that are better adapted to new host species (11). Additionally, and in common with other viral replicases, it represents a promising drug target due to its activities that are distinct from those found in the host cell (7). Yet despite its biological interest and medical importance, the absence of detailed structural information on the polymerase has limited our mechanistic understanding and our ability to design better drugs. The main reason for this absence is an overwhelming difficulty in producing purified polymerase proteins in sufficient quantities for study. Since 2007, because of heightened effort and advances in expression and crystallization technologies, a series of x-ray and NMR structures of domains from the PA, PB1, and PB2 subunits that cover approximately half of the trimeric complex have been determined. Here, we review this structural progress and discuss our improved understanding of the intrinsic polymerase mechanism, as well as its role in adaptation to the cellular environments of different host species.
Genome Replication and Transcription Activities of the Polymerase
In the intact viral particle, each RNP contains a single polymerase complex associated with the conserved 5′- and 3′-ends of each vRNA segment. The vRNA is complexed with NP, with each protomer contacting 24 nucleotides (12). Direct protein contacts also occur between NP and the polymerase, as suggested by several biochemical studies (13–17) and visualized recently in a high resolution cryo-electron microscopy reconstruction comprising a synthetic minimal vRNA, nine NP monomers, and a polymerase trimer (18). The pre-existence of a functional polymerase trimer in the infective particle is necessary to initiate the first transcription and RNA replication cycles because the negative-sense vRNA cannot be directly translated into protein. Once de novo polymerase subunits are synthesized in the cytoplasm, they are transported back into the nucleus for assembly into trimers (19), thus allowing further transcription and genome replication cycles.
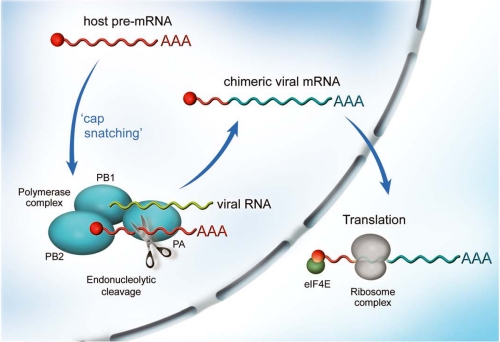
Replication of the viral genome is catalyzed by the polymerase via a cRNA intermediate. This is then copied back into vRNA segments that are used as templates for transcription and further replication in the nucleus and are exported as RNPs to the cytoplasm, where they are sorted into new viral particles via recognition of terminal packaging sequences (20, 21). In transcription, mRNA is generated by directly copying from the vRNA template, with polyadenylation occurring via a stuttering mechanism on a templated oligo(U) terminal sequence. Unlike RNA replication, the process is primer-dependent, and the virus obtains the 5′-primer by a mechanism shared with other segmented RNA viruses (e.g. bunyaviruses) called “cap snatching” (Fig. 1) (22). The polymerase binds the 5′,7-methylguanosine cap of a nuclear pre-mRNA and cleaves it 9–15 nucleotides downstream. The resulting RNA oligonucleotide is used to initiate transcription from the vRNAs, resulting in capped, polyadenylated, positive-sense mRNAs that resemble host cell messages. These are exported from the nucleus for translation in the cytoplasm, possibly via a CRM1-independent pathway (23).
FIGURE 1.
Cap-snatching transcription mechanism of influenza polymerase. The PA-PB1-PB2 complex is localized in the nucleus of the infected cell. During transcription, the PB2 subunit binds the 5′,7-methylguanosine cap of a host pre-mRNA molecule (red), which is subsequently cleaved 10–15 nucleotides downstream by the PA endonuclease. The resulting short capped RNA primer is used to initiate polymerization by the RNA-dependent RNA polymerase of the PB1 subunit using 5′- and 3′-bound vRNA (green) as template, resulting in capped, polyadenylated, chimeric mRNA molecules (red and blue) that are exported to the cytoplasm for translation into viral proteins.
PA Subunit
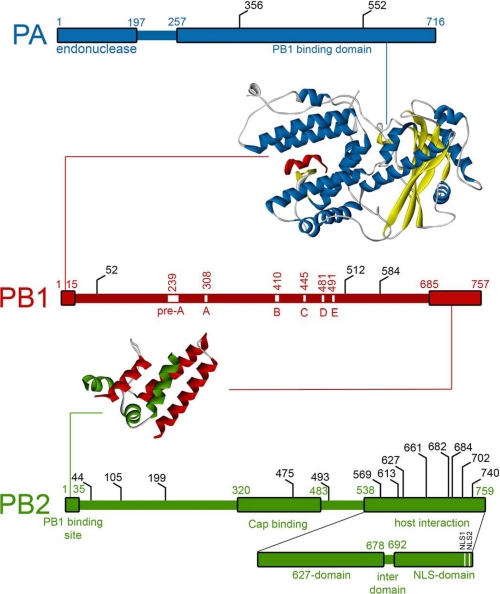
The PA subunit has no significant homology to other proteins, and for a long time, the function was unclear. Various functions were proposed, including a chymotrypsin-like serine protease (24–26) and various aspects of RNA replication (27–30). A soluble subunit could be expressed in insect cells for limited proteolysis studies, revealing cleavage into N-terminal 25-kDa and C-terminal 55-kDa fragments, indicative of stable domains (31, 32). The structure of the C-terminal domain complexed with a short PB1 N-terminal peptide was determined by two groups (33, 34), showing how the PB1 fragment is gripped in a highly conserved cleft resembling “jaws” in the “head of a dragon” (Fig. 2) (33). The essential nature of this interaction was demonstrated by mutagenesis of the interface (34), and sequence analysis revealed how the interfacial residues were resistant to mutational drift, probably because multiple compensatory mutations in both PA and PB1 subunits would be required. This interface has thus been proposed as possibly druggable, and a 25-residue PB1-derived peptide has been shown to inhibit polymerase assembly and virus replication in influenza A and B strains (35, 36).
FIGURE 2.
Features of the polymerase subunits. Linear representations of the three polymerase subunits are annotated to show the structurally characterized domains (large boxes). Residues implicated in host adaptation (67) are shown in black. X-ray crystal structures of interaction regions of PA-PB1 (33, 34) and PB1-PB2 (48) are presented with helix colors as in the linear representations.
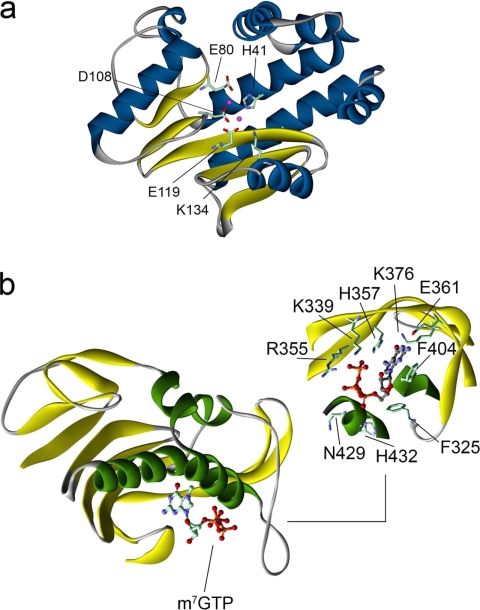
Two groups recently determined the structure of the PA N-terminal domain (37, 38). Only at this point did it become clear that the endonuclease activity was present in this subunit and not PB1 as suggested previously from work on purified polymerase complexes (39). The domain fold and active-site arrangement is similar to that of the PD-(D/E)XK family of nucleases (Fig. 3a). That the crystallized domain possessed the true endonuclease activity was confirmed by the hydrolysis of both single-stranded RNA and single-stranded DNA substrates (37), as observed previously with purified RNPs (40). Structure-based mutagenesis of key PA active-site residues in reconstituted polymerase trimer showed that endonuclease-independent RNA replication was maintained, whereas endonuclease-dependent transcription was abolished (38). A known specific inhibitor of influenza endonuclease activity in intact polymerase, dioxo-4-phenylbutanoic acid (41), strongly stabilized the purified domain in thermal shift assays and inhibited single-stranded RNA hydrolysis (37). The identity of the coordinated metal ions differs between the two structures, with one structure containing a single Mg2+ ion (38) and the other containing two Mn2+ ions coordinated in adjacent positions. The difference between the two observations is due in part to the addition of MnCl2 to the crystallization medium (37), with Mn2+ preferentially binding to the site that includes a histidine ligand. This was deliberate and followed the observation of strong thermal stabilization and enhancement of endonuclease activity in the presence of Mn2+ or Co2+; both the identity of these ions and the presence of two metal-binding sites accord with a cooperative two-metal ion mechanism measured on purified RNPs (42).
FIGURE 3.
X-ray structures of the cap-snatching domains. a, the PA endonuclease domain (37) exhibits a fold similar to members of the PD-(D/E)XK family of nucleases. The key active-site residues and two manganese ions (pink) are shown. b, the PB2 cap-binding domain complexed with m7GTP (55) shows a novel fold but binds the positively charged base through a sandwich of aromatic residues in a manner similar to other cap-binding proteins. The binding site is shown with ligand-contacting residues.
PB1 Subunit
In contrast to the progress on PA and PB2, PB1 remains poorly structurally characterized. The central location of the polymerase domain is predicted from the presence of conserved motifs characteristic of segmented negative-strand RNA-dependent polymerases (43, 44) and would suggest that it has a classic polymerase fold. However, expression of the full-length subunit has not yielded soluble material, and the domain-by-domain approach used successfully for the other subunits has not yet led to expression of an isolated polymerase domain at levels compatible with structural studies. Previous studies demonstrated that the N-terminal region of PB1 bound the C terminus of PA (35, 45, 46), and this was borne out in the PA C-terminal domain structures, where short N-terminal PB1 peptides were co-crystallized (Fig. 2) (33, 34).
At the other end of PB1, the interaction of its C-terminal region with the N terminus of PB2 has also been described biochemically (45, 47). A recently determined x-ray structure of an 86-amino acid (aa) C-terminal fragment of PB1 with a 37-aa N-terminal peptide from PB2 showed how these α-helical subunit termini tightly co-fold (Fig. 2) (48). Despite its small size, this interaction interface is completely conserved across different avian and human influenza strains, and the absolute requirement of the 250-kDa trimer in this interaction for function suggests that this interaction, like that of PB1 and PA, could be a possible drug target.
PB2 Subunit
The PB2 subunit was initially identified as the site of cap binding through cross-linking studies (49, 50), with further experiments suggesting regions 242–252 and 533–577 as interaction sites (39, 51). Mutagenesis later confirmed this role of the PB2 subunit but isolated it to central residues Phe-363 and Phe-404 (52), apparently contradicting the location determined from cross-linking data. Other information was available on this subunit relating to its nucleocytoplasmic trafficking that was proposed to occur via multiple nuclear localization signals both internally and C-terminally located (53). In common with PA, the protein sequence of PB2 is unlike any other, preventing the use of sequence alignments in identifying structural domains. Unlike PA, full-length PB2 cannot be expressed solubly, precluding the use of limited proteolysis. Therefore, studies on this subunit were effectively blocked until the development of ESPRIT, a robotic random library-based construct screening process (54) that was used to systematically screen almost 90,000 random fragments of the pb2 gene, identifying a series of Escherichia coli-expressible soluble fragments for structural studies (55–57).
The first fragment identified was obtained from a 5′-pb2 deletion library and comprised a highly soluble, overexpressing C-terminal domain (aa 678–759) (Fig. 2) (56). Analysis of the NMR solution structure revealed a well folded domain fused to a relatively mobile region, with a sequence suggestive of a classic bipartite nuclear localization signal (NLS; 737RKRX12KRIR755) that was previously thought to be monopartite (53). Nuclear translocation assays with the green fluorescent protein-fused “NLS domain” (also referred to as DPDE) and full-length PB2 in transfected cells confirmed this activity. Although the NLS domain did not crystallize independently, co-crystallization with the nuclear import receptor importin α5 yielded an x-ray structure detailing the bipartite nature of the NLS and illustrating how the NLS region becomes fully unfurled during importin binding. The clear assignment of importin-dependent binding to the C-terminal region put into question the internal NLS (residues 449–495) identified through gene deletion and cellular localization studies (53). Later structural data precisely defined the location and structure of the overlapping cap-binding domain (aa 318–483; discussed below), suggesting that aberrant nuclear transfer of the Δ449–495 construct may have been the consequence of PB2 structural destabilization.
Also identified by ESPRIT was a soluble central region with constructs spanning aa 241–483 (Fig. 2) (55). Because this contained residues implicated in cap binding (52), the purified fragments were assayed with m7GTP-Sepharose, confirming an intrinsic cap-binding activity. A proteolytically stable subfragment (aa 318–483) was observed during purification, and it was this fragment that ultimately crystallized in the presence of m7GTP. The resulting structure exhibited a new fold, consistent with the absence of homology to other proteins (Fig. 3b). Despite this, the methylated guanine base bound in a mode that is commonly observed in cap-binding proteins (e.g. cap-binding complex (58) and eIF4E (59)) whereby aromatic side chains sandwich the positively charged aromatic ring of the base. The interacting residues are completely conserved in all influenza strains, and their mutation abolished cap-dependent transcription but not cap-independent replication in recombinant mini-RNPs (55). The x-ray structures and availability of active E. coli-expressible PB2 cap-binding and PA endonuclease domains open the way to the development of drugs that target the endonuclease activity (7), a strategy that was pursued previously by pharmaceutical companies (41, 60) but that proved unsuccessful due to the need to work without structures on low abundance whole RNPs purified from cells.
Host Adaptation and Role of Polymerase Subunits, Notably PB2
The World Health Organization maintains a global surveillance program that generates large amounts of sequence data from wild-type viral isolates (61), made accessible via databases at the National Center for Biotechnology Information (NCBI) and the Global Initiative on Sharing All Influenza Data (GISAID) (62, 63). Comparative sequence analyses have identified numerous variations, among which must be those responsible for antigenic drift, shift, and host adaptation. The major roles of HA and NA in host specificity are well established, and the x-ray structures of these proteins have contributed a valuable molecular level understanding of their variants (4). Several studies have also identified host signatures in polymerase subunits that are correlated with adaptation of avian viruses to mammalian hosts (Refs. 11 and 64–67; reviewed in Ref. 68). Although sequence analyses highlight host-related differences, it is not always clear which are responsible for the host shift event rather than being the result of neutral drift (i.e. are passenger mutations alongside those that are functional), hence the importance of experimental testing of hypotheses. One recent computational study (67) used a statistical method that compensated for biases from neutral drift and the underlying phylogenetic relationships between sequences in the dataset. In common with previous observations (68), putative host-adaptive mutations were found with high confidence in the polymerase subunits: 2 in PA, 3 in PB1, and 13 in PB2. The abundance of such mutations in the PB2 subunit was also predicted previously (64–66, 69). For most of these mutations, little is known about their mode of action. A notable exception is PB2 627, which is almost invariably Glu in avian viruses and Lys in human viruses. A single E627K mutation in an otherwise non-human-infective avian virus is sufficient to confer host adaptation (66, 70). Many studies (reviewed in Ref. 68) have sought to explain this observation, with a major hypothesis being that the E627K mutation confers improved replication in mammalian cells, particularly at lower temperatures (71), in accordance with the mammalian upper respiratory tract being significantly cooler (33–35 °C) than the avian intestinal tract (37–40 °C). There is currently little understanding of the mechanism by which E627K exerts its effect, although it appears to affect the PB2-NP interaction in a host cell-dependent manner (14, 15, 17).
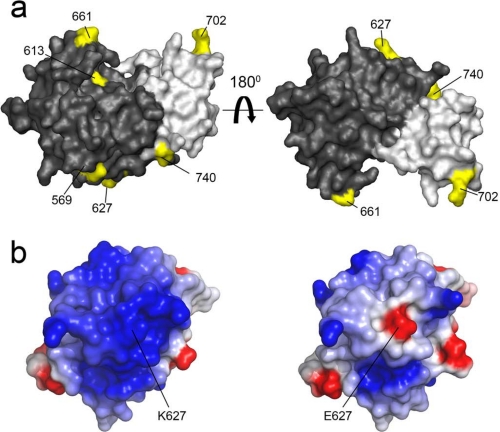
A systematic E. coli expression screen of the PB2 subunit using the ESPRIT method resulted in isolation of a large C-terminal soluble fragment containing 8 of the 13 predicted host determinant residues (67) and the previously characterized NLS domain (Fig. 2) (56). This crystallized to reveal two domains packed tightly together via a polar interface (Fig. 4a) (57). A very similar structure, also from a human virus, was solved by a second group (72). The upstream domain, termed the 627 domain because it contains this major host determinant residue, was also crystallized, revealing the side chain to be solvent-exposed in both Lys and Glu forms (Fig. 4b) (57). The 627 domain has a highly basic surface patch that was disrupted in an avian-like glutamic acid mutant (57). The function of this 627-NLS double domain remains unclear despite these structures, although a possible role in RNA binding has been proposed (72). Interestingly, the pandemic H1N1 virus has Glu-627 but is highly transmissible between humans. Using the crystal structures of the 627 domain, this was explained by a compensating double mutation, G590S and Q591R, in which Arg-591 effectively shielded the negative charge of Glu-627, re-establishing the basic surface patch that seems to characterize human-infective viruses (73). Further mutation at position 627 of this pandemic virus has not occurred to any serious degree, and a laboratory-engineered variant containing both G590S and Q591R and Lys-627 showed no obvious replicative advantage, suggesting that little further selective advantage to the virus is provided (74).
FIGURE 4.
Residues at the C terminus of PB2 implicated in host adaptation. a, the PB2 627-NLS domain comprises two independently folded domains, the 627 domain (dark gray) and the NLS domain (light gray), interacting via a polar interface (57). Residues implicated in host adaptation (67) are shown in yellow. b, the crystal structures show electrostatic surface potentials of the human 627 domain with Lys-627 (left) and a point mutant with the avian-like Glu-627 residue (right). Formation of a large basic surface occurs upon host adaptation that is mediated almost universally by an E627K mutation.
A good correlation is observed between the putative host-adaptive mutations identified from sequence analysis studies of PA and PB2 and their location on the crystal structures; most are solvent-exposed surface residues (Fig. 4). This is consistent with the hypothesis that such mutations could mediate interactions with host cell factors necessary for more efficient polymerase activity or disrupt those with host restricting factors of the innate immune system. Such has been suggested for PB2, where replication of polymerases with avian Glu-627 is inhibited by a host restricting factor present in human cells (17), although it has not yet been identified. The interaction with NP is also implicated (14, 15, 17), but it remains to be seen if it is NP itself that is the 627-dependent interactor or a host molecule.
A number of interactions between RNPs and specific host factors have been described (reviewed in Ref. 68), although it is not known if they are involved in host range determination. For example, the transcription activator hCLE (75) and the RNA polymerase II C-terminal domain (76) have been demonstrated to bind the polymerase. Perhaps best understood is the association of PB2 with members of the nuclear import receptor family, importin α. The co-crystal structure of the PB2 C-terminal NLS domain with human importin α5, together with nuclear transport assays, revealed the mechanism by which this subunit was imported into the nucleus (56). PB2 also binds importin α1 in vitro (57) and importin α7 in human cells (77), the latter report suggesting an additional function of importins as necessary cofactors for replication activity. Previously, the PB2 mutation D701N (now known to be located in this domain) had been shown to facilitate adaptation of an avian virus to mice (78). This effect is apparently explained by the observation that D701N enhances binding to importin α1 in mammalian cells but not avian cells, with a resultant increase in PB2 nuclear accumulation (79). In vivo, D701N enhances mammalian infectivity of viruses with avian-like PB2 Glu-627, normally characteristic of low pathogenicity, both in guinea pigs (80) and in humans (81), being similar in its compensatory effect to the G590S and Q591R double mutation of the adjacent 627 domain (73). Thus, the importin-PB2 association may be one mechanism by which mutations can lead to host adaptation, by modulating either nuclear localization or polymerase activity in a host-specific manner.
In recent years, high throughput proteomic approaches and yeast two-hybrid (Y2H) and genome-wide RNA interference (RNAi) screens have increased manyfold the list of putative polymerase partners, most of which still need to be properly validated. Tandem affinity purification (TAP) strategies using polymerase (82, 83) and RNPs (83) have been employed to isolate physically interacting host factors from cells, identifying various heat shock proteins and nuclear import factors. To a similar end, individual polymerase subunit baits were screened against a high quality human ORFeome library by Y2H, resulting in the description of a wide and diverse network of interactors (84). Despite screening for direct association, many interactions identified from TAP tag and Y2H experiments are difficult to rationalize from a biological perspective. Genome-wide RNAi screens in Drosophila (85) and human (86–88) cell lines have not aimed to identify direct influenza protein interactors per se but have revealed hundreds of host cell proteins whose presence appears to be essential for virus replication. Some overlap is observed between the host factor lists from these studies, e.g. the V-ATPase ATP6AP1 and COPI vesicle transport proteins are found in all RNAi screens using human cell lines, but there is also a significant variation between the different datasets (reviewed in Ref. 89). Despite the limited agreement between the proteins identified in these high throughput experiments, it may be that a subset of the binders identified in the interaction screens (TAP tag and Y2H) or the virus-associated host factors from the genome-wide RNAi screens might bind directly to the polymerase subunits in a manner modulated by their surface-exposed host-adaptive residues. The availability of well behaved, purifiable domains and their structures should help validate these interactions and explain at a molecular level the effects of polymerase-induced genetic mutations on polymerase interactions in the cellular environments of different host species.
Acknowledgments
We thank our colleagues working on influenza for many interesting discussions and P. Riedinger for assistance with figures.
This work was supported by European Union FLUPOL Contract SP5B-CT-2007-044263, Agence Nationale de Research FLU INTERPOL Contract ANR-06-MIME-014-02, and the FINOVI Foundation. This is the second article in the Thematic Minireview Series on Influenza Virus. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- vRNA
- viral RNA
- NP
- nucleoprotein
- RNP
- ribonucleoprotein
- HA
- hemagglutinin
- NA
- neuraminidase
- aa
- amino acid(s)
- NLS
- nuclear localization signal
- Y2H
- yeast two-hybrid
- RNAi
- RNA interference
- TAP
- tandem affinity purification.
REFERENCES
- 1.Noah D. L., Krug R. M. (2005) Adv. Virus Res. 65, 121–145 [DOI] [PubMed] [Google Scholar]
- 2.Wise H. M., Foeglein A., Sun J., Dalton R. M., Patel S., Howard W., Anderson E. C., Barclay W. S., Digard P. (2009) J. Virol. 83, 8021–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McAuley J. L., Zhang K., McCullers J. A. (2010) J. Virol. 84, 558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skehel J. (2009) Biologicals 37, 177–178 [DOI] [PubMed] [Google Scholar]
- 5.Iwatsuki-Horimoto K., Horimoto T., Fujii Y., Kawaoka Y. (2004) J. Virol. 78, 10149–10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake J. W. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4171–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nistal-Villán E., García-Sastre A. (2009) Nat. Med. 15, 1253–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuta Y., Takahashi K., Shiraki K., Sakamoto K., Smee D. F., Barnard D. L., Gowen B. B., Julander J. G., Morrey J. D. (2009) Antiviral Res. 82, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Hoeven N., Pappas C., Belser J. A., Maines T. R., Zeng H., García-Sastre A., Sasisekharan R., Katz J. M., Tumpey T. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann G., Noda T., Kawaoka Y. (2009) Nature 459, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miotto O., Heiny A. T., Albrecht R., García-Sastre A., Tan T. W., August J. T., Brusic V. (2010) PLoS One 5, e9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ortega J., Martín-Benito J., Zürcher T., Valpuesta J. M., Carrascosa J. L., Ortín J. (2000) J. Virol. 74, 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newcomb L. L., Kuo R. L., Ye Q., Jiang Y., Tao Y. J., Krug R. M. (2009) J. Virol. 83, 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rameix-Welti M. A., Tomoiu A., Dos Santos Afonso E., van der Werf S., Naffakh N. (2009) J. Virol. 83, 1320–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labadie K., Dos Santos Afonso E., Rameix-Welti M. A., van der Werf S., Naffakh N. (2007) Virology 362, 271–282 [DOI] [PubMed] [Google Scholar]
- 16.Poole E., Elton D., Medcalf L., Digard P. (2004) Virology 321, 120–133 [DOI] [PubMed] [Google Scholar]
- 17.Mehle A., Doudna J. A. (2008) Cell Host Microbe 4, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coloma R., Valpuesta J. M., Arranz R., Carrascosa J. L., Ortín J., Martín-Benito J. (2009) PLoS Pathog. 5, e1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huet S., Avilov S. V., Ferbitz L., Daigle N., Cusack S., Ellenberg J. (2010) J. Virol. 84, 1254–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchinson E. C., von Kirchbach J. C., Gog J. R., Digard P. (2010) J. Gen. Virol. 91, 313–328 [DOI] [PubMed] [Google Scholar]
- 21.Noda T., Sagara H., Yen A., Takada A., Kida H., Cheng R. H., Kawaoka Y. (2006) Nature 439, 490–492 [DOI] [PubMed] [Google Scholar]
- 22.Plotch S. J., Bouloy M., Ulmanen I., Krug R. M. (1981) Cell 23, 847–858 [DOI] [PubMed] [Google Scholar]
- 23.Wang W., Cui Z. Q., Han H., Zhang Z. P., Wei H. P., Zhou Y. F., Chen Z., Zhang X. E. (2008) Nucleic Acids Res. 36, 4913–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara K., Shiota M., Kido H., Ohtsu Y., Kashiwagi T., Iwahashi J., Hamada N., Mizoue K., Tsumura N., Kato H., Toyoda T. (2001) Genes Cells 6, 87–97 [DOI] [PubMed] [Google Scholar]
- 25.Sanz-Ezquerro J. J., de la Luna S., Ortín J., Nieto A. (1995) J. Virol. 69, 2420–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez A., Pérez-González A., Nieto A. (2007) J. Virol. 81, 5315–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fodor E., Crow M., Mingay L. J., Deng T., Sharps J., Fechter P., Brownlee G. G. (2002) J. Virol. 76, 8989–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fodor E., Mingay L. J., Crow M., Deng T., Brownlee G. G. (2003) J. Virol. 77, 5017–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huarte M., Falcón A., Nakaya Y., Ortín J., García-Sastre A., Nieto A. (2003) J. Virol. 77, 6007–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawaguchi A., Naito T., Nagata K. (2005) J. Virol. 79, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guu T. S., Dong L., Wittung-Stafshede P., Tao Y. J. (2008) Virology 379, 135–142 [DOI] [PubMed] [Google Scholar]
- 32.Hara K., Schmidt F. I., Crow M., Brownlee G. G. (2006) J. Virol. 80, 7789–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He X., Zhou J., Bartlam M., Zhang R., Ma J., Lou Z., Li X., Li J., Joachimiak A., Zeng Z., Ge R., Rao Z., Liu Y. (2008) Nature 454, 1123–1126 [DOI] [PubMed] [Google Scholar]
- 34.Obayashi E., Yoshida H., Kawai F., Shibayama N., Kawaguchi A., Nagata K., Tame J. R., Park S. Y. (2008) Nature 454, 1127–1131 [DOI] [PubMed] [Google Scholar]
- 35.Ghanem A., Mayer D., Chase G., Tegge W., Frank R., Kochs G., García-Sastre A., Schwemmle M. (2007) J. Virol. 81, 7801–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wunderlich K., Mayer D., Ranadheera C., Holler A. S., Mänz B., Martin A., Chase G., Tegge W., Frank R., Kessler U., Schwemmle M. (2009) PLoS One 4, e7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dias A., Bouvier D., Crépin T., McCarthy A. A., Hart D. J., Baudin F., Cusack S., Ruigrok R. W. (2009) Nature 458, 914–918 [DOI] [PubMed] [Google Scholar]
- 38.Yuan P., Bartlam M., Lou Z., Chen S., Zhou J., He X., Lv Z., Ge R., Li X., Deng T., Fodor E., Rao Z., Liu Y. (2009) Nature 458, 909–913 [DOI] [PubMed] [Google Scholar]
- 39.Li M. L., Rao P., Krug R. M. (2001) EMBO J. 20, 2078–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klumpp K., Doan L., Roberts N. A., Handa B. (2000) J. Biol. Chem. 275, 6181–6188 [DOI] [PubMed] [Google Scholar]
- 41.Tomassini J., Selnick H., Davies M. E., Armstrong M. E., Baldwin J., Bourgeois M., Hastings J., Hazuda D., Lewis J., McClements W. (1994) Antimicrob. Agents Chemother. 38, 2827–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doan L., Handa B., Roberts N. A., Klumpp K. (1999) Biochemistry 38, 5612–5619 [DOI] [PubMed] [Google Scholar]
- 43.Poch O., Sauvaget I., Delarue M., Tordo N. (1989) EMBO J. 8, 3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller R., Poch O., Delarue M., Bishop D. H., Bouloy M. (1994) J. Gen. Virol. 75, 1345–1352 [DOI] [PubMed] [Google Scholar]
- 45.González S., Zürcher T., Ortín J. (1996) Nucleic Acids Res. 24, 4456–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez D. R., Donis R. O. (2001) J. Virol. 75, 8127–8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poole E. L., Medcalf L., Elton D., Digard P. (2007) FEBS Lett. 581, 5300–5306 [DOI] [PubMed] [Google Scholar]
- 48.Sugiyama K., Obayashi E., Kawaguchi A., Suzuki Y., Tame J. R., Nagata K., Park S. Y. (2009) EMBO J. 28, 1803–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blaas D., Patzelt E., Kuechler E. (1982) Nucleic Acids Res. 10, 4803–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulmanen I., Broni B. A., Krug R. M. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 7355–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Honda A., Mizumoto K., Ishihama A. (1999) Genes Cells 4, 475–485 [DOI] [PubMed] [Google Scholar]
- 52.Fechter P., Mingay L., Sharps J., Chambers A., Fodor E., Brownlee G. G. (2003) J. Biol. Chem. 278, 20381–20388 [DOI] [PubMed] [Google Scholar]
- 53.Mukaigawa J., Nayak D. P. (1991) J. Virol. 65, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Angelini A., Tosi T., Mas P., Acajjaoui S., Zanotti G., Terradot L., Hart D. J. (2009) FEBS J. 276, 816–824 [DOI] [PubMed] [Google Scholar]
- 55.Guilligay D., Tarendeau F., Resa-Infante P., Coloma R., Crepin T., Sehr P., Lewis J., Ruigrok R. W., Ortin J., Hart D. J., Cusack S. (2008) Nat. Struct. Mol. Biol. 15, 500–506 [DOI] [PubMed] [Google Scholar]
- 56.Tarendeau F., Boudet J., Guilligay D., Mas P. J., Bougault C. M., Boulo S., Baudin F., Ruigrok R. W., Daigle N., Ellenberg J., Cusack S., Simorre J. P., Hart D. J. (2007) Nat. Struct. Mol. Biol. 14, 229–233 [DOI] [PubMed] [Google Scholar]
- 57.Tarendeau F., Crepin T., Guilligay D., Ruigrok R. W., Cusack S., Hart D. J. (2008) PLoS Pathog. 4, e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazza C., Segref A., Mattaj I. W., Cusack S. (2002) EMBO J. 21, 5548–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marcotrigiano J., Gingras A. C., Sonenberg N., Burley S. K. (1997) Cell 89, 951–961 [DOI] [PubMed] [Google Scholar]
- 60.Parkes K. E., Ermert P., Fässler J., Ives J., Martin J. A., Merrett J. H., Obrecht D., Williams G., Klumpp K. (2003) J. Med. Chem. 46, 1153–1164 [DOI] [PubMed] [Google Scholar]
- 61.McHardy A. C., Adams B. (2009) PLoS Pathog. 5, e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bao Y., Bolotov P., Dernovoy D., Kiryutin B., Zaslavsky L., Tatusova T., Ostell J., Lipman D. (2008) J. Virol. 82, 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Enserink M. (2007) Science 315, 923. [DOI] [PubMed] [Google Scholar]
- 64.Chen G. W., Chang S. C., Mok C. K., Lo Y. L., Kung Y. N., Huang J. H., Shih Y. H., Wang J. Y., Chiang C., Chen C. J., Shih S. R. (2006) Emerg. Infect. Dis. 12, 1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finkelstein D. B., Mukatira S., Mehta P. K., Obenauer J. C., Su X., Webster R. G., Naeve C. W. (2007) J. Virol. 81, 10292–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naffakh N., Massin P., Escriou N., Crescenzo-Chaigne B., van der Werf S. (2000) J. Gen. Virol. 81, 1283–1291 [DOI] [PubMed] [Google Scholar]
- 67.Tamuri A. U., Dos Reis M., Hay A. J., Goldstein R. A. (2009) PLoS Comput. Biol. 5, e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naffakh N., Tomoiu A., Rameix-Welti M. A., van der Werf S. (2008) Annu. Rev. Microbiol. 62, 403–424 [DOI] [PubMed] [Google Scholar]
- 69.Miotto O., Heiny A., Tan T. W., August J. T., Brusic V. (2008) BMC Bioinformatics 9, Suppl. 1, S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Subbarao E. K., London W., Murphy B. R. (1993) J. Virol. 67, 1761–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Massin P., van der Werf S., Naffakh N. (2001) J. Virol. 75, 5398–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuzuhara T., Kise D., Yoshida H., Horita T., Murazaki Y., Nishimura A., Echigo N., Utsunomiya H., Tsuge H. (2009) J. Biol. Chem. 284, 6855–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mehle A., Doudna J. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21312–21316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herfst S., Chutinimitkul S., Ye J., de Wit E., Munster V. J., Schrauwen E. J., Bestebroer T. M., Jonges M., Meijer A., Koopmans M., Rimmelzwaan G. F., Osterhaus A. D., Perez D. R., Fouchier R. A. (2010) J. Virol. 84, 3752–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huarte M., Sanz-Ezquerro J. J., Roncal F., Ortín J., Nieto A. (2001) J. Virol. 75, 8597–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Engelhardt O. G., Smith M., Fodor E. (2005) J. Virol. 79, 5812–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Resa-Infante P., Jorba N., Zamarreño N., Fernández Y., Juárez S., Ortín J. (2008) PLoS One 3, e3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gabriel G., Dauber B., Wolff T., Planz O., Klenk H. D., Stech J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18590–18595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gabriel G., Herwig A., Klenk H. D. (2008) PLoS Pathog. 4, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steel J., Lowen A. C., Mubareka S., Palese P. (2009) PLoS Pathog. 5, e1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Jong M. D., Simmons C. P., Thanh T. T., Hien V. M., Smith G. J., Chau T. N., Hoang D. M., Chau N. V., Khanh T. H., Dong V. C., Qui P. T., Cam B. V., Ha do Q., Guan Y., Peiris J. S., Chinh N. T., Hien T. T., Farrar J. (2006) Nat. Med. 12, 1203–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jorba N., Juarez S., Torreira E., Gastaminza P., Zamarreño N., Albar J. P., Ortín J. (2008) Proteomics 8, 2077–2088 [DOI] [PubMed] [Google Scholar]
- 83.Mayer D., Molawi K., Martínez-Sobrido L., Ghanem A., Thomas S., Baginsky S., Grossmann J., García-Sastre A., Schwemmle M. (2007) J. Proteome Res. 6, 672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shapira S. D., Gat-Viks I., Shum B. O., Dricot A., de Grace M. M., Wu L., Gupta P. B., Hao T., Silver S. J., Root D. E., Hill D. E., Regev A., Hacohen N. (2009) Cell 139, 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hao L., Sakurai A., Watanabe T., Sorensen E., Nidom C. A., Newton M. A., Ahlquist P., Kawaoka Y. (2008) Nature 454, 890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.König R., Stertz S., Zhou Y., Inoue A., Hoffmann H. H., Bhattacharyya S., Alamares J. G., Tscherne D. M., Ortigoza M. B., Liang Y., Gao Q., Andrews S. E., Bandyopadhyay S., De Jesus P., Tu B. P., Pache L., Shih C., Orth A., Bonamy G., Miraglia L., Ideker T., García-Sastre A., Young J. A., Palese P., Shaw M. L., Chanda S. K. (2010) Nature 463, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karlas A., Machuy N., Shin Y., Pleissner K. P., Artarini A., Heuer D., Becker D., Khalil H., Ogilvie L. A., Hess S., Mäurer A. P., Müller E., Wolff T., Rudel T., Meyer T. F. (2010) Nature 463, 818–822 [DOI] [PubMed] [Google Scholar]
- 88.Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., Elledge S. J. (2009) Cell 139, 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mehle A., Doudna J. A. (2010) Viruses 2, 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]