Abstract
Hematopoiesis is the process by which hemocytes mature and subsequently enter the circulation. Vertebrate prokineticins (PKs) are known to take part in this process, as are the invertebrate prokineticin domain proteins, astakines. In Pacifastacus leniusculus, astakine 1 is essential for the release of new hemocytes into the open circulatory system of these animals. In addition to astakine 1, we have now cloned a homologue of astakine 1 with an insert of 13 amino acids, named as astakine 2. Both crustacean astakines lack the N-terminal AVIT motif, which is present in vertebrate PKs, and hence receptor binding differs from that of vertebrate PKs. We have found astakine-like sequences in 19 different invertebrate species, and the sequences show that some motifs are conserved among invertebrate groups. Previously we showed that astakine 1 is directly involved in hematopoiesis, and now we show that astakine 1 and astakine 2 have different roles in hemocyte lineage differentiation. Astakine 1 can stimulate proliferation of hematopoietic tissue (Hpt) cells (precursor of hemocytes) as well as specifically induce differentiation of Hpt cells along the semigranular cell lineage, whereas astakine 2 plays a role in granular cell differentiation. Moreover, we discuss the impact of the putative structures of different astakines in comparison with the vertebrate prokineticins.
Keywords: Cytokine, Differentiation, Growth Factors, Hematopoiesis, Immunology, Astakine, Hemocyte, Invertebrate, Prokineticin, Proliferation
Introduction
Crustaceans, as other invertebrates, rely on innate immunity for the protection of the animal against invading microorganisms. The circulating hemocytes are crucial in these processes by participating in recognition, phagocytosis, melanization, and cytotoxicity (1). Thus, the regulation of hemocyte number is critical for the health of the animal, and the continuous formation of new hemocytes (hematopoiesis) is tightly regulated by factors released from circulating hemocytes (2). In crayfish, the hematopoietic tissue (Hpt)3 is a separate organ, containing five distinct types of Hpt cells, developing into two lineages of hemocytes: semigranular cells (SGC), and granular cells (GC) (3). The molecular mechanism by which Hpt cells are induced to proliferate and differentiate into hemocytes is starting to be understood in some invertebrates, including crustaceans. Recently, a cytokine, astakine, has been shown to be important in this process (2–6). Astakine, a homologue to vertebrate prokineticins, was first identified in Pacifastacus leniusculus and was found to be necessary for new hemocyte synthesis and release (3).
In vivo, when crayfish was injected with either native or recombinant astakine, the total hemocyte number was significantly increased, and astakine RNAi resulted in low hemocyte recovery after LPS stimulation. In vitro, when a primary cell culture of crayfish Hpt cells was incubated with astakine, proliferation was stimulated. Moreover, the addition of astakine results in a decrease of activity of an extracellular transglutaminase (TGase). This loss of TGase activity enables migration of the cells out of the hematopoietic tissue (4). Furthermore, we also showed that ATP synthase is present on the surface of a subpopulation of the Hpt cells and that it serves as a receptor for astakine. Surface ATP synthase activity contributes to about 40% of the extracellular ATP formation on the surface of the Hpt cells, and astakine was found to interact with the β-subunit of ATP synthase, which resulted in a block of the ATP formation by this enzyme (5).
The molecular mechanism of hematopoiesis in invertebrates has been mainly studied in Drosophila melanogaster and P. leniusculus. Several transcription factors have been characterized as lineage-specific markers in the fruit fly, and they are conserved across taxonomic groups from flies to mammals (7–9). Accordingly, the importance of GATA transcription factors as well as a Runt protein homologue during hematopoiesis has been shown in P. leniusculus (4). The importance of different signaling pathways, such as Toll/NF-κB, and JAK/STAT during embryonic and larval hematopoiesis has been studied in detail in Drosophila (9). A clear role for cytokines, such as the PDGF/VEGF-related PVF1-PVF3 and Upd (a ligand for the JAK/STAT pathway), in Drosophila hematopoiesis needs further detailed studies (10, 11). Interestingly, no astakine or prokineticin homologue is present in the genome of Drosophila or other dipterans, although such sequences are present in some other insect orders, as well as in several other arthropods.
Recently, specific marker proteins for Hpt cells, SGC, and GC were identified in P. leniusculus (6). One specific Kazal proteinase inhibitor (KPI) was found to be exclusively expressed in SGC, a superoxide dismutase (SOD) is transcribed only in GC, and a proliferating cell nuclear antigen (PCNA) is only expressed in proliferating cells and was accordingly detected in P. leniusculus Hpt cells. Because crayfish hemocytes do not divide in the circulatory system, this transcript can serve as a good marker for Hpt cells. These three marker proteins make it possible for us to study the mechanism by which astakine affects hematopoiesis in P. leniusculus. Therefore, P. leniusculus is a good model to study the role of these cytokines in blood cell development in invertebrates. In the current study, we provide direct evidence from in vitro Hpt cell culture and in vivo experiments for different functions of two related astakines during hematopoiesis.
EXPERIMENTAL PROCEDURES
Experimental Animals
Freshwater crayfish, P. leniusculus, purchased from Nils Fors (Askersund, Lake Vättern, Sweden) were maintained in tanks with aerated tap water at 10 °C. Only intermolt animals were used in the experiment.
Molecular Cloning of Astakine 2
For cDNA cloning, nested degenerate primers were designed according to the amino acid sequence GMCPCR (5′-ICK RCA IGG RCA CAT ICC-3′) and TCQLP (5′-IGG IAR YTG RCA IGT-3′) that are the homologue region in P. leniusculus astakine (hereafter named astakine 1) and Penaeus monodon astakine. Rapid amplification of 5′-complementary DNA ends (5′-RACE) was used to clone a new possible astakine from crayfish hemocytes. To complete the final gene cloning, rapid amplification of 3′-complementary DNA ends (3′-RACE) was performed, and gene-specific primers (5′-ACC CCA ACG ATC TCC AGA TCA AG-3′ and 5′-GAG GAC GGG TAC TTG GGC ATG TGC-3′) for the 3′-RACE were designed based on the partial sequence of crayfish astakine 2 from the result of 5′-RACE. The cDNA obtained is named astakine 2.
Tissue Distribution of Astakine 1 and Astakine 2 by RT-PCR
Total RNA was extracted from different crayfish tissues, including muscle, hepatopancreas, hematopoietic tissue, hemocytes, heart, intestine, nerve cord, and testis, by using the Gene Elute total mammalian RNA extraction kit (Sigma), followed by RNase-free DNase I (Ambion) treatment. Equal amounts of total RNA were used for cDNA synthesis with ThermoScriptTM (Invitrogen) according to the manufacturer's instructions and analyzed for expression of crayfish astakine 1 and astakine 2 by RT-PCR. The primers 5′-ATGCGAGGAGTTAGTGTG-3′ and 5′-CTAGTAGTAGGAGTCGAGCGTGTTGTC-3′ were used for astakine 1; primers 5′-ATGCTGGAGCGCAGTGGAGTGATGGTG-3′ and 5′-AGTGAATGATGCCCAGAGTGTTGTC-3′ were used for astakine 2. The PCR program used was as follows: 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 40 s. The transcription of a 40 S ribosomal protein was used as an internal control. All PCR products were analyzed on 1.5% agarose gel stained with ethidium bromide.
Expression of Astakine in Escherichia coli
The coding sequence of astakine 1 without signal peptide was cloned into the bacterial expression vector pet32a (Novagen) at the BamHI and SalI cleavage sites and subsequently transformed into E. coli cells Rosette-gami B (DE3), and a single colony was grown in LB medium containing 100 μg/ml ampicillin, 20 μg/ml kanamycin, and 10 μg/ml tetracycline to A600 = 0.5 and induced with 0.02 μm isopropyl 1-thio-β-d-galactopyranoside overnight at 20 °C. The recombinant protein was purified by HisTrap FF column chromatography (GE Healthcare), and the thioredoxin (Trx) tag combined with S tag and His tag was removed in the column by incubation with enterokinase (New England Biolabs) at 4 °C in cleavage buffer (20 mm Tris-HCl, 50 mm NaCl, 2 mm CaCl2, pH 7.2) overnight. Free astakine 1 was eluted with elution buffer (20 mm Tris-HCl, 50 mm NaCl, 5 mm EDTA, pH 7.2) from the column, and the enterokinase was irreversibly blocked by 4 mm Pefabloc SC (Roche Applied Science).
For astakine 2, the coding region without signal peptide was cloned into the same vector at the EcoRI and SalI cleavage sites, and furthermore one Factor Xa site was introduced in the N terminus. Thus, the tag of recombinant astakine 2 was removed on the column by incubation with Factor Xa, which generates an identical N-terminal in the recombinant astakine 2 as in native astakine 2. Factor Xa activity was irreversibly blocked by 2 μm dansyl-Glu-Gly-Arg-chloromethyl ketone (catalog no. 251700, Calbiochem).
Expression of Astakine in Spodoptera frugiperda (Sf9) Cells
The cDNA encoding the astakine 1 was cloned into pFastBac1 vector (Invitrogen) at the BamHI and SalI cleavage sites. The recombinant virus for astakine 1 expression was generated according to the manufacturer's instruction manual (Bac-to-Bac baculovirus expression system, Invitrogen). To express the protein, Sf9 cells were seeded in 6-well plates at a density of 2 × 106 cells/well and cultured in Sf-900 II medium (Invitrogen) supplemented with 5% FBS (Sigma). The Sf9 cells were lysed in lysis buffer (50 mm sodium acetate, 1% Triton, pH 3.8) containing 1× protease inhibitor mixture (Roche Applied Science) at 72–96 h postinfection of recombinant virus. The cell lysates were then centrifuged at 20,000 × g at 4 °C for 15 min, and the supernatants were collected and used for Western blot and protein purification. Western blot against astakine 1 was performed as described previously (5). Briefly, the lysates were subjected to 12.5% SDS-PAGE under both reducing and non-reducing conditions and then electrotransferred to PVDF membrane (GE Healthcare). The membrane was subsequently blocked in TBST (1% Tween 20 in 20 mm Tris-HCl, 150 mm NaCl, pH 7.5) containing 5% BSA for 1 h and washed briefly three times and then incubated with anti-astakine 1 antibody (0.2 ng/ml in blocking buffer) overnight at 4 °C or 1 h at room temperature. Then the membrane was washed with TBST for 20 min, a procedure that was repeated three times. The ECL anti-rabbit IgG peroxidase-linked, species-specific whole antibody from donkey (Amersham Biosciences) diluted 1:7500 with TBST was incubated for 1 h and washed with TBST for 20 min three times. For detection, the ECL Western blotting reagent kit (Amersham Biosciences) was used, according to the manufacturer's instructions. For purification, the supernatants were applied to a 1-ml HiTrap SP HP column (0.7 × 2.5 cm; GE Healthcare), pre-equilibrated with 50 mm sodium acetate buffer, pH 3.8, and then this column was washed with the same buffer until no material appeared in the effluent and then gradient eluted with 50 mm sodium acetate, 1 m NaCl, pH 3.8 (10 ml, 0–10%) at a flow rate of 60 ml/h. Fractions of 1 ml were collected, and the purity of astakine 1 was subjected to 12.5% SDS-PAGE and confirmed by Western blot with astakine 1 antibody. The protein concentration was determined by the Bradford protein assay (Coomassie Plus, Thermo), using BSA as a standard.
Hpt Cell Culture
Hpt cells were isolated as described previously (2) with minor modification. Briefly, the hematopoietic tissue was dissected from the dorsal side of the stomach and was incubated in 700 μl of 0.1% collagenase (Type I), 0.1% collagenase (Type IV) in crayfish phosphate-buffered saline (CPBS) (10 mm Na2HPO4, 10 mm KH2PO4, 0.15 m NaCl, 10 μm CaCl2, 10 μm MnCl2, pH 6.8) at room temperature for 40 min. After collagenase treatment, the tissue was centrifuged at 2000 × g for 5 min to remove the collagenase solution. The pellet was washed twice with 1 ml of CPBS, the undigested tissues were removed, and the isolated Hpt cells were resuspended in L-15 medium (Sigma) supplemented with 5 μm 2-mercaptoethanol, 1 μm phenylthiourea, 60 μg/ml penicillin, 50 μg/ml streptomycin, 50 μg/ml gentamycin, and 2 mm l-glutamine (3) and subsequently seeded in 96- or 6-well plates at a density of 5 × 105 cells/ml. One-third of the medium was changed every second day.
Activity of Recombinant Astakine 1 (r-astakine 1) and Recombinant Astakine 2 (r-astakine 2) in Vitro
About 100 ng (final concentration ∼5 nm) of r-astakine 1 or r-astakine 2 (quantified by a Bradford assay after purity confirmation by SDS-PAGE) was supplemented in the cell culture every second day. After 1 week, total RNA was extracted from Hpt cells, and RT-PCR was conducted to monitor the transcript of crayfish hemocyte marker genes (6). The following primers were used: for PCNA, 5′-CCTTCGCCTGTCGGTATCTCA-3′ and 5′-CCTAACTTGTCACTATCACCCAA-3′; for KPI, 5′-GTGCAACTCTCCTGCTGG-3′ and 5′-GCATTCACCGTTATAGGCA-3′; and for SOD, 5′-ATGGTGAACATGACTCTCCC-3′ and 5′-GTTGTACGTCCTCTGGTACTG-3′.
Activity of r-astakine 1 and r-astakine 2 in Vivo
Crayfish were injected at the base of a walking leg with r-astakine 1, r-astakine 2, or crayfish saline as described previously (2). The dose is about 0.02–0.06 μg/g weight of crayfish. After injection, the crayfish were kept in tanks with aerated running water at 10 °C as described above. After 24 h, blood samples were taken, and the hemocyte count was determined as described previously (2). Relative hemocyte count was calculated as the total number of hemocytes divided by the total number of hemocytes in the animal before injection.
Gel Filtration
In order to separate the oligomer form and monomer form of r-astakine, HiPrep 16/60 Sephacryl S-200 HR chromatography (GE Healthcare) was performed, and apoferritin (443 kDa; Sigma), alcohol dehydrogenase (150 kDa; Sigma), and BSA (66 kDa; Sigma) were used as standard protein to calibrate the column in order to calculate the molecular weight of the oligomer form of r-astakine 1.
Site-directed Mutagenesis
Two cysteines (Cys78 and Cys91) in astakine 1 were mutated into glycines according to the manual instructions of the QuikChange multisite-directed mutagenesis kit (Stratagene). Briefly, the plasmid pet32a-astakine 1 was used as a template for the mutagenesis, and the mutagenesis primers were designed as follows: C78G, 5′- TACATCGGAATGTGTCCTGGCCGAGCTGGGCTG-3′;C91G, 5′-ACCTTCAGCCACCGGCCAGCTGCCTTC-3′. The PCR was done as follows: 95 °C for 1 min, and 30 cycles of 95 °C for 1 min, 55 °C for 1 min, and 65 °C for 12 min. After the reaction, 1 μl of DpnI restriction enzyme (10 units/μl) was added directly to the amplification reaction, and then the reactions were incubated at 37 °C for 1 h to digest the parental (non-mutated) double-stranded DNA. The reaction mixture, enriched for mutated single-stranded DNA, was transformed into XL10-Gold ultracompetent cells. The correctly mutated plasmids, which were confirmed by sequencing, were chosen to prepare the mutant astakine 1 (r-astakine 1Mut) as described before.
For mutagenesis of astakine 2, two amino acids (Leu72 and Tyr74) in astakine 2 were mutated into glycines according to the manual instructions of the QuikChange site-directed mutagenesis kit (Stratagene). Briefly, the plasmid pet32a-astakine 2 was used as a template for the mutagenesis, and the mutagenesis primers were designed as follows: ask2m1, 5′-GCTGAGTCCCAGACGACCGGGAATGGCCCCAACGATCTCCAGATCAAG-3′; ask2m2, 5′-CTTGATCTGGGATCGTTGGGGCCATTCCCGGTCATGTCCTGGGACTCAGC-3′. The PCR was done as follows: 95 °C for 1 min and 18 cycles of 95 °C for 1 min, 55 °C for 1 min, 68 °C for 12 min. After the reaction, 1 μl of DpnI restriction enzyme (10 units/μl) was added directly to the amplification reaction, and then the reactions were incubated at 37 °C for 1 h to digest the parental (non-mutated) double-stranded DNA. The reaction mixture, enriched for mutated single-stranded DNA, was transformed into XL10-Gold ultracompetent cells. The correctly mutated plasmids, which were confirmed by sequencing, were chosen to prepare the mutant astakine 2 (r-astakine 2Mut) as described before.
Inhibition of ATP Synthase
Inhibition of ATP synthase was performed according to Lin et al. (5). Briefly, ATP was measured by using the ATP biomass kit HS (BioThema), and the light emission was detected with a luminometer (LKB Wallac 1250) according to the manufacturer's instructions. First, the ATP content of the reaction buffer (10 mm Hepes, 150 mm NaCl, 100 μm ADP, 20 mm potassium phosphate, 2 mm MgCl2, pH 7.4) was measured without Hpt cells. Then Hpt cells (3 × 105) were seeded to a 20-mm2 part near the edge of a tissue culture dish (35 × 100 mm). The Hpt cells were washed twice with 500 μl of Hepes buffer (10 mm Hepes, 150 mm NaCl, pH 7.4) after 2–5 h of incubation. Each dish of Hpt cells was first incubated with 150 μl of reaction buffer for 3 min, the formation of extracellular ATP was assayed, and the value was set as the individual control value for each dish. The cells were then washed twice with 150 μl of Hepes buffer and incubated with r-astakine 1 or r-astakine 2 for 60 min, followed by incubation with 150 μl of reaction buffer for 3 min, and the formation of ATP was calculated and compared with the control value.
Identification of Astakine-like cDNAs and Phylogenetic Analysis
The sequences of P. leniusculus astakine 1 and astakine 2 as well as B. variegata Bv8 were used as queries in a TBLASTN search against the NCBI non-redundant data base, NCBI genomes, and “non-human, non-mouse” expressed sequence tags (EST_others). In total, 23 different astakine-like sequences were detected in arthropod phyla. These sequences were aligned using ClustalW2 (see the EMBL-EBI Web site) using default alignment parameters and were manually edited, and the phylogenetic unrooted tree was constructed using the Phylip 3.69 package (27).
Homology Modeling
Homology models of Pacifastacus astakine 1 and astakine 2 were built using the three-dimensional structure of mamba toxin (Protein Data Bank entry 1IMT) (12). The LSQMAN program (13) was used to select the average from 39 models in 1IMT. An alignment of these two proteins was carried out using the PROBCONS Web service (14) and manually edited. The MODELLER program version 9.7 (15) was used to build three-dimensional models of Pacifastacus astakine 1 and astakine 2 proteins. The qualities of the final models were checked by WhatCheck programs (16).
RESULTS
Two Different Astakines Are Found in Crayfish and Have Different Roles in Crayfish Hematopoiesis
Two isoforms of a second astakine were cloned from a crayfish hemocyte cDNA, and they were named astakine 2a and 2b, differing only in their 3′-UTR (GenBankTM accession numbers EF568370 and EF568371). Our new astakine 2a and 2b, both contain an insert of 13 amino acid residues as compared with the crayfish astakine 1 molecule (3) (Fig. 1). We have also identified and cloned an astakine from shrimp P. monodon (AY787657) (3). This shrimp astakine contains a 13-amino acid insert at the same site as astakine 2a and 2b. One putative motif GX2RYSX(P/R)C is present in all of these three astakines (underlined in Fig. 1) that is not present in the vertebrate PKs.
FIGURE 1.
Two different astakines are present in P. leniusculus. A new astakine in crayfish (pl-astakine 2) is more similar to the previously cloned astakine (pm-astakine 2) from shrimp; both of them have an insert of 13 amino acid residues (boxed) when compared with crayfish astakine (pl-astakine 1). The conserved GX2RYSX(P/R)XC motif and LXYP motif are underlined.
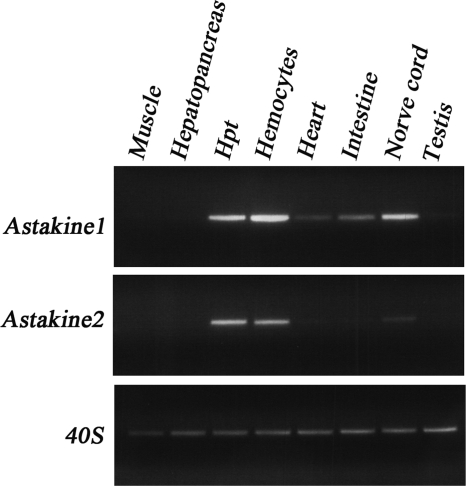
As analyzed by semiquantitative RT-PCR and illustrated in Fig. 2, astakine 1 is highly expressed in hemocytes, the Hpt, and nerve tissue. Very weak bands could be detected in intestine and heart, and these are most likely due to contaminating hemocytes infiltrating these tissues. Astakine 2 shows a similar expression pattern, although the expression level is much lower in general, and in nerve tissue, it is barely detectable (Fig. 2).
FIGURE 2.
Astakine 1 and astakine 2 share a similar tissue expression pattern. Semiquantitative RT-PCR was used for this expression analysis. Lane 1, muscle; lane 2, hepatopancreas; lane 3, Hpt; lane 4, hemocytes; lane 5, heart; lane 6, intestine; lane 7, nerve cord; lane 8, testis.
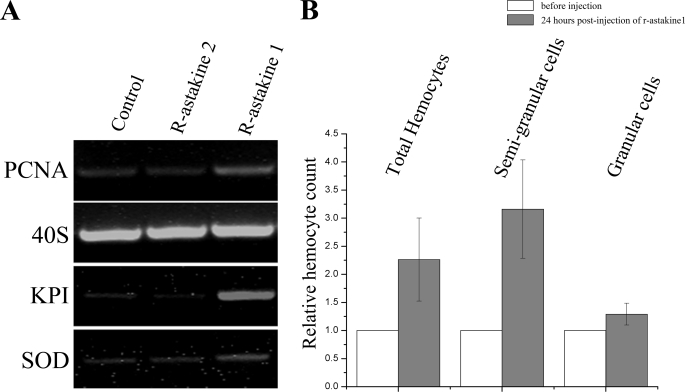
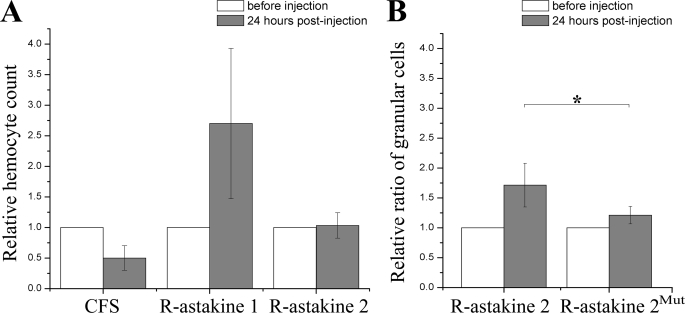
Recently, we have detected a KPI that is specific for SGC, whereas a SOD is expressed exclusively in GC (6). By using these transcripts as markers for differentiation, we can now conclude that astakine 1 (at concentrations of 4–100 nm) promotes proliferation of Hpt cells in vitro as well as induces differentiation into the SGC lineage because the expression of PCNA and the semigranular specific KPI were induced by the addition of r-astakine 1 (Fig. 3A). This result was also confirmed by the analysis of differential hemocyte counts in vivo, showing an increase in the number of SGC after injection of r-astakine 1 into live crayfish (Fig. 3B). Recombinant astakine 2 (r-astakine 2), in contrast, did not induce proliferation in the Hpt cell culture (Fig. 3A), but a significant increase in total hemocyte count was found after in vivo injection (Fig. 4A) compared with control injection with crayfish saline. In addition, r-astakine 2 injection was followed by a concomitant increase in the proportion of granular hemocytes, suggesting that astakine 2 is involved in the maturation of granular cells (Fig. 4B). The insert present in all astakine 2 molecules and displayed in Table 1 varies in size from 13 amino acid residues in P. leniusculus and P. monodon to 15 residues in A. variegatum and Daphnia pulex. Nearly all inserts contain a conserved LXYP motif, indicating an important role for these amino acids, and this was confirmed by our mutagenesis experiment. When leucine (Leu72) and tyrosine (Tyr74) in this conserved motif were mutated into glycines in r-astakine 2Mut, the activity of astakine 2 in promoting the maturation of granular cells was abolished (Fig. 4B).
FIGURE 3.
Astakine 1 stimulates proliferation and differentiation into the semigranular cell lineage of Hpt cells both in vitro and in vivo. A, Hpt cell cultures were supplemented with r-astakine 1 or r-astakine 2 every second day. After 1 week, RT-PCR was conducted to monitor the expression of PCNA (proliferation marker of Hpt cells), KPI (gene marker for semigranular cells), and SOD (gene marker for granular cells). The addition of astakine 1 could stimulate transcription of PCNA and KPI significantly. The effect of r-astakine 1 and r-astakine 2 is detectable from ∼3 nm (4.5 ng/well) and is optimal at a concentration of ∼65 nm (∼100 ng/well), whereas a toxic effect to the cells is obtained at ≥120 nm. B, r-astakine 1 mainly stimulates the synthesis of semigranular cells in vivo. Relative hemocyte counts for total hemocytes, semigranular cells, or granular cells were calculated as hemocyte number 24 h postinjection/initial hemocyte number. r-astakine 1 injection induced a significant increase in the number of total hemocytes, mainly due to an increase of semigranular cells. The columns represent the mean of three separate experiments; the error bars indicate S.D. values.
FIGURE 4.
Astakine 2 promotes maturation of granular cells in vivo. A, r-astakine 2 injection slightly stimulated the total number of hemocytes in vivo. Relative hemocyte counts were calculated as the total number of hemocytes after injection of r-astakine 1 or r-astakine 2/initial total number of hemocytes. Crayfish saline (CFS) was used for control injection. B, r-astakine 2 injection stimulated the maturation of granular cells in vivo. The relative ratio of granular cells was calculated as the ratio of granular cells after 24 h post-treatment/initial ratio of granular cells. r-astakine 2 can increase the ratio of granular cells in vivo by stimulating the maturation of granular cells, whereas the mutated r-astakine 2Mut showed no such activity. The columns represent the mean of six separate experiments, and error bars represent S.D. values. *, t test, p < 0.01.
TABLE 1.
Arthropod astakines or astakine-like transcripts found in GenBankTM
| Subphylum | Species | Accession number |
|---|---|---|
| Astakine 1 | ||
| Chelicerata | Acanthoscurria gomesiana | DR447331 |
| Ornithoctonus huwena | ABY77690 | |
| Crustacea | Pacifastacus leniusculus | AY787656 |
| Astakine 2 | ||
| Chelicerata | Rhipicephalus appendiculatus | CD794853 |
| Ixodes scapularis | EW845057 | |
| Amblyomma variegatum | BM292046 | |
| Acanthoscurria gomesiana | DR445103 | |
| Dysdera erythrina | CV178181 | |
| Mesobuthus gibbosus | CB333881 | |
| Crustacea | Pacifastacus leniusculus | EF568370–EF568371 |
| Penaeus monodon | AY787657 | |
| Litopenaeus vannamei | FE148214 | |
| Homarus americanus | FE535609 | |
| Carcinus maenas | DW585080 | |
| Daphnia pulex | FE329237 | |
| Insecta, Hymenoptera | Nasonia vitripennis | XM_001605610 |
| Solenopsis invicta | EE139835 | |
| Insecta, Blattodea | Blattella germanica | FG125716 |
| Insecta, Hemiptera | Homalodisca vitripennis | CO639882 |
| Acyrthosiphon pisum | XM_001945710 | |
| Insecta, Pterygota | Pediculus humanus corporis | CB887301, XP_002431167 |
The Two Astakines Differ in Putative Structure from Each Other and from Vertebrate Prokineticins
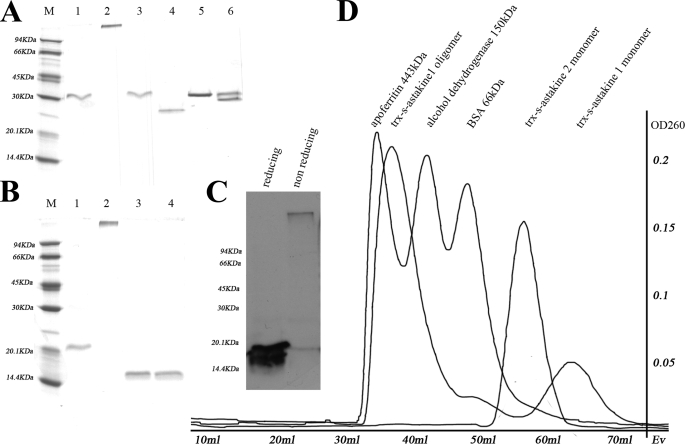
During preparation of r-astakine 1 and r-astakine 2, we found that these two different astakines moved differently in SDS-PAGE. Recombinant astakine 1 fusion protein containing the Trx-S tag as well as without the tag formed a high molecular weight oligomer under non-reducing conditions (Fig. 5, A and B). In contrast, r-astakine 2 was only found to be present as a monomer (Fig. 5A). Similarly, r-astakine 1 expressed in an insect-baculovirus system also showed a high oligomer in SDS-PAGE under non-reducing conditions, which was confirmed by Western blot using the astakine 1 antibody (Fig. 5C). Moreover, when hemocyte-free plasma was analyzed by Western blot under non-reducing conditions, a high molecular mass oligomer of astakine 1 as well as a monomeric astakine 1 was identified by astakine 1 antibodies (supplemental Fig. 1), indicating that this oligomerization of astakine 1 also occurs in the native protein. Gel filtration analysis using Sephacryl S-200 revealed that the molecular mass for the recombinant fusion astakine 1 oligomer is ∼370 kDa (Fig. 5D).
FIGURE 5.
Astakine 1 and astakine 2 move differently in SDS-PAGE. A, recombinant astakine 1 shows an oligomer and a monomer, but recombinant astakine 2 only runs as a monomer. Lane 1, r-astakine 1 (Trx-S tag-astakine 1) in reducing conditions (molecular mass is about 30 kDa); lane 2, r-askakine1 (Trx-S tag-astakine 1) in non-reducing conditions (molecular mass is about 370 kDa, which was measured by Gel filtration); lane 3, monomer of r-astakine 1 (Trx-S tag-astakine 1) in reducing conditions (purified by gel filtration); lane 4, monomer of r-astakine 1 (Trx-S tag-astakine 1) in non-reducing conditions (purified by gel filtration); lane 5, r-astakine 2 (Trx-S tag-astakine 2) in reducing conditions; lane 6, r-astakine 2 (Trx-S tag-astakine 2) in non-reducing conditions. B, oligomer formation of r-astakine 1 is independent of the fusion tag. The oligomer form of r-astakine 1 was treated with the protease thrombin to remove the fusion tag Trx. Lane 1, r-astakine 1 without Trx tag in reducing conditions; lane 2, r-astakine 1 without Trx tag in non-reducing conditions; lane 3, free Trx in reducing conditions; lane 4, free Trx in non-reducing conditions. C, r-astakine 1 expressed in the insect cell-baculovirus expression system similarly forms oligomers. Lysates of Sf9 cells infected with recombinant baculovirus containing the ORF of astakine 1 were used for Western blot using astakine 1 antibody. D, in order to separate the oligomer and monomer form of recombinant fusion astakine 1, HiPrep 16/60 Sephacryl S-200 HR column (GE Healthcare) gel filtration was performed. Apoferritin (443 kDa), alcohol dehydrogenase (150 kDa), and BSA (66 kDa) were used as standard proteins to calibrate the column.
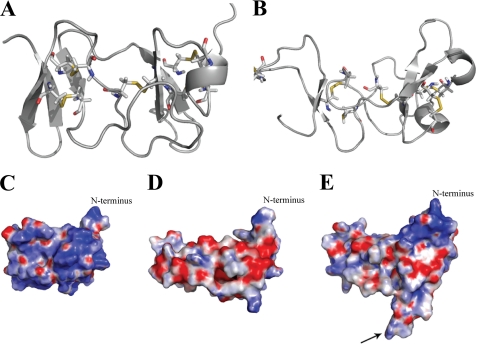
Astakine 1 is involved in the hematopoietic process in crayfish, and similar stimulation of blood cell formation has been shown to be one of the functions for vertebrate prokineticins (17). However, although these molecules share a similar function, their receptors are different (5), and the conserved vertebrate AVIT motif is lacking in astakine 1. Due to this difference and because of the observed oligomeric structure of astakine 1, we decided to further elucidate the similarities and differences between astakines and prokineticins by building homology models of astakines.
The model structure of astakine 1 shows that it may have only four cysteine bridges compared with mamba toxin (a vertebrate PK used as template for modeling) that is stabilized by five bridges (Fig. 6, A and B). A similar structure has also been shown for Dkk (Dickkopf proteins) and colipase (18). Both astakine structures are more negatively charged than mamba toxin or Dkk, and there is a large charge difference at the N terminus between astakine 1 and astakine 2 (Fig. 6, C–E). Moreover, astakine 2 has a 13-amino acid insert (loop) compared with astakine 1, and the conserved tyrosine-proline residues in the astakine 2 loop are most likely exposed on the surface of the protein (Fig. 6E). The mutation of tyrosine (Tyr74) located on the tip of the loop resulted in loss of astakine 2 activity (Fig. 4B). Thus, this tyrosine may participate in the interaction with its receptor or some other important co-factor.
FIGURE 6.
Homology modeling of crayfish P. leniusculus astakine 1 and surface charge prediction of astakine 1 and astakine 2, compared with the template of black mamba intestinal toxin (MIT1). A, model of MIT1 (Protein Data Bank entry 1IMT) showing the predicted five cysteine bridges. B, homology model of astakine 1, predicted cysteine bridges, and free cysteines are shown. C, surface charge calculation of 1IMT. The N terminus is indicated. D, surface charge prediction of P. leniusculus astakine 1. The N terminus is indicated. E, surface charge prediction of P. leniusculus astakine 2. The N terminus is indicated, and the conserved tyrosine-proline residues in the astakine 2 loop are indicated by an arrow.
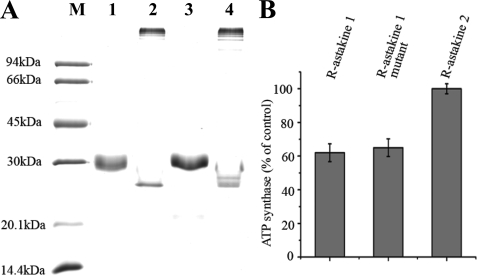
Site-directed Mutagenesis of Two Cysteines Did Not Affect Astakine 1 Activity
Because the homology modeling using MIT1 as a template indicates that only four cysteine bridges are present in astakine 1, compared with five in vertebrate PKs (as well as in the C-terminal sequence of the Dickkopf proteins), we decided to investigate whether the two free cysteines are involved in oligomer formation as indicated in Fig. 5. Therefore, we performed site-directed mutagenesis of Cys78 and Cys91 (according to Fig. 1) and produced recombinant astakine 1Mut. We then analyzed this mutated astakine using SDS-PAGE, but there was no significantly difference in the performance between the mutated and the wild type form of astakine 1 (Fig. 7A). We then tested whether the mutated r-astakine 1Mut exhibits activity similar to that of astakine 1 in the ATP synthase assay, and as clearly shown in Fig. 7B, there is no difference in inhibitory activity between the two astakines, indicating that the two mutated cysteines are involved neither in the formation of high molecular form of astakine 1 nor in receptor binding. Although r-astakine 1Mut can interact with ATP synthase and inhibit its ATP synthase activity, r-astakine 2 did not interact with ATP synthase on the surface of Hpt cells. This result supports our previous finding that astakine 1 and astakine 2 have different functions.
FIGURE 7.
Mutation of two free cysteines in astakine 1 to glycines does not affect its properties. A, purified recombinant astakine 1 and mutant of astakine 1 were separated by 12.5% SDS-PAGE. Lane 1, r-astakine 1 (Trx-S tag-astakine 1) in reducing conditions; lane 2, r-astakine 1 (Trx-S tag-astakine 1) in non-reducing conditions; lane 3, mutant of r-astakine 1 (Trx-S tag-astakine 1Mut) in reducing conditions; lane 4, mutant of r-astakine 1 (Trx-S tag-astakine 1Mut) in non-reducing conditions. B, effects of r-astakine 1, and mutation of r-astakine 1 and r-astakine 2 on extracellular ATP synthesis by Hpt cells. 3 × 105 Hpt cells were seeded to tissue culture dishes, and each dish of Hpt cells was first incubated with reaction buffer, and the formation of extracellular ATP was assayed and used as the individual control value for each dish. Then the cells were washed with Hepes buffer, incubated with 0.2 μm recombinant protein for 60 min, and subsequently incubated with reaction buffer. The formation of ATP was calculated, and the remaining ATP formation was calculated by comparison with its individual control value. The columns represent the mean of three separate experiments, and error bars represent S.D. values.
Astakine 1 and Astakine 2 Are Conserved in Arthropods
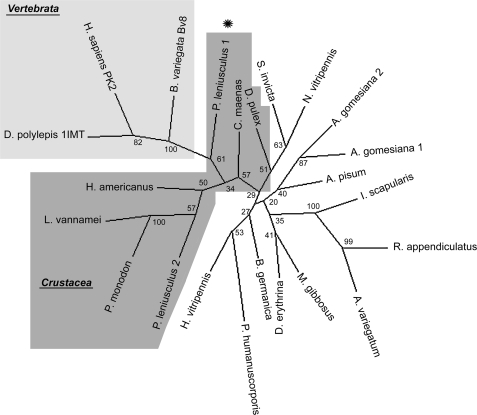
We searched available databases for astakine-like sequences among invertebrate species and could identify astakines or astakine-like sequences in several arthropod species (Table 1). Astakine 1-like sequences were only detected in crayfish P. leniusculus, the tarantula spider Acanthoscurria gomesiana, and the Chinese bird spider Ornithoctonus huwena, whereas astakine 2 was found in crustaceans, ticks, and spiders as well as in blattodean, hymenopteran, and hemipteran insects (Table 1). However, we could not find any indications of astakine-like sequences in the dipterans, showing that hematopoiesis in Drosophila may be regulated in a different manner. It is interesting to note that the insert in invertebrate astakine 2-like proteins is located in the prokineticin domain at a similar site as the arginine-rich insert found in human prokineticin 2L. This insert is encoded by exon number 3 in the human PK gene (19). The gene structure of human PK2L reveals four exons, and when comparing crustacean astakines, human exon number 2 and exon number 4 are highly similar in sequence to the corresponding location in crustacean astakine 1. Exon number 1 is different, and in humans (and possible other vertebrates), exon number 1 encodes the signal peptide and the conserved N-terminal motif (AVIT) that is missing in all invertebrate astakines known so far (19, 20). All astakines share a conserved motif in the loop between the putative third and fourth β-strands, namely GX2RYS, where the two X residues are usually Met or Gln. The corresponding amino acids in vertebrate PKs are not conserved.
A phylogenetic analysis of all arthropod astakines (Table 1) shows that despite the highly conserved motifs, the arthropod astakines are fairly distantly related. An interesting finding is that P. leniusculus astakine 1 is closer to the vertebrate PKs compared with other crustacean astakines, and similar results were obtained by using different methods (not shown). The other crustacean astakines except for D. pulex and Carcinus maenas are different. In addition, most insect astakines are clustered together, whereas the different chelicerate astakine-like proteins (which are mainly detected in venoms) are found scattered throughout the tree (Fig. 8).
FIGURE 8.
Phylogenetic analysis of invertebrate astakine and vertebrate PKs. Phylogenetic analysis of the astakine sequences in Table 1 was performed using ClustalW2 (see the EMBL-EBI Web site), and the phylogenetic unrooted tree was constructed using the Phylip 3.69 package (27).
DISCUSSION
Prokineticin-containing proteins are involved in a broad variety of biological processes in vertebrates (e.g. hematopoiesis and regulation of immune response) (17, 20). Although PKs in humans and mice modulate growth, survival and differentiation of cells of the innate immune system (17, 21), as do crustacean astakines, the mechanisms by which they do this are different.
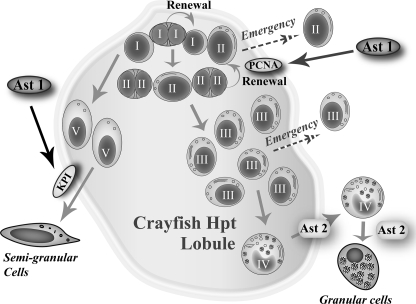
Astakine 1 is an invertebrate prokineticin, which plays a key role in the hematopoietic processes of crustaceans (3), and its significance for cell proliferation, differentiation, and release of mature hemocytes into the circulation was shown by the injection of the recombinant protein into the animal as well as by gene silencing in vivo (3). In this study, we demonstrate a specific function for astakine 1 in stimulating proliferation of Hpt cells as well as inducing differentiation along the semigranular cell lineage (Fig. 9) because the addition of astakine 1 induced expression of the proliferation marker PCNA and the SGC-specific Kazal type proteinase inhibitor (6). Rapid release of hemocytes is needed after infection or tissue damage, and astakine 1 is important for this release, as is also the case for PKs in vertebrates (21). We have recently identified TGase as an important extracellular protein that keeps the Hpt cells in an undifferentiated stage inside the hematopoietic tissue (4). If expression of TGase mRNA is silenced, the cells start to migrate out into the circulating hemolymph. Astakine 1 plays an important role in this process because astakine 1 by some unidentified mechanism reduces the extracellular TGase activity, and when TGase activity is reduced, cell migration is induced (4).
FIGURE 9.
Different roles of astakines as hematopoietic growth factors. The hypothetical model shows five types of Hpt cells and two main lineages of hemocytes: SGC and GC (adapted from Refs. 6 and 26). Astakine 1 and astakine 2 have different roles in hemocyte lineage differentiation. Astakine 1 can stimulate proliferation of Hpt cells as well as specifically induce differentiation of Hpt cells along the semigranular cell lineage, whereas astakine 2 plays a role in granular cell differentiation.
In the present work, we also identified and cloned a new crayfish astakine, astakine 2, similar to our previously described shrimp astakine (3). In a recent paper by Hsiao and Song (22), this peptide was shown to stimulate proliferation in the hematopoietic tissue of P. monodon in vivo. However, although astakine 2 has been shown to increase proliferation in Hpt cells in penaeid shrimp, its role in crayfish seems more complex. Crayfish r-astakine 2 failed to stimulate the proliferation of Hpt cells in vitro in contrast to r-astakine 1. Instead, r-astakine 2 was found to stimulate an increase of the total hemocyte number in vivo, although the stimulation was not as dramatic as that obtained with r-astakine 1, but the ratio of granular cells was always significantly increased. Therefore, astakine 2 appears to be involved in the maturation of granular cells (Fig. 9), and the conserved LXYP motif is critical for its biological activity because if Leu and Tyr were mutated into Gly, the biological activity was lost.
So far, no functional studies have been reported for any insect astakine. One study of the venom of two Australian funnel web spiders did focus on the function of astakine-like peptides, the so-called MIT-like atracotoxins (24). Like other invertebrate astakines, these peptides lack the N-terminal motif (AVIT) present in vertebrate prokineticins and accordingly did not affect smooth muscle contractility or restrain contractions induced by human PK1 in guinea pig ileum (23, 24).
We have previously reported that crayfish astakine 1 can bind to a cell surface-located ATP synthase β-subunit, indicating different binding properties of invertebrate and vertebrate prokineticin domain proteins. Consequently, we decided to study the putative structure of astakines by homology modeling and furthermore compare surface properties by calculating probable charge distributions on the surface of crayfish astakines as compared with a vertebrate prokineticin, the black mamba intestinal toxin (MIT1). MIT1 has a structure similar to those of the antagonists of the canonical Wnt pathway, Dkk and pancreatic procolipase. They all share typical ribbon architecture with a rather low proportion of secondary structure elements (less than 30%). These proteins are all stabilized by five cysteine bridges. Although there are 10 cysteines in the primary sequence of astakines, homology models of astakines show that they cannot form more than four cysteine bridges. The loss of one cysteine bridge most likely results in an increased flexibility of astakines compared with mamba toxin, Dkk, or colipase. Two free cysteines nevertheless do not effect or cause the multimerization observed by astakine 1 because mutation of both free cysteines (Cys78 and Cys91) did not prevent astakine 1 from forming multimers.
We have also compared the electrostatic potential of astakines with mamba toxin, Dkk, and colipase. Astakines are more negatively charged, which probably also should result in different substrate specificity. Astakine 1 and 2 differ by a 13-amino acid insert that might explain why astakine 2 is strictly monomeric. The differences in electrostatic potential of their N termini further indicate that these two astakines have different functions. The tyrosine-proline residues in this insert are conserved in all astakine 2 found in GenBankTM and, together with the GX2RYSX(P/R)XC motif, seem to be vital for astakines in general.
Vertebrate PKs are released from damaged tissues and act as regulators of inflammatory responses, including recruitment of new blood cells (21). Prokineticins act by binding to two closely related G-protein-coupled receptors (25), and although it has been shown that receptor activation mediates proliferation and differentiation through calcium mobilization (19), the downstream signaling is until now unknown. Crustaceans, as other invertebrates, do not possess erythrocytes or blood cells of the adaptive immune defense and therefore offer a simple model system to study in more detail evolutionary aspects of hematopoiesis and the function of prokineticin domain-containing proteins. The present study showing that astakine 1 alone can induce differentiation along the SGC lineage and astakine 2 can induce differentiation along the GC lineage provides us with a simple system to study astakine 1- and astakine 2-induced signaling pathways. In conclusion, our findings that astakines are key regulators of hematopoietic lineage commitment in crustaceans and release of astakines into the circulatory system after infection or wounding support an evolutionarily conserved role for the prokineticin domain-containing proteins in regulating hematopoiesis.
Supplementary Material
This work was supported by the Swedish Science Research Council and the Swedish Research Council Formas.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- Hpt
- hematopoietic tissue
- SGC
- semigranular cell(s)
- GC
- granular cell(s)
- KPI
- Kazal proteinase inhibitor
- SOD
- superoxide dismutase
- PCNA
- proliferating cell nuclear antigen
- RACE
- rapid amplification of cDNA ends
- Trx
- thioredoxin
- r-astakine
- recombinant astakine
- TGase
- transglutaminase.
REFERENCES
- 1.Jiravanichpaisal P., Lee B. L., Söderhäll K. (2006) Immunobiology 211, 213–236 [DOI] [PubMed] [Google Scholar]
- 2.Söderhäll I., Bangyeekhun E., Mayo S., Söderhäll K. (2003) Dev. Comp. Immunol. 27, 661–672 [DOI] [PubMed] [Google Scholar]
- 3.Söderhäll I., Kim Y. A., Jiravanichpaisal P., Lee S. Y., Söderhäll K. (2005) J. Immunol. 174, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 4.Lin X., Söderhäll K., Söderhäll I. (2008) BMC Immunol. 9, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin X., Kim Y. A., Lee B. L., Söderhäll K., Söderhäll I. (2009) Exp. Cell. Res. 315, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 6.Wu C., Söderhäll I., Kim Y. A., Liu H., Söderhäll K. (2008) Proteomics 8, 4226–4235 [DOI] [PubMed] [Google Scholar]
- 7.Crozatier M., Meister M. (2007) Cell. Microbiol. 9, 1117–1126 [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Agosto J. A., Mikkola H. K., Hartenstein V., Banerjee U. (2007) Genes Dev. 21, 3044–3060 [DOI] [PubMed] [Google Scholar]
- 9.Radtke F., Wilson A., MacDonald H. R. (2005) BioEssays 27, 1117–1128 [DOI] [PubMed] [Google Scholar]
- 10.Munier A. I., Doucet D., Perrodou E., Zachary D., Meister M., Hoffmann J. A., Janeway C. A., Jr., Lagueux M. (2002) EMBO Rep. 3, 1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agaisse H., Perrimon N. (2004) Immunol. Rev. 198, 72–82 [DOI] [PubMed] [Google Scholar]
- 12.Boisbouvier J., Albrand J. P., Blackledge M., Jaquinod M., Schweitz H., Lazdunski M., Marion D. (1998) J. Mol. Biol. 283, 205–219 [DOI] [PubMed] [Google Scholar]
- 13.Kleywegt G. J. (1996) Acta Crystallogr. D Biol. Crystallogr. 52, 842–857 [DOI] [PubMed] [Google Scholar]
- 14.Do C. B., Mahabhashyam M. S., Brudno M., Batzoglou S. (2005) Genome Res. 15, 330–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez R., Chinea G., Lopez N., Pons T., Vriend G. (1998) Bioinformatics 14, 523–528 [DOI] [PubMed] [Google Scholar]
- 17.LeCouter J., Zlot C., Tejada M., Peale F., Ferrara N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16813–16818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L., Wang K., Shao Y., Huang J., Li X., Shan J., Wu D., Zheng J. J. (2008) J. Biol. Chem. 283, 23364–23370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Kuei C., Sutton S., Wilson S., Yu J., Kamme F., Mazur C., Lovenberg T., Liu C. (2005) Mol. Pharmacol. 67, 2070–2076 [DOI] [PubMed] [Google Scholar]
- 20.Urayama K., Guilini C., Messaddeq N., Hu K., Steenman M., Kurose H., Ert G., Nebigil C. G. (2007) FASEB J. 21, 2980–2993 [DOI] [PubMed] [Google Scholar]
- 21.Dorsch M., Qiu Y., Soler D., Frank N., Duong T., Goodearl A., O'Neil S., Lora J., Fraser C. C. (2005) J. Leukoc. Biol. 78, 426–434 [DOI] [PubMed] [Google Scholar]
- 22.Hsiao C. Y., Song Y. L. (2010) Fish Shellfish Immunol. 28, 77–86 [DOI] [PubMed] [Google Scholar]
- 23.Wen S., Wilson D. T., Kuruppu S., Korsinczky M. L., Hedrick J., Pang L., Szeto T., Hodgson W. C., Alewood P. F., Nicholson G. M. (2005) Peptides 26, 2412–2426 [DOI] [PubMed] [Google Scholar]
- 24.Negri L., Lattanzi R., Giannini E., Melchiorri P. (2007) Life Sci. 81, 1103–1116 [DOI] [PubMed] [Google Scholar]
- 25.Kaser A., Winklmayr M., Lepperdinger G., Kreil G. (2003) EMBO Rep. 4, 469–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaga O., Lignell M., Söderhäll K. (1995) Anim. Biol. 4, 59–70 [Google Scholar]
- 27.Felsenstein J. (1989) Cladistics 5, 164–166 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.