Abstract
The CCR4-CAF1-NOT complex is a major cytoplasmic deadenylation complex in yeast and mammals. This complex associates with RNA-binding proteins and microRNAs to repress translation of target mRNAs. We sought to determine how CCR4 and CAF1 participate in repression and control of maternal mRNAs using Xenopus laevis oocytes. We show that Xenopus CCR4 and CAF1 enzymes are active deadenylases and repress translation of an adenylated mRNA. CAF1 also represses translation independent of deadenylation. The deadenylation-independent repression requires a 5′ cap structure on the mRNA; however, deadenylation does not. We suggest that mere recruitment of CAF1 is sufficient for repression, independent of deadenylation.
Keywords: MicroRNA, Repressor Protein, RNA, Translation Control, Xenopus, CAF1/POP2, Deadenylases, mRNA Decay, Poly(A), Translational Repression
Introduction
Regulation of RNA localization, translation, and decay determine when, where, and how much protein is produced from an mRNA (1–3). The 3′-UTR is pivotal in these controls (1, 2). Regulatory proteins and microRNAs bind specific sequence elements in the 3′-UTR to trigger repression or decay (1–6). To do so, the 3′-UTR complexes recruit enzymes that promote removal of the poly(A) tail (7–14). This process, termed deadenylation, can cause repression (4, 5, 15).
Multiple enzymes remove poly(A) in vivo and can be grouped into families based on sequence relatedness (5). CCR4 is a member of the exonuclease-endonuclease-phosphatase superfamily, whereas CAF1 (also known as POP2, CNOT7, and CNOT 8) belongs to the DEDD superfamily (5, 16, 17). CCR4 and CAF1 proteins interact directly with one another and are part of a larger CCR4-CAF1-NOT complex (18–21). Many regulatory proteins and miRNAs2 recruit this multi-component deadenylation complex (7–14, 22–26). For example, PUF proteins recruit deadenylases to specific mRNAs (8), as do microRNAs (12, 13). The wide spectrum of mRNAs controlled in this fashion underlies the broad biological functions of CCR4 and CAF1, which range from control of the cell cycle, early development, and fertility (5, 20, 27–36).
The role of CAF1 and CCR4 enzymes in regulation appears to be complex (5). In some systems, deadenylation is required for repression, yet in others it is dispensable (5, 6, 37). For example, in budding yeast, CAF1 is required for repression of mRNAs targeted by the regulatory protein, PUF5, even though deadenylation of the target is not (24). Similarly, miRNA complexes elicit deadenylation via the CCR4-CAF1 complex, yet deadenylation is not essential for repression (13, 22, 37–39). These findings suggest that the deadenylase enzymes CCR4 and CAF1 may possess a second, deadenylation-independent role in translational repression.
Control of mRNAs is pervasive during early development and often is mediated by changes in poly(A) length (1, 3). Maternal mRNAs direct oogenesis and embryogenesis until zygotic transcription begins (1). The temporal control of deadenylation in Xenopus oocytes and embryos is highly regulated; specific mRNAs are deadenylated at precise times throughout the early period of development and lose poly(A) to characteristic extents (9, 40–42). In Xenopus, the PARN deadenylase, recruited by the 3′-UTR binding protein, CPEB, removes poly(A) from target mRNAs (e.g. cyclin B1) prior to maturation (9). It dissociates once oocyte maturation begins, allowing polyadenylation and translation (9). Other deadenylases, including CCR4 and CAF1, likely facilitate the intricacies of deadenylation. The CCR4-NOT complex catalyzes deadenylation of multiple mRNAs in Drosophila embryos and again is recruited by specific regulatory proteins, such as Smaug (7, 11).
We sought to determine how CAF1 and CCR4 contribute to repression and participate in the control of maternal mRNAs, focusing on the Xenopus oocyte. Our data reveal that the CAF1 enzyme possesses an intrinsic repression activity, independent of its ability to deadenylate the mRNA. This activity requires the mRNA 5′ cap structure. We show that mere recruitment of CAF1, even without deadenylation, is sufficient to repress a target mRNA.
EXPERIMENTAL PROCEDURES
DNA Constructs
With pCS2+3HA:MS2, three HA tags were inserted in the BamHI/EcoRI restriction sites of pCS2+ vector (addGene). An NcoI restriction site was introduced before the EcoRI site that is in-frame with the HA tags (pCS2+3HA). The MS2 fragment from pET-MS2 was ligated into the NcoI/StuI restriction sites of pCS2+3HA, creating pCS2+3HA:MS2 (47). This vector contains the Sp6 promoter followed by three HA tags, the MS2 coat protein, and the MCS. pCS2+3HA:MS2+PL. Additional restriction sites were added to the MCS of pCS2+3HA:MS2. The vector was cleaved with StuI/XhoI. AP005 (sense, cctggacccatcgatgaaggaagatcttcctagactagtctagaac) and AP006 (antisense, tcgagttctagactagtctaggaagatcttccttcatcgatgggtccagg) were annealed, kinased, and inserted into the StuI/XhoI restriction sites creating pCS2+3HA:MS2+PL vector. This vector contains the same features as pCS2+3HA:MS2 with ClaI, BglII, and SpeI added in the MCS between StuI/XhoI.
MS2 Fusion Proteins
Full-length cDNA clones for CCR4a (BC091632.1), CCR4b (BC084200.1), CAF1a (BC106339.1), and CAF1b (BC041239) were ordered from Open Biosystems. These clones were used to amplify full-length PCR templates of each protein for cloning into the pCS2+3HA:MS2 vector: CCR4a, CAF1a and CAF1b were cloned into the StuI/XhoI restriction sites, and CCR4a was cloned into the StuI/XbaI restriction sites. GLD-2 D242A was PCR-amplified from the pLW073 plasmid and cloned into the BglII/XhoI restriction sites of pCS2+3HA:MS2+PL vector (42). Xp54 was PCR-amplified from the MS2Xp54 3′-UTR+MSP plasmid and cloned into the StuI/BglII restriction sites of pCS2+3HA:MS2+PL vector (43).
Each plasmid carrying an MS2 fusion contains the SP6 promoter, followed by three HA tags, one copy of the MS2 coat protein, and the protein to be tested. All of the plasmids were linearized with NotI before in vitro transcription.
mRNA Reporter Plasmids
pLG-MS2 (firefly luciferase), pSP65 ren (Renilla luciferase), and pLGMS2-LucHS plasmids have been described (48–50). The pCSFV-Luc-MS2 plasmid was supplied by Nicola Gray and has been described (46). pLG-MS2+A39 was made by inserting 39 adenosines between the BglII/BamHI restriction sites of pLG-MS2. To make pLGMS2+A39-LucHS, pLGMS2+A39 was cleaved with HindIII and SpeI to excise the luciferase gene fragment; ends were filled in and relegated. To make pLG-SL-MS2+A39, pLG-MS2+A39 was cleaved with HindIII; primers ac242 (agcttggccggccggccggccggccggccgtaaggccggccggccggccggccggcca) andac243 (agcttggccggccggccggccggccggccttacggccggccggccggccggccggcca) were annealed, kinased, and inserted into the HindIII site.
pLG-MS2 and pLGMS2-LucHS were linearized with BglII and transcribed with T7 RNA polymerase. pLG-MS2+A39 and pLGMS2+A39-LucHS were linearized with BamHI and transcribed with T7 RNA polymerase. pJSP65 was linearized with SalI and transcribed with Sp6 RNA polymerase. pCSFV-Luc-MS2 was linearized with BglII and transcribed with T7 RNA polymerase.
In Vitro Transcription
Plasmids were linearized with the restriction enzymes indicated above. All of the in vitro transcriptions were described by Kwak et al. (49).
Western Blotting
All Western blotting was done as described by Kwak et al. (49) with the following modifications. The oocytes were lysed in 10 μl of PBS plus protease inhibitors (Roche Applied Science) centrifuged at 3000 rpm for 10 min at 4 °C, and the supernatants were collected. The lysates from two oocytes were loaded on to 8–16% SDS/PAGE gels (Lonza). The proteins were analyzed by Western blotting using mouse monoclonal anti-HA tag antibody (HA11 1:1000 dilution, from Covance, Princeton) and anti-actin antibody (actin 1:40,000 dilution, from MP Biomedicals).
Tethered Function Assays
Oocyte injections were performed as described (47, 48). The oocytes were collected at the indicated time points.
RNA Extraction and Gel Electrophoresis
Oocyte RNA was prepared by using TRI reagent, following the manufacturer's instructions (Sigma). RNAs were separated on a 6% polyacrylamide gel (short RNAs) and analyzed by autoradiography.
Northern Blots
10 μg of total mRNA was separated on a 1% agarose, formaldehyde, 1× MOPS gel. The RNA was transferred by capillary action to activated nylon membrane (Millipore) using 10× SSC. The blots were hybridized in ULTRAhybTM (Ambion) and washed according to the manufacturer's specifications. The firefly luciferase mRNA was detected with a probe antisense to nucleotides 1131–1640. The probe was transcribed using the T7 megascript kit from Ambion and was internally labeled with [α-32P]UTP. Northern blots were exposed to a phosphorimaging screen for 1 day. The screens were scanned on a Strom phosphorimaging (Molecular Dynamics).
RESULTS
Domain Structure of Xenopus CCR4 and CAF1 Homologs
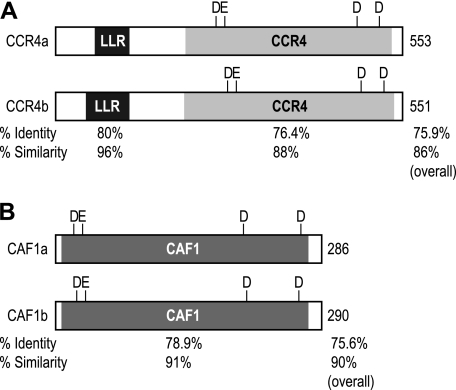
The Xenopus laevis genome is predicted to encode four CCR4 and two CAF1 homologs. Expressed sequence tag data indicate that each mRNA is expressed in stage VI oocytes. CCR4a and CCR4b contain leucine-rich repeats and a CCR4-deadenylase domain. The Xenopus CCR4 proteins are in the exonuclease-endonuclease phosphatase family and include four amino acids (DEDD) necessary for catalytic activity (Fig. 1A) (5, 16). Based on conservation (Fig. 1A), CCR4a and CCR4b appear to be functionally distinct (supplemental Fig. S1 and Fig. 1A). The two CAF1 homologs, CAF1a and CAF1b, belong to the RNase D/DEDD family of endonucleases and possess the characteristic four amino acids necessary for catalytic activity (Fig. 1B) (5, 17).
FIGURE 1.
Domain structure of Xenopus CCR4 and CAF1 homologs. A, domain structure for two of four Xenopus CCR4 homologs (CCR4a and CCR4b). Depicted are the leucine-rich repeat (LRR) motif and the CCR4 deadenylase domain; the catalytic residues are indicated. The percentages of identity and similarity are indicated below. B, domain structure for the two Xenopus CAF1 homologs. Depicted is the CAF1 domain; the catalytic residues are indicated. The percentages of identity and similarity are indicated below.
Tethered CCR4 and CAF1 Repress
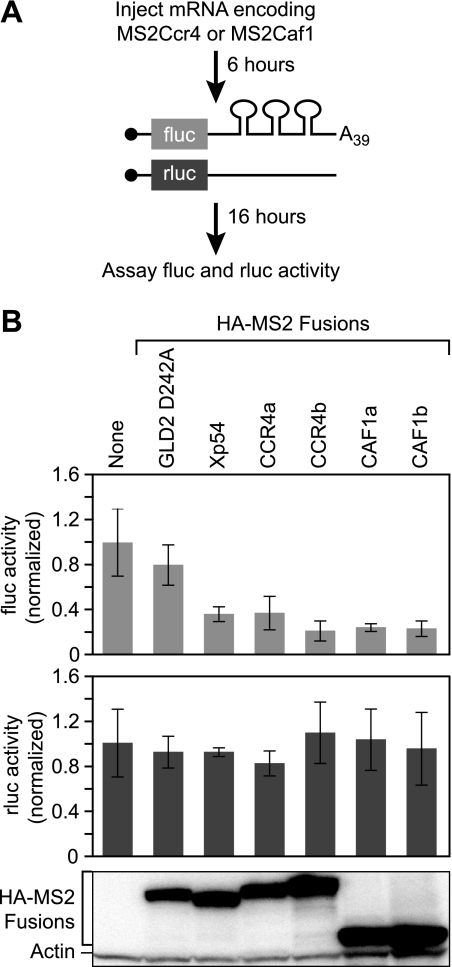
To test the activities of Xenopus CCR4 and CAF1, CCR4 (CCR4a and CCR4b) or CAF1 (CAF1a and CAF1b) was tethered to firefly luciferase reporter RNAs using the MS2 coat protein (Fig. 2A). Xenopus oocytes first were injected with mRNAs encoding HA-MS2-CCR4 or HA-MS2-CAF1. These proteins possessed three HA tags followed by the MS2 coat protein and the protein of interest. The oocytes were left for 6 h to allow accumulation of the fusion proteins and then were co-injected with two reporter RNAs: a firefly luciferase mRNA with three MS2 coat protein-binding sites in its 3′-UTR and, as a control, a Renilla luciferase mRNA without MS2-binding sites. The firefly luciferase mRNA possessed a 39-nucleotide poly(A) tail. After 16 h, the firefly and Renilla luciferase levels were determined.
FIGURE 2.
Tethered Xenopus CCR4 and CAF1 repressed translation of an adenylated luciferase mRNA. A, mRNA encoding MS2 fusion proteins of interest were injected into oocytes. The oocytes were incubated for 6 h to allow translation of the fusion protein, followed by co-injection with two reporter mRNAs. The firefly luciferase reporter mRNA contained three MS2-binding sites in the 3′-UTR and a poly(A) tail of 39 adenosines; the Renilla luciferase reporter mRNA lacked the MS2-binding sites and was used as a control. After 16 h of incubation, the levels of firefly and Renilla luciferase were measured. B, top panel, relative repression of firefly luciferase translation by each protein. The values were normalized to firefly luciferase activity without an MS2 fusion protein expressed. The error bars were derived from analysis of four sets of four oocytes within the same experiment. Middle panel, relative translational activity of Renilla mRNA when expressed with each fusion protein. Renilla and firefly luciferase levels were normalized the same way. Bottom panel, lysates equivalent to two oocytes were loaded in each lane and analyzed by Western blotting with α-HA-11 and α-actin.
HA-MS2-CCR4 and HA-MS2-CAF1 proteins reduced the level of firefly luciferase 3–4-fold (Fig. 2B, top panel). The reduction was specific, because it required MS2-binding sites; the level of Renilla luciferase, which lacked the MS2-binding sites was unaffected by any of the proteins (Fig. 2B, bottom panel). Similarly, tethered GLD2 D242A, a protein that lacks repression activity, had no effect (Fig. 2B) (42). The magnitude of decrease with HA-MS2-CCR4 and HA-MS2-CAF1 was similar to that observed with HA-MS2-Xp54, a known translational repressor (Fig. 2B) (43). All of the MS2 fusion proteins were expressed at similar levels (Fig. 2B).
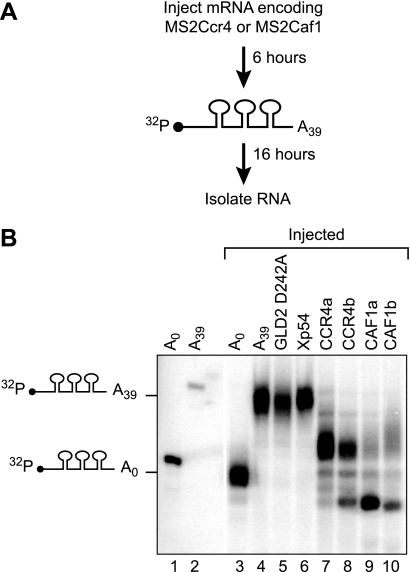
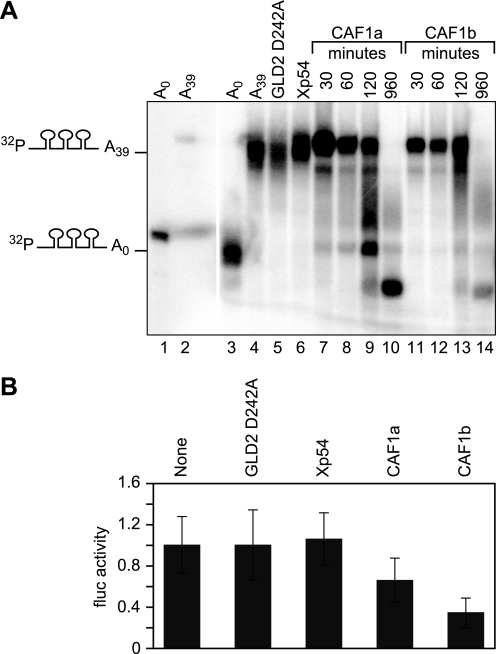
To determine whether CCR4 or CAF1 could catalyze deadenylation in oocytes, we injected radiolabeled (130 nucleotides) RNAs with MS2-binding sites and an A39 tail (Fig. 3A). These RNAs were deadenylated by MS2-CCR4 and MS2-CAF1 but not by MS2-GLD2 D242A or MS2-Xp54 (Fig. 3B). For example, 89% of the RNA tethered to XlCAF1b was fully or partially deadenylated (supplemental Fig. S2). The deadenylases also excise several nucleotides into the RNA “body.”3 We conclude that CCR4 and CAF1 can repress translation of an adenylated mRNA and are active deadenylases.
FIGURE 3.
Xenopus CCR4 and CAF1 homologs are active deadenylases. A, repeated scheme as in Fig. 1A, except a short radiolabeled RNA was injected instead of reporter mRNAs. The radiolabeled RNA was internally labeled with [α-32P]UTP and contained three MS2-binding sites and a poly(A) tail of 39 adenosines. RNA was extracted after 16 h of incubation to determine the length of the poly(A) tail with each MS2 fusion protein. B, poly(A) status of reporter RNA. Lanes 1 and 3 show the 32P RNA without a poly(A) before and after injection into oocytes, respectively. Lanes 2 and 4 show the 32P RNA with a poly(A) tail before and after injection into oocytes, respectively. Lanes 5–10 indicate poly(A) tail length of the 32P RNA when tethered to the indicated MS2 fusion protein. Tethered CCR4 and CAF1 excised about 10 nucleotides from the RNA body.3
Tethered CAF1 Proteins Repress mRNAs without Poly(A)
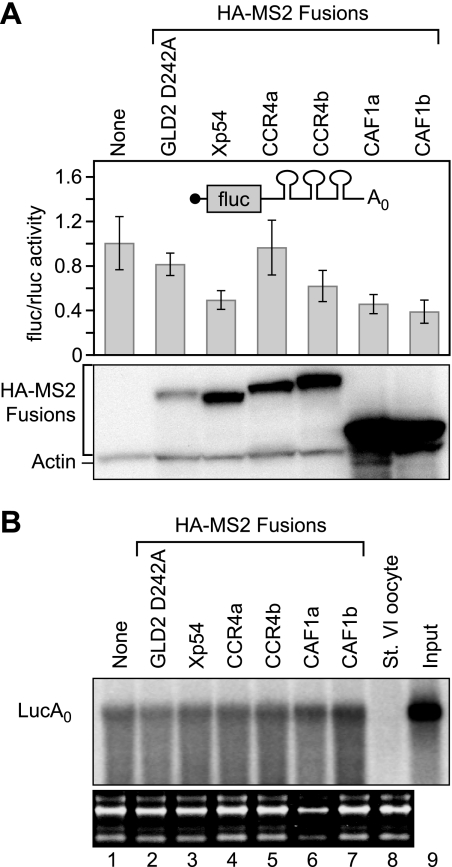
To test whether CCR4 and CAF1 repress translation independent of deadenylation, we next assayed their activities on a reporter RNA without a poly(A) tail. Repression activity was expressed as a ratio of firefly to Renilla luciferase activities. CCR4a did not repress firefly expression, whereas CCR4b moderately repressed expression of a nonadenylated firefly mRNA (Fig. 4A). The moderate repression by CCR4b was statistically significant (p value < 0.002, eight experiments). Surprisingly, both CAF1 homologs repressed the same reporter mRNA by 2–3-fold (Fig. 4A). The reduction was specific; GLD2 D242A had no significant effect (Fig. 4A), and the levels of Renilla luciferase, produced from the mRNA without MS2-binding sites, were unaffected (data not shown). The MS2:CAF1 proteins were similar in activity to MS2:Xp54 (Fig. 4A) (43), and were expressed at comparable levels. The reporter mRNAs were not destabilized by the tethered CAF1 proteins or the controls (Fig. 4B). We conclude that the XlCAF1 can repress translation of a nonadenylated RNA. Repression by XlCAF1 proteins was more robust then by XlCCR4b; therefore, hereafter, we focus on the XlCAF1 proteins.
FIGURE 4.
Tethered Xenopus CAF1 homologs repressed translation of a nonadenylated luciferase mRNA, while not destabilizing the mRNA. A, top panel, the relative translation of luciferase activity in response to each protein was quantified as the ratio of firefly to Renilla luciferase activity. The firefly luciferase mRNA contained three MS2-binding sites in the 3′-UTR and no poly(A) tail. The values were normalized to the ratio of luciferase activity without an MS2 fusion protein expressed. The error bars were derived from analysis of four sets of four oocytes within the same experiment. Bottom panel, lysates equivalent to two oocytes were loaded in each lane and analyzed by Western blotting with α-HA-11 and α-actin. B, lanes 1–7 indicate the stability of the reporter mRNAs when tethered to each protein. The reporter mRNA with and without a poly(A) tail are indicated. Blots show rRNA below each panel used as a loading control. Lane 8 shows signal from an uninjected oocyte. Lane 9 shows the reporter RNA before injection.
CAF1 Repression Is Deadenylation-independent
If Xenopus CAF1 proteins repress translation independent of deadenylation, then mutations in the catalytic domain of CAF1 should not disrupt repression. To test this prediction, we generated missense mutations in the first two amino acids required for catalysis in other CAF1 proteins (Fig. 1B) (31).
Both CAF1 mutants CAF1a and CAF1b repressed expression of a nonadenylated mRNA (Fig. 5, A and B, top panels) and an adenylated mRNA (Fig. 5, A and B, bottom panels). Surprisingly, the CAF1a mutant repressed a mRNA with a poly(A) tail to the same extent as the wild type protein (Fig. 5A, bottom panel). As expected, the missense mutations abolish catalytic activity of the proteins (Fig. 5, C and D, compare lanes 5 and 6), and the two proteins were expressed at similar levels (Fig. 5, A and B).
FIGURE 5.
Mutations in catalytic amino acids of Xenopus CAF1 abolish deadenylase activity but not translational repression. A and B, the relative translation of luciferase activity in response to HA:MS2:CAF1a (A) or HA:MS2:CAF1b (B) wild type or catalytic mutant was quantified as the ratio of firefly to Renilla luciferase activity. The reporter used in the experiment is indicated. Normalization and error bars were determined as for Fig. 1B. Bottom panel, lysates equivalent to two oocytes were loaded in each lane and analyzed by Western blotting with α-HA-11 and α-actin. C and D, poly(A) status of reporter RNA. Lanes 1 and 3 show the 32P RNA without a poly(A) before and after injection into oocytes, respectively. Lanes 2 and 4 show the 32P RNA with a poly(A) tail before and after injection into oocytes, respectively. Lanes 5 and 6 indicate poly(A) tail length of the 32P RNA when tethered to HA:MS2:CAF1a (C) or HA:MS2:CAF1b (D) wild type or catalytic mutant, respectively. A t test was performed on at least four replicate experiments in A and B. No statistical difference was found for any of the values except CAF1b wild type and mutant forms with the adenylated reporter mRNA (p value < 0.005).
The results of XlCAF1b were similar to those of XlCAF1a but differ in one important respect. Like the XlCAF1a mutant, the XlCAF1b mutant repressed translation with or without a poly(A) tail, was catalytically inactive, and was expressed well (Fig. 5, B and D). However, the XlCAF1b mutant relieved repression by 2-fold on an adenylated mRNA (Fig. 5B, bottom panel; p value < 0.005, seven experiments). We conclude that both proteins repress translation through a poly(A)- and deadenylation-independent mechanism. XlCAF1b also represses through deadenylation, whereas XlCAF1a does not.
Human CAF1 Proteins Repress Nonadenylated mRNAs
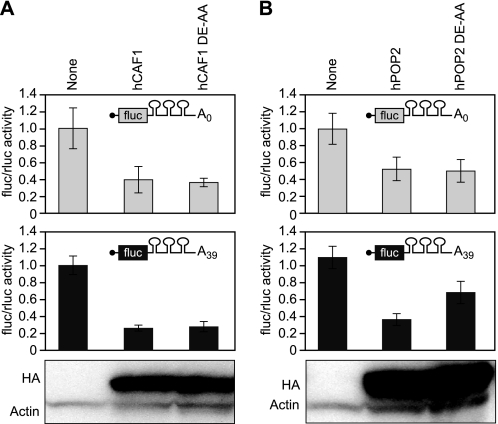
Next, we sought to determine whether the poly(A)- and deadenylation-independent mechanism of repression was specific to Xenopus. To test this, we tethered human CAF1 (hCAF1 and hPOP2). hCAF1 and hPOP2 repressed translation independent of a poly(A) tail (Fig. 6). Similarly, the hCAF1 catalytic mutant repressed an adenylated mRNA to the same extent as the wild type hCAF1 protein (Fig. 6A).
FIGURE 6.
hCAF1 represses translation independent of deadenylation. A and B, the relative translation of luciferase activity in response to HA:MS2:hCAF1 (A) or HA:MS2:hCAF1 (B) wild type or catalytic mutant was quantified as the ratio of firefly to Renilla luciferase activity. The reporter used in the experiment is indicated. Normalization and error bars were determined as for Fig. 1B. Bottom panel, lysates equivalent to two oocytes were loaded in each lane and analyzed by Western blotting with α-HA-11 and α-actin.
We conclude that the poly(A)- and deadenylation-independent mechanism of repression is not specific to XlCAF1 proteins. Strikingly, hCAF1 does not appear to utilize deadenylation akin to XlCAF1a; however, similar to XlCAF1b, hPOP2 can repress through deadenylation.
Deadenylation Rates and Repression
To examine the kinetics of deadenylation, we analyzed labeled RNAs at various times after they had been injected into oocytes expressing XlCAF1a or XlCAF1b proteins. One hour after injection, the majority of RNA remained adenylated (Fig. 7A, lanes 7, 8, 11, and 12). By 2 h, deadenylation had begun (Fig. 7A, lanes 9 and 13). For example, after 1 h, only 73% of the RNA tethered to XlCAF1a was fully or partially deadenylated; whereas after 2 h, 47% of the RNA was deadenylated or partially deadenylated (data not shown). The rates of deadenylation, as tethered proteins, are low (∼0.5 nucleotide/min), as is the default deadenylation pathway during oocyte maturation (40).
FIGURE 7.
XlCAF1 deadenylation rates and translational repression. A, poly(A) status of reporter RNA. Lanes 1 and 3 show the 32P RNA without a poly(A) before and after injection into oocytes, respectively. Lanes 2 and 4 show the 32P RNA with a poly(A) tail before and after injection into oocytes, respectively. Lanes 5 and 6 indicate poly(A) tail length of the 32P RNA when tethered to the control HA:MS2 fusion protein. Lanes 7–10 and 11–14 indicate the poly(A) tail length of the 32P RNA when tethered HA:MS2:CAF1a or HA:MS2:b for designated lengths of time, respectively. B, the relative translation of firefly luciferase activity 1 h post injection of the reporter mRNA in response to each protein. The luciferase mRNA contained three MS2-binding sites in the 3′-UTR and a poly(A) tail of 39 adenosines. Normalization and error bars were determined as for Fig. 1B.
The kinetic analysis prompted an additional test of the deadenylation independence of repression activity. If XlCAF1b repressed through a deadenylation-independent mechanism, then an mRNA could be repressed before it was deadenylated. To test this, we measured the level of firefly luciferase 1 h after injection of the mRNA in oocytes expressing XlCAF1b. We chose 1 h after injection because in Fig. 7A the majority (86%, data not shown) of the RNA was adenylated. XlCAF1b reduced the expression at the 1-h time point after the firefly luciferase mRNA was injected (Fig. 7B). The reduction was specific because GLD2 D242A, Xp54, and XlCAF1a did not reduce the level of firefly luciferase (Fig. 7B). However, as seen before, after 16 h Xp54, XlCAF1a, and XlCAF1b reduced the level of firefly luciferase (data not shown).
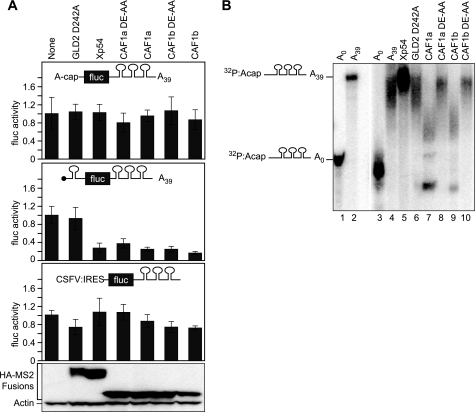
The 7mGpppG Cap Is Required for CAF1 Repression
To assess whether the cap structure was necessary for repression, we used a firefly luciferase mRNA bearing a 5′ cap structure of ApppG. As expected, the level of translation from an ApppG capped mRNA was reduced by 97% (supplemental Fig. S3A). Strikingly, XlCAF1 proteins did not repress translation of an ApppG capped adenylated mRNA (Fig. 8A, top panel; p value > 0.5; three experiments). However, the XlCAF1 proteins repressed a reporter mRNA with a 7-methyl GpppG cap (Figs. 2B and 4A; p value of less then or equal to 1.8 × 10−6; five and eight experiments, respectively). As seen before, Xp54 did not repress the mRNA that contained an ApppG (Fig. 8A) (44). The results were the same with an ApppG capped nonadenylated mRNA (supplemental Fig. S3B).
FIGURE 8.
XlCAF1 required the 7-methyl GpppG cap for translational repression but not deadenylation. A, the relative translation of firefly luciferase activity in response to each protein is shown. The luciferase mRNAs used contained a ApppG cap (top) or a 7-methyl GpppG cap and stem-loop in the 5′-UTR (middle) or a ApppG cap and a CSFV IRES in the 5′ UTR (bottom). The ApppG and stem-loop reporters contained a poly(A) tail of 39 adenosines. The IRES reporter does not contain a poly(A) tail. Normalization and error bars were determined as for Fig. 1B. Bottom panel, lysates equivalent to two oocytes were loaded in each lane and analyzed by Western blotting with α-HA-11 and α-actin. B, poly(A) status of reporter RNA with an ApppG cap. Lanes 1 and 3 show the 32P RNA without a poly(A) before and after injection into oocytes, respectively. Lanes 2 and 4 show the 32P RNA with a poly(A) tail before and after injection into oocytes, respectively. Lanes 5–10 indicate poly(A) tail length of the 32P RNA when tethered to the indicated MS2 fusion protein.
We next tested whether the reduced translation rate of the ApppG mRNA was enough to alleviate repression by XlCAF1. To do so, we introduced a stem-loop before the translational start of a 7-methyl GpppG mRNA (Fig. 8A). As with the ApppG capped mRNA, the stem-loop reduced translation by 98% (supplemental Fig. S3A). However, unlike the ApppG capped mRNA, the translation of the stem-loop mRNA with the 7-methyl GpppG cap was repressed by the XlCAF1 proteins (Fig. 8A, middle panel). Moreover, the missense XlCAF1 mutants repressed the stem-loop mRNA as well (Fig. 8A, middle panel). This reduction was specific because GLD2 D242A does not repress translation (Fig. 8A, middle panel). We conclude that XlCAF1 represses translation at or before recognition of the 7-methyl GpppG cap.
To further probe the mechanism of repression, we used an IRES derived from classical swine flu virus (CSFV). Initiation from the CSFV IRES does not require eIF4F or eIF3 (45, 46). We prepared a luciferase mRNA with an IRES and an ApppG cap to avoid degradation when injected into oocytes. XlCAF1 did not repress translation of the IRES-containing reporter (Fig. 8A, bottom panel). As seen previously, Xp54 did not repress the reporter mRNA with the IRES (Fig. 8A, bottom panel) (44). The IRES was functional, because that mRNA was translated more than 75-fold more efficiently than a replica mRNA lacking the CSFV IRES (data not shown). We conclude that translational repression by XlCAF1 proteins requires the 7-methyl GpppG cap and, likely, the translation initiation factors eIF4F and eIF3.
To determine whether XlCAF1 required the cap to catalyze deadenylation, we injected ApppG-capped radiolabeled RNAs with an A39 tail (Fig. 8B). As expected, these RNAs were deadenylated by XlCAF1b, but not by GLD2 D242A or Xp54 or XlCAF1 DE-AA (Fig. 8B). Therefore, repression by CAF1 is deadenylation-independent.
DISCUSSION
CAF1 proteins possess an intrinsic repression activity independent of deadenylation. By “intrinsic,” we imply that the mere presence of a CAF1 protein on an mRNA is sufficient to elicit repression, whether or not deadenylation occurs. Several lines of evidence support this central conclusion. CAF1 proteins repress mRNAs without poly(A) tails, catalytically dead enzymes retain repression activity, and wild type CAF1 represses prior to deadenylation.
Deadenylation-independent repression by CAF1 proteins requires the cap, because it is blocked by a nonfunctional cap analog, ApppG. Repression also appears to require eIF4F and/or eIF3, because it is alleviated by use of the CSFV IRES. CAF1 may interfere with translation by contacting the cap and initiation factors directly or by acting through protein intermediaries. For simplicity, we will refer to CAF1 as a translational repressor, without implying its mechanism of action.
CAF1b possesses a low level of deadenylation-dependent repression activity, whereas CAF1a does not. Human POP2 also shows some deadenylation-dependent repression activity, whereas human CAF1 does not, demonstrating that the difference between CAF1a and CAF1b is conserved. Thus regulatory proteins and miRNAs might differ in activity depending on which isoform they recruit.
In vivo, regulatory factors, including both proteins and miRNAs, recruit CAF1 to mRNAs (7, 8, 11–14, 22). We suggest that repression induced by RNA-binding proteins may be due in part to recruitment of CAF1 per se, independent of deadenylation. 3′-UTR-bound regulatory proteins, such as the PUF proteins, recruit CAF1 and other components of the NOT-CCR4-CAF1 complex (8). Similarly, Ago proteins, which associate with mRNAs via miRNAs, recruit CAF1 and several other components of the complex (12). miRNAs often (though not always) elicit deadenylation of their targets (37), as do many regulatory proteins (5). Although deadenylation often is required for full repression, that is not universally the case (37). The repression activity of CAF1 may be responsible.
The two activities of CAF1 proteins diversify mechanisms of repression by 3′-UTR-binding proteins. For example, repression by a yeast PUF protein, PUF5, requires CAF1 but not deadenylation (8, 24, 25), whereas repression by a closely related protein, PUF4, requires both CAF1 and deadenylation (8, 24, 25). miRNAs recruit CCR4 and CAF1 deadenylases and induce deadenylation, yet deadenylation is not invariably required for repression (37). Similarly, miRNA-mediated translational repression precedes deadenylation in vitro, yet is alleviated by CAF1 depletion in vitro (12). We suggest that deadenylation-independent repression relies on the intrinsic repression activity of CAF1.
Mere recruitment of CAF1 causes translational repression. In that sense, CAF1 can be viewed as a repressor, independent of its enzymatic activity. From that perspective, our findings reveal an unanticipated mode of mRNA control.
Supplementary Material
Acknowledgments
We thank members of the Wickens laboratory, especially Aaron Caesar Goldstrohm, for discussions and suggestions. We also thank Olivia for catalyzing organization and L. Vanderploeg for help preparing figures. We thank Nancy Standart and Nicola Gray for plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants GM31892 and GM50942 (to M. W.). This work was also supported by an National Institutes of Health Molecular Biosciences Training Grant (to A. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Tethered CAF1 generated RNAs that are 9–12 nucleotides shorter and 10–15 nucleotides longer than the A0 marker. Previous work shows that CAF1 can degrade non-adenosine nucleotides (51, 52) and that deadenylation in oocytes results in removal of a few non-adenosine nucleotides from the end of the RNA (40). Partially deadenylated RNAs with oligo(A) tracts of 10–15 nucleotides have been reported in previous studies (8, 24, 25, 53).
- miRNA
- microRNA
- CSFV
- classical swine flu virus.
REFERENCES
- 1.Wickens M., Goodwin E. B., Kimble J., Strickland S., Hentze M. W. (2000) Translational Control, pp. 295–370, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 2.Wickens M., Bernstein D. S., Kimble J., Parker R. (2002) Trends Genet 18, 150–157 [DOI] [PubMed] [Google Scholar]
- 3.Gebauer F., Hentze M. W. (2004) Nat. Rev. Mol. Cell Biol. 5, 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickens M., Goldstrohm A. (2003) Science 300, 753–755 [DOI] [PubMed] [Google Scholar]
- 5.Goldstrohm A. C., Wickens M. (2008) Nat. Rev. Mol. Cell Biol. 9, 337–344 [DOI] [PubMed] [Google Scholar]
- 6.Omer A. D., Janas M. M., Novina C. D. (2009) Mol Cell 35, 739–740 [DOI] [PubMed] [Google Scholar]
- 7.Semotok J. L., Cooperstock R. L., Pinder B. D., Vari H. K., Lipshitz H. D., Smibert C. A. (2005) Curr. Biol. 15, 284–294 [DOI] [PubMed] [Google Scholar]
- 8.Goldstrohm A. C., Hook B. A., Seay D. J., Wickens M. (2006) Nat. Struct. Mol. Biol. 13, 533–539 [DOI] [PubMed] [Google Scholar]
- 9.Kim J. H., Richter J. D. (2006) Mol. Cell 24, 173–183 [DOI] [PubMed] [Google Scholar]
- 10.Moraes K. C., Wilusz C. J., Wilusz J. (2006) RNA 12, 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadyrova L. Y., Habara Y., Lee T. H., Wharton R. P. (2007) Development 134, 1519–1527 [DOI] [PubMed] [Google Scholar]
- 12.Fabian M. R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J. T., Svitkin Y. V., Rivas F., Jinek M., Wohlschlegel J., Doudna J. A., Chen C. Y., Shyu A. B., Yates J. R., 3rd, Hannon G. J., Filipowicz W., Duchaine T. F., Sonenberg N. (2009) Mol. Cell 35, 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piao X., Zhang X., Wu L., Belasco J. G. (2010) Mol. Cell. Biol. 30, 1486–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki A., Igarashi K., Aisaki K., Kanno J., Saga Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3594–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilusz C. J., Wormington M., Peltz S. W. (2001) Nat. Rev. Mol. Cell Biol. 2, 237–246 [DOI] [PubMed] [Google Scholar]
- 16.Dlakić M. (2000) Trends Biochem. Sci. 25, 272–273 [DOI] [PubMed] [Google Scholar]
- 17.Zuo Y., Deutscher M. P. (2001) Nucleic Acids Res. 29, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J., Rappsilber J., Chiang Y. C., Russell P., Mann M., Denis C. L. (2001) J. Mol. Biol. 314, 683–694 [DOI] [PubMed] [Google Scholar]
- 19.Tucker M., Valencia-Sanchez M. A., Staples R. R., Chen J., Denis C. L., Parker R. (2001) Cell 104, 377–386 [DOI] [PubMed] [Google Scholar]
- 20.Denis C. L., Chen J. (2003) Prog. Nucleic Acid Res. Mol. Biol. 73, 221–250 [DOI] [PubMed] [Google Scholar]
- 21.Wagner E., Clement S. L., Lykke-Andersen J. (2007) Mol. Cell. Biol. 27, 1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. (2006) Genes Dev. 20, 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ezzeddine N., Chang T. C., Zhu W., Yamashita A., Chen C. Y., Zhong Z., Yamashita Y., Zheng D., Shyu A. B. (2007) Mol. Cell. Biol. 27, 7791–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstrohm A. C., Seay D. J., Hook B. A., Wickens M. (2007) J. Biol. Chem. 282, 109–114 [DOI] [PubMed] [Google Scholar]
- 25.Hook B. A., Goldstrohm A. C., Seay D. J., Wickens M. (2007) J. Biol. Chem. 282, 15430–15438 [DOI] [PubMed] [Google Scholar]
- 26.Standart N., Jackson R. J. (2007) Genes Dev. 21, 1975–1982 [DOI] [PubMed] [Google Scholar]
- 27.Körner C. G., Wormington M., Muckenthaler M., Schneider S., Dehlin E., Wahle E. (1998) EMBO J. 17, 5427–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berthet C., Morera A. M., Asensio M. J., Chauvin M. A., Morel A. P., Dijoud F., Magaud J. P., Durand P., Rouault J. P. (2004) Mol. Cell. Biol. 24, 5808–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura T., Yao R., Ogawa T., Suzuki T., Ito C., Tsunekawa N., Inoue K., Ajima R., Miyasaka T., Yoshida Y., Ogura A., Toshimori K., Noce T., Yamamoto T., Noda T. (2004) Nat. Genet. 36, 528–533 [DOI] [PubMed] [Google Scholar]
- 30.Temme C., Zaessinger S., Meyer S., Simonelig M., Wahle E. (2004) EMBO J. 23, 2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viswanathan P., Ohn T., Chiang Y. C., Chen J., Denis C. L. (2004) J. Biol. Chem. 279, 23988–23995 [DOI] [PubMed] [Google Scholar]
- 32.Westmoreland T. J., Marks J. R., Olson J. A., Jr., Thompson E. M., Resnick M. A., Bennett C. B. (2004) Eukaryot. Cell 3, 430–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianchin C., Mauxion F., Sentis S., Séraphin B., Corbo L. (2005) RNA 11, 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molin L., Puisieux A. (2005) Gene 358, 73–81 [DOI] [PubMed] [Google Scholar]
- 35.Morris J. Z., Hong A., Lilly M. A., Lehmann R. (2005) Development 132, 1165–1174 [DOI] [PubMed] [Google Scholar]
- 36.Morita M., Suzuki T., Nakamura T., Yokoyama K., Miyasaka T., Yamamoto T. (2007) Mol. Cell. Biol. 27, 4980–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E. (2009) RNA 15, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beilharz T. H., Humphreys D. T., Clancy J. L., Thermann R., Martin D. I., Hentze M. W., Preiss T. (2009) PLoS One 4, e6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wakiyama M., Yokoyama S. (2010) Prog. Mol. Subcell. Biol. 50, 85–97 [DOI] [PubMed] [Google Scholar]
- 40.Fox C. A., Sheets M. D., Wickens M. P. (1989) Genes Dev. 3, 2151–2162 [DOI] [PubMed] [Google Scholar]
- 41.Barkoff A. F., Dickson K. S., Gray N. K., Wickens M. (2000) Dev. Biol. 220, 97–109 [DOI] [PubMed] [Google Scholar]
- 42.Rouhana L., Wang L., Buter N., Kwak J. E., Schiltz C. A., Gonzalez T., Kelley A. E., Landry C. F., Wickens M. (2005) RNA 11, 1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minshall N., Thom G., Standart N. (2001) RNA 7, 1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minshall N., Reiter M. H., Weil D., Standart N. (2007) J. Biol. Chem. 282, 37389–37401 [DOI] [PubMed] [Google Scholar]
- 45.Pestova T. V., Shatsky I. N., Fletcher S. P., Jackson R. J., Hellen C. U. (1998) Genes Dev. 12, 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorgoni B., Andrews S., Schaller A., Schümperli D., Gray N. K., Müller B. (2005) RNA 11, 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gray N. K., Coller J. M., Dickson K. S., Wickens M. (2000) EMBO J. 19, 4723–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dickson K. S., Thompson S. R., Gray N. K., Wickens M. (2001) J. Biol. Chem. 276, 41810–41816 [DOI] [PubMed] [Google Scholar]
- 49.Kwak J. E., Wang L., Ballantyne S., Kimble J., Wickens M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4407–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chritton J. J., Wickens M. (2010) RNA 16, 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daugeron M. C., Mauxion F., Séraphin B. (2001) Nucleic Acids Res. 29, 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finoux A. L., Séraphin B. (2006) J. Biol. Chem. 281, 25940–25947 [DOI] [PubMed] [Google Scholar]
- 53.Thompson S. R., Goodwin E. B., Wickens M. (2000) Mol. Cell Biol. 20, 2129–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.