Abstract
Ferlins are an ancient family of C2 domain-containing proteins, with emerging roles in vesicular trafficking and human disease. Dysferlin mutations cause inherited muscular dystrophy, and dysferlin also shows abnormal plasma membrane expression in other forms of muscular dystrophy. We establish dysferlin as a short-lived (protein half-life ∼4–6 h) and transitory transmembrane protein (plasma membrane half-life ∼3 h), with a propensity for rapid endocytosis when mutated, and an association with a syntaxin-4 endocytic route. Dysferlin plasma membrane expression and endocytic rate is regulated by the C2B-FerI-C2C motif, with a critical role identified for C2C. Disruption of C2C dramatically reduces plasma membrane dysferlin (by 2.5-fold), due largely to accelerated endocytosis (by 2.5-fold). These properties of reduced efficiency of plasma membrane expression due to accelerated endocytosis are also a feature of patient missense mutant L344P (within FerI, adjacent to C2C). Importantly, dysferlin mutants that demonstrate accelerated endocytosis also display increased protein lability via endosomal proteolysis, implicating endosomal-mediated proteolytic degradation as a novel basis for dysferlin-deficiency in patients with single missense mutations. Vesicular labeling studies establish that dysferlin mutants rapidly transit from EEA1-positive early endosomes through to dextran-positive lysosomes, co-labeled by syntaxin-4 at multiple stages of endosomal transit. In summary, our studies define a transient biology for dysferlin, relevant to emerging patient therapeutics targeting dysferlin replacement. We introduce accelerated endosomal-directed degradation as a basis for lability of dysferlin missense mutants in dysferlinopathy, and show that dysferlin and syntaxin-4 similarly transit a common endosomal pathway in skeletal muscle cells.
Keywords: Endocytosis, Intracellular Trafficking, Membrane Proteins, Muscular Dystrophy, Skeletal Muscle, Dysferlin, Syntaxin-4
Introduction
Dysferlin belongs to the ferlin family of proteins, an ancient family with emerging roles in cellular trafficking and vesicle fusion. Caenorhabditis elegans FER-1 has been proposed as a calcium sensor or regulatory protein involved in the process of fusion of membranous organelles to the plasma membrane of spermatozoa (1). Impaired calcium-regulated fusion of vesicular organelles as seen in FER-1 mutants (1) is a theme that unites ferlin-related disease in humans.
Dysferlin was identified through linkage analyses of two families with an inherited form of muscular dystrophy (2, 3). Dysferlin is ubiquitously expressed, but with predominant expression in striated muscle. Dysferlin-deficient murine skeletal muscle fibers exhibit impaired calcium-mediated membrane repair (4), a process thought to involve calcium-activated vesicular exocytosis (5, 6) and/or endocytosis (7). Otoferlin is expressed in the brain, vestibular system, and cochlea, and underlies a form of human deafness (8, 9). Otoferlin-null mice are profoundly deaf, and exhibit a primary defect in calcium-mediated fusion of neurotransmitter-containing vesicles at the specialized ribbon synapse of cochlear inner hair cells (10). Myoferlin is broadly expressed in many tissues, with high expression in skeletal muscle during development and in regeneration (11). Myoferlin has not yet been implicated in human disease but has been shown to be important for myoblast fusion and myotube formation (12) and has an emerging role as a regulator of endocytosis in epithelial cells (13, 14). Mammalian Fer-1 L4, L5, and L6 GenBankTM sequences have been identified, but these proteins have not yet been characterized.
Ferlin proteins contain multiple C2 domains and have structural homology to the synaptotagmins. The synaptotagmins possess two calcium-binding C2 domains that lie on the cytoplasmic face of a lipid bilayer, anchored by an N-terminal transmembrane domain. The two C2 domains of synaptatogmin exhibit complex calcium-regulated binding to both SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) protein machinery and membrane phospholipids, to regulate vesicular fusion (15). The ferlin proteins possess between four and seven tandem C2 domains within their cytoplasmic domain and are also anchored to lipid bilayers via their C-terminal transmembrane domain.
We are seeking to better understand dysferlin cell biology and its molecular role in membrane resealing. Defective membrane resealing is likely to be widely implicated in the muscular dystrophies, supported by recent findings of membrane resealing defects in mouse models of caveolinopathy (16). Caveolin-3 interacts with dysferlin (17) and regulates its plasma membrane expression (18) and rate of endocytosis (18, 19). Consequently, it is likely that other proteins that influence the plasma membrane expression of dysferlin, or interact with dysferlin in membrane repair or vesicular trafficking pathways, may represent additional disease candidates for muscular dystrophy.
Although the cellular function of dysferlin has not been fully defined, data from other ferlin proteins suggest a trafficking role for dysferlin, which may underpin its involvement or requirement for effective membrane resealing. Primary dysferlinopathies are associated with complete deficiency of dysferlin in patient skeletal muscle by immunostaining and Western blot. It remains enigmatic why many dysferlinopathy patients bearing only single missense substitutions should display dysferlin deficiency, with one previous study implicating a role for protein misfolding and endoplasmic reticulum-associated degradation pathways (ERAD) with one mutation (20). We (21) and others (22) have documented marked abnormalities in dysferlin plasma membrane localization in the muscle biopsies of many patients with muscular dystrophy without mutations in the dysferlin gene. This led us to hypothesize that dysferlin may be a protein only transiently expressed at the muscle plasma membrane, and recruited to intracellular membrane compartments under certain conditions. Therefore, we have defined the basic cell biology properties of dysferlin relating to protein trafficking, half-life, and membrane dynamics. We establish that dysferlin is a transitory plasma membrane protein, whose plasma membrane expression is regulated by its endocytosis via a syntaxin-4-associated pathway.
EXPERIMENTAL PROCEDURES
Patients and Ethics
We identified the L344P mutation in two sisters referred to our Neuromuscular Clinic and laboratory. The elder sister first presented in her early twenties, with elevated CK (not precisely documented) and mild limb girdle weakness on examination. Muscle biopsy histology demonstrated variation in fiber size, several degenerate and necrotic fibers, and a moderate increase in lymphocytes and macrophages. The younger sister presented at age 16, with an elevated CK level of up to 4000 units/liter on occasion (normal range <230 units/liter), and with a painful swollen calf muscle, which was enlarged on MRI. She also presented with knee, hip, and shoulder pain and scoliosis. Routine muscular dystrophy screening performed in our laboratory revealed the absence of dysferlin by Western blot, with normal protein levels of dystrophin, calpain, lamin A/C, emerin, and caveolin-3. cDNA sequencing identified two mutations in the dysferlin gene; the L344P mutation in exon 11 and a mutation in the splice acceptor site preceding exon 49, which gives rise to two alternate transcripts; one with a 36-bp intronic insertion and one which skips exon 49, depending on the alternate splice site used by the splicing machinery. The unaffected parents were confirmed to carry one of these mutations each. Approval for these studies was obtained from the Human Ethics Committees of The Children's Hospital at Westmead and the University of Sydney, with patients providing informed consent.
Antibodies and Fluorescent Reagents
Antibodies to the following proteins were used: dysferlin NCL-Hamlet (Novocastra, Leica Microsystems, Australia); anti-Myc (Santa Cruz Biotechnology); anti-MycAlexa555 (Millipore); anti-His (Abcam, Cambridge, UK); anti-HisAlexa647 (Qiagen Pty Limited); anti-HA, anti-EEA1 (BD Biosciences); anti-Syntaxin-4 (Synaptic Systems); Alexa Fluor®-conjugated secondary antibodies (Invitrogen); Cy3-conjugated secondary antibodies (Jackson Immunoresearch Laboratories); HRP-conjugated secondary antibodies (Amersham Biosciences, Buckinghamshire, UK). Other fluorescent reagents were used: Cholera toxinAlexa488 and low density lipoprotein conjugated to lipophilic carbocyanine DiI (LDLdil) (Molecular Probes, Invitrogen). Flow cytometry isotype control staining was performed using LEAFTM Purified Mouse IgG1 (κ Isotype, clone MG1-45, BioLegend).
Cell Culture
C2C12 cells (P22-P29) were provided by Dr Edna Hardeman, Children's Medical Research Institute, Sydney. All culture media and reagents were obtained from Invitrogen unless otherwise specified. Cells were grown in a humidified incubator containing 5% CO2 and maintained at 37 °C. C2C12 cells were maintained in 1:1 F-12/DMEM containing 15% heat-inactivated fetal bovine serum (FBS) and 50 μg/ml gentamycin.
Dysferlin Expression Constructs
The dysferlin expression construct pcDNA4-EGFPdysferlinMycHis was kindly provided by Prof. Kate Bushby (23). ΔC2C*B (Dysferlin ΔMet303-Ala497) was derived through ligation of the dysferlin 5′-fragment EcoRI/BspEI (Klenow fill-in to blunt), into the plasmid backbone pcDNA4-EGFPdysferlinMycHis restricted with EcoRI/Sbf I (T4 blunt), resulting in deletion of the cDNA sequence between BspEI and Sbf I. ΔC2BC (Dysferlin ΔVal140-Ala497) was derived through Sbf I restriction, and ligation of annealed oligos (5′-AGCGCTTGCA-3′, 5′-AGCGCTTGCA-3′) to restore frame and introduce an Afe I site. ΔC2BCD (Dysferlin ΔVal140-Pro1327) was derived through Afe I digestion of ΔC2BC, and re-ligation. C2EF-TM contains the C-terminal dysferlin fragment from Asp1475 derived through EcoRI/ClaI restriction, Klenow fill-in to blunt ends, and re-ligation. C2F-TM contains the C-terminal dysferlin fragment from Iso1738 and was derived through ligation of a BamHI (Klenow blunted)/NotI fragment of pcDNA4-EGFPdysferlinMycHis religated into EcoRI (Klenow blunted)/NotI-digested vector backbone. His-only and Myc-only constructs were derived via NotI digestion and ligation of oligos (His: 5′-GGCCGCGAACAAAAACTCATCTCAGAAGAGGATCTGTAAGCTAGC-3′; 5′-GGCCGCTAGCTTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGC-3′; Myc 5′-GGCCGCGAACAAAAACTCATCTCAGAAGAGGATCTGTAAGCTAGC-3′; 5′-GGCCGCTAGCTTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGC-3′). −EGFP constructs were derived through KpnI (T4 blunt) and BsrG1 (T4 blunt) digestion and re-ligation of pcDNA4-EGFPdysferlinMycHis. The L344P construct was derived by amplification of the patient cDNA (Forward primer: 5′-ACCTTTCCGAGCCCTCTTTG-3′ Reverse primer: 5′-CCAAGTCTGAGGCTTTCGAAG-3′) followed by digestion of this fragment by Sbf I and ligation into the pcDNA4-EGFP full-length (FL)3 plasmid backbone digested with Sbf I and de-phosphorylated. The syntaxin-4-HA construct (pcDNA3-Syn4HA) was kindly provided by Prof. David James, Garvan Institute, Sydney.
Transfection
C2C12 cells were transfected overnight or for 24 h at 70–80% confluence with GeneJuiceTM (Merck, Darmstadt, Germany), using 0.5 μg DNA/1.5 μl GeneJuiceTM per 2 cm2 culture area in 0.5 ml culture media. For FL-dysferlin and C2F-TM or FL dysferlin and syntaxin 4 -HA co-expression studies, plasmids were used at a ratio of FL: C2F-TM at 2:1 or FL: Syn4-HA at 2:1. For FL-dysferlin and ΔC2C co-expression, plasmids were used at a ratio of 1:1. Twenty-four hours post-transfection, cells were switched to low serum medium to induce differentiation (1:1 F12/DMEM, 3% horse serum, 50 μg/ml gentamycin).
Immunocytochemistry and Microscopy
Cells grown on ThermanoxTM coverslips (Nunc) were transfected and differentiated as described above. On the third day of differentiation, intact, live cells were immunolabeled at 4 °C with anti-MycAlexa555 (1:100) and/or anti-HisAlexa647 (1:100) diluted in Hank's-buffered salt solution (HBSS) containing 20% FBS. Cells were washed in PBS and coverslips mounted on 22 × 50 mm glass coverslips using FluorsaveTM mounting reagent (Merck, Darmstadt, Germany). Confocal microscopy was performed using a 100× HCX Plan Apo oil immersion lens (1.4×) by imaging through the glass coverslip, as described (29), using a Leica SP2 Scanning Laser Confocal Microscope. Images (1024 × 1024) were captured without subtraction of glow-under. Background was subtracted postcapture using Adobe Photoshop software, through a single adjustment of the levels histogram. Deconvolution images were obtained on an inverted Olympus IX-71 microscope (Deltavision Image Restoration Microscope; Applied Precision/Olympus) and a Photometrics CoolSnap QE camera with a 100 × 1.4 NA oil immersion lens. Images were de-convolved and overlaid using DeltaVision SoftWoRx software version 3.7.1, with a single adjustment of the levels histogram.
Antibody and Fluorescent Reagent Uptake
C2C12 cells were grown on thermanox coverslips, transfected, and differentiated as above. For dysferlin endocytosis studies, myotubes differentiated for 2 or 3 days were incubated with anti-MycAlexa555 (1:100), anti-His Alexa647(1:100), and/or anti-FLAG (1:100) in the culture medium at 37 °C. Coverslips were removed after 15 min or 3 h, acid washed (0.5 m glycine, pH 2.2) to remove surface-bound antibody, washed briefly in HBSS, fixed in 3% PFA (paraformaldehyde), and mounted for confocal microscopy. Similarly, for studies using un-conjugated antibodies, cells were incubated in primary antibodies for 90 min or 3 h at 37 °C, acid washed, fixed in 3% PFA, permeabilized with 0.15% saponin, and incubated with an appropriate species-specific secondary antibody as described in the legend. For cholera toxin uptake, cells were incubated in medium containing cholera toxinAlexa488 (1:100) and anti-MycAlexa555 (1:100) for 2 h at 37 °C, then acid washed, fixed, and mounted as above. For LDL uptake, transfected differentiated C2C12 myotubes were serum starved overnight, then incubated in LDLdil (33 μg/ml) for 30 min at 37 °C, acid washed, fixed, and mounted as above. For dextran loading, cells were incubated in fluorescent dextran overnight (∼16 h), rinsed, and incubated with anti-Myc antibody for 90 min at 37 °C before acid washing, fixing, and secondary antibodies as above.
Cycloheximide, MG132, Brefeldin-A, and Bafilomycin Treatment
C2C12 cells were grown in 12-well plates, transfected, and differentiated as above. Cells were either harvested at t0 or incubated at 37 °C in medium containing 100 μg/ml cycloheximide (Sigma-Aldrich) and/or 5 μg/ml brefeldin A (Sigma-Aldrich) and/or 5 μm MG132 (Merck, Darmstadt, Germany) and/or 10 μm bafilomycin (Sigma-Aldrich).
Cell Lysis
Cells were washed with cold PBS, and incubated in RIPA buffer (50 mm Tris pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, and 1 mm EDTA) containing freshly added protease inhibitors (Sigma-Aldrich) for 30 min, rocking gently, on ice. Samples were collected and centrifuged for 5 min at 4 °C, 18,000 × g to pellet cell debris. Supernatants were removed and stored at −80 °C for Western blotting.
Western Blotting, Densitometry, and Half-life Calculations
Protein concentration was determined with a BCA protein assay (Pierce, Thermo Fisher). 50 μg of protein of FL and 10 μg of C2F-TM were separated by SDS-PAGE using NuPAGE 3–8% gradient gels (Invitrogen). Proteins were transferred to PVDF membranes, blocked (PBS containing 5% skim milk powder, 0.01% Tween) and incubated in NCL-Hamlet (1:500) or anti-Myc (1:500) overnight at 4 °C with rotation. After washing, membranes were incubated in anti-mouseHRP secondary antibody (1:2000) at room temperature for two hours and developed using ECL detection reagents (GE Healthcare Biosciences, Buckinghamshire, UK). Densitometry was performed with a Bio-Rad GS-800 calibrated densitometer using Quantity One software. For half-life calculations, experiments with multiple time points were plotted on a log density versus time graph, and half-life derived from a line of best fit using the equation: half-life = log(2)/gradient. For experiments involving replicates at a single time point, the half-life equation: half-life = elapsed time × [log(2)]/log(beginning density/ending density) was used.
Flow Cytometry
Transfected C2C12 myotubes on the third day of differentiation were treated with or without 5 μg/ml brefeldin A prior to harvesting. Cells were dissociated from the plate through incubation for 15 min at 37 °C in Versene (0.48 mm EDTA·4Na in PBS), with or without supplementation with brefeldin A. Cells were then cooled rapidly in ice-cold Ca/Mg-free PBS, triturated to a single cell suspension using a pipette tip precoated with Ca/Mg-free PBS containing 1% BSA, and pelleted by centrifugation at 250 × g for 5 min at 6 °C. Cells were resuspended in Ca/Mg-free PBS containing 1% BSA with anti-His (1:200) for 1 h at 4 °C, washed twice, pelleted as before, then resuspended in buffer containing donkey anti-mouse IgGAlexa647 (1:200) for 1 h at 4 °C. Cells were then washed twice and resuspended in Ca/Mg-free PBS. Cells with compromised membranes were identified through addition of 10 μl (0.5 μg) 7-amino-actinomycin D (7-AAD, BD Biosciences) to 300 μl of cell suspension. Flow cytometry was performed using a BD LSRII cytometer equipped with FACSDivaTM software (BD Biosciences). Postacquisition analysis was performed using FlowJo software (version 7.2.5) (Tree Star Inc.).
RESULTS
The Effect of Dysferlin Domain Deletions upon Plasma Membrane Targeting and Vesicular Localization
Dysferlin's modular structure, with seven putative cytoplasmic C2 domains, led us to consider the potential for specific roles of particular domains upon dysferlin trafficking and plasma membrane biology. We utilized an expression construct bearing an N-terminal EGFP dysferlin fusion protein with extracellular C-terminal Myc- and His-epitope tags (23), from which we derived a panel of dysferlin deletion mutants (Fig. 1a). Cell surface labeling of live cells using the extracellular epitope tag was used to assess plasma membrane targeting by confocal microscopy (Fig. 2a) and quantified by flow cytometry (Fig. 2b). Previous use of flow cytometry to analyze isolated myofibres (24) initiated our development of this quantitative approach, analyzing thousands of cells to produce a normal distribution of cell surface labeling, effectively normalizing cell-to-cell variability. All cells within a population of differentiated myotubes successfully transited the flow cytometer in hydrodynamic flow, producing a broad scatter profile of cells of increasing size with differentiation.
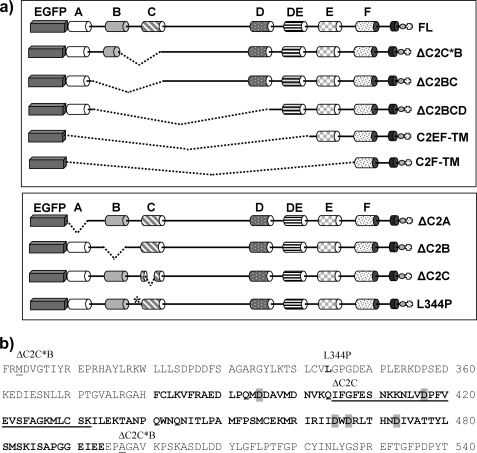
FIGURE 1.
Dysferlin deletion constructs. a, schematic of the dysferlin deletion constructs highlighting the domain deletions for each expression construct. Cylinders A → F mark C2 domains, TM denotes the transmembrane domain, and the circle and cross symbolize Myc and His epitope tags. b, dysferlin amino acid sequence surrounding the C2C domain. Predicted residues constituting the eight β-sheets within the C2C domain are in bold typeface, with the putative aspartic acid residues implicated in calcium-binding (based on homology and alignment with synaptotagmin C2A) highlighted in gray. The 28-residue microdeletion of expression construct ΔC2C is underlined. The first and last residues of the larger deletion of ΔC2C*B are annotated and underlined. The patient missense mutant L344P is annotated and in bold typeface.
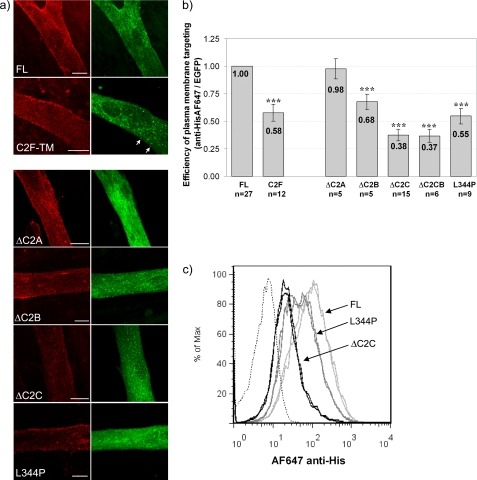
FIGURE 2.
Efficiency of plasma membrane targeting of dysferlin deletion constructs. a, confocal analysis of surface anti-MycAlexa555 and cellular EGFP fluorescence. Transfected and differentiated C2C12 myotubes were labeled with anti-Mycalexa555 at 4 °C to specifically label plasma membrane dysferlin (red) with absent cytoplasmic immunoreactivity to surface applied antibodies confirming maintenance of an intact plasma membrane (not shown). Striking EGFP vesicles are apparent for C2F-TM (green, C2F-TM, arrows), and are also clearly observed with ΔC2B, ΔC2C, and patient missense mutant L344P. Scale bars, 10 μm. b, quantification of cell surface dysferlin by flow cytometry. Populations of transfected and differentiated C2C12 myotubes were surface labeled with anti-Hisalexa647 and subject to flow cytometry. A histogram represents cumulative data of the relative efficiency of plasma membrane targeting for each dysferlin deletion construct over many experiments. For each experiment, transfected cells were selectively analyzed (see supplemental Fig. S1), and levels of surface dysferlin labeled by anti-HisAlexa647 versus levels of total EGFP fluorescence quantified (see “Experimental Procedures”). For each construct, results are presented relative to results observed for FL dysferlin within the same experiment. A highly significant reduction in plasma membrane dysferlin is observed for C2F-TM, ΔC2B, ΔC2C, ΔC2C*B and patient missense mutant L344P. Statistical significance was assessed using a Mann Whitney non-parametric test, with p < 0.001 represented by three stars (***). The error bars represent the 95% confidence interval. c, representative flow cytometry data derived from duplicates within a single experiment, assessing plasma membrane expression of FL dysferlin (light gray), ΔC2C (dark gray), and patient missense mutant L344P (black). Nonspecific labeling was determined using an IgG Alexa647 isotype control (black dotted line). A clear reduction in the surface anti-HisAlexa647 labeling is observed for cells expressing ΔC2C and L344P dysferlin constructs, relative to that observed for the reference FL dysferlin construct. Levels of total EGFP fluorescence were comparable for each construct. Separately transfected experimental duplicates were highly consistent, as shown, and data presented are derived from n = 4000–6000 cells for each construct in each experiment.
All dysferlin deletion constructs targeted the plasma membrane, with varying efficiencies (Fig. 2, a and b). C2C12 myotubes transfected with FL dysferlin showed punctate labeling of the surface membrane (Fig. 2a, FL, red), and a mixed pattern of cellular EGFP fluorescence, labeling intracellular networks, surface membranes (filapodia and lamellapodia), and a population of small vesicles (Fig. 2a, FL, green). The vesicle population was noted due to the striking abundance of vesicular labeling with several dysferlin expression constructs, particularly truncated mutants C2EF-TM and C2F-TM (shown for C2F-TM, Fig. 2a, arrows). Strong vesicular labeling for C2F-TM was similarly observed in both brightly fluorescent cells, and weakly fluorescent cells, suggesting that vesicular localization was not determined by protein overexpression. Prominent vesicle labeling was also observed with three constructs bearing different internal deletions within the dysferlin N-terminal domain (not shown); ΔC2BC dysferlin, ΔC2C*B dysferlin, and ΔC2BCD dysferlin (see Fig. 1a, upper panel), united by disruption of the region encompassing C2B and C2C. Thus, we derived four more expression constructs (see Fig. 1, a, lower panel, and b); three constructs bearing a specific microdeletion of C2A, C2B, or C2C, and one construct bearing missense mutation L344P, based on a patient from our Australian dysferlinopathy cohort (Fig. 1b). Leu344 is a highly conserved residue within the Fer1 domain, sandwiched between C2 domains C2B and C2C.
The C2B-FerI-C2C Motif Critically Regulates Dysferlin Plasma Membrane Expression
Confocal microscopy revealed the prominent vesicular labeling observed with C2F-TM was recapitulated by any construct with a disruption within the C2B-FerI-C2C motif (Fig. 2a, ΔC2B, ΔC2C, L344P). ΔC2C showed the most consistently profound vesicular localization (Fig. 2a, ΔC2C, green), and was also accompanied by the greatest reduction in plasma membrane expression levels (2.5-fold, Fig. 2b). Reduced plasma membrane expression levels (Fig. 2b), and prominent vesicular labeling (Fig. 2a) was also obvious, but less consistently dramatic, with L344P and ΔC2B. In contrast, ΔC2A showed normal plasma membrane expression levels without overt intracellular vesicular labeling. The trend for lower levels of plasma membrane expression correlating with increased vesicular labeling was also observed with severely truncated C2F-TM, though only to 60% levels of FL dysferlin (discussed in more detail in Fig. 7).
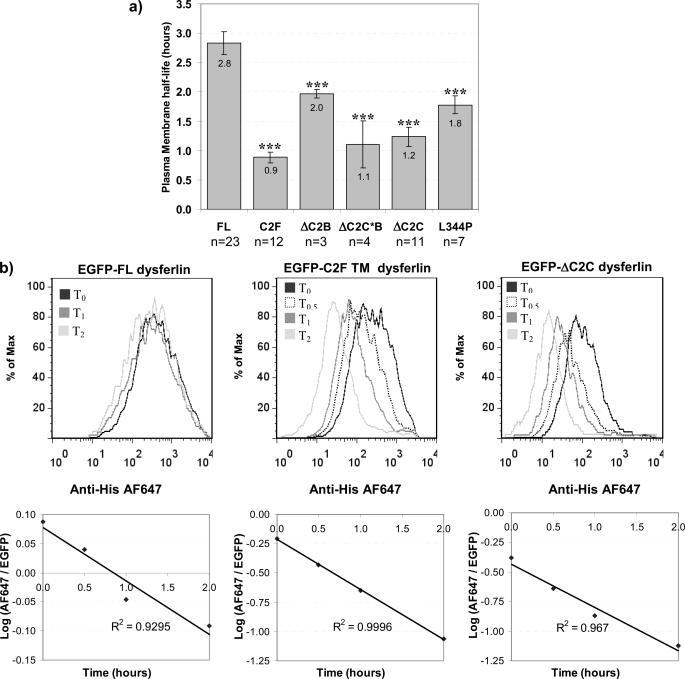
FIGURE 7.
Determination of the plasma membrane half-life of dysferlin. Transfected and differentiated C2C12 myotubes (on the third day of differentiation) were incubated with or without brefeldin A for the times indicated. Cells were harvested and labeled with anti-HisAlexa647 as described under “Experimental Procedures.” Immediately prior to flow cytometry analysis, cells were incubated in the presence of actinomycin D (7-AAD), to identify any cells with compromised membranes. Cells labeled by 7-AAD were excluded and cells positive for EGFP fluorescence analyzed. a, histogram representing cumulative data of the calculated plasma membrane half-life for dysferlin expression constructs. Plasma membrane half-lives were calculated using the geometric mean of the normal distribution of the anti-HisAlexa647 fluorescence, relative to the geometric mean of the normal distribution of the EGFP fluorescence, for each cell population, at each time point. Data from between 3 and 23 experiments were averaged, as indicated. Error bars are 95% confidence intervals. Statistical significance was assessed using a Mann Whitney non-parametric test, with p < 0.001 represented by three stars (***). b, representative graph of flow cytometry data demonstrating surface labeling for transfected cells expressing EGFP-FL dysferlin, C2F-TM dysferlin, and ΔC2C dysferlin, as annotated. Note the incremental decline in plasma membrane expression for expressed dysferlin constructs with increasing periods of brefeldin A treatment. The Alexa647/EGFP fluorescent ratio for each time point was plotted on a semi-log plot as described under “Experimental Procedures.” Only experiments with a line-of-best-fit of r2 ≥ 0.9 were included.
Of note, discreet disruption of ΔC2C (28 amino acid deletion) induced the same degree of vesicular labeling and reduction in plasma membrane expression as did ΔC2C*B (both by 2.5-fold, Fig. 2b), which bears a much larger deletion of 204 amino acids encompassing C2C, FerI and perhaps also disrupting C2B (depending on exact domain boundaries, see Fig. 1b). These results suggest a critical requirement for C2C, and the C2B-Fer1-C2C motif generally, for normal plasma membrane expression of dysferlin.
Intracellular Vesicles for C2F-TM Are Endocytic
The correlation between prominent vesicular labeling and reduced levels of dysferlin plasma membrane expression raised the possibility of endosomal internalization of cell surface dysferlin. To address this, we performed antibody uptake assays in C2C12 myotubes transfected with either FL dysferlin or severely truncated C2F-TM, to determine if intracellular vesicles observed at steady state for C2F-TM were endocytic. After a three hour antibody uptake, all intracellular vesicles for both FL and C2F-TM dysferlin were co-labeled with endocytosed anti-Myc555 (red) and EGFP, indicating EGFP-dysferlin labeled vesicles were endosomal (Fig. 3a). When antibody uptake was limited to 15 min, considerably more anti-Myc555 uptake was observed for C2F-TM, relative to FL dysferlin (supplemental Fig. S2). Together, these results suggest truncated C2F-TM is more rapidly endocytosed following export to the plasma membrane, than is FL dysferlin.
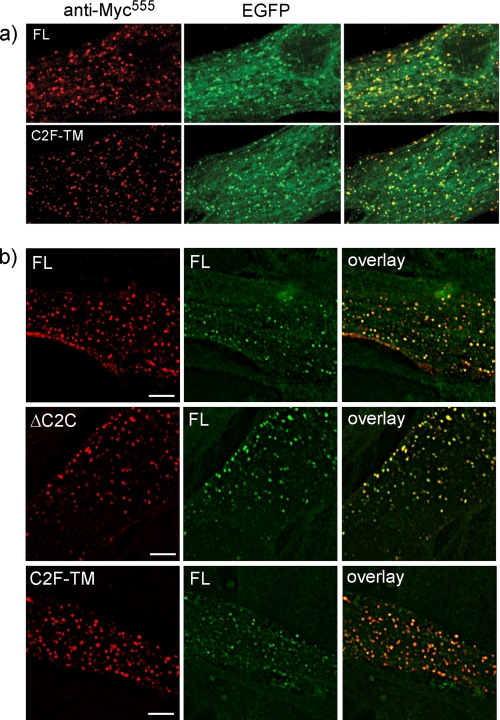
FIGURE 3.
Dysferlin vesicles are endocytic and C2F-TM and ΔC2C co-endocytose with full-length dysferlin. a, transfected and differentiated myotubes were incubated in anti-Mycalexa555 for 3 h at 37 °C showing endocytosed anti-Mycalexa555 antibody (red) and cellular EGFP (green) indicating vesicles formed by dysferlin expression are endocytic. b, cells co-transfected with FLmyc and FLhis (top row) or FLflag and ΔC2Cmychis (middle row) or FLhis and C2Fmyc (third row). Antibody uptake was performed using rabbit α-Myc and mouse α-His or mouse α-Flag with differently fluorescent secondary antibodies (rabbit Alexa-647 and mouse Alexa-555) to show the presence of both dysferlin species in the same vesicle, marked by arrows (deconvolved Deltavision images). Alexa647-labeled vesicles have been false colored green (from blue) for best reproduction. Scale bars, 10 μm.
C2F-TM and ΔC2C Co-endocytose with Full-length Dysferlin within Endosomal Compartments Labeled by Syntaxin-4 and Cholera Toxin
To determine whether dysferlin deletion constructs C2F-TM and ΔC2C highlight trafficking pathways also employed by FL dysferlin, we derived uniquely tagged expression constructs and performed pairwise co-transfections (i.e. C2F-TMMyc with FLHis, ΔC2CMycHis and FLFlag, FLMycHis, and FLFlag). Antibody uptake assays were performed using two antibodies conjugated with different colored fluorophores, to uniquely label each dysferlin species within a pairwise co-transfection (Fig. 3b). FL dysferlin is co-endocytosed with ΔC2C and C2F-TM, within the same endocytic vesicle, as shown by dual labeling of endocytic vesicles with both red and blue antibody fluorophores (Fig. 3b, arrows). Thus, rapid endocytosis of mutant ΔC2C and C2F-TM does not appear due to abnormal targeting to atypical plasma membrane microdomains, nor usage of an alternate endosomal trafficking route. Co-endocytosis of FL dysferlin with ΔC2C or C2F-TM was similarly observed using FL dysferlin lacking the EGFP fusion protein, and using different combinations of extracellular epitope tags (not shown).
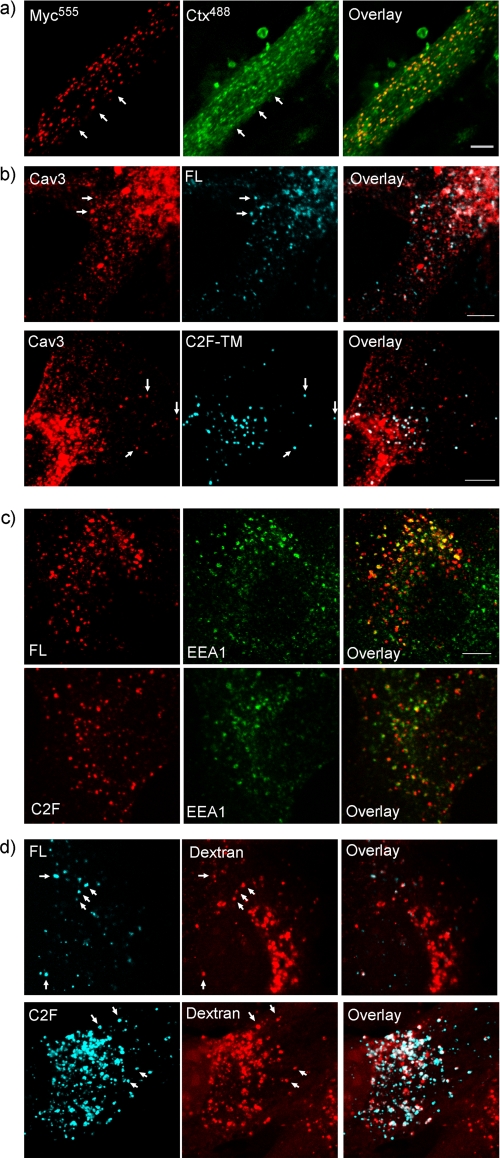
To define the endocytic route of dysferlin, we used characterized markers of various endocytic pathways. Dysferlin endocytic vesicles co-labeled with fluorescent cholera toxin, known to bind a specific ganglioside of lipid rafts (Fig. 4a), and with caveolin-3 (Fig. 4b), but did not show significant co-labeling with transferrin (not shown). These results are in agreement with previous studies demonstrating caveolin-dependent, cholera toxin-associated, but transferrin-independent dysferlin endocytosis in mouse embryonic fibroblasts (18).
FIGURE 4.
Dysferlin endocytic vesicles label with cholera toxin and caveolin-3 and transit from early endosomes to lysomes in ∼1.5 h. Co-labeling of dysferlin vesicles with various markers of endocytic compartments. a, differentiated myotubes transfected with FL dysferlin lacking the EGFP tag were incubated with α-MycAlexa555 with cholera toxinAlexa488 at 37 °C, with co-labeled vesicles marked by white arrows. b, dysferlin vesicles marked by uptake of anti-Mycalexa647 (blue) also labeled with caveolin-3 (red) in cells transfected with both FL (upper row) and C2F-TM dysferlin (lower row). c, endocytic vesicles produced by expression of either FL (top row) or C2F-TM dysferlin (bottom row) and marked by a 1.5 h anti-MycCy3 antibody uptake (red) also labeled with EEA1alexa647, a marker of early endosomes (green) labeled with arrows. EEA1alexa647 staining has been false-colored green (from blue) for best reproduction. d, endocytosed dysferlin (myc uptake, blue) is seen in vesicular compartments labeled by overnight fluorescent dextran loading (red, see “Experimental Procedures”), indicating that a proportion of dysferlin endocytosed within this time frame has reached lysosomes (overlay, white, arrows). Scale bars, 5 μm.
We also tracked dysferlin endocytosis from EEA1-labeled (Fig. 4c) or Rab-5 labeled (not shown) early endosomes, through to dextran-loaded lysosomes (Fig. 4d), using a 90 min anti-His647antibody uptake. Dysferlin transit to the late endosomal/lysosomal compartment was also confirmed using fluorescently conjugated low density lipoprotein (LDL, supplemental Fig. S3), a marker of the late endosomal/lysosomal pathway. More dextran and LDL positive vesicles were observed for C2F-TM compared with FL dysferlin (Fig. 4d). Thus, within 90 min, a large proportion of C2F-TM dysferlin transits from the early endosomal compartment through to late endosomes/lysosomes, suggesting that lysosomal degradation may be an end point for truncated C2F-TM following endocytosis.
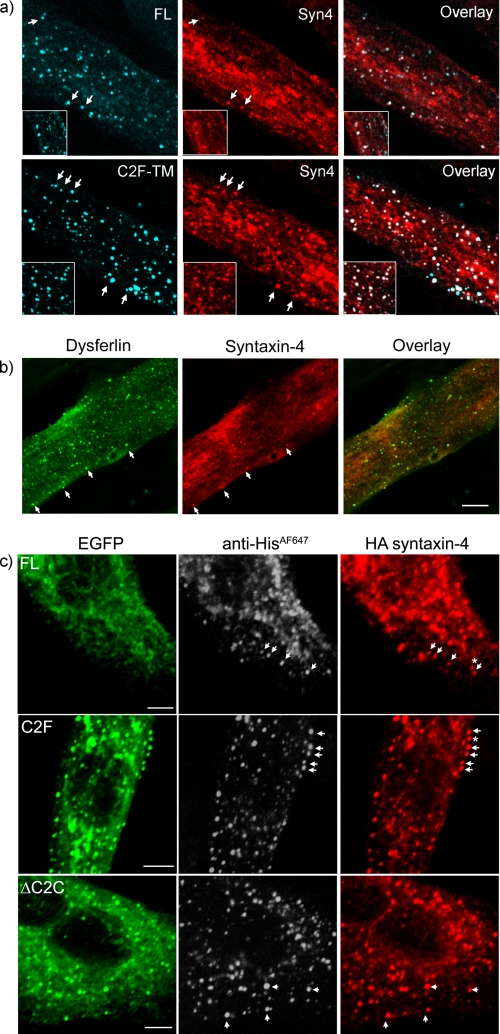
Given the recent study associating otoferlin with syntaxin-1 (25), we explored the relationship between dysferlin and syntaxin-4, which localizes to the plasma membrane and is highly expressed in skeletal muscle (26). Antibodies recognizing endogenously expressed syntaxin-4 co-labeled FL and C2F-TM dysferlin endocytic vesicles in transfected myotubes (Fig. 5a). We also observed evidence for syntaxin-4/dysferlin co-labeled vesicles in untransfected myotubes (Fig. 5b); though in this case we could not confirm their endosomal origin via antibody uptake (an antibody raised to the short extracellular tail of dysferlin was tried unsuccessfully). To specifically examine endocytosed syntaxin 4, we co-transfected dysferlin with syntaxin-4-HA, which bears an extracellular HA tag (27). Antibody uptake experiments using anti-His and anti-HA antibodies revealed a striking co-labeling of dysferlin- and syntaxin-4-positive endosomal compartments (Fig. 5c), observed with all dysferlin mutants (including ΔC2C and patient missense L344P), and also with wild-type dysferlin lacking the N-terminal EGFP fusion protein (not shown). For truncated C2F-TM following a 90 min anti-His647 antibody uptake (where all endosomal compartments from early endosomes through to late endosomes/lysosomes are labeled by anti-His647), virtually every endosomal vesicle showed labeling for both dysferlin and syntaxin-4 (Fig. 5c, middle row), suggesting these two proteins similarly transit all maturation stages of this endosomal pathway.
FIGURE 5.
Dysferlin endocytic vesicles contain syntaxin-4. a, confocal analysis of differentiated myotubes transfected with FL (upper row) or C2F-TM dysferlin (lower row). Endocytic vesicles labeled with anti-Mycalexa647 (blue) also labeled with endogenous syntaxin-4 (red). b, vesicles observed in untransfected differentiated myotubes labeled with both dysferlin (NCL-hamlet 1, green) and syntaxin-4 (red). c, cells transfected with dysferlin and syntaxin 4. Endocytic dysferlin vesicles (shown with GFP, green, and HisAlexa647 uptake, gray scale) formed by expression of FL (top row), ΔC2C (middle row), or C2F-TM (third row) also labeled positively with syntaxin 4 (HA staining, red). Scale bars, 5 μm.
Accelerated Endocytosis of C2F-TM Increases Protein Turnover and Lability
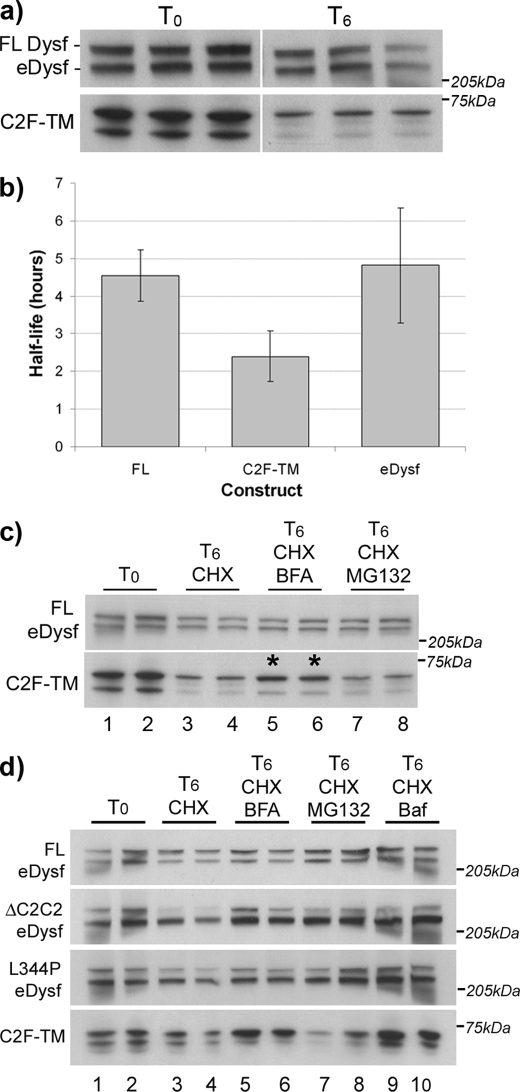
Protein half-life studies were used to assess whether the rapid transit of C2F to lysosomal compartments reflected greater protein lability, by using cycloheximide to block cytoplasmic protein synthesis. Studies of dysferlin protein decay over a time-course of cycloheximide treatment showed FL dysferlin to have an average protein half-life of 4.6 ± 0.7 h (Fig. 6, a and b), similar to endogenously expressed murine dysferlin (t½ = 4.8 ± 1.5 h). In contrast, more rapid decay was observed for C2F-TM (t½ = 2.4 ± 0.7 h), that could be blocked by preventing secretion of C2F-TM to the plasma membrane using brefeldin A (which collapses the Golgi complex and prevents export from the endoplasmic reticulum to the plasma membrane), and using bafilomycin to prevent endosomal acidification (a specific blocker of vacuolar ATPase). In contrast, MG132, a specific inhibitor of the ubiquitin proteosome (targeting misfolded proteins from the endoplasmic reticulum), did not protect C2F-TM from degradation. Therefore, rapid protein turnover of C2F-TM appears largely due to endosomal proteolysis, without clear evidence for misfolding and degradation from the endoplasmic reticulum (ERAD) (Fig. 6, c and d).
FIGURE 6.
Half-life analysis of dysferlin expression constructs. Transfected and differentiated C2C12 myotubes (on the third day of differentiation) were incubated with or without cycloheximide (100 μg/ml) and other drugs for the time periods indicated. Duplicate or triplicate wells for each condition were harvested in RIPA buffer and 50 μg (for FL, DC2C2, and L344P) or 10 μg of total protein (for C2F-TM) was separated on a 3–8% gradient gel and Western blotted. Blots were probed with NCL-Hamlet, and levels of dysferlin expression assessed through densitometry. a, treated cells were harvested at t0, t2, t6, t12 (t6 shown). eDysf represents endogenous dysferlin, detected by both Hamlet I and Hamlet II antibodies (not shown). eDysf has a lower apparent molecular mass than transfected FL-dysferlin due to conjugation of the 29 kDa EGFP tag. The doublet band observed for C2F-TM is presumed to be a breakdown product. b, graphed data represents averaged half-life calculations based on four or five separate experiments. For each individual experiment, density readings for duplicates or triplicates were averaged, and half-life calculations were performed as described under “Experimental Procedures.” c, brefeldin A protects C2F-TM from degradation (asterisks), whereas MG132 has little effect on protein levels. d, ΔC2C2, L344P and C2F-TM dysferlin are all more labile then FL dysferlin after 6 h of cycloheximide treatment (lanes 3 and 4), with partial protection of degradation by brefeldin A (lanes 5 and 6) and bafilomycin (lanes 9 and 10). MG132 treatment of cells transfected with ΔC2C2 and L344P dysferlin also affords partial protection from cycloheximide-induced protein degradation (lanes 7 and 8).
ΔC2C and L344P showed greater protein lability than FL dysferlin, and were also partially protected from degradation by incubation with brefeldin A and bafilomycin. Interestingly, MG132 also partially protected ΔC2C and L344P from degradation, suggesting a role for the ubiquitin proteosome in their proteolysis. This contrasts with truncated mutant C2F-TM, unaffected by MG132, perhaps reflecting greater folding constraints of the much larger full-sized dysferlin mutants (Fig. 6d).
Disruption of the C2B-FerI-C2C Motif Reduces Dysferlin Plasma Membrane Expression via Accelerated Endocytosis
We sought to quantify the plasma membrane half-life of dysferlin, and the effect of our specific mutations on accelerating dysferlin endocytosis. Previous studies with brefeldin A had demonstrated that dysferlin relied upon an intact Golgi apparatus for cell surface export (supplemental Fig. S4), and thus we employed a time course of brefeldin A treatment to examine the time-dependent loss of dysferlin from the cell surface across a large cell population using flow cytometry analysis. This approach presents the significant advantage of examining the rate of endocytosis, independently from antibody-induced or ligand-induced endocytosis, and independently of endosomal recycling (which requires an intact Golgi network).
Fig. 7a depicts a histogram of cumulative plasma membrane half-life data for each dysferlin expression construct over many experiments. Fig. 7b shows representative data from a single experiment examining the plasma membrane half-life of FL dysferlin, and deletion mutants ΔC2C and C2F-TM. The upper panels of Fig. 7b show the normal distributions of cell surface dysferlin labeling, which incrementally decrease as a function of time (t0, t30 min, t1 h, t2 h), once all further export and recycling of dysferlin is blocked by treatment with brefeldin A. Normalizing cell surface dysferlin (anti-His647) relative to total dysferlin (EGFP) over time was then used to derive the plasma membrane half-life (half-life = log(2)/gradient, see “Experimental Procedures”).
These analyses were consistent and reproducible, and estimated the plasma membrane half-life (PM t½) of FL dysferlin to be ∼3 h (PM t½ = 2.8 ± 0.2 h). In contrast, C2F-TM was endocytosed 3-fold faster (PM t½ = 0.9 ± 0.1 h) and ΔC2C 2.5-fold faster (PM t½ = 1.2 ± 0.2 h). Again, ΔC2C (28 amino acid microdeletion) and ΔC2C*B (206 amino acid deletion) behaved similarly, supporting a crucial role for the C2C domain for plasma membrane stability. ΔC2B and patient missense mutant L344P also showed a significant, but less marked reduction in plasma membrane half-life (PM t½ = 1.8 ± 0.2 h) relative to ΔC2C, supporting a general requirement for an intact C2B-FerI-C2C domains for normal regulation of dysferlin plasma membrane expression.
Endocytic Rate Is a Major Determinant of Dysferlin Plasma Membrane Expression Levels
When we compared the levels of plasma membrane dysferlin (Fig. 2), with the plasma membrane half-life (Fig. 7), we observed a striking correlation between the fold-acceleration in endocytic rate, and the fold-reduction in plasma membrane expression levels (Table 1). This relationship bore out for all full-sized dysferlin constructs, and suggests that even with some degree of misfolding (as suggested in Fig. 6d by partial MG132-mediated protection of ΔC2C and L344P proteolysis), endocytosis remains a primary regulator of dysferlin plasma membrane expression. This correlation did not hold true for truncated C2F-TM, where marked acceleration of endocytosis (3-fold) was not reflected in a similar fold-reduction in plasma membrane expression levels (reduced by only 40%). These results may yet be reasonable, whereby enhanced biosynthesis and export of much smaller C2F-TM assists plasma membrane expression levels, despite its extremely transient plasma membrane half-life.
TABLE 1.
Endocytic rate is a primary regulator of dysferlin surface expression
| Dysferlin expression construct | Predicted steady-state plasma membrane dysferlin (based on half-life) | Actual steady-state plasma membrane dysferlin |
|---|---|---|
| EGFP-FL dysferlin | 1.00 (PM t½ = 2.8 ± 0.2 h) | 1.00 |
| EGFP-C2F dysferlin | 0.32 (PM t½ = 0.9 ± 0.1 h) | 0.58 |
| EGFP-ΔC2B dysferlin | 0.71 (PM t½ = 2.0 ± 0.1 h) | 0.68 |
| EGFP-ΔC2C dysferlin | 0.43 (PM t½ = 1.2 ± 0.2 h) | 0.38 |
| EGFP-ΔC2C*B dysferlin | 0.40 (PM t½ = 1.1 ± 0.4 h) | 0.37 |
| EGFP-L344P dysferlin | 0.64 (PM t½ = 1.8 ± 0.2 h) | 0.55 |
Our results do not support that accelerated endocytosis results from excess plasma membrane dysferlin (because of overexpression); in this case, plasma membrane expression levels for each of the different constructs would be expected to be more broadly similar. Instead, each mutant construct consistently showed a similar fold reduction in efficiency of plasma membrane expression relative to wild-type dysferlin, over many experiments (see Fig. 2). Overall, the correlation between endocytic rate and plasma membrane expression levels for full-sized dysferlin proteins is striking and leads us to propose endocytosis as a major regulator of dysferlin plasma membrane expression.
DISCUSSION
Dysferlin is a tail-anchored protein, with multiple tandem C2 domains that are likely to underlie its role in calcium-dependent and vesicle-mediated membrane resealing. We sought to further define the basic cell biology of dysferlin, as a reference to study the consequences of patient-derived mutations, and to define the roles of calcium and C2 domains upon dysferlin trafficking and membrane biology. The ferlins are an ancient family of proteins,4 and it is likely that they possess primordial functions in vesicular trafficking that can be revealed through studies of dysfunction in human disease. Synaptotagmin 1 possesses two C2 domains, with each showing distinct calcium-regulated binding to different phospholipid membranes (28, 29), and different SNARE proteins (30, 31). Dysferlin has seven putative C2 domains, of which four show potential calcium sensitivity (32), suggesting even more modular complexity.
Our studies reveal dysferlin to be a transitory transmembrane protein in cultured myotubes, with an average plasma membrane half-life of ∼3 h, and a reasonably short-lived protein half-life of ∼4–6 h. Transfected myotubes provide a good myogenic model to derive baseline information of dysferlin cell biology. This information is highly relevant for the development of gene replacement therapies to treat muscular dystrophy caused by loss of dysferlin, and perhaps also relevant to otoferlin replacement strategies in human deafness. However, it is important to acknowledge that dysferlin behavior may be differentially regulated in different cellular compartments, and in different cell types.
To study domain-specific roles for dysferlin, we constructed a panel of dysferlin deletion constructs. We identified a requirement for the dysferlin C2B-Fer1-C2C region for stable plasma membrane expression. Disruption or mutation of each domain within the C2B-Fer1-C2C motif resulted in reduced plasma membrane expression due to accelerated endocytosis, and, increased protein lability from endosomally driven proteolysis. Targeted disruption of C2C-destabilized dysferlin plasma membrane expression most profoundly accelerating dysferlin endocytic rate by 2.5-fold. Our previous collaborative studies have shown that caveolin deficiency results in more rapid dysferlin endocytosis via a caveolin-independent pathway in mouse embryonic fibroblasts (18, 19). In our current study, rapidly endocytosed dysferlin mutants were shown to maintain an association with caveolin-3-positive endosomes in cultured myotubes. Thus, employing a caveolin-independent endocytic route does not easily explain our findings, though altered dysferlin/caveolin-3 endocytic regulation cannot be excluded. Importantly, we show that a patient missense mutation also displays reduced plasma membrane expression due to accelerated endocytosis, which also increases protein lability. This is the first evidence for endosomally driven proteolysis of dysferlin as the mechanism underlying dysferlin lability in dysferlinopathy. We are now exploring whether accelerated endocytosis is a feature of other patient missense mutants, particularly those affecting the C2B-FerI-C2C motif.
On the basis of an identified association between otoferlin and syntaxin I in cochlear inner hair cells (25), we explored a relationship between dysferlin and the muscle counterpart of syntaxin-1, syntaxin-4. Syntaxin-4 positively labeled dysferlin endocytic vesicles, from early endosomes to late endosomes/lysosomes, suggesting both proteins similarly transit a common endosomal pathway. This dysferlin/syntaxin-4 endocytic pathway may be relevant to studies of known syntaxin-4-dependent cargo; syntaxin-4 is involved in the trafficking of GLUT4 receptors in response to insulin (33–35), and has also been associated with cytokine release in macrophages (36). This inflammatory association is particularly interesting given the marked inflammatory response in dysferlinopathy (37) and the abnormal immune signaling in the dysferlin-deficient mouse (38–41). Of note, the dysferlin/syntaxin-4 endosomal association is maintained even when dysferlin is severely truncated (C2F-TM). The syntaxin-1 interactive domain of otoferlin was mapped to C2F (25), highly conserved between paralogues, and our ongoing studies are exploring the potential of a direct interaction mediated by dysferlin C2F and syntaxin-4.
In summary, our data establish dysferlin as a transient plasma membrane protein subject to endocytic regulation of cell surface expression levels relevant to a potential plasma membrane- or vesicle-based role in membrane resealing, and a likely basis for abnormal dysferlin localization in muscle of patients with muscular dystrophy (21, 22). We identify a key role for the C2B-FerI-C2C motif for dysferlin plasma membrane expression, and propose that increased protein lability due to accelerated endocytosis may underpin disease pathogenesis in some cases of dysferlinopathy. Significantly, we identify a dysferlin and syntaxin-4 endocytic pathway, relevant for known syntaxin-4 dependent cargo, and perhaps also more widely significant in the pathology of dysferlinopathy. Our results raise questions as to whether accelerated endocytosis of dysferlin mutants reflects abnormal exocytic delivery of dysferlin-rich vesicles that are then endocytosed, active removal of malfunctioning dysferlin, or dysregulation of a primary endocytic role that may also influence other endocytic cargo. Distinguishing between these possibilities is now essential to understanding the molecular role dysferlin plays in membrane resealing and the pathology of dysferlinopathy.
Supplementary Material
Acknowledgments
We thank the patients and their families for participation and support, and the clinicians involved in their care, especially Dr. Lesley McGregor for referring the patients in this study to our laboratory. We thank Prof. Kate Bushby and Prof. David James for kindly distributing expression constructs.
This work was supported by the Muscular Dystrophy Association, USA (STC Development Grant), the NSW Muscular Dystrophy Association (to S. T. C. and K. N. N.), the Australian National Health and Medical Research Council (301946 to K. N. N. and S. T. C.), the Brain Foundation Australia (to S. T. C.), and the Jain Foundation (to S. T. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
A. Lek, M. Lek, K. N. North, and S. T. Cooper, submitted manuscript.
- FL
- full-length.
REFERENCES
- 1.Washington N. L., Ward S. (2006) J. Cell. Sci. 119, 2552–2562 [DOI] [PubMed] [Google Scholar]
- 2.Bashir R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z., Sadeh M., Mahjneh I., Marconi G., Passos-Bueno M. R., Moreira Ede S., Zatz M., Beckmann J. S., Bushby K. (1998) Nat. Genet. 20, 37–42 [DOI] [PubMed] [Google Scholar]
- 3.Liu J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J. A., Hentati F., Hamida M. B., Bohlega S., Culper E. J., Amato A. A., Bossie K., Oeltjen J., Bejaoui K., McKenna-Yasek D., Hosler B. A., Schurr E., Arahata K., de Jong P. J., Brown R. H., Jr. (1998) Nat. Genet. 20, 31–36 [DOI] [PubMed] [Google Scholar]
- 4.Bansal D., Miyake K., Vogel S. S., Groh S., Chen C. C., Williamson R., McNeil P. L., Campbell K. P. (2003) Nature 423, 168–172 [DOI] [PubMed] [Google Scholar]
- 5.McNeil P. L., Vogel S. S., Miyake K., Terasaki M. (2000) J. Cell. Sci. 113, 1891–1902 [DOI] [PubMed] [Google Scholar]
- 6.Gerasimenko J. V., Gerasimenko O. V., Petersen O. H. (2001) Curr. Biol. 11, R971–R974 [DOI] [PubMed] [Google Scholar]
- 7.Idone V., Tam C., Goss J. W., Toomre D., Pypaert M., Andrews N. W. (2008) J. Cell. Biol. 180, 905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasunaga S., Grati M., Cohen-Salmon M., El-Amraoui A., Mustapha M., Salem N., El-Zir E., Loiselet J., Petit C. (1999) Nat. Genet. 21, 363–369 [DOI] [PubMed] [Google Scholar]
- 9.Yasunaga S., Grati M., Chardenoux S., Smith T. N., Friedman T. B., Lalwani A. K., Wilcox E. R., Petit C. (2000) Am. J. Hum. Genet. 67, 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roux I., Safieddine S., Nouvian R., Grati M., Simmler M. C., Bahloul A., Perfettini I., Le Gall M., Rostaing P., Hamard G., Triller A., Avan P., Moser T., Petit C. (2006) Cell 127, 277–289 [DOI] [PubMed] [Google Scholar]
- 11.Britton S., Freeman T., Vafiadaki E., Keers S., Harrison R., Bushby K., Bashir R. (2000) Genomics 68, 313–321 [DOI] [PubMed] [Google Scholar]
- 12.Doherty K. R., Cave A., Davis D. B., Delmonte A. J., Posey A., Earley J. U., Hadhazy M., McNally E. M. (2005) Development 132, 5565–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernatchez P. N., Sharma A., Kodaman P., Sessa W. C. (2009) Am. J. Physiol. Cell. Physiol 297, C484–C492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernatchez P. N., Acevedo L., Fernandez-Hernando C., Murata T., Chalouni C., Kim J., Erdjument-Bromage H., Shah V., Gratton J. P., McNally E. M., Tempst P., Sessa W. C. (2007) J. Biol. Chem. 282, 30745–30753 [DOI] [PubMed] [Google Scholar]
- 15.Südhof T. C. (2002) J. Biol. Chem. 277, 7629–7632 [DOI] [PubMed] [Google Scholar]
- 16.Cai C., Weisleder N., Ko J. K., Komazaki S., Sunada Y., Nishi M., Takeshima H., Ma J. (2009) J. Biol. Chem. 284, 15894–15902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda C., Hayashi Y. K., Ogawa M., Aoki M., Murayama K., Nishino I., Nonaka I., Arahata K., Brown R. H., Jr. (2001) Hum. Mol. Genet. 10, 1761–1766 [DOI] [PubMed] [Google Scholar]
- 18.Hernández-Deviez D. J., Martin S., Laval S. H., Lo H. P., Cooper S. T., North K. N., Bushby K., Parton R. G. (2006) Hum. Mol. Genet. 15, 129–142 [DOI] [PubMed] [Google Scholar]
- 19.Hernández-Deviez D. J., Howes M. T., Laval S. H., Bushby K., Hancock J. F., Parton R. G. (2008) J. Biol. Chem. 283, 6476–6488 [DOI] [PubMed] [Google Scholar]
- 20.Fujita E., Kouroku Y., Isoai A., Kumagai H., Misutani A., Matsuda C., Hayashi Y. K., Momoi T. (2007) Hum. Mol. Genet. 16, 618–629 [DOI] [PubMed] [Google Scholar]
- 21.Lo H. P., Cooper S. T., Evesson F. J., Seto J. T., Chiotis M., Tay V., Compton A. G., Cairns A. G., Corbett A., MacArthur D. G., Yang N., Reardon K., North K. N. (2008) Neuromuscul. Disord. 18, 34–44 [DOI] [PubMed] [Google Scholar]
- 22.Piccolo F., Moore S. A., Ford G. C., Campbell K. P. (2000) Ann. Neurol. 48, 902–912 [PubMed] [Google Scholar]
- 23.Klinge L., Laval S., Keers S., Haldane F., Straub V., Barresi R., Bushby K. (2007) Faseb. J. 21, 1768–1776 [DOI] [PubMed] [Google Scholar]
- 24.Jackaman C., Nowak K. J., Ravenscroft G., Lim E. M., Clément S., Laing N. G. (2007) Cell. Motility Cytoskel. 64, 914–925 [DOI] [PubMed] [Google Scholar]
- 25.Ramakrishnan N. A., Drescher M. J., Drescher D. G. (2009) J. Biol. Chem. 284, 1364–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumitani S., Ramlal T., Liu Z., Klip A. (1995) Biochem. Biophys. Res. Commun. 213, 462–468 [DOI] [PubMed] [Google Scholar]
- 27.Latham C. F., Lopez J. A., Hu S. H., Gee C. L., Westbury E., Blair D. H., Armishaw C. J., Alewood P. F., Bryant N. J., James D. E., Martin J. L. (2006) Traffic 7, 1408–1419 [DOI] [PubMed] [Google Scholar]
- 28.Schiavo G., Gu Q. M., Prestwich G. D., Söllner T. H., Rothman J. E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13327–13332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chae Y. K., Abildgaard F., Chapman E. R., Markley J. L. (1998) J. Biol. Chem. 273, 25659–25663 [DOI] [PubMed] [Google Scholar]
- 30.Schiavo G., Stenbeck G., Rothman J. E., Söllner T. H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett M. K., Calakos N., Scheller R. H. (1992) Science 257, 255–259 [DOI] [PubMed] [Google Scholar]
- 32.Therrien C., Di Fulvio S., Pickles S., Sinnreich M. (2009) Biochemistry 48, 2377–2384 [DOI] [PubMed] [Google Scholar]
- 33.Volchuk A., Wang Q., Ewart H. S., Liu Z., He L., Bennett M. K., Klip A. (1996) Mol. Biol. Cell. 7, 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jagadish M. N., Fernandez C. S., Hewish D. R., Macaulay S. L., Gough K. H., Grusovin J., Verkuylen A., Cosgrove L., Alafaci A., Frenkel M. J., Ward C. W. (1996) Biochem. J. 317, 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tellam J. T., Macaulay S. L., McIntosh S., Hewish D. R., Ward C. W., James D. E. (1997) J. Biol. Chem. 272, 6179–6186 [DOI] [PubMed] [Google Scholar]
- 36.Pagan J. K., Wylie F. G., Joseph S., Widberg C., Bryant N. J., James D. E., Stow J. L. (2003) Curr. Biol. 13, 156–160 [DOI] [PubMed] [Google Scholar]
- 37.McNally E. M., Ly C. T., Rosenmann H., Mitrani Rosenbaum S., Jiang W., Anderson L. V., Soffer D., Argov Z. (2000) Am. J. Med. Genet. 91, 305–312 [DOI] [PubMed] [Google Scholar]
- 38.Roche J. A., Lovering R. M., Roche R., Ru L. W., Reed P. W., Bloch R. J. (2010) Am. J. Physiol. Cell. Physiol. 298, C298–C312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagaraju K., Rawat R., Veszelovszky E., Thapliyal R., Kesari A., Sparks S., Raben N., Plotz P., Hoffman E. P. (2008) Am. J. Pathol. 172, 774–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiu Y. H., Hornsey M. A., Klinge L., Jørgensen L. H., Laval S. H., Charlton R., Barresi R., Straub V., Lochmüller H., Bushby K. (2009) Hum. Mol. Genet. 18, 1976–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawat R., Cohen T. V., Ampong B., Francia D., Henriques-Pons A., Hoffman E. P., Nagaraju K. (2010) Am J. Pathol, 176, 2891–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.