Abstract
Alzheimer disease (AD) is a devastating neurodegenerative disease with no cure. The pathogenesis of AD is believed to be driven primarily by amyloid-β (Aβ), the principal component of senile plaques. Aβ is an ∼4-kDa peptide generated via cleavage of the amyloid-β precursor protein (APP). Curcumin is a compound in the widely used culinary spice, turmeric, which possesses potent and broad biological activities, including anti-inflammatory and antioxidant activities, chemopreventative effects, and effects on protein trafficking. Recent in vivo studies indicate that curcumin is able to reduce Aβ-related pathology in transgenic AD mouse models via unknown molecular mechanisms. Here, we investigated the effects of curcumin on Aβ levels and APP processing in various cell lines and mouse primary cortical neurons. We show for the first time that curcumin potently lowers Aβ levels by attenuating the maturation of APP in the secretory pathway. These data provide a mechanism of action for the ability of curcumin to attenuate amyloid-β pathology.
Keywords: Alzheimer Disease, Amyloid, Endoplasmic Reticulum (ER), Neurodegeneration, Protein Processing
Introduction
Alzheimer disease (AD)2 is a devastating neurodegenerative disorder and the primary cause of dementia in the elderly (1). Genetic studies of the disease have revealed a complex and strong genetic etiology. Four genes have been established to either cause early-onset familial AD with complete penetrance (amyloid-β (A4) protein precursor (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) or to increase susceptibility for late-onset AD with partial penetrance (APOE) (2–4). Despite its heterogeneous inheritance, the functional neuropathology of AD is represented commonly by disruption in neural circuits, such as loss of neurons and synapses primarily in the neocortex and hippocampus (5, 6). The pathophysiology of AD is characterized by two distinctive features: amyloid plaques comprised primarily of a small peptide named Aβ and neurofibrillary tangles composed of hyperphosphorylated Tau (2, 6, 7). Whereas Aβ42 and Aβ40 are the two primary Aβ species, Aβ42 is more prevalent than Aβ40 in amyloid plaques. Mounting genetic, biochemical, and molecular biological evidence suggests that the excessive accumulation of Aβ is the primary pathological event leading to AD (3, 6, 7). Aβ is produced by a sequential proteolytic cleavage of the type I transmembrane protein, amyloid-β (A4) precursor protein (APP) (8). The initial cleavage of APP can be mediated by α- or β-secretase (or BACE1). α-Secretase cleavage produces sAPPα and α C-terminal fragment (or C83); β-secretase cleavage produces sAPPβ and β C-terminal fragment (C99). C83 and C99 can be further cleaved by γ-secretase to produce P3 or Aβ (2, 7).
For more than a decade after the discovery of Aβ and the establishment of the Aβ hypothesis, one essential strategy for AD therapeutics has focused on modulating APP processing and decreasing Aβ levels (2, 9). Recently, considerable effort has been concentrated on identifying natural dietary supplements that can prevent, inhibit, or reverse Aβ accumulation or aggregation (10). Emerging evidence supports a use for curcumin in AD therapeutics. Curcumin (diferulomethane) is a yellow pigment in turmeric (or curcuma longa), the widely used spice, and a food additive used primarily in Indian culinary preparations (11). Curcumin is a low molecular weight molecule with broad and beneficial biological activities including potent antioxidant, anti-inflammatory, and chemo-preventative effects (12–15) with a favorable toxicity profile (13, 14). Epidemiological studies have suggested that curcumin contributes to the reported 4.4-fold reduced (age-adjusted) prevalence of AD in India compared with the United States (16). Both in vitro and in vivo studies have shown that curcumin can bind to amyloid and inhibit Aβ aggregation (14, 17), as well as fibril and oligomer formation (14). In vivo studies have shown that dietary curcumin can cross the blood-brain barrier and significantly decrease Aβ deposition and plaque burden in AD transgenic mice (14, 15, 18, 19), markedly inhibit Tau phosphorylation (20), and attenuate inflammation and reduce oxidative damage (14, 18), and reduce genomic instability events (21).
These findings support a beneficial role for the use of curcumin in treating AD. However, to date, the effects of curcumin on APP metabolism have not been elucidated. Here, we investigated the effects of curcumin on Aβ levels and APP processing in mouse primary cortical neurons and various cell lines. Curcumin potently lowered Aβ levels and hindered APP maturation. Curcumin also markedly attenuated the effects of brefeldin A (BFA), a specific Golgi-disrupting agent, on APP maturation and trafficking. Taken together, we have elucidated a novel curcumin-dependent mechanism for lowering Aβ levels via attenuation of APP maturation.
EXPERIMENTAL PROCEDURES
Cell Culture and Mouse Primary Cortical Neuron Culture
Human neuroglioma H4 cells that stably overexpress human APP751 (H4-APP751) cells have been described previously (22, 23), as has the rat neuroblastoma cell line (B104-APP751) stably overexpressing human APP751, and the Chinese hamster ovary cell line (CHO-APP751) stably overexpressing human APP751 (24, 25). These cell lines were cultured on regular tissue culture plates in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 200 μg/ml G418. Mouse primary cortical neurons were from BrainBits (E18) and were cultured on poly-d-lysine-coated plates in B27/neurobasal medium supplemented with 1× GlutaMAX (Invitrogen).
Chemicals and Antibodies
Curcumin was purchased from Sigma (catalogue no. C7727). The APP C-terminal antibody (targeting the last 19 amino acids of APP; 1:1000) was purchased from Sigma (catalogue no. A8717) and used to detect the full-length APP and APP C-terminal fragment. The anti-APP antibody (antibody 6E10) was purchased from Covance and utilized for detection of sAPPα (1:1000). The APLP2 antibody was a gift from Dr. W. Wasco (26), and the ADAM10 antibody was purchased from Sigma (catalogue no. A2726; 1:1000). The β-actin antibody (1:10,000) was purchased from Sigma. The HRP-conjugated secondary antibodies (anti-mouse and anti-rabbit; 1:10,000) were purchased from Pierce.
Aβ Measurement
Aβ measurement was following the manufacturer's suggested protocols and was described previously (27). In brief, Aβ40 and Aβ42 levels (pg/ml) were quantified using a sandwich enzyme-linked immunosorbent assay (ELISA) from Wako (catalogue no. 292-62301/294-62501 to detect Aβ40; and catalogue no. 298-62401/290-62601 to detect Aβ42). Aβ levels were further normalized to protein concentration from the same cell lysates. Normalized Aβ40 and Aβ42 levels of the curcumin treatment were compared and normalized to those levels of control treatment.
Cell Lysis and Protein Amount Quantification
Cells were lysed in M-PER (mammalian protein extraction reagent, Thermo Fisher Scientific) with 1× Halt protease inhibitor mixture (Thermo Fisher Scientific). The lysates were collected, centrifuged at 10,000 rpm using a microcentrifuge from Eppendorf (model 5417) for 20 min, pellets were discarded, and supernatants were transferred to a new Eppendorf tube. Total protein was quantified using the BCA protein assay kit (Pierce) (28, 29).
Western Blotting Analysis
Western blotting analysis was carried out by the method described previously (27, 28). Briefly, after centrifugation and protein concentration measurement, an equal amount of protein was applied to electrophoresis, followed by membrane transfer, antibody incubation, and signal development. β-Actin was used as an internal control. We used the VersaDoc imaging system (Bio-Rad) to develop the blots and the software Quantity One (Bio-Rad) to quantify the proteins of interest, following the protocols described previously (27, 28).
Cell Surface Biotinylation
Cell surface biotinylation was carried out using a protocol reported previously(27). Briefly, stable H4-APP751 and CHO-APP751 cells were preincubated in cold Mg2+/Ca2+ containing PBS for 20 min and then incubated with 0.5 mg/ml sulfo-NHS-LC-biotin (Pierce) for 30 min with gentle rocking at room temperature. Excess biotin was quenched with 0.1 m glycine for 20 min. Cells were then lysed in M-PER lysis buffer and immunoprecipitated with streptavidin beads (Pierce) overnight at 4 °C. Samples were boiled at 95 °C for 5 min then applied to Western blotting analysis.
alamarBlue Analysis
The alamarBlue assay was a noninvasive way to assess cell viability and proliferation rate and has been reported previously (30). The alamarBlue agent was purchased from Invitrogen, and the assay was performed according to the manufacturer's recommended protocol. Briefly, the alamarBlue agent was added to culture medium at a final concentration of 10% (v/v), medium was collected after 6 h of incubation, and the fluorescence intensity was read on the Criterion Analyst AD high throughput fluorescence detection system (Molecular Devices) using a 550-nm excitation wavelength and a 590-nm emission wavelength. The fluorescence readouts from the treatments with curcumin of different concentrations were compared with the readout from the treatment of control (0 μm curcumin; dimethyl sulfoxide).
Data Analysis
β-Actin was used in the Western blotting analysis to account for any differences in loading. The levels of various proteins, e.g. full-length APP, mature and immature APP, C99, and C83 were normalized to the β-actin values from the same lane or sample. sAPPα levels were normalized to the cell lysates protein concentration and then compared with full-length APP levels from each sample. All results were demonstrated as means ± S.E. from at least three independent experiments. We used the two-tailed Student's t test, as appropriate, to reveal the differences between the experimental groups. The Bonferroni correction analysis was used to correct for multiple comparisons within a single experiment. p values < 0.05 were considered statistically significant.
RESULTS
Curcumin Decreases Aβ Levels and Attenuates APP Maturation in Mouse Primary Cortical Neurons
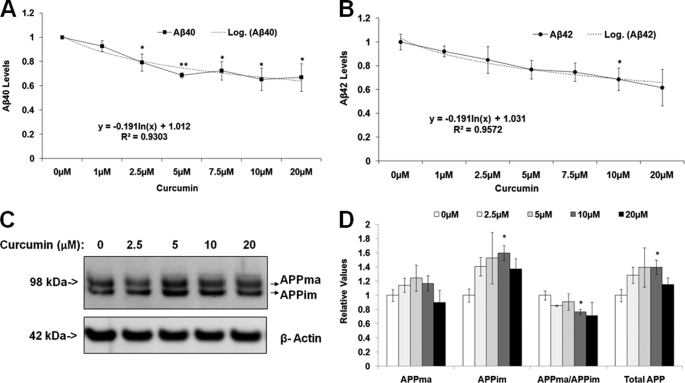
We first tested whether curcumin affects Aβ levels and/or APP processing and metabolism in mouse primary neuronal cells. Mouse primary neurons (E18) were prepared on poly-d-lysine-coated plates and treated with curcumin (0, 1, 2.5, 7.5, 10, and 20 μm). Cells were harvested after 24 h of treatment. Conditioned medium was applied to ELISA analysis to measure the Aβ40 and Aβ42 levels, which were normalized to cell number. Curcumin treatment decreased both Aβ40 and Aβ42 levels in a dose-dependent manner (Fig. 1, A and B). For example, 20 μm curcumin decreased Aβ40 and Aβ42 levels by 38.4% (p < 0.01), and 43.9% (p < 0.05), respectively, compared with the control treatment (0 μm) (Fig. 1, A and B).
FIGURE 1.
Curcumin significantly decreases Aβ levels and the ratio of APPma:APPim in mouse primary cortical neurons in a dose-dependent manner. A and B, curcumin significantly decreases both Aβ40 and Aβ42 levels. Mouse primary cortical neurons (E18) were treated with different concentrations of curcumin and harvested after 24 h. Conditioned medium was used in ELISA analysis to detect the Aβ40 and Aβ42 levels, which were normalized to cell numbers. C and D, the curcumin treatment altered APP levels and decreased the ratio of APPma:APPim. In the Western blotting analysis, cell lysates were probed with the APP8717 antibody to reveal APP. β-Actin was used as the loading control. C, a representative gel showing full-length APP and β-actin. D, densitometry of C (n = 3 for each treatment group). Mean ± S.E. *, p < 0.05; **, p < 0.01.
Next, we studied the effects of curcumin on APP metabolism and processing. Cell lysates from the previous experiment were subjected to quantitative Western blotting analysis with antibody, APP8717, targeted at the C terminus of APP. β-Actin was used to normalize loading variation between gel lanes. We assessed the effects of curcumin treatment on levels of total APP (APPma and APPim), mature and immature APP, as well as the APPma:APPim ratio. Curcumin did not significantly alter the levels of mature APP but increased levels of immature APP and total APP in comparison to control. For example, 10 μm curcumin treatment increased immature APP levels by 59.7% and increased total APP levels by 41.6% versus control (p < 0.05; Fig. 1, C and D). In addition, curcumin decreased the ratio of mature APP to immature APP compared with control. For example, 10 μm curcumin treatment decreased the ratio by 23.3% (p < 0.05) (Fig. 1, C and D). Overall, these data suggest that curcumin decreases Aβ levels by retarding APP maturation.
Curcumin Significantly Alters APP Maturation and Processing in Other Cell Types
Next, we asked whether curcumin impairs APP maturation in other cell types. For this purpose, we used stably transfected rat neuroblastoma B104-APP751 cells, which were treated with different concentrations of curcumin for 24 h and then collected for Western blotting analysis with the antibody APP8717. In addition, the media was probed with antibody 6E10 to measure sAPPα levels. Curcumin significantly increased levels of mature APP at doses of 5 and 10 μm (by 60.0%; p < 0.05, compared with control) but revealed a trend toward decreased levels at 15 and 20 μm compared with control (Fig. 2, A and C). The level of immature APP was increased significantly by curcumin treatment (e.g. increased by 279.1% at 20 μm; p < 0.01). Additionally, curcumin treatment increased markedly the ratio of APPma/APPim at lower concentrations, but significantly decreased the ratio with increasing concentrations (Fig. 2, A and C). Specifically, the doses of 5 and 10 μm curcumin decreased the ratio by 56.3% (p < 0.01) and 82.1% (p < 0.01), respectively, compared with the control, whereas 15 and 20 μm curcumin markedly decreased the APPma/APPim ratio by 55.8% (p < 0.01) and 84.4% (p < 0.01), respectively, compared with the control. Finally, curcumin treatment significantly increased total APP levels in comparison to control treatment (e.g. increased by 137.7% at 20 μm; p < 0.01) (Fig. 2C). In contrast to what was observed in the primary neurons, lower doses of curcumin, e.g. 5 and 10 μm, increased APP maturation. However, at higher doses, e.g. 15 and 20 μm, these data recapitulate the data from the primary neurons showing that curcumin treatment retards APP maturation in rat neuroblastoma B104-APP751 cells.
FIGURE 2.
Curcumin significantly modulates APP processing in B104-APP751 cells in a dose-dependent manner. Stable rat neuroblastoma B104-APP751 cells were treated with different concentrations of curcumin for 24 h and were collected for Western blotting analysis. Cell lysates were probed with the APP8717 antibody to reveal APP. β-Actin was used as the loading control. The cell medium was probed with 6E10 to reveal sAPPα. A and B, a representative gel showing full-length APP, APP-C83, and sAPPα. C, densitometry of A. Curcumin significantly increased the levels of immature APP and total APP. It also markedly increased and then decreased the level of mature APP, as well as the ratio of APPma:APPim with increasing curcumin concentration. D, curcumin treatment significantly decreased the ratio of C83/APPtotal. E, curcumin treatment significantly decreased the ratio of sAPPα/APPtotal compared with control (n = 3 for each treatment group). Mean ± S.E.; *, p < 0.05; **, p < 0.01.
In this same experiment, we also assessed the effects of curcumin on APP processing by measuring levels of the APP proteolytic products, sAPPα and C83, the products of α-secretase cleavage of APP. We observed a trend toward decreased levels of C83 and sAPPα (Fig. 2, A and C–E) with curcumin treatment. We also observed a significant decrease in the ratio of C83/total APP as well as in the ratio of sAPPα/total APP, compared with the control treatment (Fig. 2, D–E), following curcumin treatment. For example, 20 μm curcumin decreased the ratio of C83/APPtotal by 74.3% (p < 0.01) (Fig. 2D) and decreased the ratio of sAPPα/total APP by 57.1% in comparison to control (p < 0.05) (Fig. 2E). Collectively, these data show that curcumin treatment attenuates α-secretase processing of APP in rat neuroblastoma B104-APP751 cells, consistent with attenuation of APP maturation.
Next, we tested the effects of curcumin on APP metabolism and processing in human H4 neuroglioma cells stably transfected with APP751 (H4-APP751) and in Chinese hamster ovary cells stably transfected with APP751 (CHO-APP751). As observed with the rat neuroblastoma B104-APP751 cells, curcumin treatment led to an increase (trend) and then a decrease in the level of mature APP with increasing concentrations (p > 0.05) (Fig. 3A) in H4-APP751 cells. Curcumin treatment also significantly increased levels of immature APP compared with the control treatment (0 μm) (Fig. 3B). 20 μm curcumin markedly increased immature APP levels by 79.1% (p < 0.01; versus control) (Fig. 3B). Additionally, curcumin significantly decreased the ratio of mature APP to immature APP with increasing doses, compared with control. 20 μm curcumin markedly decreased the ratio of APPma:APPim by 42.2% (p < 0.01; compared with control). Curcumin treatment significantly increased levels of total APP (APPma and APPim) with increased concentration. 20 μm curcumin markedly increased total APP levels by 44.3% (p < 0.01; compared with control) (Fig. 3B). Curcumin treatment significantly increased levels of mature APP and immature APP, as well as total APP compared with the control in CHO-APP751 cells (Fig. 3, C–D). 20 μm curcumin elevated the level of immature APP by 65.0% (p < 0.05) and increased the levels of total APP by 59.2% (p < 0.05). Curcumin treatment also revealed a trend toward decrease in the ratio of APPma:APPim compared with control (p > 0.05) (Fig. 3D). We also measured the Aβ levels from curcumin treated samples in these three cell models. We found that curcumin significantly decreased both Aβ40 and Aβ42 levels in all cell models (supplemental Fig. 1, A–C). Collectively, these data recapitulated the effects of curcumin treatment on APP metabolism and processing in several different cell types.
FIGURE 3.
Curcumin treatment alters APP metabolism in H4-APP751 cells and CHO-APP751 cells in a dose-dependent manner. Various cell models were treated with different concentrations of curcumin for 24 h and collected for Western blotting analysis. Cell lysates were probed with the APP8717 antibody to reveal APP. β-Actin was used as the loading control. A and B, the effects of curcumin treatment on H4-APP751 cells. Curcumin treatment had a trend to increase the level of mature APP, significantly increased the levels of immature APP and total APP, and markedly decreased the ratio of APPma/APPim. C and D, the effects of curcumin on CHO-APP751 cells. Curcumin significantly increased the levels of mature APP, immature APP, and total APP and had a trend to decrease the ratio of APPma:APPim with increasing concentration (n = 3 for each treatment group). Mean ± S.E.; *, p < 0.05; **, p < 0.01.
Curcumin Alters the Turnover of Mature and Immature APP
In the next step, we assessed the effects of curcumin on the rate of APP turnover using cycloheximide treatment. H4-APP751 cells were treated with 20 μm curcumin for 24 h and then treated with 40 μg/ml cycloheximide for various time intervals (0, 0.5, 1.5, and 3 h). Cell lysates were collected and utilized for quantitative Western blotting analysis. Curcumin markedly decreased the half-life of mature APP from 2.03 to 1.14 h (changing by 43.4%), increased the half-life of immature APP from 1.86 to 2.36 h (changing by 26.0%), and shortened total APP half-life from 1.94 to 1.87 h (Fig. 4, A–D). Curcumin also markedly decreased the ratio of APPma/APPim (e.g. decreased by 67.6% at 0.5 h; p < 0.05) (Fig. 4E). Thus, curcumin increased the stability of immature APP while decreasing the stability of mature APP, consistent with attenuated APP maturation.
FIGURE 4.
Curcumin treatment alters the turnover rate of both mature and immature APP in H4-APP751 cells. H4-APP751 cells were treated with 20 μm curcumin for 24 h and then treated with 40 μg/ml cycloheximide for different time (0, 0.5, 1.5, and 3 h). Cell lysates were collected and utilized for Western blotting analysis. Cell lysates were probed with the APP8717 antibody to reveal APP. β-Actin was used as the loading control. A, a representative gel revealing the cycloheximide treatment of different time points with or without curcumin treatment. B and C, quantitative Western blot analysis for mature (B) and immature (C) APP levels. Immature APP levels in curcumin treatment were significantly higher at 0.5, 1.5, and 3 h, compared with the corresponding control treatment. D, quantitative Western blot analysis for total APP levels. E, the ratio of APPma:APPim was decreased in curcumin treatment compared with control at cycloheximide treatment of 0, 0.5, 1.5, and 3 h (n = 3 for each treatment group). Mean ± S.E. *, p < 0.05; **, p < 0.01.
Curcumin Increases the Level of Plasma Membrane APP
Next, we tested the effects of curcumin on plasma membrane levels of APP. Stable H4-APP751 and CHO-APP751 cells were treated with 20 μm curcumin for 24 h and then subjected to biotinylation analysis to assess the levels of cell surface APP. Cell lysates were collected and utilized for Western blotting analysis. Curcumin significantly increased the levels of cell surface APP by 28.1% in H4-APP751 cells (p < 0.05) (Fig. 5, A and B) and markedly increased the level of cell surface APP by 133.7% in CHO-APP751 cells (p < 0.01) (Fig. 5, C and D), in comparison with control. We also studied whether curcumin may alter plasma membrane levels of APLP2, an APP homologue protein. We did not find any differences in the levels of APLP2 (amyloid precursor-like protein 2) between cells treated with curcumin versus control samples.3 In combination with prior observations of attenuated maturation of APP following treatment with curcumin, these data suggest that curcumin treatment may also lead to decreased endocytosis of APP, consistent with decreased Aβ levels.
FIGURE 5.
Curcumin treatment significantly increases cell surface APP levels in both H4-APP751 and CHO-APP751 cells. Cells were treated with 20 μm curcumin for 24 h and then subjected to biotinylation analysis to assess cell surface APP. Cell lysates were collected and utilized for Western blotting analysis. A and B, curcumin treatment markedly increased the level of cell surface APP in H4-APP751 cells compared with control treatment. C and D, curcumin treatment significantly increased the level of cell surface APP in CHO-APP751 cells compared with control (n = 3 for each treatment group). Mean ± S.E. *, p < 0.05; **, p < 0.01.
Curcumin Decreases the Levels of Intermediate APP Induced by Brefeldin A
Next, we investigated the mechanism by which curcumin may increase immature APP levels. Previous reports have shown that curcumin is a sarcoplasmic/endoplasmic reticulum calcium-ATPase inhibitor (31). It potentially may affect the functions of ER lumen chaperones, which are calcium binding proteins (32). We hypothesized that curcumin may affect APP metabolism at the level of the endoplasmic reticulum. BFA is an agent that disassembles the Golgi complex and redistributes proteins into the ER. BFA treatment has been shown to induce the buildup of an intermediate APP isoform (33). Thus, if curcumin decreased levels of intermediate APP induced by BFA, it would strongly suggest that the curcumin acts before the Golgi complex, likely at the ER.
First, H4-APP751 cells were treated with 5 μg/ml BFA for 5 or 30 min and then harvested and subjected to Western blotting analysis. As expected, 5 μg/ml and 10 μg/ml BFA for 30 min induced the generation of intermediate APP (Fig. 6A). Then, H4-APP751 cells were treated with 20 μm curcumin ± 5 μg/ml BFA for 0.5 or 3 h. BFA markedly induced the generation of the intermediate APP at both 0.5 and 3 h, compared with control (Fig. 6, B–C). BFA-induced generation of intermediate APP was attenuated significantly in the presence of curcumin (at both 0.5 and 3 h; Fig. 6, B and C). These data suggest that curcumin affects APP metabolism in the secretory pathway at the level of the endoplasmic reticulum.
FIGURE 6.
Curcumin treatment affects APP metabolism at the endoplasmic reticulum. A, BFA disrupts APP maturation process at the Golgi complex and induces the generation of the intermediate APP in H4-APP751 cells. H4-APP751 cells were treated with or without 5 and 10 μg/ml BFA for 5 or 30 min. Cell lysates were collected and prepared for Western blot analysis. B and C, curcumin treatment markedly decreased the level of intermediate APP in the presence of BFA. H4-APP751 cells were treated with 5 μg/ml BFA in the presence or absence of 20 μm curcumin for 5 or 30 min (n = 3 for each treatment group). Cell lysates were collected and prepared for Western blot analysis, as described under “Experimental Procedures.”
Curcumin Does Not Alter APLP2 Levels
To assess whether the effects of curcumin are specific to APP, we next measured the levels of APP homologue APLP2 following curcumin treatment. Stable B104-APP751 cells and H4-APP751 cells were treated with different concentrations of curcumin for 24 h, and cell lysates were collected and utilized for Western blotting analysis. APLP2 antibody has been reported previously(26, 27). β-Actin was used as the loading control. Curcumin treatment did not alter the levels of full length APLP2 in either B104-APP751 cells (p > 0.05) (Fig. 7, A and B) or H4-APP751 cells (p > 0.05) and did not alter levels of mature versus immature forms of APLP2 (Fig. 7, C and D). In addition, we found that curcumin treatment does not alter the levels of ADAM10 in mouse primary cortical neurons (supplemental Fig. 2). Thus, these data suggest that the effects of curcumin on APP levels and maturation are specific.
FIGURE 7.
Curcumin treatment does not alter APLP2 protein levels. B104-APP751 and H4-APP751 cells were treated with different concentrations of curcumin for 24 h, and cell lysates were collected and utilized for Western blot analysis. Cell lysates were probed with the APLP2 antibody to reveal APLP2. β-Actin was used as the loading control. A and B, the effects of curcumin treatment on APLP2 in B104-APP751 cells. Curcumin treatment did not alter full-length APLP2 levels and did not alter levels of mature versus immature forms of APLP2. C and D, the effects of curcumin treatment on APLP2 in H4-APP751 cells. Curcumin treatment did not alter full-length APLP2 levels (n = 3 for each treatment group). Mean ± S.E. p > 0.05.
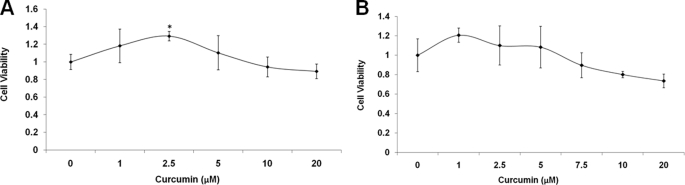
Curcumin Does Not Decrease Cell Viability
Finally, we confirmed that the treatment of curcumin did not decrease cell viability, which could, otherwise, alter Aβ levels via apoptotic pathways (34). H4-APP751 cells and mouse primary cortical neurons were treated with varying concentrations of curcumin for 24 h and were then treated with the alamarBlue for 6 h. Curcumin did not decrease cell viability in either cell type compared with control (0 μm) (Fig. 8, A and B). In fact, 2.5 μm curcumin significantly increased cell viability by 29.3% in H4-APP751 cells compared with control (0 μm) (p < 0.05; Fig. 8A).
FIGURE 8.
Curcumin treatment does not decrease cell viability. A and B, H4-APP751 cells (A) or mouse primary cortical neurons (B) were treated with different concentrations of curcumin for 24 h and then subjected to alamarBlue analysis as described under “Experimental Procedures.” Curcumin treatment did not decrease cell viability in either cell type, compared with control (0 μm). The dose of 2.5 μm curcumin significantly increased cell viability by 29.3% in H4-APP751 cells compared with control (A) (p < 0.05) (n = 3 in each treatment group). Mean ± S.E. *, p < 0.05.
DISCUSSION
We have described a possible cellular mechanism underlying in vivo observations of decreased Aβ deposition and plaque burden in AD transgenic mice following treatment of curcumin (14, 15, 18, 19). We have shown that curcumin decreases Aβ levels by retarding APP maturation and possibly, impairing endocytosis from the plasma membrane. Immature APP (or N-APP) is N-glycosylated in the ER, and a fraction of these molecules exit the ER and undergo O-glycosylation in the Golgi complex to become mature APP (or N,O-APP) (35). Mature APP is then sorted onto the plasma membrane after which it can undergo endocytosis via clathrin-coated pits (36). Disruption of APP trafficking and sorting has been proposed to underlie the pathogenesis of AD (33, 35, 37). Our data show that curcumin significantly alters the ratio of APPma/APPim and markedly decreases the level of intermediate APP induced by BFA, an agent that disrupts the Golgi complex. Additionally, curcumin has been shown to be an inhibitor of the sarcoplasmic/endoplasmic reticulum calcium ATPase pump (31, 32). Collectively, these findings suggest that curcumin may affect APP metabolism at the level of the ER. Though the precise underlying mechanism requires further elucidation, curcumin may delay the exit of immature APP from the ER, thereby increasing the stability of immature APP at the ER. Curcumin also may affect the endocytosis of APP from the cell surface. The cumulative effect would be a significant decrease in both Aβ40 and Aβ42 levels.
Curcumin has been reported to modulate trafficking and maturation of other proteins, MLC1 (megalencephalic leukoencephalopathy with subcortical cysts) (38) and ΔF508 cystic fibrosis transmembrane conductance regulator (31). MLC1 encodes a plasma membrane protein, MLC1, which is responsible for a severe autosomal recessive clinical disorder, MLC, characterized by macrocephaly, deterioration in motor functions, cerebellar ataxia, and mental decline. Levels of MLC1 have been shown to be significantly decreased in cells expressing the mutant form of the gene.
Curcumin has been shown to target dozens of proteins (39), and it may act on different biological pathways with distinct mechanisms at different dosages. It has been shown that curcumin has a dose-dependent effect on Aβ levels and inflammatory reactions in vivo (14, 15). Treatment with curcumin in vivo for 5–6 months potently attenuated proteins oxidization and interleukin-1β generation in the brain (15, 16) and reduced Aβ levels and plaque burden (14, 15). Here, we treated mouse primary cortical neurons with different concentrations of curcumin (1–20 μm) for 24 h and found that both Aβ40 and Aβ42 levels significantly decreased compared with control.
Collectively, our data suggest that the cellular mechanism by which curcumin decreases Aβ levels is via the modulation of APP levels in the secretory and, possibly, endocytic pathways. Curcumin treatment significantly increased the retention of immature APP in the ER and simultaneously attenuated APP endocytosis from the plasma membrane. Collectively, our data, together with the previously published in vivo data, suggest that curcumin and its derivatives may prove useful in the search for small molecule pharmacological agents for the effective treatment and prevention of AD-related β-amyloid pathology.
Supplementary Material
This work was supported by the Cure Alzheimer's Fund, the Funds for Medical Discovery from Massachusetts General Hospital, and a Neurodegenerative Disease Pilot Study Grant from the Harvard NeuroDiscovery Center and Massachusetts Alzheimer's Disease Research Center.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
C. Zhang, A. Browne, D. Child, and R. E. Tanzi, unpublished data.
- AD
- Alzheimer disease
- APP
- amyloid-β precursor protein
- Aβ
- amyloid-β peptide
- APPma
- mature APP
- APPim
- immature APP
- sAPPα
- α-secretase-soluble amyloid-β precursor protein.
REFERENCES
- 1.Cummings J. L. (2004) N. Engl. J. Med. 351, 56–67 [DOI] [PubMed] [Google Scholar]
- 2.Bertram L., Tanzi R. E. (2008) Nat. Rev. Neurosci. 9, 768–778 [DOI] [PubMed] [Google Scholar]
- 3.Tanzi R. E., Bertram L. (2005) Cell 120, 545–555 [DOI] [PubMed] [Google Scholar]
- 4.Tanzi R. E., Gusella J. F., Watkins P. C., Bruns G. A., St George-Hyslop P., Van Keuren M. L., Patterson D., Pagan S., Kurnit D. M., Neve R. L. (1987) Science 235, 880–884 [DOI] [PubMed] [Google Scholar]
- 5.Selkoe D. J. (2002) Science 298, 789–791 [DOI] [PubMed] [Google Scholar]
- 6.Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 7.Gandy S. (2005) J. Clin. Invest. 115, 1121–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C., Saunders A. J. (2007) Discov. Med. 7, 113–117 [PubMed] [Google Scholar]
- 9.Selkoe D. J. (2001) Physiol. Rev. 81, 741–766 [DOI] [PubMed] [Google Scholar]
- 10.Calabrese V., Scapagnini G., Colombrita C., Ravagna A., Pennisi G., Giuffrida Stella A. M., Galli F., Butterfield D. A. (2003) Amino Acids 25, 437–444 [DOI] [PubMed] [Google Scholar]
- 11.Begum A. N., Jones M. R., Lim G. P., Morihara T., Kim P., Heath D. D., Rock C. L., Pruitt M. A., Yang F., Hudspeth B., Hu S., Faull K. F., Teter B., Cole G. M., Frautschy S. A. (2008) J. Pharmacol. Exp. Ther. 326, 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimmyo Y., Kihara T., Akaike A., Niidome T., Sugimoto H. (2008) Neuroreport 19, 1329–1333 [DOI] [PubMed] [Google Scholar]
- 13.Kelloff G. J., Boone C. W., Crowell J. A., Nayfield S. G., Hawk E. T., Steele V. E., Lubet R. A., Sigman C. C. (1997) Prog. Clin. Biol. Res. 396, 159–183 [PubMed] [Google Scholar]
- 14.Yang F., Lim G. P., Begum A. N., Ubeda O. J., Simmons M. R., Ambegaokar S. S., Chen P. P., Kayed R., Glabe C. G., Frautschy S. A., Cole G. M. (2005) J. Biol. Chem. 280, 5892–5901 [DOI] [PubMed] [Google Scholar]
- 15.Lim G. P., Chu T., Yang F., Beech W., Frautschy S. A., Cole G. M. (2001) J. Neurosci. 21, 8370–8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganguli M., Chandra V., Kamboh M. I., Johnston J. M., Dodge H. H., Thelma B. K., Juyal R. C., Pandav R., Belle S. H., DeKosky S. T. (2000) Arch. Neurol. 57, 824–830 [DOI] [PubMed] [Google Scholar]
- 17.Hong H. S., Rana S., Barrigan L., Shi A., Zhang Y., Zhou F., Jin L. W., Hua D. H. (2009) J. Neurochem. 108, 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y. J., Thomas P., Zhong J. H., Bi F. F., Kosaraju S., Pollard A., Fenech M., Zhou X. F. (2009) Neurotox. Res. 15, 3–14 [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Alloza M., Borrelli L. A., Rozkalne A., Hyman B. T., Bacskai B. J. (2007) J. Neurochem. 102, 1095–1104 [DOI] [PubMed] [Google Scholar]
- 20.Ma Q. L., Yang F., Rosario E. R., Ubeda O. J., Beech W., Gant D. J., Chen P. P., Hudspeth B., Chen C., Zhao Y., Vinters H. V., Frautschy S. A., Cole G. M. (2009) J. Neurosci. 29, 9078–9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas P., Wang Y. J., Zhong J. H., Kosaraju S., O'Callaghan N. J., Zhou X. F., Fenech M. (2009) Mutat. Res. 661, 25–34 [DOI] [PubMed] [Google Scholar]
- 22.Xie Z., Dong Y., Maeda U., Xia W., Tanzi R. E. (2007) J. Biol. Chem. 282, 4318–4325 [DOI] [PubMed] [Google Scholar]
- 23.Xie Z., Romano D. M., Tanzi R. E. (2005) J. Biol. Chem. 280, 15413–15421 [DOI] [PubMed] [Google Scholar]
- 24.Huttunen H. J., Guénette S. Y., Peach C., Greco C., Xia W., Kim D. Y., Barren C., Tanzi R. E., Kovacs D. M. (2007) J. Biol. Chem. 282, 28285–28295 [DOI] [PubMed] [Google Scholar]
- 25.Zhang C., Browne A., Kim D. Y., Tanzi R. E.Curr. Alzheimer Res. 7, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh D. M., Fadeeva J. V., LaVoie M. J., Paliga K., Eggert S., Kimberly W. T., Wasco W., Selkoe D. J. (2003) Biochemistry 42, 6664–6673 [DOI] [PubMed] [Google Scholar]
- 27.Hiltunen M., Lu A., Thomas A. V., Romano D. M., Kim M., Jones P. B., Xie Z., Kounnas M. Z., Wagner S. L., Berezovska O., Hyman B. T., Tesco G., Bertram L., Tanzi R. E. (2006) J. Biol. Chem. 281, 32240–32253 [DOI] [PubMed] [Google Scholar]
- 28.Zhang C., Khandelwal P. J., Chakraborty R., Cuellar T. L., Sarangi S., Patel S. A., Cosentino C. P., O'Connor M., Lee J. C., Tanzi R. E., Saunders A. J. (2007) Mol. Neurodegener. 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C., Browne A., Child D., Divito J. R., Stevenson J. A., Tanzi R. E.J. Biol. Chem. 285, 8515–8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antczak C., Shum D., Radu C., Seshan V. E., Djaballah H. (2009) Comb. Chem. High Throughput Screen. 12, 258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egan M. E., Pearson M., Weiner S. A., Rajendran V., Rubin D., Glöckner-Pagel J., Canny S., Du K., Lukacs G. L., Caplan M. J. (2004) Science 304, 600–602 [DOI] [PubMed] [Google Scholar]
- 32.Egan M. E., Glöckner-Pagel J., Ambrose C., Cahill P. A., Pappoe L., Balamuth N., Cho E., Canny S., Wagner C. A., Geibel J., Caplan M. J. (2002) Nat. Med. 8, 485–492 [DOI] [PubMed] [Google Scholar]
- 33.Haass C., Lemere C. A., Capell A., Citron M., Seubert P., Schenk D., Lannfelt L., Selkoe D. J. (1995) Nature Medicine 1, 1291–1296 [DOI] [PubMed] [Google Scholar]
- 34.Tesco G., Koh Y. H., Tanzi R. E. (2003) J. Biol. Chem. 278, 46074–46080 [DOI] [PubMed] [Google Scholar]
- 35.Xia W., Zhang J., Kholodenko D., Citron M., Podlisny M. B., Teplow D. B., Haass C., Seubert P., Koo E. H., Selkoe D. J. (1997) J. Biol. Chem. 272, 7977–7982 [DOI] [PubMed] [Google Scholar]
- 36.Small S. A., Gandy S. (2006) Neuron 52, 15–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandy S., Zhang Y. W., Ikin A., Schmidt S. D., Bogush A., Levy E., Sheffield R., Nixon R. A., Liao F. F., Mathews P. M., Xu H., Ehrlich M. E. (2007) J. Neurochem. 102, 619–626 [DOI] [PubMed] [Google Scholar]
- 38.Teijido O., Martínez A., Pusch M., Zorzano A., Soriano E., Del Río J. A., Palacín M., Estévez R. (2004) Human Molecular Genetics 13, 2581–2594 [DOI] [PubMed] [Google Scholar]
- 39.Aggarwal B. B., Sundaram C., Malani N., Ichikawa H. (2007) Adv. Exp. Med. Biol. 595, 1–75 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.