Abstract
Dendritic growth is essential for the establishment of a functional nervous system. Among extrinsic signals that control dendritic development, substantial evidence indicates that BDNF regulates dendritic morphology. However, little is known about the underlying mechanisms by which BDNF controls dendritic growth. In this study, we show that the MAPK signaling pathway and the transcription factor cAMP response element-binding protein (CREB) mediate the effects of BDNF on dendritic length and complexity. However, phosphorylation of CREB alone is not sufficient for the stimulation of dendritic growth by BDNF. Thus, using a mutant form of CREB unable to bind CREB-regulated transcription coactivator (CRTC1), we demonstrate that this effect also requires a functional interaction between CREB and CRTC1. Moreover, inhibition of CRTC1 expression by shRNA-mediated knockdown abolished BDNF-induced dendritic growth of cortical neurons. Interestingly, we found that nuclear translocation of CRTC1 results from activation of NMDA receptors by glutamate, a process that is essential for the effects of BDNF on dendritic development. Together, these data identify a previously unrecognized mechanism by which CREB and the coactivator CRTC1 mediate the effects of BDNF on dendritic growth.
Keywords: Calcineurin; CREB; Glutamate; Ionotropic Glutamate Receptors (AMPA, NMDA); MAP Kinases (MAPKs); Neurotrophin; Transcription Coactivators; BDNF; CRTC1; Dendrite
Introduction
Dendrites are the primary sites where neurons receive and integrate information from a vast number of synaptic inputs. The specific branching pattern of dendrites determines the number and type of synapses received by a neuron (1). Dendritic development is essential for the formation of neuronal circuits. Indeed, defects in dendritic growth are associated with some forms of mental retardation, including Down syndrome and fragile X syndrome (2, 3). Dendritic arbor development is characterized by extension and retraction of dendritic branches, followed by stabilization and growth of these branches (4, 5). This multistep process is regulated both by intrinsic genetic programs that are capable of generating a basic dendritic arborization and by external signals such as neuronal activity, guidance molecules, and growth factors that are essential for sculpting dendrites to their final form (1, 6, 7). There is compelling in vitro and in vivo evidence that BDNF, a member of the neurotrophin family, regulates dendritic morphology. In particular, BDNF plays an important role in controlling the dendritic length and branching of pyramidal neurons in the developing visual cortex (8, 9). In addition, BDNF overexpression in pyramidal neurons induces sprouting of basal dendrites (10), and release of BDNF from single cells elicits local dendritic growth in nearby neurons (11). Despite these findings, the underlying mechanisms by which BDNF exerts its effects on dendritic growth remain largely unknown.
Neurotrophins trigger a variety of biological responses by activating Trk receptor tyrosine kinases (12). Binding of neurotrophins to Trk receptors leads to the activation of three major intracellular signaling pathways, including MAPK, PI3K, and phospholipase Cγ1 (PLCγ1)2 (12). Neurotrophin signaling through the MAPK, PI3K, and PLCγ1 pathways regulates neuronal differentiation, neuronal survival, and synaptic plasticity, respectively (12). Trk-mediated signaling can propagate to the nucleus to regulate gene transcription through the activation of several transcription factors (12). Of particular interest, the transcription factor cAMP response element-binding protein (CREB) is activated by BDNF (13) and plays a key role in mediating dendritic development in response to neuronal activity (14). Although CREB activation requires phosphorylation of serine 133, there is evidence that phosphorylation of CREB is not always sufficient to initiate gene transcription (15, 16). These observations suggest that additional factors such as CREB-regulated transcription coactivators (CRTCs), also known as transducers of regulated CREB activity (17), may control CREB-mediated gene transcription. CRTCs are latent cytoplasmic coactivators that shuttle to the nucleus in response to increased levels of calcium and cAMP (18, 19). After translocation into the nucleus, CRTCs associate with the basic leucine zipper domain of CREB independently of its phosphorylation status and increase CREB transcriptional activity (17, 20). Among CRTC family members, CRTC1 is primarily expressed in the brain and is involved in activity-dependent transcription of BDNF and in late-phase long-term potentiation (21, 22). Although there is compelling evidence supporting a critical role of BDNF in regulating dendritic morphology, the signaling pathways and downstream effectors necessary for BDNF to promote dendritic development of cortical neurons remain to be identified.
In this study, we show that activation of MAPK, CREB, and CRTC1 mediates BDNF-induced changes in cortical dendritic morphology. We provide evidence that nuclear translocation of CRTC1 results from NMDA receptor-mediated activation of calcineurin and is essential for the regulation of cortical dendritic development by BDNF.
EXPERIMENTAL PROCEDURES
Cortical Neuron Culture
All experiments were performed in accordance with the European Communities Council Directive regarding the care and use of animals for experimental procedures. Cerebral cortices from Day 18 Sprague-Dawley rat embryos were isolated in modified Hanks' balanced salt solution (1.26 mm CaCl2, 0.5 mm MgCl2, 0.4 mm MgSO4, 5.33 mm KCl, 0.44 mm KH2PO4, 145 mm NaCl, 0.34 mm Na2HPO4, 5.56 mm d-glucose, and 10 mm HEPES, pH 7.4) and incubated at 37 °C for 30 min in modified Hanks' balanced salt solution containing 0.12 mg/ml l-cysteine (Sigma), 20 units/ml papain (Worthington), and 1 unit/ml DNase I (Worthington). Dissociated cells were then cultured in NeurobasalTM medium (Invitrogen) supplemented with B27 (Invitrogen), 0.5 mm glutamine (Sigma), and 2 μm glutamate (Sigma). Cells were usually plated at a density of 60,000 cells/cm2 on poly-d-lysine (Sigma)-coated dishes or glass coverslips and maintained for 4 days at 37 °C in a humidified atmosphere of 95% air and 5% CO2. For experiments performed in the absence of glutamate, cells were plated at a density of 2000 cells/cm2 to minimize the accumulation of extracellular glutamate released from cortical neurons. In addition, the culture medium was replaced during the stimulation period with medium lacking glutamine and glutamate.
HEK293 Cell Culture
HEK293 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (HyClone), 100 units/ml penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen) at 37 °C in a humidified atmosphere of 95% air and 5% CO2. The day before transfection, cells were plated at a density of 50,000 cells/cm2 in the above fresh medium lacking antibiotics.
Site-directed Mutagenesis and RNA Interference
An shRNA- resistant CRTC1 mutant (pcDNA3-*CRTC1-Myc) was generated with the QuikChange Lightning site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The expression plasmid pcDNA3-CRTC1-Myc (21) was used as template, and four silent mutations, engineered in the middle of the CRTC1 sequence targeted by CRTC1 shRNA, were introduced with the following set of primers: 5′-CCATGGCGACTTCGAACAACCCCAGAAAATTTAGCGAGAAGATCGC-3′ and 5′-GCGATCTTCTCGCTAAATTTTCTGGGGTTGTTCGAAGTCGCCATGG-3′. All mutations were confirmed by sequencing. pRNAT-CRTC1-shRNA-GFP and pRNAT-Ctrl-shRNA-GFP were constructed by cloning, into pRNAT-U6.3-cGFP (GenScript), a nucleotide sequence targeted against CRTC1 (21) and a nucleotide sequence with no significant homology to any mammalian gene sequence (21), respectively.
Transfection
Transfection of plasmids was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Neurons were transfected 5 h after plating with the following plasmids: pRc/CMV-FLAG-A-CREB (A-CREB) (23), pRc/RSV-FLAG-R314A-CREB (CREB R314A) (21), pRc/RSV-CREB-M1 (CREB S133A) (24), pRNAT-Ctrl-shRNA-GFP (Ctrl shRNA), pRNAT-CRTC1-shRNA-GFP (CRTC1 shRNA), pcDNA3-CRTC1-Myc (CRTC1) (21), and pcDNA3-*CRTC1-Myc (*CRTC1). Plasmids coding for Myc-CMV5-ERK2-MEK1-LA (CA ERK2) (25) and pcDNA3- Myc-PI3K* (CA PI3K) (26) were transfected 3 days after plating for 24 h. Neurons were always cotransfected with the enhanced GFP expression vector pCAG-EGFP (Addgene), except when neurons were transfected with pRNAT-Ctrl-shRNA-GFP or pRNAT-CRTC1-shRNA-GFP. Confluent HEK293 cells were transfected for 48 h with the Ctrl shRNA, CRTC1 shRNA, CRTC1, or *CRTC1 plasmid.
Cell Stimulation
For dendritic analysis, 3-day in vitro cortical neurons were stimulated with 10 ng/ml BDNF (Alomone Labs) for 24 h. When indicated, neurons were treated 30 min before stimulation with 50 μm U0126 (Calbiochem), 10 μm LY294002 (Calbiochem), 0.5 μm U73122 (Calbiochem), 5 μm actinomycin D (Sigma), 1 μm MK-801 (Tocris Bioscience), 5 μm FK506 (LC Laboratories), or 1 μm tetrodotoxin (Latoxan). For Western blot experiments, neurons were treated with 10 ng/ml BDNF for 30 min (except for time course analysis) in the presence of 50 μm U0126 or 10 μm LY294002. For CRTC1 translocation experiments, neurons were exposed for 6 h to 1 μm MK-801, 5 μm FK506, 10 μm nifedipine (Sigma), or 40 μm 6-cyano-7-nitroquinoxaline-2,3-dione (Sigma). Neurons were also treated with 10 ng/ml BDNF for various periods of time in the presence or absence of 1 μm MK-801. To examine the effect of glutamate on CRTC1 translocation, cortical neurons were cultured for 12 h in Neurobasal/B27TM not supplemented with glutamate and glutamine and then stimulated with 10 μm glutamate for 6 h.
Immunocytochemistry
After stimulation, cortical neurons were processed for immunostaining as described previously (27). The antibodies used were as follows: rabbit anti-phospho-Thr202/Tyr204 p44/42 MAPK (1:50), rabbit anti-phospho-Ser473 Akt (1:50), mouse anti-Myc tag (9B11; 1:1000), and rabbit anti-CRTC1 (C71D11; 1:1500) (Cell Signaling Technology); mouse anti-MAP2 (clone HM-2; 1:200; Sigma), and rabbit or mouse anti-GFP (1:500; Invitrogen). Alexa Fluor 488- and 546-labeled goat anti-mouse or anti-rabbit IgG (1:1000; Invitrogen) was used as a secondary antibody. Glass coverslips were mounted with ProLong Gold antifade reagent (Invitrogen) on microscope slides after DAPI counterstaining (1:4000; Invitrogen) and analyzed with an Axioplan 2 fluorescence microscope (Carl Zeiss).
Image Analysis
Analysis of dendritic morphology was performed with Neurolucida software (MBF Bioscience). CRTC1 immunofluorescence intensity was analyzed with ImageJ software (National Institutes of Health) in the cytoplasmic and nuclear compartments of cortical neurons, which were delineated using MAP2 immunofluorescence staining and DAPI counterstaining, respectively. Data are shown as the ratio of CRTC1 immunofluorescence intensity in the nucleus to the cytoplasm. To examine the knockdown efficiency of endogenous CRTC1 expression, cortical neurons were transfected with CRTC1 shRNA or Ctrl shRNA, and the ratio of CRTC1 immunofluorescence intensity in the nucleus of the transfected neuron to that in neighboring non-transfected neurons was determined using ImageJ software.
Western Blotting
Western blotting was performed as described previously (27) with rabbit anti-phospho-Thr202/Tyr204 p44/42 MAPK (1:1000), rabbit anti-phospho-Ser473 Akt (1:1000), rabbit anti-phospho-Ser133 CREB (1:1000; Cell Signaling Technology), mouse anti-Myc tag (9B11; 1:2000), rabbit anti-CRTC1 (C71D11; 1:1500), and mouse anti-β-tubulin (clone tub 2.1; 1:1000; Sigma) antibodies. ECL horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibody (1:10,000; Amersham Biosciences) was used as a secondary antibody. Results were quantified with ImageJ software, and densitometric values were normalized to corresponding β-tubulin levels.
Statistical Analysis
Data were analyzed for statistical significance using one-way analysis of variance, followed either by Bonferroni's post hoc test for analysis of dendritic morphology or by Dunnett's post hoc test for CRTC1 translocation experiments. Data from CA ERK2 and CA PI3K experiments were analyzed using unpaired Student's t test.
RESULTS
Activation of the MAPK Signaling Pathway Is Required for the Effects of BDNF on Dendritic Development
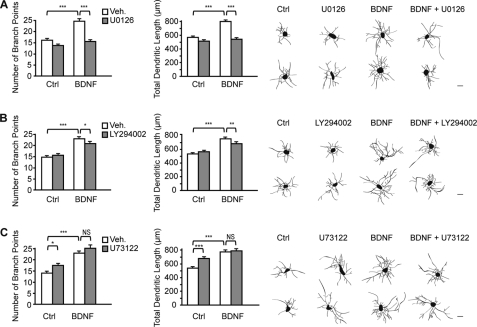
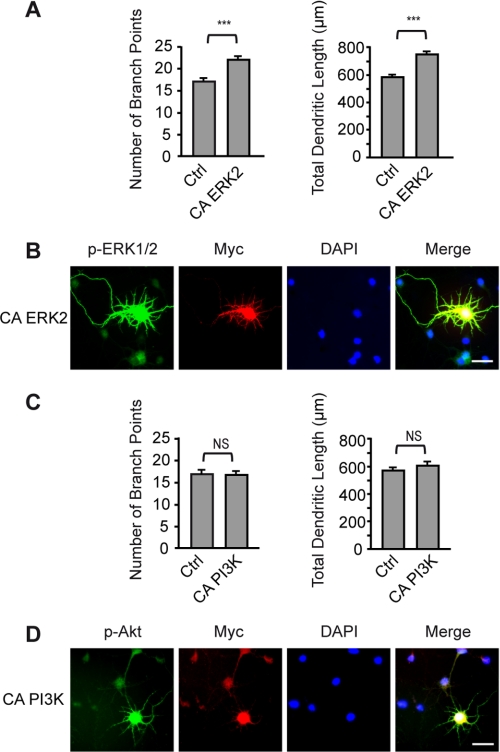
BDNF exerts its cellular effects through activation of the TrkB receptor tyrosine kinase. Binding of BDNF to TrkB produces biological responses via activation of three well described intracellular pathways, including MAPK, PI3K, and PLCγ1 (12). To identify which signaling pathway mediates the effects of BDNF on dendritic growth, cortical neurons were stimulated by BDNF in the presence or absence of selective pharmacological inhibitors of the MAPK, PI3K, and PLC pathways, and the dendritic morphology was analyzed. Compared with unstimulated cultures, neurons exposed to BDNF exhibited an increased number of branch points and total dendritic length, an effect that was completely abolished by the MEK inhibitor U0126 (Fig. 1A). In contrast, blockade of the PI3K pathway by LY294002 only attenuated but did not prevent the effects of BDNF on dendritic morphology (Fig. 1B). The partial inhibitory effect of LY294002 on BDNF-induced dendritic growth does not result from an inhibitory effect of LY294002 on the activation of ERK1/2 by BDNF, as shown by Western blot analysis (supplemental Fig. S1). Inhibition of PLCγ1 by U73122 did not affect BDNF-induced increases in dendritic length and branching (Fig. 1C). These results indicate that activation of the MAPK signaling pathway is required for mediating the effects of BDNF on cortical dendritic morphology. These findings led us to investigate whether activation of MAPK is sufficient to promote dendritic growth of cortical neurons. Expression of a constitutively active form of the MAPK ERK2 (25) was sufficient to increase the total dendritic length and branch point number of cortical neurons (Fig. 2, A and B), whereas transfection of cortical neurons with a constitutively active form of PI3K (26) did not enhance the extent and complexity of dendritic processes (Fig. 2, C and D). These data indicate that activation of the MAPK signaling cascade, but not of the PI3K pathway, is sufficient to increase dendritic length and arborization of cortical neurons.
FIGURE 1.
Activation of the MAPK signaling pathway mediates BDNF-induced dendritic growth. Cortical neurons were stimulated with BDNF for 24 h in the presence or absence of the MEK inhibitor U0126 (A), the PI3K inhibitor LY294002 (B), or the PLC inhibitor U73122 (C). In the bar charts, the number of branch points and the total dendritic length of cortical neurons were quantified for each condition. The right panels show reconstructions of representative neurons for each condition. Scale bars = 20 μm. Results represent the mean ± S.E. of 160 neurons from four independent experiments. ***, p < 0.001; **, p < 0.01; *, p < 0.05; NS, not significantly different. Ctrl, control; Veh, vehicle.
FIGURE 2.
Expression of a constitutively active form of MAPK is sufficient to increase dendritic growth of cortical neurons. A, cortical neurons were transfected with a plasmid encoding a constitutively active form of MAPK (CA ERK2). The number of branch points and the total dendritic length were quantified 24 h after transfection. Ctrl, control. B, immunofluorescence staining of phosphorylated ERK1/2 (p-ERK1/2) and Myc showed a pronounced increase in phosphorylated ERK1/2 expression in transfected neurons compared with untransfected neurons. C, cortical neurons were transfected with a plasmid encoding a constitutively active form of PI3K (CA PI3K). The number of branch points and the total dendritic length were quantified 24 h after transfection. D, immunofluorescence staining of phosphorylated Akt (p-Akt) and Myc showed a marked enhancement of phosphorylated Akt expression in transfected neurons compared with untransfected neurons. Scale bars = 20 μm. Data in A and C are the mean ± S.E. of at least 180 neurons from five independent experiments. ***, p < 0.001; NS, not significantly different.
Role of CREB in the Control of Dendritic Growth by BDNF
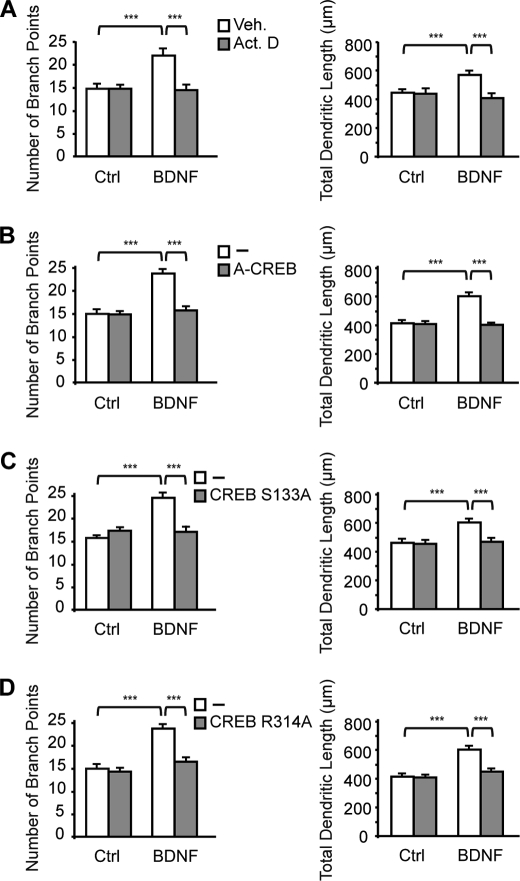
Activation of MAPK signaling by neurotrophins can propagate to the nucleus to regulate gene transcription via specific transcription factors (12). This led us to examine whether BDNF-induced dendritic development is dependent on gene transcription. Treatment of cortical neurons with the transcription inhibitor actinomycin D suppressed the effects of BDNF on dendritic morphology (see Fig. 4A), indicating that activation of gene transcription mediates BDNF-induced increases in dendritic length and complexity.
FIGURE 4.
CREB and its interaction with CRTC1 are essential for the stimulation of dendritic growth by BDNF. Cortical neurons were treated with BDNF for 24 h in the presence of actinomycin D (Act. D) (A) or after transfection with plasmids encoding a dominant-negative form of CREB (A-CREB) (B), a mutant form of CREB that lacks the protein kinase A phosphorylation site (CREB S133A) (C), or a mutant form of CREB defective in CRTC1 binding (CREB R314A) (D). The number of branch points and the total dendritic length of cortical neurons were quantified for each condition. Results represent the mean ± S.E. of 120 neurons from three independent experiments. ***, p < 0.001. Ctrl, control; Veh, vehicle.
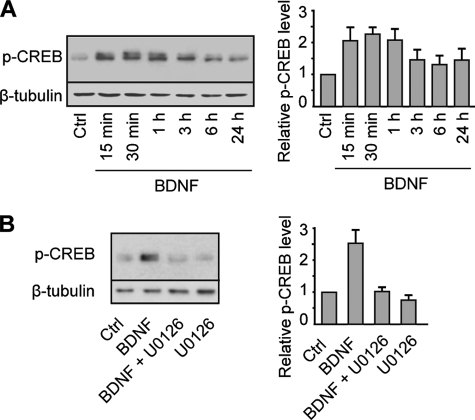
Among transcription factors activated by neurotrophins via the MAPK pathway, CREB plays a central role in mediating neurotrophin responses (13). We therefore investigated the role of CREB in the control of dendritic development by BDNF. Because phosphorylation of CREB at serine 133 is critical for CREB-mediated gene transcription (28), we first examined whether BDNF enhances CREB phosphorylation. Exposure of cortical neurons to BDNF induced a sustained phosphorylation of CREB that was suppressed by the MEK inhibitor U0126 (Fig. 3, A and B). We next determined whether CREB is necessary to induce dendritic growth by BDNF. To this end, we examined the consequences of expressing A-CREB, a dominant-negative form of CREB that prevents the basic region of wild-type CREB from binding to DNA (23), on BDNF-induced dendritic growth. Expression of A-CREB in cortical neurons suppressed BDNF-induced increases in dendritic length and branching (Fig. 4B), indicating that the regulation of dendritic development by BDNF requires CREB-mediated gene transcription. On the basis of this observation, we determined whether phosphorylation of CREB at serine 133 is required for the stimulation of dendritic growth by BDNF. Expression of a dominant-negative form of CREB in which serine 133 was mutated to alanine (CREB S133A) (24) abolished BDNF-induced increases in dendritic length and branching (Fig. 4C), indicating that phosphorylation of CREB at serine 133 is required for the stimulation of dendritic growth by BDNF. Although phosphorylation of serine 133 is critical for the activation of CREB, there is evidence that CREB phosphorylation initiated by different stimuli, including neurotrophins, is not always sufficient to activate CREB-dependent gene transcription (15). This suggests that cooperative interactions between CREB and other transcription factors or coactivators are necessary to activate CREB-mediated gene expression (15). Recently, a new family of CREB coactivators called CRTCs has been shown to dramatically increase CREB-mediated transcriptional activity independently of CREB serine 133 phosphorylation (17, 20). Characterization of CREB-CRTC interaction revealed that arginine 314 in the CREB basic leucine zipper domain is essential for the binding of CRTC to CREB (19, 29). To test whether the interaction between CREB and CRTC contributes to the regulation of dendritic morphology by BDNF, cortical neurons were transfected with a CREB construct containing an R314A mutation (21) and treated with BDNF. Analysis of dendritic morphology revealed that the R314A mutation in CREB suppressed BDNF-induced increases in dendritic length and branching (Fig. 4D), indicating that the interaction between CREB and CRTC is essential for the stimulation of dendritic growth by BDNF. In this context, it is important to note that the R314A mutation in CREB was previously shown not to affect the dimerization or DNA binding properties of the CREB basic leucine zipper domain, indicating that suppression of BDNF-induced dendritic growth by expression of CREB R314A in cortical neurons reflects a direct effect on formation of the CREB·CRTC complex and not on CREB occupancy (19).
FIGURE 3.
BDNF stimulates CREB phosphorylation via the MAPK signaling pathway. A, left panel, Western blot analysis of phospho-Ser133 CREB (p-CREB) expression in cortical neurons stimulated with BDNF for various periods of time. Right panel, quantitative analysis. Values represent the mean ± S.E. of five independent experiments and are shown as phospho-CREB expression levels relative to the control (Ctrl) value. B, left panel, Western blot analysis of phospho-CREB expression in cortical neurons stimulated with BDNF for 30 min in the presence or absence of the MEK inhibitor U0126. Right panel, quantitative analysis. Values represent the mean ± S.E. of three independent experiments and are shown as phospho-CREB expression levels relative to the control value.
CRTC1 Mediates the Effects of BDNF on Dendritic Development
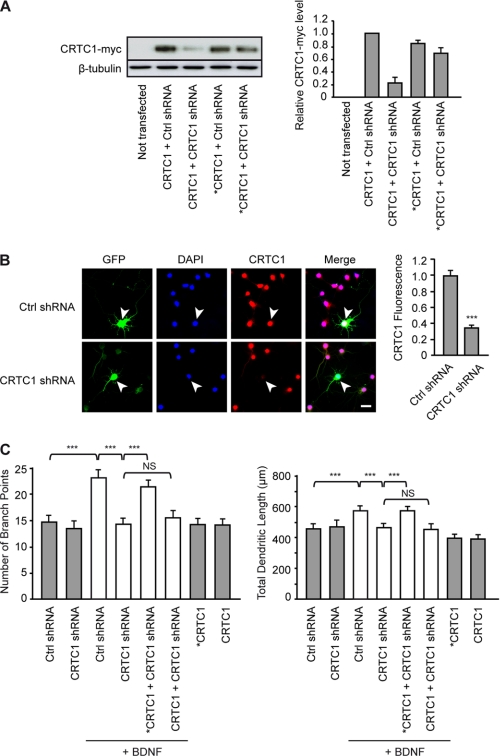
Among CRTC family members, CRTC1 is highly expressed in the rodent cerebral cortex, hippocampus, striatum, and cerebellum (21, 22). These observations led us to examine whether knockdown of CRTC1 with shRNA inhibits BDNF-induced increases in dendritic length and branching. Transfection of HEK293 cells with a CRTC1 shRNA-producing plasmid (21) markedly inhibited exogenous expression of CRTC1 protein, whereas it did not significantly reduce expression of a mutant form of CRTC1 resistant to shRNA silencing (Fig. 5A). We confirmed the efficacy of CRTC1 shRNA in cortical neurons. Thus, transfection of cortical neurons with CRTC1 shRNA markedly inhibited the endogenous expression of CRTC1 (Fig. 5B), indicating that the shRNA against CRTC1 efficiently decreased CRTC1 expression. Inhibition of CRTC1 expression by shRNA- mediated knockdown abolished BDNF-induced dendritic growth of cortical neurons (Fig. 5C). Blockade of BDNF-induced increases in dendritic length and branching by CRTC1 shRNA was prevented by coexpression of shRNA-resistant but not wild-type CRTC1 (Fig. 5C). Together, these data indicate that CRTC1 is a critical mediator of the effects of BDNF on cortical dendritic development.
FIGURE 5.
CRTC1 is essential for mediating the effects of BDNF on dendritic development. A, knockdown efficiency of shRNA against CRTC1. Left panel, Western blot analysis of CRTC1-Myc expression in HEK293 cells transfected with plasmids encoding CRTC1-Myc (CRTC1) or a mutant form of CRTC1-Myc resistant to shRNA silencing (*CRTC1) together with plasmids producing either non-silencing shRNA (Ctrl shRNA) or shRNA targeted against CRTC1 (CRTC1 shRNA). Right panel, quantitative analysis. Values are the mean ± S.E. of three independent experiments and are shown as the levels of CRTC1-Myc in cells cotransfected with CRTC1 and Ctrl shRNA plasmids. B, left panel, representative images of cortical neurons transfected with the Ctrl shRNA or CRTC1 shRNA plasmid and immunostained for GFP and CRTC1, followed by DAPI counterstaining. White arrowheads point to the transfected neuron. Scale bar = 20 μm. Right panel, quantitative analysis of the ratio of CRTC1 immunofluorescence intensity in the nucleus of transfected neurons to that of neighboring non-transfected neurons. Results are the mean ± S.E. of 40 neurons from three independent experiments. ***, p < 0.001. C, cortical neurons were transfected with the Ctrl shRNA or CRTC1 shRNA plasmid alone or along with the CRTC1 or *CRTC1 plasmid and stimulated with BDNF for 24 h. The number of branch points and the total dendritic length of cortical neurons were quantified for each condition. Data are the mean ± S.E. of 120 neurons from three independent experiments. ***, p < 0.001; NS, not significantly different.
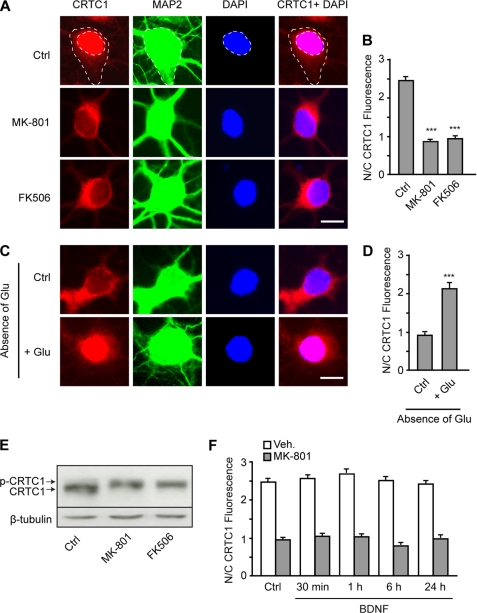
Nuclear Translocation of CRTC1 Is Triggered by NMDA Receptor-mediated Activation of Calcineurin
To stimulate CREB-dependent transcription, CRTC1 must undergo dephosphorylation and subsequent translocation into the nucleus (19). On the basis of this observation, we examined whether BDNF triggers nuclear translocation of CRTC1. To our surprise, immunofluorescence studies revealed that CRTC1 protein was already confined to the nucleus of cortical neurons not treated with BDNF, whereas only low levels of CRTC1 were localized in the cytoplasm (Fig. 6, A and B). Increases in intracellular calcium levels have been shown to induce nuclear translocation of CRTC proteins (18, 19). Because calcium entry through NMDA receptors and L-type voltage-sensitive calcium channels plays an essential role in the control of dendritic growth (14, 30), we examined whether activation of these calcium channels contributes to the nuclear localization of CRTC1. Treatment of cortical neurons with the NMDA receptor antagonist MK-801 prevented nuclear accumulation of CRTC1 (Fig. 6, A and B), whereas the L-type calcium channel blocker nifedipine did not reverse nuclear localization of CRTC1 (nuclear/cytoplasmic fluorescence ratio, control = 2.48 ± 0.07 and nifedipine = 2.40 ± 0.08; n = 250). In contrast to the effects of MK-801, blockade of AMPA/kainate glutamate receptors by 6-cyano-7-nitroquinoxaline-2,3-dione did not affect nuclear accumulation of CRTC1 (nuclear/cytoplasmic fluorescence ratio, control = 2.41 ± 0.08 and 6-cyano-7-nitroquinoxaline-2,3-dione = 2.43 ± 0.13; n = 120). In agreement with these findings, cortical neurons exposed to a medium lacking glutamate exhibited cytoplasmic localization of CRTC1 similar to that observed in neurons treated with MK-801, whereas the addition of glutamate induced nuclear translocation of CRTC1 (Fig. 6, C and D). The data from these experiments show that translocation of CRTC1 from the cytoplasm to the nucleus of cortical neurons results from the activation of NMDA receptors by glutamate. Previous studies have demonstrated that nuclear transport of CRTC1 in response to increased intracellular calcium is mediated by the calcium/calmodulin- dependent protein phosphatase calcineurin (18, 19). In agreement with this observation, inhibition of calcineurin by FK506 prevented nuclear accumulation of CRTC1 in cortical neurons (Fig. 6, A and B). In addition, treatment of cortical neurons with MK-801 or FK506 increased CRTC1 phosphorylation (Fig. 6E), which is consistent with their inhibitory effect on the nuclear accumulation of CRTC1 (Fig. 6, A and B). Taken together, these data indicate that nuclear translocation of CRTC1 is triggered by NMDA receptor-mediated activation of calcineurin. Interestingly, stimulation of cortical neurons with BDNF did not affect the nuclear/cytoplasmic ratio of CRTC1 expression even after exposure to MK-801 (Fig. 6F), a treatment that reverses the nuclear localization of CRTC1, indicating that BDNF does not regulate nuclear transport of CRTC1.
FIGURE 6.
Nuclear localization of CRTC1 results from the activation of NMDA receptors and calcineurin. A, representative images of neurons treated with MK-801 or FK506 for 6 h and immunostained for CRTC1 and MAP2, followed by DAPI counterstaining. Dashed lines represent the cytoplasmic and nuclear compartments, which were delineated using MAP2 immunofluorescence staining and DAPI counterstaining, respectively. Scale bar = 10 μm. Ctrl, control. B, quantitative analysis of the nuclear/cytoplasmic ratio (N/C) of CRTC1 immunofluorescence in neurons treated with MK-801 or FK506. C, representative images of neurons exposed to a culture medium lacking glutamate for 12 h and then stimulated or not with 10 μm glutamate for 6 h. Cortical neurons were then immunostained for CRTC1 and MAP2, followed by DAPI counterstaining. D, quantitative analysis of the nuclear/cytoplasmic ratio of CRTC1 immunofluorescence in neurons treated as described for C. E, Western blot analysis of CRTC1 expression in cortical neurons treated with MK-801 or FK506 for 6 h. The upper and lower bands correspond to the phosphorylated (p-CRTC1) and non-phosphorylated forms of CRTC1, respectively. F, quantitative analysis of the nuclear/cytoplasmic ratio of CRTC1 immunofluorescence in neurons treated with BDNF for various periods of time in the presence or not of MK-801. Results shown in B, D, and F represent the mean ± S.E. of at least 140 neurons from three independent experiments. ***, p < 0.001. Veh, vehicle.
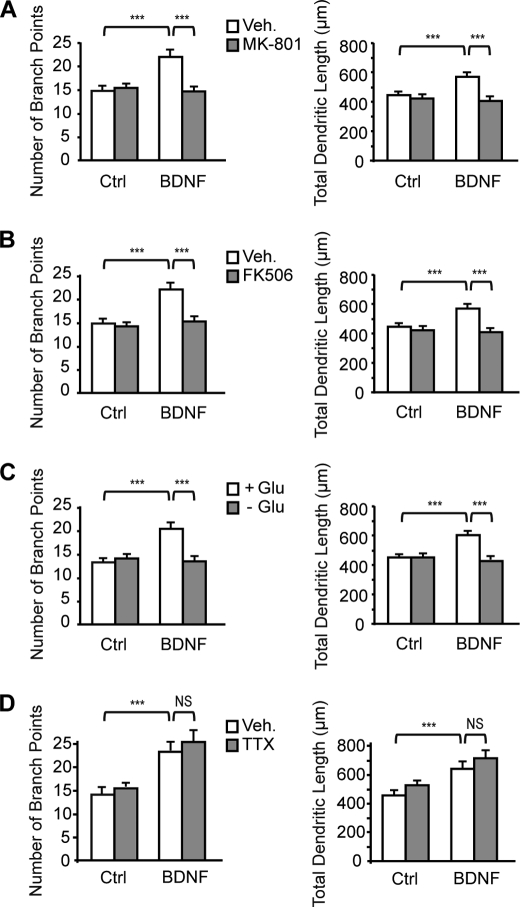
Increases in Dendritic Growth by BDNF Require NMDA Receptor-mediated Activation of Calcineurin by Glutamate
Finally, we examined whether nuclear translocation of CRTC1 by NMDA receptor-mediated activation of calcineurin contributes to the effects of BDNF on dendritic growth. Although exposure of cortical neurons to MK-801 or FK506 did not affect basal dendritic growth, they both abolished BDNF-induced increases in dendritic length and complexity (Fig. 7, A and B), indicating that activation of NMDA receptors and subsequently of calcineurin mediates the effects of BDNF on dendritic development. In agreement with these findings, stimulation of cortical neurons with BDNF in the absence of glutamate prevented BDNF-induced increases in dendritic length and branching (Fig. 7C). Although there is evidence that BDNF enhances glutamate release from cortical neurons through a tetrodotoxin-sensitive mechanism (31), treatment of cortical neurons with tetrodotoxin did not reduce the effects of BDNF on dendritic length and complexity (Fig. 7D), indicating that BDNF does not act by increasing glutamate release from cortical neurons. Together, these data support the conclusion that stimulation of dendritic growth by BDNF requires NMDA receptor-mediated activation of calcineurin by glutamate.
FIGURE 7.
Activation of NMDA receptors and calcineurin mediates the effects of BDNF on dendritic development. Cortical neurons were stimulated with BDNF for 24 h in the presence or absence of MK-801 (A), FK506 (B), glutamate (C), or tetrodotoxin (TTX) (D). The number of branch points and the total dendritic length of cortical neurons were quantified for each condition. Data are the mean ± S.E. of 120 neurons from three independent experiments. ***, p < 0.001; NS, not significantly different. Ctrl, control; Veh, vehicle.
DISCUSSION
There is considerable evidence that BDNF regulates dendritic growth during development, but the underlying mechanisms remain unclear. In this study, we have shown that activation of MAPK, CREB, and CRTC1 plays a critical role in mediating the effects of BDNF on dendritic growth. In addition, our data revealed that nuclear translocation of CRTC1 results from NMDA receptor-mediated activation of calcineurin. Finally, we found that activation of CRTC1 by glutamate via stimulation of NMDA receptors and calcineurin is essential for the effects of BDNF on dendritic development.
Characterization of the signaling cascade by which BDNF regulates dendritic development revealed that activation of the MAPK signaling pathway mediates BDNF-induced dendritic growth and branching (Fig. 1A). In line with these findings, activation of MAPK was found to be sufficient to increase the total dendritic length and branch point number of cortical neurons (Fig. 2, A and B). Interestingly, previous studies have provided evidence that BDNF induces primary dendrite formation in cortical neurons via activation of the PI3K and MAPK signaling pathways (6). However, it is important to note that the effects of BDNF on primary dendrite formation were independent of new protein synthesis (6), whereas increases in dendritic length and complexity by BDNF, described in the present study, were mediated by transcription-dependent mechanisms, as shown by the complete inhibition of BDNF-induced dendritic growth by actinomycin D (Fig. 4A). Inhibition of CREB phosphorylation by expression of CREB S133A suppressed BDNF-induced increases in dendritic length and branching (Fig. 4C), indicating that phosphorylation of CREB is required for the stimulation of dendritic growth by BDNF. In line with these observations, BDNF induced a sustained phosphorylation of CREB that was abolished by inhibition of the MAPK pathway (Fig. 3, A and B). Although CREB phosphorylation is critical for the regulation of cAMP response element-mediated gene expression (28), there is evidence that phosphorylation of CREB at serine 133 is not sufficient to activate gene transcription (15, 16). In this context, members of the CRTC family potently enhance CREB-dependent transcription via a phosphorylation-independent interaction with the basic leucine zipper DNA-binding/dimerization domain of CREB (17, 20). Among CRTC family members, CRTC1 is abundantly expressed in the brain and is involved in activity-dependent dendritic growth and hippocampal long-term potentiation (21, 22, 32). Expression of a CREB construct bearing a point mutation in the CRTC1-binding domain, as well as depletion of CRTC1 expression by shRNA-mediated knockdown, abolished BDNF-induced increases in dendritic length and branching (Figs. 4D and 5C), indicating that the interaction of CREB with CRTC1 is required for the effects of BDNF on dendritic growth.
Under resting conditions, CRTC1 is sequestered in the cytoplasm through a phosphorylation-dependent interaction with 14-3-3 proteins (19). Increases in intracellular calcium levels have been shown to trigger the dephosphorylation and nuclear translocation of CRTC1 via activation of the calcium/calmodulin- dependent protein phosphatase calcineurin (18, 19). In our study, cortical neurons exhibited a strong nuclear accumulation of CRTC1 (Fig. 6, A and B) that was not further increased by exposure of cortical neurons to BDNF (Fig. 6F). In this context, it is interesting to note that previous studies have shown that stimulation of cortical neurons by BDNF induces calcineurin activity and nuclear translocation of NFATc, a transcription factor that regulates embryonic axon outgrowth (33). The marked nuclear accumulation of CRTC1 exhibited by cortical neurons in our study was prevented in the absence of glutamate or in the presence of the NMDA receptor antagonist MK-801 (Fig. 6, A--D), indicating that nuclear translocation of CRTC1 results from the activation of NMDA receptors. In agreement with these data, recent studies have provided evidence that activation of NMDA receptors by intravitreous injection of NMDA triggers nuclear accumulation of CRTC1 in retinal ganglion cells (34). Pharmacological inhibition of calcineurin prevented the dephosphorylation and nuclear accumulation of CRTC1 (Fig. 6, A, B, and E), which is consistent with previous findings (18, 19). Together, these data demonstrate that nuclear translocation of CRTC1 results from NMDA receptor-mediated activation of calcineurin. Finally and most importantly, nuclear translocation of CRTC1 by NMDA receptor-mediated activation of calcineurin was shown to be essential for the effects of BDNF on dendritic growth of cortical neurons. Thus, prevention of nuclear localization of CRTC1 by MK-801 or FK506 or by stimulating cortical neurons in the absence of glutamate suppressed BDNF-induced increases in dendritic length and complexity (Fig. 7, A–C), indicating that regulation of dendritic growth by BDNF requires glutamate-mediated activation of NMDA receptors and calcineurin.
Previous studies in ferret cortical brain slices have demonstrated that BDNF increases dendritic growth of layer 4 pyramidal neurons (9). Interestingly, application of the competitive NMDA receptor antagonist 2-amino-5-phosphonovaleric acid prevented BDNF from increasing the complexity of basal and apical dendrites (9). These observations support our findings and indicate that regulation of dendritic growth of cortical neurons by BDNF requires activation of NMDA receptors. However, the cellular mechanisms underlying the cooperative actions of BDNF and glutamate in the regulation of dendritic development were not known. Here, we identified the cellular mechanisms underlying these concerted actions by showing that activation of MAPK and phosphorylation of CREB by BDNF are necessary but not sufficient to mediate the effects of BDNF on dendritic development. Indeed, inhibition of NMDA receptors or stimulation of cortical neurons in the absence of glutamate prevented the increased dendritic growth by BDNF (Fig. 7, A and C). Further analysis revealed that activation of NMDA receptors triggered the nuclear translocation of CRTC1 (Fig. 6, A and B), whose interaction with CREB was shown to be essential for the regulation of cortical dendritic growth by BDNF (Fig. 4D).
Together, these results support the conclusion that regulation of dendritic growth by BDNF requires both the stimulation of CREB phosphorylation by BDNF and the induction of CRTC1 nuclear translocation by glutamate through NMDA receptor activation. These data are consistent with previous studies demonstrating that phosphorylation of CREB at serine 133 is necessary but not always sufficient for CREB-dependent transcription (15). For instance, in PC12 cells that extend neurites upon exposure to the neurotrophin NGF, it has been shown that NGF induces the phosphorylation of CREB but is unable to activate CREB-mediated transcription (15). These observations suggest that, in addition to phosphorylation of CREB at serine 133, the cooperation of CREB with transcription coactivators may be required to effectively stimulate CREB-dependent transcription in response to NGF. Our data support this hypothesis by revealing a previously unrecognized mechanism by which CREB and the coactivator CRTC1 mediate the effects of BDNF on dendritic length and complexity.
There is compelling evidence supporting functional and cooperative interactions between BDNF and glutamate. Thus, in developing cortical and hippocampal neurons, glutamate and BDNF co-regulate one another such that glutamate increases the expression and secretion of BDNF (35, 36), and conversely, BDNF enhances glutamate release (31). Moreover, BDNF enhances glutamatergic synaptic transmission through pre- and postsynaptic mechanisms (37). Data from the present study provide evidence for a novel cooperative interaction between BDNF- and glutamate-mediated signaling that converges on CREB to regulate the expression of target genes involved in dendritic development.
A role for activity in shaping dendritic morphology is well established (4, 38, 39). For instance, visual stimulation of Xenopus laevis tadpoles, which enhances synaptic activity on tectal neurons, promotes dendritic growth in an NMDA receptor-dependent manner (38). Because excitatory activity regulates the expression and secretion of BDNF, predominantly by glutamate signaling, this suggests that the cooperative interaction between BDNF- and glutamate-mediated signaling, described in this study, may have important implications in activity-dependent dendritic growth by facilitating the transcription of CREB target genes that contribute to the development of dendritic morphology.
Supplementary Material
Acknowledgments
We thank Charles Vinson for the pRc/CMV-FLAG-A-CREB plasmid, Melanie Cobb and Nathalie Ahn for the ERK2-MEK1-LA plasmid, and Wen-Cheng Xiong and Bai Lu for the pcDNA3-Myc-PI3K* plasmid.
This work was supported by Swiss National Science Foundation Grants 31003A-124783 (to J.-L. M.) and 3100A0–120699 (to J.-R. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PLCγ1
- phospholipase Cγ1
- CREB
- cAMP response element-binding protein
- CRTC
- CREB-regulated transcription coactivator.
REFERENCES
- 1.Whitford K. L., Dijkhuizen P., Polleux F., Ghosh A. (2002) Annu. Rev. Neurosci. 25, 127–149 [DOI] [PubMed] [Google Scholar]
- 2.Benavides-Piccione R., Ballesteros-Yáñez I., de Lagrán M. M., Elston G., Estivill X., Fillat C., Defelipe J., Dierssen M. (2004) Prog. Neurobiol. 74, 111–126 [DOI] [PubMed] [Google Scholar]
- 3.O'Donnell W. T., Warren S. T. (2002) Annu. Rev. Neurosci. 25, 315–338 [DOI] [PubMed] [Google Scholar]
- 4.Cline H. T. (2001) Curr. Opin. Neurobiol. 11, 118–126 [DOI] [PubMed] [Google Scholar]
- 5.Jan Y. N., Jan L. Y. (2001) Genes Dev. 15, 2627–2641 [DOI] [PubMed] [Google Scholar]
- 6.Dijkhuizen P. A., Ghosh A. (2005) J. Neurobiol. 62, 278–288 [DOI] [PubMed] [Google Scholar]
- 7.Miller F. D., Kaplan D. R. (2003) Curr. Opin. Neurobiol. 13, 391–398 [DOI] [PubMed] [Google Scholar]
- 8.McAllister A. K., Lo D. C., Katz L. C. (1995) Neuron 15, 791–803 [DOI] [PubMed] [Google Scholar]
- 9.McAllister A. K., Katz L. C., Lo D. C. (1996) Neuron 17, 1057–1064 [DOI] [PubMed] [Google Scholar]
- 10.Horch H. W., Krüttgen A., Portbury S. D., Katz L. C. (1999) Neuron 23, 353–364 [DOI] [PubMed] [Google Scholar]
- 11.Horch H. W., Katz L. C. (2002) Nat. Neurosci. 5, 1177–1184 [DOI] [PubMed] [Google Scholar]
- 12.Reichardt L. F. (2006) Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1545–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkbeiner S., Tavazoie S. F., Maloratsky A., Jacobs K. M., Harris K. M., Greenberg M. E. (1997) Neuron 19, 1031–1047 [DOI] [PubMed] [Google Scholar]
- 14.Redmond L., Ghosh A. (2005) Cell Calcium 37, 411–416 [DOI] [PubMed] [Google Scholar]
- 15.Bonni A., Ginty D. D., Dudek H., Greenberg M. E. (1995) Mol. Cell. Neurosci. 6, 168–183 [DOI] [PubMed] [Google Scholar]
- 16.Ravnskjaer K., Kester H., Liu Y., Zhang X., Lee D., Yates J. R., 3rd, Montminy M. (2007) EMBO J. 26, 2880–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conkright M. D., Canettieri G., Screaton R., Guzman E., Miraglia L., Hogenesch J. B., Montminy M. (2003) Mol. Cell 12, 413–423 [DOI] [PubMed] [Google Scholar]
- 18.Bittinger M. A., McWhinnie E., Meltzer J., Iourgenko V., Latario B., Liu X., Chen C. H., Song C., Garza D., Labow M. (2004) Curr. Biol. 14, 2156–2161 [DOI] [PubMed] [Google Scholar]
- 19.Screaton R. A., Conkright M. D., Katoh Y., Best J. L., Canettieri G., Jeffries S., Guzman E., Niessen S., Yates J. R., 3rd, Takemori H., Okamoto M., Montminy M. (2004) Cell 119, 61–74 [DOI] [PubMed] [Google Scholar]
- 20.Iourgenko V., Zhang W., Mickanin C., Daly I., Jiang C., Hexham J. M., Orth A. P., Miraglia L., Meltzer J., Garza D., Chirn G. W., McWhinnie E., Cohen D., Skelton J., Terry R., Yu Y., Bodian D., Buxton F. P., Zhu J., Song C., Labow M. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12147–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovács K. A., Steullet P., Steinmann M., Do K. Q., Magistretti P. J., Halfon O., Cardinaux J. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4700–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Wu H., Li S., Chen Q., Cheng X. W., Zheng J., Takemori H., Xiong Z. Q. (2006) PLoS ONE 1, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn S., Olive M., Aggarwal S., Krylov D., Ginty D. D., Vinson C. (1998) Mol. Cell. Biol. 18, 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loriaux M. M., Rehfuss R. P., Brennan R. G., Goodman R. H. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9046–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson M. J., Stippec S. A., Goldsmith E., White M. A., Cobb M. H. (1998) Curr. Biol. 8, 1141–1150 [DOI] [PubMed] [Google Scholar]
- 26.Yang F., He X., Feng L., Mizuno K., Liu X. W., Russell J., Xiong W. C., Lu B. (2001) Nat. Neurosci. 4, 19–28 [DOI] [PubMed] [Google Scholar]
- 27.Burkhalter J., Fiumelli H., Erickson J. D., Martin J. L. (2007) J. Biol. Chem. 282, 5152–5159 [DOI] [PubMed] [Google Scholar]
- 28.Ginty D. D., Bonni A., Greenberg M. E. (1994) Cell 77, 713–725 [DOI] [PubMed] [Google Scholar]
- 29.Xu W., Kasper L. H., Lerach S., Jeevan T., Brindle P. K. (2007) EMBO J. 26, 2890–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konur S., Ghosh A. (2005) Neuron 46, 401–405 [DOI] [PubMed] [Google Scholar]
- 31.Takei N., Numakawa T., Kozaki S., Sakai N., Endo Y., Takahashi M., Hatanaka H. (1998) J. Biol. Chem. 273, 27620–27624 [DOI] [PubMed] [Google Scholar]
- 32.Li S., Zhang C., Takemori H., Zhou Y., Xiong Z. Q. (2009) J. Neurosci. 29, 2334–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graef I. A., Wang F., Charron F., Chen L., Neilson J., Tessier-Lavigne M., Crabtree G. R. (2003) Cell 113, 657–670 [DOI] [PubMed] [Google Scholar]
- 34.Deng J., Zhang X. L., Wang J. W., Teng L. L., Ge J., Takemori H., Xiong Z. Q., Zhou Y. (2009) Neuroscience 159, 1023–1031 [DOI] [PubMed] [Google Scholar]
- 35.Lessmann V., Gottmann K., Malcangio M. (2003) Prog. Neurobiol. 69, 341–374 [DOI] [PubMed] [Google Scholar]
- 36.Zafra F., Castrén E., Thoenen H., Lindholm D. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicoll R. A., Schmitz D. (2005) Nat. Rev. Neurosci. 6, 863–876 [DOI] [PubMed] [Google Scholar]
- 38.Sin W. C., Haas K., Ruthazer E. S., Cline H. T. (2002) Nature 419, 475–480 [DOI] [PubMed] [Google Scholar]
- 39.Wong R. O., Ghosh A. (2002) Nat. Rev. Neurosci. 3, 803–812 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.