Abstract
Reconstitution of eukaryotic Okazaki fragment processing implicates both one- and two-nuclease pathways for processing flap intermediates. In most cases, FEN1 (flap endonuclease 1) is able to efficiently cleave short flaps as they form. However, flaps escaping cleavage bind replication protein A (RPA) inhibiting FEN1. The flaps must then be cleaved by Dna2 nuclease/helicase before FEN1 can act. Pif1 helicase aids creation of long flaps. The pathways were considered connected only in that the products of Dna2 cleavage are substrates for FEN1. However, results presented here show that Dna2, Pif1, and RPA, the unique proteins of the two-nuclease pathway from Saccharomyces cerevisiae, all stimulate FEN1 acting in the one-nuclease pathway. Stimulation is observed on RNA flaps representing the initial displacement and on short DNA flaps, subsequently displaced. Neither the RNA nor the short DNA flaps can bind the two-nuclease pathway proteins. Instead, direct interactions between FEN1 and the two-nuclease pathway proteins have been detected. These results suggest that the proteins are either part of a complex or interact successively with FEN1 because the level of stimulation would be similar either way. Proteins bound to FEN1 could be tethered to the flap base by the interaction of FEN1 with PCNA, potentially improving their availability when flaps become long. These findings also support a model in which cleavage by FEN1 alone is the preferred pathway, with the first opportunity to complete cleavage, and is stimulated by components of the backup pathway.
Keywords: DNA, DNA-binding Protein, DNA Enzymes, DNA Helicase, DNA Replication, One-nuclease Pathway, Two-nuclease Pathway
Introduction
Double-stranded DNA in eukaryotes is synthesized through elongation of leading and lagging strands copied using the original DNA as a template. The leading strand DNA is made in a continuous manner by DNA polymerase ϵ, which adds nucleotides in the same direction of the opening replication fork (1). The lagging strand, which grows in the opposite direction, must be made discontinuously, through the creation of short (∼150-nt) oligonucleotides known as Okazaki fragments (2). This process begins with DNA polymerase α/primase (pol α),2 which makes a mixed primer initiating with 10–12 nt of RNA, to which 10–20 nt of DNA is added (3). The primer is then lengthened by DNA polymerase δ (pol δ). After adding deoxynucleotides to make the bulk of the fragment, pol δ encounters the adjacent downstream Okazaki fragment. At this point, pol δ will displace the downstream fragment into a flap while continuing to synthesize DNA, a process called strand displacement synthesis (4, 5). These flaps are cleaved by specific endonucleases, creating a nick that will then be sealed by DNA ligase I. The ligation event finishes the production of the complete DNA strand.
Two pathways have been suggested for flap processing. The first pathway, proposed by Burgers and co-workers (6–8) based on reconstitutions in vitro, is referred to as the FEN1-only pathway, or “one-nuclease pathway.” As the name suggests, in this pathway, the only nuclease involved in flap cleavage is FEN1 (flap endonuclease 1). FEN1 is an endonuclease with a preferred substrate having a 5′-flap with a 1-nucleotide-long 3′-flap. After pol δ displaces a short flap of only a few nucleotides, FEN1 is able to bind to the 5′-end of the flap and track to its base, where it proceeds to cleave off the entire flap, leaving behind only a nick (4, 9, 10).
Reconstitutions of Okazaki fragment processing in vitro suggest that FEN1 cleaves most flaps when they are only a few nucleotides long. However, in these reactions, a small fraction of flaps escape FEN1 cleavage and achieve greater lengths (11, 12). Once this occurs, the flaps can be bound by replication protein A (RPA), the eukaryotic single-stranded binding protein. RPA is able to stably bind flaps greater than 22 nt in length, and doing so has been shown to inhibit FEN1 cleavage (13, 14).
Bae and Seo (15) proposed a second pathway through which they suggested that flaps are cleaved in vivo. In the two-nuclease pathway, cleavage by the endonuclease Dna2 is promoted on RPA-coated flaps (14). Dna2 exhibits both a directional 5′–3′ endonuclease activity and a 5′–3′ helicase activity that is specific for forked substrates (15, 16). Like FEN1, Dna2 enters flaps from the 5′-end. However, whereas FEN1 cleaves a single time, Dna2 cleaves multiple times while tracking on the flap. Also, unlike FEN1, Dna2 is not able to cleave the flap entirely but leaves a terminal flap of 5–7 nt in length (17). RPA is no longer able to bind these short flaps, allowing FEN1 to freely bind and cleave the remainder of the flap. Although the two-nuclease pathway was originally proposed to predominate, later reconstitutions suggest that its role is to complete processing of the small fraction of flaps that are missed by FEN1 (11, 12, 15).
An additional protein, Pif1, has been implicated in lagging strand synthesis. Pif1 is a 5′–3′ helicase that has been shown to have an important role in telomere and mitochondrial DNA maintenance (18). Interestingly, Pif1 has been shown to have a genetic interaction with Dna2; deletion of PIF1 rescues the lethality created by a Dna2 nuclease-deficient mutant in yeast (19). Previous work from our research group has also suggested that Pif1 plays a role in the two-nuclease pathway by aiding the creation of longer flaps during Okazaki fragment processing (12, 20).
Although originally formulated as two independent pathways, some evidence suggests that the one- and two-nuclease pathways have relevant interactions. Previous characterization of Dna2 suggests that it is able to stimulate FEN1 although indirectly. By cleaving long flaps bound by RPA, Dna2 not only removes the RPA block but also leaves short flaps that are more readily processed by FEN1 (21, 22).
We therefore questioned here whether the two pathways truly operate independently (i.e. whether the protein components of the two-nuclease pathway performed no function until a flap grew to the length that would serve as a substrate for the two-nuclease pathway). In such a model, relative processing of substrates through either of the two pathways would depend solely on the fraction of flaps missed for cleavage by FEN1. Alternatively, the two pathways might be interactive, with components of one affecting the efficiency of the other. For example, the components of the two-nuclease pathway might interfere with FEN1 cleavage, helping to generate more substrate for their pathway. To test this concept, we measured the effect of the unique two-nuclease protein components on the ability of FEN1 to cleave substrates of the one-nuclease pathway. Surprising results showed that the two-nuclease proteins all stimulate FEN1 activity in a way that suppresses formation of the substrate for their own pathway.
EXPERIMENTAL PROCEDURES
Materials
[α-32P]dCTP and [γ-32P]ATP were obtained from PerkinElmer Life Sciences. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). T4 polynucleotide kinase and Escherichia coli DNA polymerase I Klenow were obtained from Roche Applied Science. Other reagents were of the best grade commercially available.
Protein Expression and Purification
Saccharomyces cerevisiae Rad27 (FEN1) was cloned into the T7 expression vector pET-24b (Novagen/EMD Biosciences, Madison, WI), expressed in E. coli BL21(DE3) codon plus strain (Stratagene), and purified as described previously (23). S. cerevisiae Pif1 and Pif1 K264A were cloned into the pET-28b bacterial expression vector (Novagen/EMD Biosciences), expressed in the E. coli Rosetta strain (Novagen/EMD Biosciences), and purified as described previously (24). S. cerevisiae RPA was overexpressed and purified from E. coli as previously described (25). S. cerevisiae Dna2 E675A was produced by site-directed mutagenesis as previously described. Wild-type Dna2 and Dna2 E675A were overexpressed and purified from baculovirus High Five cells as previously described (26).
Oligonucleotide Substrates
Oligonucleotides were used to design substrates that simulate Okazaki fragment processing intermediates. Downstream primers of 28, 32, and 56 nt in length were annealed at their 3′-ends to a 20-nt labeling template with a 5′-GCTA overhang and radiolabeled using [α-32P]dCTP and Klenow polymerase. For 5′ labeling, the downstream primer of 28 nt was labeled with [γ-32P]ATP using polynucleotide kinase. Radiolabeled primers were separated by electrophoresis on a 15%, 7 m urea polyacrylamide gel and then gel-purified. Substrates were then created by annealing primer components in annealing buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm dithiothreitol), heating at 95 °C for 5 min, transferring to 70 °C, and slowly cooling to room temperature. The annealing ratio was 1:2:4 (downstream primer/template/upstream primer). Each downstream primer contains a region of 26 nt complementary to the template, resulting in 2-, 6-, and 30-nt unannealed 5′-flaps, respectively. The template itself is 51 nt in length. The upstream primer is 26 nt in length, containing a 25-nt region complementary to the template, leaving a 1-nt 3′-flap. These substrates were designed to allow measurement of cleavage by FEN1. Additionally, an RNA-DNA substrate was created utilizing the above procedure. However, for the RNA-DNA substrate, the 32-nt-long downstream primer contained 12 nt of RNA in the 5′-region. Annealing resulted in a substrate with a 6-nt 5′ RNA flap and 6 nt of RNA annealed to the template. Specific substrates used in each figure are indicated in the legends and pictured at the top of the figures. The location of the radiolabel on the downstream primer is indicated by an asterisk in the respective figures.
Cleavage Assays
For the fixed flap cleavage reactions, one or more of the proteins, FEN1, Pif1, Dna2, or RPA, were mixed together in reaction buffer containing 30 mm HEPES, 40 mm KCl, 4 mm MgCl2, 0.01% Nonidet P-40, 0.5% inositol, 0.1 mg/ml bovine serum albumin, 1 mm dithiothreitol, 0.5 mm ATP, and 5% glycerol. To start the reactions, 5 fmol of 3′-radiolabeled substrate was added for a total volume of 20 μl. Reactions were run for 10 min at 37 °C. Reactions were stopped by adding 20 μl of 2× termination dye (90% formamide (v/v), 10 mm EDTA, 0.01% bromphenol blue, and xylene cyanol), followed by heating at 95 °C for 5 min. Products were separated by electrophoresis on a 22.5%, 7 m urea polyacrylamide gel for 1 h and 40 min at 70 watts. The gel was placed on filter paper and vacuum-dried on a gel drier (Bio-Rad). Dried gels were exposed to a phosphor screen, which was scanned using a GE Healthcare PhosphorImager and analyzed with ImageQuant version 5.0 software.
Electrophoretic Mobility Shift Assay (EMSA)
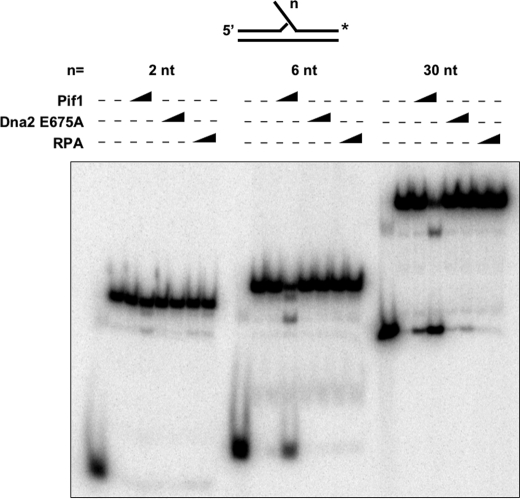
Binding efficiency of Pif1, RPA, or Dna2 to a substrate with a flap of either 2, 6, or 30 nt was assessed using EMSA. For clearer visualization, the 2-nt flap substrate was 5′-labeled in this experiment. Five fmol of substrate was incubated with increasing concentrations of the specific protein at either 50 or 500 fmol and incubated for 10 min at 37 °C in the same reaction buffer as described above but lacking ATP and MgCl2. The reactions were loaded on prerun, non-denaturing 12% polyacrylamide gels in 1× TBE. Gels were subjected to electrophoresis for 1.5 h at a constant 250 V.
Helicase Assay
To measure the helicase activity of Pif1 or Dna2 or the strand melting activity of RPA, the respective protein was added to a reaction containing a flap substrate of either 2, 6, or 30 nt in the reaction buffer utilized for our cleavage reactions. For clearer visualization, the 2-nt flap substrate was 5′-labeled in this experiment. Five fmol of substrate was incubated with concentrations of protein of 50 or 500 fmol and incubated for 10 min at 37 °C. The reactions were terminated using a helicase dye consisting of 30% glycerol, 50 mm EDTA, 0.9% SDS, 0.125% bromphenol, and 0.125% xylene cyanol. The reactions were loaded on prerun, non-denaturing 12% polyacrylamide gels in 1× TBE. Gels were subjected to electrophoresis for 1.5 h at a constant 250 V.
Binding Assay
Purified S. cerevisiae FEN1 (1 ng) and 1 ng of a purified two-nuclease protein (S. cerevisiae Dna2, Pif1, or RPA) were allowed to bind together in a coupling buffer consisting of 25 mm HEPES (pH 7.5), 100 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, and 10% glycerol for 2 h at 4 °C (in a 1:1 ratio). When three proteins were bound to FEN1, 2 ng of the competing proteins were added first and allowed to bind for 2 h. Then 1 ng of the antibody-specific protein was added and incubated with the mixture for an additional 2 h. Antibody to either Dna2, prepared as previously described (27), Pif1 (sc-48377 (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), or RPA70 (kindly provided by Dr. Marc Wold) or control IgG (sc-2027, Santa Cruz Biotechnology, Inc.) were prebound to protein A-agarose for 1 h at room temperature and washed twice with phosphate-buffered saline. The bound proteins were then added to the washed protein A-agarose-antibody complex and incubated overnight at 4 °C with end-over mixing. The following day, the proteins were released from the protein A-agarose using elution buffer. The immunoprecipitates were separated on precast 4–20% gels (Bio-Rad). Western blot analysis was performed with anti-FEN1 polyclonal antibody (ab17993, Abcam). Cross-reactivity of mouse monoclonal Pif1 antibody with purified S. cerevisiae Pif1 was tested and confirmed before the immunoprecipitation.
The amount of each protein used in each experiment is given in the appropriate figure legend. All enzyme assays were repeated at least in triplicate with a representative gel shown in each figure.
RESULTS
Dna2 and RPA have been implicated in the two-nuclease pathway, important for the proper cleavage of long flaps formed during eukaryotic Okazaki fragment processing. Previous work has also shown that Pif1 helicase promotes the long flap pathway. Although the two-nuclease and FEN1-only pathways are generally represented as occurring separately from one another, we considered the possibility that the pathways have evolved to be interactive. Specifically, we set out to examine whether the components of the two-nuclease pathway influence the efficiency of the one-nuclease pathway.
Substrate Design
For our experiments, we created three substrates with flap lengths of 2, 6, or 30 nt. The 2-nt flap represents an intermediate formed during the beginning of strand displacement synthesis, which we would expect to have no affinity for Dna2, RPA, or Pif1. The 6-nt flap represents an intermediate considered at the borderline for detectable binding by the three two-nuclease pathway proteins. The 30-nt flap serves as a positive control to which all three proteins should be able to bind with maximum affinity. FEN1 is able to bind and cleave all of these flaps. By utilizing these three different lengths, we have been able to distinguish the influence of each protein on flap cleavage by FEN1 and whether that influence depends on binding of each protein to the flap.
Because our experimental protocol utilized fixed flap substrates and because Dna2 and Pif1 are helicases, the potential existed that the helicase functions of these proteins would disrupt the substrates. The concern was that the helicases would partially displace the downstream primer, creating a gap. This would alter the double flap configuration favored by FEN1, decreasing FEN1 cleavage activity and masking any stimulatory effects. This is not an issue in vivo because elongation of the upstream primer by pol δ would continuously renew the favored double flap structure. To ensure that we did not experience interference, we chose a buffer system in which the helicase functions would cause minimal disruption of our substrates.
Effect of Dna2 on FEN1 Cleavage
Dna2 was found to stimulate FEN1 cleavage on long flaps. It has been proposed that the ability of Dna2 to cleave long 5′-flaps, terminating in a minimum flap length of about 6 nt, promotes FEN1 to cleave the remainder of the flap more efficiently. The action of Dna2 would remove regions of secondary structure in the flap that might interfere with tracking of FEN1. Moreover, Dna2 would also reduce the flap length so that RPA could not bind to interfere with FEN1 tracking. However, we were interested in determining whether Dna2 is able to directly stimulate FEN1 on the short flaps cleaved in the one-nuclease pathway.
To measure whether Dna2 influences FEN1 activity, we first performed a cleavage assay. For this experiment, amounts of Dna2 ranging from 0 to 50 fmol were added to reactions containing FEN1 and varying flap length substrates. We observed a dose-dependent stimulation of FEN1 cleavage by Dna2 (Fig. 1A). Interestingly, in addition to measuring stimulation on the 30-nt flap substrate, to which Dna2 is expected to bind avidly, we also measured significant stimulation on the 6- and 2-nt flaps, implying that the ability of Dna2 to stimulate FEN1 is independent of Dna2 binding to the substrate. After adding 50 fmol of Dna2 to the reaction in the presence of ATP, we measured a stimulation of FEN1 cleavage of 12.5-fold on the 2-nt flap substrate, 44.2-fold on the 6-nt flap substrate, and 15.9-fold on the 30-nt flap substrate.
FIGURE 1.
Dna2 stimulates FEN1 cleavage of 2-, 6-, and 30-nt flap substrates. A, cleavage by FEN1 (2 fmol) was assayed on a 2-, 6-, and 30-nt flap substrate in the presence or absence of ATP and varying amounts of Dna2 (5, 15, 25, or 50 fmol) as indicated in the figure and described under “Experimental Procedures.” The substrate is depicted at the top with the location of the radiolabel indicated by an asterisk. The length of the flap utilized is denoted above the corresponding lanes. The presence (+) and absence (−) of components are indicated. B, Dna2 binding activity (50 or 500 fmol) was measured on a 2-, 6-, and 30-nt flap substrate by non-denaturing PAGE.
Additionally, this experiment was performed in the absence of ATP. This was done to determine whether the helicase function of Dna2 is responsible for its ability to stimulate FEN1. The outcome was similar, showing that Dna2 is able to stimulate FEN1 cleavage even in the absence of helicase function.
To confirm the expected binding characteristics of Dna2 to the different length flap substrates, we performed an electrophoretic mobility shift assay (EMSA) utilizing all three substrates (Fig. 1B). For these EMSA measurements, we used protein levels of 50 and 500 fmol. Whereas 50 fmol of Dna2 represents the highest amount used in our cleavage assays, to observe high efficiency binding of the protein for an EMSA, a 10-fold increase in protein (i.e. 500 fmol) was needed. In the EMSA reactions, we utilized a buffer lacking both ATP and Mg2+ to prevent unwinding or, in the case of Dna2, cleavage. At 50 fmol of Dna2, we detected no binding to the 2- and 6-nt flap substrates, and a small amount of Dna2 bound to the 30-nt flap substrate. At 500 fmol, there was little binding to the 2-nt flap substrate. More binding was observed on the 6-nt flap substrate, with the most binding occurring on the 30-nt flap substrate in the presence of 500 fmol of Dna2. This is consistent with the binding properties expected from Dna2.
Finally, we performed a helicase assay utilizing the nuclease-deficient mutant of Dna2. Although the absence of ATP negated the helicase activity of Dna2 in the cleavage assay, the helicase assay was performed in conditions that allowed for maximal helicase activity by the nuclease-deficient Dna2 mutant. Although Dna2 did not show stable binding to the short flap substrates in our EMSA study, we envisioned that there could be transient binding of Dna2 to these substrates, allowing for some helicase activity. However, at 50 fmol of Dna2, the maximum amount used in our cleavage assay, we saw only minimal unwinding and only on the 30-nt flap (Fig. 2). Even at 500 fmol of Dna2, unwinding of the downstream primer could only be observed on the 30-nt flap, suggesting that robust helicase activity by Dna2 requires a longer single-stranded DNA region for it to track and unwind the substrate. This confirms that although Dna2 can stimulate FEN1 cleavage on short flaps, it cannot perform its helicase or endonuclease functions.
FIGURE 2.
Substrate unwinding by Pif1, Dna2, and RPA. Unwinding of a 2-, 6-, and 30-nt substrate by Pif1, Dna2 E675A, or RPA was assessed by non-denaturing PAGE using 50 or 500 fmol of protein, as indicated in the figure and described under “Experimental Procedures.” Substrate depictions and designations are as in Fig. 1.
Effect of Pif1 on FEN1 Cleavage
Pif1 has previously been shown to promote utilization of the two-nuclease pathway in reconstitution reactions. Because it appears to be a two-nuclease pathway component, we assessed the ability of Pif1 to stimulate FEN1 on short flaps.
As with Dna2, we first performed a cleavage assay, now utilizing FEN1 and Pif1. Varying amounts of Pif1, ranging from 0 to 50 fmol, were added to reactions containing FEN1. Results revealed a dose-dependent stimulation of FEN1 cleavage by Pif1 (Fig. 3A). As with Dna2, stimulation of FEN1 occurred not only for cleavage of the 30-nt flap substrate but also with the 6- and 2-nt flap substrates. This implies that the ability of Pif1 to stimulate FEN1 is not dependent on Pif1 binding to the flap. The addition of 50 fmol of Pif1 to the reaction resulted in a stimulation of FEN1 cleavage by 16.6-fold on the 2-nt flap substrate, 10.5-fold on the 6-nt flap substrate, and 9.4-fold on the 30-nt flap substrate. A similar experiment was also performed in the absence of ATP to determine whether the helicase function of Pif1 is in any way responsible for its ability to stimulate FEN1. We observed that, like Dna2, Pif1 is able to stimulate FEN1 cleavage even in the absence of ATP. A Pif1 helicase-deficient mutant was also utilized to further confirm that the stimulation of FEN1 by Pif1 does not require its helicase function (data not shown).
FIGURE 3.
Pif1 stimulates FEN1 cleavage of 2-, 6-, and 30-nt flap substrates. A, cleavage by FEN1 (2 fmol) was assayed on a 2-, 6-, and 30-nt flap substrate in the presence or absence of ATP and varying amounts of Pif1 (5, 15, 25, or 50 fmol) as indicated in the figure and described under “Experimental Procedures.” Substrate depictions and designations are as in Fig. 1. B, Pif1 binding activity (50 or 500 fmol) was measured on a 2-, 6-, and 30-nt flap substrate by non-denaturing PAGE.
We expected that Pif1 would exhibit weak binding to a 2- or 6-nt flap but would bind avidly to a 30-nt flap. To confirm, we performed an EMSA utilizing all three substrates in the presence of Pif1 (Fig. 3B). We observed a very low level of binding on the 2-nt flap, even at 500 fmol of protein. As expected, the amount of binding increased on the 6-nt flap, although the amount of Pif1 bound was still low. On the 30-nt flap, binding was evident even at 50 fmol of Pif1 and even more efficient at 500 fmol of Pif1. This result is consistent with our expectation that Pif1 would bind poorly to the short flap substrates, if at all, but readily to the 30-nt flap substrate.
In order to determine how the helicase of Pif1 is functioning on the different length flaps utilized in our previous assays, we performed a helicase assay. In addition to characterizing the helicase ability of Pif1, we also sought to confirm that Pif1 was not disrupting our substrates by removing the downstream primer. At 50 fmol of Pif1, the maximum amount used in our cleavage assay, we saw minimal unwinding of the downstream primer and only on the 30-nt flap (Fig. 2). This level of Pif1 was chosen intentionally with the expectation that it would not disrupt the substrate. At the higher concentration of 500 fmol, however, Pif1 displayed helicase activity, most effective on the 30-nt flap substrate, with some unwinding on the 6-nt flap. This confirms what we expected; there is no helicase activity of Pif1 directly on the 2-nt flap and minimal activity on the 6-nt flap, and Pif1 is able to freely interact with the 30-nt flap. This pattern of unwinding is consistent with our hypothesis that Pif1, on short flaps, is at first working to stimulate FEN1 cleavage. However, after a critical flap length is reached, Pif1 is able to bind to the flap, allowing its helicase function to act. This allows Pif1 to lengthen the flap, thereby biasing the processing of the flap toward the two-nuclease pathway.
Effect of RPA on FEN1 Cleavage
RPA is a key component of the Okazaki fragment maturation pathway, responsible for switching processing from the one-nuclease pathway to the two-nuclease pathway via binding of the flaps. To better understand these actions of RPA, we measured regulation of FEN1 by RPA on the same substrates that revealed stimulation of FEN1 by Dna2 and Pif1.
Amounts of RPA ranging from 0 to 100 fmol were added to reactions containing FEN1. This resulted in a dose-dependent stimulation of FEN1 cleavage by RPA (Fig. 4A). On the 2- and 6-nt flap substrates, to which RPA cannot stably bind, FEN1 cleavage was increased by 9.3- and 8.3-fold, respectively, at 100 fmol of RPA. For the 30-nt flap substrate, we included two additional, higher amounts of RPA, 200 and 300 fmol. This allowed us to measure the expected inhibition of FEN1 by RPA on long flaps. At lower levels, RPA was indeed able to stimulate FEN1 cleavage; there was a 2-fold stimulation of FEN1 cleavage at 100 fmol of RPA. However, at 300 fmol of RPA, at which we expected stable coating of the flap, FEN1 cleavage was inhibited to 25% of its basal level. As with our previous assays, this experiment was also performed in the absence of ATP. We observed a generally similar stimulation/inhibition pattern irrespective of inclusion of ATP, although inhibition of FEN1 on the 30-nt flap substrate occurred at lower concentrations in the absence of ATP. This could simply be a result of a lower ionic strength of our buffer without the ATP present.
FIGURE 4.
RPA stimulates FEN1 cleavage of 2-, 6-, and 30-nt flap substrates. A, cleavage by FEN1 (2 fmol) was assayed on a 2-, 6-, and 30-nt flap substrate in the presence or absence of ATP and varying amounts of RPA (15, 25, 50, or 100 fmol, with two additional reactions containing 200 and 300 fmol on the 30 nt flap substrate) as indicated in the figure and described under “Experimental Procedures.” Substrate depictions and designations are as in Fig. 1. B, RPA binding activity (50 or 500 fmol) was measured on a 2-, 6-, and 30-nt flap substrate by non-denaturing PAGE.
To confirm expected binding characteristics of RPA, we performed an EMSA utilizing all three substrates in the presence of RPA (Fig. 4B). There was little binding of RPA on the 2- and 6-nt flaps at 50 fmol of protein, but there was substantial binding at 500 fmol. RPA is expected to exhibit a low affinity binding mode on shorter flaps, but once bound, it may encourage local melting until the flap is long enough for more stable interaction. On the 30-nt flap, substantial binding was evident at 50 fmol of RPA, with a complete substrate shift at 500 fmol.
Finally, we performed a melting assay for RPA. Although RPA does not have helicase activity, high levels of RPA have been show to melt double-stranded DNA. Therefore, we intended to confirm that RPA was not removing either our upstream or downstream primers. At 50 fmol of RPA, we saw no removal of either primer, and even at 500 fmol of RPA, removal of either primer from the template was not observed (Fig. 2).
RPA has previously been reported to stimulate FEN1 cleavage. In 2003, Chai et al. (28) observed that on a 9- or 1-nt flap substrate, the presence of RPA would stimulate FEN1 cleavage. Biswas et al. (29) also noticed a similar stimulation followed by inhibition of FEN1 as RPA is titrated onto a 16-nt flap substrate. The detailed mechanism of RPA stimulation of FEN1 and whether it involved RPA interaction with the substrate was not explored. Based on our results, stimulation of FEN1 by RPA does not appear to derive from an interaction of RPA with the flap substrate but rather a direct interaction with FEN1.
After observing stimulation of FEN1 cleavage by Dna2, Pif1, and RPA, we also performed several negative controls. We showed that boiled Dna2, Pif1, and RPA do not possess the ability to stimulate FEN1 cleavage (data not shown). Additionally, we noted that stimulation of FEN1 cleavage was species-specific because human Dna2 and RPA failed to stimulate S. cerevisiae FEN1 (data not shown).
Additionally, to confirm that the stimulation of FEN1 by our proteins did not result from a contaminant in our protein preparations, Dna2, Pif1, and RPA (each containing a His6 tag) were allowed to adsorb to an Ni2+-NTA-agarose column. Following protein sequestration, the flow-through supernatant fraction was added to FEN1 and produced no stimulation of cleavage. Following elution of each protein from the nickel-agarose column, it stimulated FEN1 cleavage (data not shown).
Effect of Dna2, Pif1, or RPA on FEN1 Cleavage of an RNA Flap
Flaps are created when pol δ displaces the primer laid down by pol α and then extended by pol δ, which would generate an RNA segment of 10–12 nt on the 5′-end, followed by DNA. However, neither Dna2, Pif1, nor RPA is known to bind to single-stranded RNA. Therefore, it was important to determine whether protein components of the two-nuclease pathway are capable of stimulating FEN1 on a flap with a 5′ RNA segment.
For these experiments, we utilized a substrate containing a 6-nt-long 5′-flap composed entirely of RNA. At the base of the flap, there are 6 additional nucleotides of RNA annealed to the DNA template. This represents the 12 nt of RNA anticipated to be laid down by pol α. We then repeated the titrations of Pif1, Dna2, and RPA that we performed previously, utilizing the DNA flap substrates.
In each case, we observed stimulation of FEN1 cleavage. There was an approximately 12-fold stimulation at 50 fmol of Dna2 (Fig. 5A) and an approximately 13-fold stimulation at 50 fmol of Pif1 (Fig. 5B). Moreover, at 50 fmol of RPA, FEN1 cleavage was stimulated 8-fold (Fig. 5C). These values are consistent with the level of stimulation observed with a DNA flap substrate. We noted that with the RNA substrate, FEN1 cleavage produced two bands. This is not surprising because FEN1 sometimes will move 1 nt further into the downstream primer, where it then can cleave a second time. Therefore, for analysis of these results, FEN1 cleavage was assessed as a combination of the two cleavage products.
FIGURE 5.
Two-nuclease pathway proteins stimulate FEN1 on an RNA flap substrate. A, cleavage by FEN1 (2 fmol) was assayed on a 6-nt RNA flap substrate in the presence of varying amounts of Dna2 (5, 15, 25, or 50 fmol) as indicated in the figure and described under “Experimental Procedures.” The substrate is depicted at the top with the location of the radiolabel indicated by an asterisk. The dotted line represents the RNA region of the substrate. The presence (+) and absence (−) of components are indicated. B, cleavage by FEN1 (2 fmol) on a 6-nt RNA flap substrate in the presence of varying amounts of Pif1 (5, 15, 25, or 50 fmol). C, cleavage by FEN1 (2 fmol) on a 6-nt RNA flap substrate in the presence of varying amounts of RPA (5, 15, 25, or 50 fmol).
Effect of the Combination of Dna2, Pif1, and RPA on FEN1 Cleavage
After observing the ability of Dna2, Pif1, and RPA to stimulate FEN1 individually, we further explored the combined effect of all three proteins on FEN1 activity. One could imagine a situation in which each protein stimulates FEN1 in a different way, perhaps by inducing different conformational changes within FEN1. In that case, we would expect that adding different proteins would produce an additive stimulation of FEN1. Alternatively, if each protein component induced the same structural change in FEN1, then we would expect that there would be a maximal level of stimulation that one protein could impose, which could not be exceeded by the addition of one or more additional stimulatory proteins.
First, we determined the amounts of either Dna2, RPA, or Pif1 necessary to maximally stimulate a very low level (0.2 fmol) of FEN1 on a 6-nt flap. The low level ensured that additive stimulations could be readily detected. The maximum stimulatory amounts were determined to be 400 fmol of Dna2, 400 fmol of RPA, and 300 fmol of Pif1 (data not shown). Using these concentrations, we then added Dna2, RPA, and Pif1 to FEN1 in different combinations (Fig. 6). FEN1 controls of 10 and 2 fmol were also included. The 10 fmol of FEN1 was used to demonstrate that FEN1 can cleave 100% of our substrate. 2 fmol (a 10-fold increase over the 0.2 fmol control) was used to more easily visualize a basal level of FEN1 cleavage. We observed that the highest stimulation of FEN1 cleavage by a single protein resulted from the addition of Dna2 (42.9% cleavage). When adding all three proteins together, there was little difference in the amount of FEN1 cleavage compared with only adding Dna2 (46.8%). Combining two proteins had little effect on the maximal cleavage observed as well. Rather, although each protein stimulated to a somewhat different maximum, the various combinations of proteins stimulated to a maximum that was not substantially different from the stimulation level with Dna2 alone. These results demonstrate that the two-nuclease pathway proteins do not exhibit an additive stimulatory effect and that the proteins are each not stimulating FEN1 in a unique manner.
FIGURE 6.
Individual proteins are able maximize stimulation of cleavage by FEN1. Cleavage by FEN1 (0.2 fmol) was assayed on a 6-nt flap substrate in the presence of combinations of Dna2 (400 fmol), RPA (400 fmol), and Pif1 (300 fmol) as indicated in the figure and described under “Experimental Procedures.” Control reactions assessing cleavage at 2 and 10 fmol FEN1 are shown. Substrate depictions and designations are as in Fig. 1.
Protein Components of the Two-nuclease Pathway Are Present in a Complex with FEN1
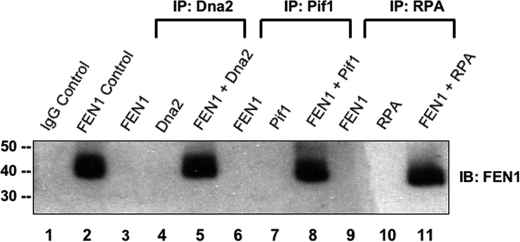
Stimulation of FEN1 by each protein component of the two-nuclease pathway even when those proteins were not expected to interact with the substrate suggested direct interactions of Dna2, Pif1, and RPA with FEN1. Pif1 and Dna2 are known to genetically interact (19), and direct associations between human FEN1 and human Dna2 and S. cerevisiae Dna2 and S. cerevisiae RPA70 have also been shown (30, 31). Binding assays were performed to clarify the direct interactions of S. cerevisiae FEN1 with S. cerevisiae Dna2, S. cerevisiae Pif1, and S. cerevisiae RPA in vitro. Purified recombinant yeast proteins were bound to FEN1 overnight and were immunoprecipitated the following day with antibodies to Dna2, Pif1, or RPA70. The immunoprecipitates were separated on a SDS-polyacrylamide gel, and Western blotting was performed using antibody against FEN1. The nonspecific IgG control showed no contamination with FEN1 (Fig. 7, lane 1). Lane 2 served as a positive control for FEN1. Direct interaction of Dna2 and FEN1 was detected and confirmed as previously reported (Fig. 7, lane 5). Pif1 (Fig. 7, lane 8) and RPA (Fig. 7, lane 11) also associated with FEN1 in vitro. Lanes 4, 7, and 10 serve as controls to demonstrate that Dna2, Pif1, and RPA, respectively, are not contaminated with FEN1 and thus stimulation by these proteins is not a result of additional FEN1 being included with these proteins. Lanes 3, 6, and 9 are controls to show that the Dna2, Pif1, and RPA antibodies, respectively, do not bind FEN1.
FIGURE 7.
Dna2, Pif1, and RPA each directly bind FEN1. Lane 1, a nonspecific IgG control. Lane 2, a positive control for 200 pg of purified FEN1. Lanes 3 (containing 1 ng of purified FEN1), 4 (containing 1 ng of purified Dna2), and 5 (containing 1 ng of purified FEN1 and 1 ng of purified Dna2) were immunoprecipitated (IP) with antibody to Dna2. Lanes 6 (containing 1 ng of purified FEN1), 7 (containing 1 ng of purified Pif1), and 8 (containing 1 ng of purified FEN1 and 1 ng of purified Pif1) were immunoprecipitated with antibody to Pif1. Lanes 9 (containing 1 ng of purified FEN1), 10 (containing 1 ng of purified RPA) and 11 (containing 1 ng of purified FEN1 and 1 ng of purified RPA) were immunoprecipitated with antibody to RPA. The immunoprecipitates were separated on an SDS-polyacrylamide gel, and Western blotting (IB) was performed using antibody against FEN1.
Our results suggest that each protein component of the two-nuclease pathway is able to directly interact with and stimulate FEN1 in vitro. When all of the protein components of the two-nuclease pathway were simultaneously allowed to bind FEN1 and then individually immunoprecipitated with antibodies specific for either Dna2, Pif1, or RPA and finally immunoblotted for FEN1, we still observed interaction between individual proteins and FEN1 (Fig. 8). However, the quantity of each protein bound to FEN1 was lower in the presence of the other proteins. Possibly, the presence of other proteins in a complex partially occluded the antibody interaction sites, decreasing the efficiency of immunoprecipitation. Alternatively, this could be an indication of partial or even complete overlap of protein binding sites, such that the proteins competed for binding FEN1. Additionally, interactions of the proteins among themselves might have altered the efficiency of their binding to FEN1. Our experiments are not able to distinguish among these possibilities. Results are consistent with formation of a complex containing all of the proteins but also with complexes that allow just one or two proteins at a time to bind FEN1.
FIGURE 8.
Binding of Dna2, Pif1, RPA, and FEN1. Lane 1, a nonspecific IgG control; lane 2, a positive control for 200 pg of purified FEN1. Lanes 3 (containing 1 ng of purified FEN1 and 1 ng of purified Dna2), and 4 (containing 2 ng of purified Pif1, 2 ng of purified RPA, 1 ng of purified FEN1, and 1 ng of purified Dna2) were immunoprecipitated (IP) with antibody to Dna2. Lanes 5 (containing 1 ng of purified FEN1 and 1 ng of purified Pif1), and 6 (containing 2 ng of purified Dna2, 2 ng of purified RPA, 1 ng of purified FEN1, and 1 ng of purified Pif1) were immunoprecipitated with antibody to Pif1. Lanes 7 (containing 1 ng of purified FEN1 and 1 ng of purified RPA) and 8 (containing 2 ng of purified Dna2, 2 ng of purified Pif1, with 1 ng of purified FEN1 and 1 ng of purified RPA) were immunoprecipitated with antibody to RPA. The immunoprecipitates were separated on a SDS-polyacrylamide gel, and Western blotting (IB) was performed using antibody against FEN1.
DISCUSSION
Current models of eukaryotic Okazaki fragment processing envision that it occurs by two enzymatic pathways. The one-nuclease pathway is proposed to convert the large majority of short flaps to nicks for ligation through the nuclease action of FEN1. The two-nuclease pathway is thought to process those flaps that escape FEN1 cleavage to become long. The actions of these pathways until now have been viewed as sequential and functionally independent. They also may be viewed as unidirectionally competitive, in the sense that the one-nuclease pathway reduces substrate available for two-nuclease processing. We demonstrate here that all of the unique protein components proposed to participate in the two-nuclease pathway, Dna2, Pif1, and RPA, stimulate the cleavage activity of FEN1 via direct interaction. This stimulation is ATP-independent and occurs on substrates with flaps that are too short to bind these two-nuclease pathway proteins. Additionally, although Pif1 and RPA cannot bind to RNA, and Dna2 cannot cleave RNA, all three proteins are able to stimulate FEN1 on a flap substrate containing a 5′ RNA region.
Relative use of the two pathways has been inferred from reconstitution of Okazaki fragment processing using oligonucleotide substrates and purified enzymes in vitro (12, 20). In the reconstituted system, instead of using a preformed flap substrate, pol δ actively creates a flap. Pol δ, complexed with PCNA, extends an upstream primer into a downstream primer, generating a flap by strand displacement synthesis, the mechanism of flap creation in Okazaki fragment processing. In the presence of FEN1, the large majority of cleavage products were less than 8 nt long (12). These observations suggest that the natural one-nuclease pathway is highly efficient and that the two-nuclease pathway has evolved to catch a small percentage of the flaps that are missed by effective FEN1 cleavage. Although it is currently not very clear why certain flaps escape FEN1 cleavage, one can imagine the deleterious effects on genome stability if the system were not equipped with a fail-safe mechanism in the form of the two-nuclease pathway. FEN1 haploinsufficiency has been linked to genomic instability and rapid progression to cancer (32, 33). Furthermore, post-translational modification of FEN1 by either phosphorylation or acetylation inhibits its cleavage function (34–36). Uncoupling of FEN1 from the replisome or post-translational modification of the protein might drive the system to utilize the two-nuclease pathway.
Assuming that the replication machinery has evolved to maximize genome stability, it makes sense to favor processing of flaps by FEN1 while they are short because problems caused by longer flap intermediates are easily envisioned. Long flaps can fold back onto themselves, forming secondary structures anticipated to inhibit processing. Formation of secondary structures not only inhibits the one-nuclease pathway, but these structures are also refractory to action by protein components of the two-nuclease pathway thereby creating an undesirable, unresolved structure. The long flaps can also promote harmful recombination events that would disrupt the continuity of the DNA sequence. Additionally, it has been shown that repeated cleavage of short flaps by FEN1 promotes more effective strand displacement synthesis by pol δ (6, 11). Thus, quick, efficient cleavage of short flaps would help lagging strand processing to the continuous final product occur more quickly, avoiding a series of long-lived breaks in the chromosome that can also promote recombination.
Furthermore, the requirement for removal of the RNA portion of the mixed primer made by pol α highlights the need for efficiency of the one-nuclease pathway. Pif1 cannot bind to a single-stranded RNA flap (24), and RPA shows approximately 1000-fold lower binding affinity to single-stranded RNA as compared with single-stranded DNA (37, 38). Moreover, although Dna2 is able to track over an RNA flap, it cannot cleave (15, 21). FEN1, however, is similarly active on RNA or DNA flaps (39). Therefore the RNA portion of the flap must be either fully displaced or displaced and cleaved before the two-nuclease pathway is even an option. Again, this suggests that the replication system has evolved in a way that favors FEN1 cleavage by allowing what would otherwise be inactive members of the two-nuclease pathway to stimulate FEN1 cleavage even when they themselves cannot bind or cleave.
With further displacement, the region of DNA beyond the RNA begins to become single-stranded. FEN1 cleavage can now generate a nick that serves as a substrate for the ligation that completes fragment processing. Our results indicate that while the DNA flap is short, neither Pif1 nor Dna2 can exhibit helicase activity, but they can exert a stimulatory effect on FEN1. Only after additional DNA flap displacement, and only in the infrequent cases in which the flaps escape FEN1 cleavage, would the flaps achieve lengths of ∼20 nt. At this point, RPA would bind to inhibit FEN1, Pif1 would accelerate displacement, and these flaps would require a shift to the two-nuclease pathway.
To further characterize the manner in which Pif1, Dna2, and RPA stimulate FEN1, we evaluated whether these proteins could each stimulate FEN1 in an additive manner or whether the presence of any one of the three proteins was capable of driving FEN1 to maximum activity. When using low levels of FEN1 and observing stimulation by each individual protein, we note that the addition of a second or third protein has little to no effect on the ability of FEN1 to cleave over the stimulation already conferred. The highest level of stimulation from an individual protein is from Dna2, causing an increase in cleavage from 0.6% to ∼43%, a stimulation of ∼71-fold. The addition of RPA and Pif1 to the reaction only had a modest effect, increasing cleavage to about 47%.
We also demonstrated that each of the unique proteins of the two-nuclease pathway has a direct binding affinity for FEN1, independent of the presence of a DNA substrate. However, our binding measurements could not establish unambiguously whether all three proteins can bind FEN1 simultaneously or whether they have evolved for a process of sequential binding during the execution of the two-nuclease pathway. Because single interactions or complexes involving combinations of two or all three proteins are all capable of maximum stimulation of FEN1, the effect on FEN1 cleavage of short flaps should be similar whether the proteins are bound simultaneously or sequentially.
Previous work has demonstrated that another protein involved in Okazaki fragment processing, PCNA, is also able to influence FEN1 cleavage. PCNA is a central component of Okazaki fragment processing that interacts with pol δ, FEN1, and DNA ligase I. It is a processivity factor for the polymerase and a stimulator of FEN1 and the ligase. It is also thought to coordinate the sequential actions of these proteins. PCNA and its similarly structured DNA repair counterpart, the 9-1-1 checkpoint complex, both bind and stimulate FEN1. Stimulation by PCNA has been extensively investigated (40–42). To exert stimulation, PCNA must be loaded onto the double-stranded part of the FEN1 substrate, specifically upstream of the flap. Evidence suggests that it captures the FEN1 as it slides to the base of the flap and stabilizes FEN1 interaction with its cleavage site. Although PCNA may also induce a conformational change in FEN1 that increases activity, effects on tethering FEN1 to its cleavage site and induction of a more active conformation are not readily distinguished. PCNA and 9-1-1 are capable of stimulating FEN1 cleavage activity over 10-fold, similar to the stimulation factors conferred by the protein components of the two-nuclease pathway.
FEN1, like other enzymes, presumably has a stimulation maximum that cannot be exceeded. Whereas experiments analyzing FEN1 cleavage function on cognate substrates in vitro are done in the presence of either individual or a combination of stimulating proteins, in the cellular system, the modes of interaction and stimulation might vary. For example, FEN1 interaction with PCNA might primarily provide for an increase in binding to the flap, and interactions with Dna2, Pif1, and RPA may change the conformation of FEN1 in a such manner that it can optimally function on the substrate.
Based on these observations and binding studies, we know that FEN1 is capable of complexing with multiple proteins, including Dna2, Pif1, RPA, and PCNA. Moreover, it appears that only PCNA tethers the FEN1 to its cleavage site on a short flap substrate because the two-nuclease proteins do not bind short flaps. However, the two-nuclease proteins are capable of interacting with FEN1 and each other, in a way that is likely to raise their local concentrations near flaps. When the flaps grow long, the two-nuclease components can develop additional productive interactions with the substrate. These interactions would help the two-nuclease components to be in proximity to their long flap substrates as soon as they are created.
In summary, the protein components of the two-nuclease pathway for eukaryotic Okazaki fragment processing appear to interact directly with FEN1 in order to stimulate FEN1 cleavage of short flaps. We propose that these proteins form a complex with FEN1 and PCNA that involves either simultaneous or successive interactions. Moreover, if a flap becomes long enough to require the two-nuclease pathway, the presence of the complex ensures that the two-nuclease proteins are available to carry out their functions.
Acknowledgments
We thank the members of the Bambara laboratory for helpful discussions and suggestions. We thank Dr. Marc Wold for providing purified RPA.
This work was supported, in whole or in part, by National Institutes of Health Grants GM024441 (to R. A. B.) and GM087666 (to J. L. C.).
- nt
- nucleotide(s).
REFERENCES
- 1.Kunkel T. A., Burgers P. M. (2008) Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg A., Baker T. A. (1992) DNA Replication, 2nd Ed., pp. 113–195, W. H. Freeman, New York [Google Scholar]
- 3.Bambara R. A., Murante R. S., Henricksen L. A. (1997) J. Biol. Chem. 272, 4647–4650 [DOI] [PubMed] [Google Scholar]
- 4.Liu Y., Kao H. I., Bambara R. A. (2004) Annu. Rev. Biochem. 73, 589–615 [DOI] [PubMed] [Google Scholar]
- 5.Rossi M. L., Purohit V., Brandt P. D., Bambara R. A. (2006) Chem. Rev. 106, 453–473 [DOI] [PubMed] [Google Scholar]
- 6.Ayyagari R., Gomes X. V., Gordenin D. A., Burgers P. M. (2003) J. Biol. Chem. 278, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 7.Garg P., Stith C. M., Sabouri N., Johansson E., Burgers P. M. (2004) Genes Dev. 18, 2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Y. H., Ayyagari R., Resnick M. A., Gordenin D. A., Burgers P. M. (2003) J. Biol. Chem. 278, 1626–1633 [DOI] [PubMed] [Google Scholar]
- 9.Harrington J. J., Lieber M. R. (1994) EMBO J. 13, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murante R. S., Rust L., Bambara R. A. (1995) J. Biol. Chem. 270, 30377–30383 [DOI] [PubMed] [Google Scholar]
- 11.Rossi M. L., Bambara R. A. (2006) J. Biol. Chem. 281, 26051–26061 [DOI] [PubMed] [Google Scholar]
- 12.Rossi M. L., Pike J. E., Wang W., Burgers P. M., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fanning E., Klimovich V., Nager A. R. (2006) Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae S. H., Bae K. H., Kim J. A., Seo Y. S. (2001) Nature 412, 456–461 [DOI] [PubMed] [Google Scholar]
- 15.Bae S. H., Seo Y. S. (2000) J. Biol. Chem. 275, 38022–38031 [DOI] [PubMed] [Google Scholar]
- 16.Bae S. H., Choi E., Lee K. H., Park J. S., Lee S. H., Seo Y. S. (1998) J. Biol. Chem. 273, 26880–26890 [DOI] [PubMed] [Google Scholar]
- 17.Kao H. I., Veeraraghavan J., Polaczek P., Campbell J. L., Bambara R. A. (2004) J. Biol. Chem. 279, 15014–15024 [DOI] [PubMed] [Google Scholar]
- 18.Boulé J. B., Zakian V. A. (2006) Nucleic Acids Res. 34, 4147–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budd M. E., Reis C. C., Smith S., Myung K., Campbell J. L. (2006) Mol. Cell. Biol. 26, 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pike J. E., Burgers P. M., Campbell J. L., Bambara R. A. (2009) J. Biol. Chem. 284, 25170–25180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart J. A., Campbell J. L., Bambara R. A. (2006) J. Biol. Chem. 281, 38565–38572 [DOI] [PubMed] [Google Scholar]
- 22.Stewart J. A., Miller A. S., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 31356–31365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao H. I., Henricksen L. A., Liu Y., Bambara R. A. (2002) J. Biol. Chem. 277, 14379–14389 [DOI] [PubMed] [Google Scholar]
- 24.Boulé J. B., Vega L. R., Zakian V. A. (2005) Nature 438, 57–61 [DOI] [PubMed] [Google Scholar]
- 25.Sibenaller Z. A., Sorensen B. R., Wold M. S. (1998) Biochemistry 37, 12496–12506 [DOI] [PubMed] [Google Scholar]
- 26.Budd M. E., Choe W., Campbell J. L. (2000) J. Biol. Chem. 275, 16518–16529 [DOI] [PubMed] [Google Scholar]
- 27.Budd M. E., Campbell J. L. (1997) Mol. Cell. Biol. 17, 2136–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chai Q., Zheng L., Zhou M., Turchi J. J., Shen B. (2003) Biochemistry 42, 15045–15052 [DOI] [PubMed] [Google Scholar]
- 29.Biswas E. E., Zhu F. X., Biswas S. B. (1997) Biochemistry 36, 5955–5962 [DOI] [PubMed] [Google Scholar]
- 30.Balakrishnan L., Stewart J., Polaczek P., Campbell J. L., Bambara R. A. (2010) J. Biol. Chem. 285, 4398–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae K. H., Kim H. S., Bae S. H., Kang H. Y., Brill S., Seo Y. S. (2003) Nucleic Acids Res. 31, 3006–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kucherlapati M., Yang K., Kuraguchi M., Zhao J., Lia M., Heyer J., Kane M. F., Fan K., Russell R., Brown A. M., Kneitz B., Edelmann W., Kolodner R. D., Lipkin M., Kucherlapati R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9924–9929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng L., Dai H., Zhou M., Li M., Singh P., Qiu J., Tsark W., Huang Q., Kernstine K., Zhang X., Lin D., Shen B. (2007) Nat. Med. 13, 812–819 [DOI] [PubMed] [Google Scholar]
- 34.Henneke G., Koundrioukoff S., Hübscher U. (2003) Oncogene 22, 4301–4313 [DOI] [PubMed] [Google Scholar]
- 35.Hasan S., Stucki M., Hassa P. O., Imhof R., Gehrig P., Hunziker P., Hübscher U., Hottiger M. O. (2001) Mol. Cell 7, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 36.Friedrich-Heineken E., Henneke G., Ferrari E., Hübscher U. (2003) J. Mol. Biol. 328, 73–84 [DOI] [PubMed] [Google Scholar]
- 37.Wold M. S. (1997) Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 38.Kim C., Snyder R. O., Wold M. S. (1992) Mol. Cell. Biol. 12, 3050–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murante R. S., Rumbaugh J. A., Barnes C. J., Norton J. R., Bambara R. A. (1996) J. Biol. Chem. 271, 25888–25897 [DOI] [PubMed] [Google Scholar]
- 40.Jónsson Z. O., Hindges R., Hübscher U. (1998) EMBO J. 17, 2412–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X., Li J., Li X., Hsieh C. L., Burgers P. M., Lieber M. R. (1996) Nucleic Acids Res. 24, 2036–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tom S., Henricksen L. A., Bambara R. A. (2000) J. Biol. Chem. 275, 10498–10505 [DOI] [PubMed] [Google Scholar]