Abstract
Transcription activation by estrogen receptor (ER) is rapid and dynamic. How the prompt and precise ER response is established and maintained is still not fully understood. Here, we report that two boundary elements surrounding the well defined ERα target TFF1 locus are occupied by the CCCTC-binding factor (CTCF). These elements are separated by 40 kb but cluster in the nuclear space depending on CTCF but independent of estrogen and transcription. In contrast, in estrogen non-responsive breast cancer cells, the spatial proximity of these two elements is lost and the entire locus instead displays a polycomb repressive complex 2-controlled heterochromatin characteristic. We showed that CTCF acts upstream of the “pioneer” factor FOXA1 in determining the genomic response to estrogen. We propose that the CTCF-bound boundary elements demarcate active versus inactive regions, building a framework of adjacent chromosome territory that predisposes ERα-regulated transcription.
Keywords: Chromatin Remodeling, Chromatin Structure, Estrogen, Gene Regulation, Hormone Receptors
Introduction
Estrogen is a classical etiological factor for breast and endometrial cancers (1–3). Estrogen exerts its biological function via two members of the nuclear receptor family, ERα2 and ERβ, which function as ligand-dependent transcription factors (4–6), with ERα being the dominant conducer in both physiological and pathological settings. Previously, we and others showed that ERα, together with a number of cofactor proteins, rapidly associates with its genomic targets in a cyclic fashion following estrogen stimulation (7–9).
The mechanism responsible for the prompt and precise recognition of target genes in the whole genome by liganded ER is not yet fully understood. However, it is becoming clear that the stochastic collision model, which assumes the binding of a transcription factor with its cognate sequence in a random collision fashion, does not fully explain the promptness and the precision of the target gene recognition, especially considering the enormous complexity of genome organization in the nuclear space. In this regard, it is interesting to note that recent studies demonstrated that the recruitment of ERα to its target genes requires forkhead protein FOXA1 being in close proximity in the genome (10, 11); FOXA1 acts as a “pioneer factor” to read and translate histone H3 lysine 4 mono/dimethylation (H3K4M1/2) marks (12).
CCCTC-binding factor (CTCF) is the major insulator-binding protein identified in vertebrates. Together with its associated protein factors that are capable of configuring chromatin structure, CTCF endows insulators with the ability to make a gene immune from promiscuous cis effects (13, 14). Based on their biological effects, insulators are divided into two classes: enhancer-blocking insulators, which protect a promoter from communicating with an outside enhancer, and barrier insulators, which prevent the encroachment of neighboring heterochromatin (14, 15). CTCF interacts with insulators/boundary elements through its 11 zinc finger motifs (13, 16, 17) and has been reported to be a transcriptional activator, repressor, and silencer, depending on the DNA context of different gene loci (13, 18). CTCF has been shown to be essential for stabilizing distal cis regulatory element interaction and the higher-order chromatin structure to form an active chromosome hub at the mouse β-globin and Igf2/H19 loci (16, 19–21).
Here we show that two boundary elements separated by 40 kb in the ERα target TFF1 locus act as barrier insulators and demarcate the estrogen responsiveness of this region. These two elements cluster in nuclear space in a manner that is dependent on CTCF but independent of estrogen and active transcription.
EXPERIMENTAL PROCEDURES
Antibodies and Western Blotting
αFOXA1 and αH3K4M1/2 were from Abcam; αCTCF, αSUZ12, αH3K9M2/3, αH3K9Ac, and αH3K27M3 from Upstate; αEZH2 from BD Biosciences; and αERα from Santa Cruz Biotechnology. Western blottings were performed according to the procedures described elsewhere (22–27).
Chromatin Immunoprecipitation (ChIP)
ChIP experiments were performed according to the protocol described previously (22, 24, 26, 28).
Electrophoretic Mobility Shift Assay (EMSA)
Biotinylated probes and methylated cold competitors were synthesized commercially. The assay was performed with gel shift assay systems from Promega using MCF-7 and MDA-MB-231 nuclear extracts. The probe sequences used were as follows: 3′1, GTGGCCGCAAGGGGCCAGTCTGCTTCAAGG; 5′1, CCCAGCATGGATAGCAGAGGGCGCTGTGGAGC; 3′1 cold mutant probe, GTGGAAATAGCTTTTCAGTCTGCTTCAAGG; 3′1 methylated mutant probe, GTGGC(m)CGCAAGGGGCCAGTCTGCTTCAAGG.
Chromosome Conformation Capture Assay (3C)
The 3C assay was done essentially as described (29) with minor modifications. Briefly, collected cell pellets were washed with PBS buffer and then cross-linked with formaldehyde (Sigma) to achieve a final concentration of 2%. After a 10-min incubation at 37 °C, glycine (0.125 m final concentration) was added to stop the reaction. The pellet was then subjected to cold lysis buffer with protease inhibitors (Roche Applied Science), homogenized with a Dounce homogenizer on ice, and centrifuged to pellet the nuclei. To remove non-cross-linked proteins from DNA, SDS (Sigma) was added to a final concentration of 0.3%. For DpnII digestion, Triton X-100 (Sigma) was added to a final concentration of 1% to sequester excess SDS. A 5-μl aliquot of the sample was kept as an undigested genomic DNA control. DpnII (New England Biolabs) restriction enzyme was added to the remaining sample, and it was digested overnight at 37 °C. Digestion efficiency was optimized and monitored by PCR using primer pairs designed specifically for each of the DpnII restriction sites in the TFF1 locus. After complete digestion, 1.6% SDS was added for 20 min at 65 °C to inactivate DpnII, and a 5-μl aliquot of the sample was set aside as the digested genomic DNA control. The ligation reaction was performed with 400 units of T4 DNA ligase (New England Biolabs) for 4 h at 16 °C, followed by incubation for 30 min at room temperature in a total of 5-ml reaction system. Cross-linking was reversed by overnight incubation of the samples with proteinase K at 65 °C, followed by phenol-chloroform purification of DNA. Purified DNA was subjected to PCR amplification with site-specific primer pairs.
RNAi
Chemically synthesized double-stranded siRNA was used against the transcript of CTCF, ERα (28), and FOXA1 (10). Cells were transfected with 100 nm small interfering RNA oligonucleotides for 72 h using Lipofectamine 2000 (Invitrogen) (23). The siRNA sequences were as follows: siCTCF, GGAAGAUCCUAGUUGGCAA; siFOXA1, GGACUUCAAGGCAUACGAA; siERα, GCUACUGUUUGCUCCUAAC; and siNS, UUCUCCGAACGUGUCACGU.
DNA Methylation Analysis
Genomic DNA was extracted and suspended in TE buffer. For each sample, 1 μg of DNA was incubated with the restriction enzymes AciI, EaeI, and HaeIII in a total volume of 10 μl for 12 h at 37 °C. The digested DNA was PCR amplified.
RT-PCR and Real Time PCR
Total cellular RNAs were isolated with the TRIzol reagent (Invitrogen) and used for first strand cDNA synthesis with the Reverse Transcription System (Promega, A3500). Quantitation of all gene transcripts was done by qPCR using Power SYBR Green PCR Master Mix and an ABI PRISM 7300 sequence detection system (Applied Biosystems, Foster City, CA) with the expression of GAPDH as the internal control (25, 27).
RESULTS
TFF Locus Is Delimited by CTCF-bound Boundary Elements for Estrogen Responsiveness
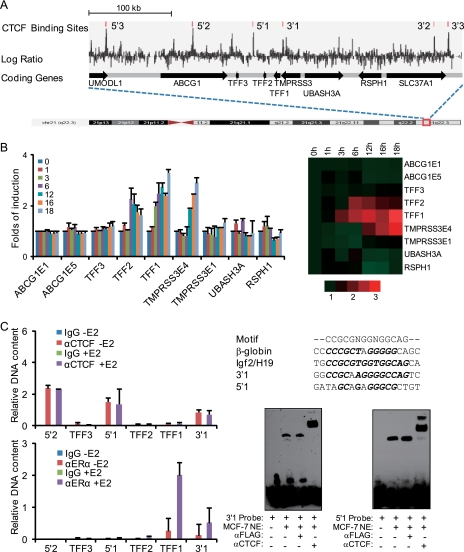
To explore the molecular basis for the efficient genomic response to estrogen, we first investigated what roles the higher-order chromatin structures, especially the genome organizers, might play in this process. We chose the TFF locus as our model system, as its responsiveness to estrogen and its regulatory elements for ERα are well documented (30, 31). Based on recent genomewide chromatin immunoprecipitation (ChIP)-on-chip and ChIP-seq results (32, 33), we identified three putative boundary elements around the TFF locus spanning 100 kb in the genome that are potentially bound by CTCF with repetitive elements masked, as repetitive sequences are stretches of DNA that repeat themselves with unknown function thus are unlikely to represent CTCF-binding sites (Fig. 1A). We denoted these three elements numerically from the 5′ to 3′ direction as 5′2, 5′1, and 3′1, with 5′2 and 3′1 residing in the introns of corresponding neighboring genes ABCG1 and TMPRSS3, respectively, whereas 5′1 is located in the intergenic region of the TFF2 and TFF3 genes.
FIGURE 1.
CTCF demarcates the estrogen response region in the TFF locus. A, the genomic organization/ideogram and CTCF-binding sites in the TFF locus. CTCF-binding sites were identified by bioinformatics analysis of the published ChIP-on-chip data (33) with repeat sequences masked, and these sites were numbered as indicated from the 5′ to 3′ direction of the chromosome. B, CTCF-bound boundary elements confine estrogen responsiveness in the TFF locus. MCF-7 cells were deprived of estrogen for 3 days followed by treatment with E2 for the indicated periods of time. The transcript expression of the indicated genes or exons (E) of genes was measured by real time RT-PCR with GAPDH mRNA as the normalizer. Each bar represents the mean ± S.D. for triplicate experiments. Right panel, a heat map based on the expression of the indicated genes. The scale at the bottom indicates fold-changes. C, binding of CTCF in the identified putative boundary elements in vivo and in vitro. Left panel, qChIP was performed in MCF-7 cells treated with E2 for 45 min for the detection of the in vivo binding of CTCF (upper) or ERα (lower) to the identified putative boundary elements and on the promoters of the indicated genes. Right panel, nucleotide alignment of the consensus CTCF-binding motif, the CTCF-binding motifs in mouse β-globin and Igf2/H19, and the predicted CTCF-binding motif of the 5′1 and 3′1 sites (upper). Italics represent conserved bases. EMSA were performed with MCF-7 nuclear extracts (NE) and biotin-labeled DNA sequences were derived from the 5′1 and the 3′1 sites to detect the binding of CTCF to these sites in vitro (lower).
To determine the estrogen responsiveness of the putative boundary element-enclosed region, we designed real time reverse transcriptase (RT) PCR primers for the three TFF genes, the element-separated exons of the ABCG1 and TMPRSS3 genes, and neighboring UBASH3A and RSPH1 genes. Cells from the estrogen responsive human breast cancer cell line MCF-7 were deprived of estrogen for at least 3 days followed by treatment with 17β-estradiol (E2) for different times. Total RNA was extracted and analyzed by real time RT-PCR for mRNA expression. As shown in Fig. 1B, the mRNA expression of TFF1 and TFF2 increased in response to E2 treatment; the level of TFF1 mRNA peaked at 3-fold and that of TFF2 mRNA peaked at ∼2-fold relative to the control after 6 h of E2 treatment, whereas the mRNA expression of TFF3, which lies outside of 5′1 and 3′1, was barely induced. Remarkably, the messenger induction of the fourth exon of TMPRSS3, which lies within the region of 5′1 and 3′1, showed analogous kinetics to that of TFF1 and TFF2, whereas the first exon of TMPRSS3, which resides outside the 5′1 and 3′1 region, were almost uninduced. Similarly, no significant induction was detected for the messenger expression of exons 1 and 5 of the ABCG1 gene and the UBASH3A and RSPH1 genes, which also lies outside the 5′1 and 3′1 boundaries. A heat map clearly illustrates that the estrogen responsive region is confined within the 5′1 and 3′1 boundaries and centered on TFF1 (Fig. 1B, right).
To determine whether the 5′1 and 3′1 sites are indeed bound by the CTCF protein, we first examined in vivo binding of CTCF to these elements by quantitative PCR-coupled ChIP (qChIP). The results revealed that, although absent at the promoter regions, CTCF binding was detected at the 5′1 and 3′1 sites as well as 5′2 site (Fig. 1C, upper panel). Moreover, the binding of CTCF to these sites was not dependent on E2, which is in contrast to the profile of E2-triggered ERα recruitment on the TFF1 promoter (Fig. 1C, lower panel).
We then performed electrophoretic mobility shift assays (EMSA) to determine whether CTCF could bind to the 5′1 and 3′1 elements in vitro. For this purpose, we searched in these putative insulator sequences for the consensus binding sites of CTCF with a score matrix produced by Ren and co-workers (34) and found that all three putative insulators contain one or two CTCF-binding motifs that scored as high as that in the FII insulator or Igf2 ICR (imprinted control region) (20). In fact, the 3′1 element contains a motif with just one biased base compared with that of the FII insulator (Fig. 1C, right panel). We then synthesized 30–40-bp double-stranded probes based on the sequences of the three putative insulators harboring the identified CTCF-binding motif and performed EMSA with MCF-7 cell nuclear extracts. The results indicated that both 5′1 and 3′1 probes were able to interact with the protein factor(s) from MCF-7 cell nuclear extracts (Fig. 1C, right panel). Incubation of MCF-7 cell nuclear extracts with anti-CTCF antibodies resulted in supershifts of the DNA-protein complex. These experiments indicate that the 5′1 and 3′1 boundary elements are able to bind CTCF and might act as intrinsic insulators for estrogen responsiveness in the TFF locus.
CTCF Is Required for the Genomic Response to Estrogen
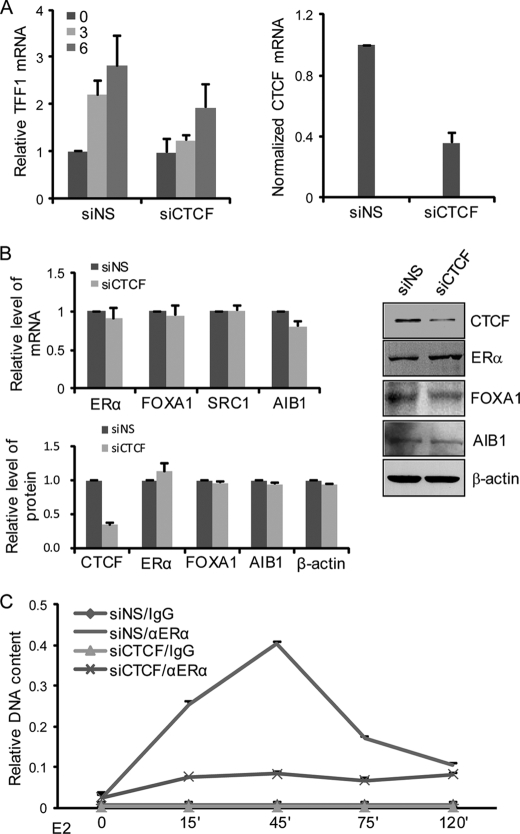
To further establish the role of the 5′1 and 3′1 boundary elements and their associated CTCFs in the genomic response to estrogen, loss-of-function experiments with CTCF were performed and their effect on ERα-mediated TFF1 transcription was evaluated. In these experiments, MCF-7 cells were transfected with either control or CTCF-specific siRNA molecules. The cells were then cultured in estrogen-deprived medium for 72 h followed by treatment with E2 for 3 and 6 h. Real time RT-PCR analysis of TFF1 expression indicated that knockdown of CTCF expression led to a significant decrease in E2-induced TFF1 mRNA expression (Fig. 2A). This effect was not due to a decrease in either the expression of ERα itself or the expression of its critical cofactors such as FOXA1 or p160 coactivators in these cells, as the expression of ERα, FOXA1, SRC-1, and AIB1 mRNAs and proteins remained unchanged in cells with CTCF knockdown (Fig. 2B). These experiments indicate that CTCF is required for ERα-mediated transcription.
FIGURE 2.
CTCF is required for estrogen-stimulated transcription. A, the effect of CTCF knockdown on estrogen-induced TFF1 expression. MCF-7 cells were transfected with either nonspecific control siRNA (siNS) or CTCF siRNA (siCTCF). These cells were deprived of estrogen for 3 days and treated with E2 for 3 or 6 h before the measurement of TFF1 expression by real time RT-PCR. The knockdown effect of CTCF was validated by real time RT-PCR measurement of CTCF mRNA expression (right). B, the effect of CTCF knockdown on estrogen-induced TFF1 expression was not due to changes in expression of ERα and its cofactors. Left panel, mRNA (upper) or protein (bottom) levels of CTCF, ERα, FOXA1, AIB1, SRC-1, and β-actin were analyzed by real time RT-PCR and/or Western blotting, in the above described MCF-7 cells after 6 h of E2 treatment. Each bar represents the mean ± S.D. for triplicate experiments. Right panel, representative Western blots of CTCF and other ERα regulators. C, the effect of CTCF knockdown on the recruitment of ERα on the TFF1 promoter. MCF-7 cells were transfected with either siNS or siCTCF. These cells were deprived of estrogen for 3 days and treated with E2 for the indicated times followed by qChIP assays with control IgG or anti-ERα. Each point represents the mean ± S.D. for triplicate experiments.
To gain a mechanistic insight into the effect of CTCF knockdown on ERα-mediated transcription, qChIP assays were performed and the recruitment dynamics of ERα on the TFF1 promoter were monitored in MCF-7 cells with CTCF knockdown. In nonspecific control siRNA-transfected MCF-7 cells, ERα was associated with the TFF1 promoter in response to E2 treatment, and the association peaked at 45 min of E2 treatment (Fig. 2C), which was consistent with the cyclic kinetics observed previously (8). However, when the expression of CTCF was silenced, the recruitment of ERα on the TFF1 promoter was severely impaired, exhibiting a flattened accumulation curve and a loss of peak occupancy (Fig. 2C). These experiments suggest that CTCF influences the ability of ERα to bind to its target gene promoters and thus affects its regulation of the transcription.
DNA Methylation Status of the 3′1 Element Influences Its Ability for CTCF Binding
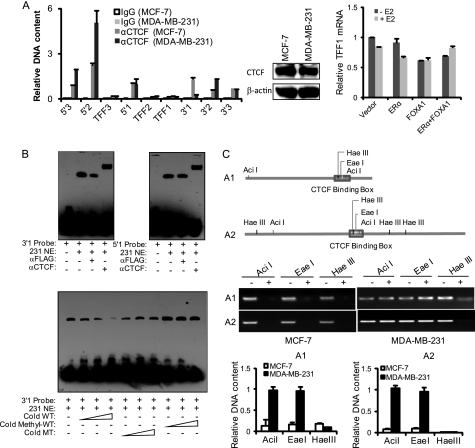
Next, we extended our study to include a total of six putative CTCF-binding sites spanning the ∼700-kb genomic sequence surrounding the TFF1 locus (Fig. 1A). We compared the CTCF binding profile in these sites between estrogen responsive MCF-7 cells and estrogen non-responsive MDA-MB-231 cells using qChIP experiments. The results indicated that, whereas CTCF binding was detected in all of the 6 putative insulators in MCF-7 cells and 5 of them in MDA-MB-231 cells (some with even elevated affinity in MDA-MB-231 cells), this binding was completely lost in the 3′1 element in estrogen non-responsive MDA-MB-231 cells, although expression of the CTCF protein was similar between these two cell lines (Fig. 3A). The correlation between CTCF binding in the 3′1 element and cellular estrogen responsiveness was also observed in estrogen responsive T47-D cells and estrogen non-responsive MDA-MB-468 cells (data not shown). Therefore, the loss of CTCF binding at the 3′1 element in estrogen non-responsive cells could also contribute, beyond the obvious contribution of lost expression of ERα, to the non-responsiveness of these cells to estrogen. In support of this notion, ectopic expression of ERα and/or FOXA1 in MDA-MB-231 cells failed to restore ERα-mediated transcription of the TFF1 gene in response to E2 treatment (Fig. 3A, right panel).
FIGURE 3.
DNA methylation status of the 3′1 site influences its ability for CTCF binding. A, the in vivo binding of CTCF to the identified putative CTCF-binding sites in estrogen-responsive and non-responsive cells. qChIP was performed and the binding of CTCF in the identified putative CTCF-binding sites in MCF-7 cells and MDA-MB-231 cells was compared (left). CTCF expression was also compared in MCF-7 and MDA-MB-231 cells by Western blotting (middle). Ectopic expression of ERα and/or FOXA1 in MDA-MB-231 cells fails to restore ERα-mediated transcription of the TFF1 gene (right). MDA-MB-231 cells were transfected with the indicated expression constructs and treated with E2 for 6 h. The expression of TFF1 was then measured by real time RT-PCR. Each bar represents the mean ± S.D. for triplicate experiments. B, EMSA were performed with MDA-MB-231 cell nuclear extracts (231 NE) and biotin-labeled DNA sequences derived from the 5′1 and 3′1 sites to detect the binding of CTCF in vitro (upper). The methylated 3′1 CTCF probe is unable to compete for the binding of CTCF to the wild-type 3′1 CTCF probe. EMSA were performed with MDA-MB-231 cell nuclear extracts (231 NE) and biotin-labeled DNA sequences derived from the 3′1 site with cold wild-type probe (WT), cold methylated wild-type probe (Methyl-WT), or cold probe mutant (MT) as competitors (lower). C, the CTCF-binding motif of the 3′1 site is hypermethylated in MDA-MB-231 cells. Genomic DNAs from MCF-7 cells or MDA-MB-231 cells were extracted and restricted with endonuclease AciI, EaeI, or HaeIII overnight. The DNAs were then amplified by PCR and separated on 1% agarose gel. The two amplicons labeled as A1 and A2 for these endonucleases in the 3′1 site are illustrated in the upper panel, the PCR products are shown in the middle, and the abundance of PCR products was also measured by real time PCR and shown at the bottom. Each bar represents the mean ± S.D. for triplicate experiments.
To investigate whether the lost binding at the 3′1 site in estrogen non-responsive cells was a result of a trans effect or a cis effect, we performed EMSA with a synthesized sequence derived from the 3′1 fragment containing the CTCF-binding motif and MDA-MB-231 cell nuclear extracts. The results showed that the CTCF binding capacity of the 3′1 fragment was retained in MDA-MB-231 cells, arguing against the possibility that a trans effect was the cause (Fig. 3B, upper panel). Therefore, we asked if the lost binding at the 3′1 site in MDA-MB-231 cells was a result of a cis effect. In this regard, it is important to note that the core cognate motif recognized by CTCF is CG-enriched (Fig. 1C), and it has been reported that, in the mouse Igf2/H19, DNA methylation repels CTCF binding and causes methylation-dependent monoallelic expression of these loci (20, 21, 35). To investigate whether DNA methylation is also a determinant of the cell-specific CTCF binding affinity at the 3′1 site of the TFF locus, we performed nucleotide endonuclease sensitivity assays with DNA methylation-sensitive AciI and EaeI and insensitive HaeIII, all of which have their restriction sites residing in the CTCF-binding motif of the 3′1 site (Fig. 3C). Genomic DNAs were extracted from MCF-7 and MDA-MB-231 cells, treated with the above listed nucleotide endonucleases overnight, and then analyzed with primers to amplify the fragment of the 3′1 element. The abundance of PCR products was indicative of the degree of DNA methylation. As shown in Fig. 3C, whereas the 3′1 DNA from both MCF-7 and MDA-MB-231 cells could be efficiently restricted by HaeIII and that from MCF-7 cells could be effectively cut by AciI and EaeI, the 3′1 DNA from MDA-MB-231 cells could not be digested by AciI or EaeI, suggesting that the 3′1 DNA is hypermethylated in MDA-MB-231 cells.
To establish a causal relationship between DNA methylation and CTCF binding at the 3′1 site, we synthesized cold probes containing methylated bases and performed competitive EMSA. The results indicated that the methylated 3′1 CTCF probe was unable to compete for protein binding in EMSA, similar to the cold probe mutants (Fig. 3B, lower panel). This observation supports a functional association between DNA methylation and CTCF binding in the 3′1 boundary in estrogen non-responsive cells.
Clustering of 5′1 and 3′1 Elements Depends on CTCF but Is Independent of Active Transcription
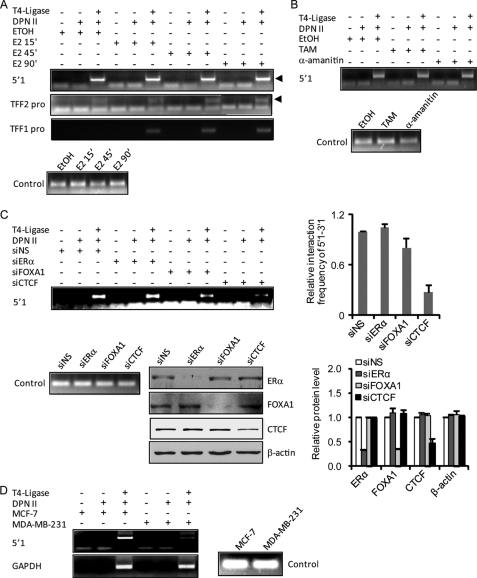
To further explore the molecular mechanism underlying the role of the boundary elements in defining estrogen responsiveness, we used chromosome conformation capture (3C) assays (29, 36, 37) to examine the potential higher-order chromatin organization between CTCF-binding sites surrounding the TFF locus. To this end, MCF-7 cells were depleted of estrogen for 3 days and then treated with E2 for different times. The genomic DNA was fixed with formaldehyde and digested with the restriction enzyme DpnII, which recognizes the frequently appearing four bases GATC. The DNA samples were subsequently ligated by limited T4 DNA ligase for maximal intermolecular joining. We used an anchoring primer located in the 3′1 boundary as prey to capture other potential interacting DNA sequences. Of all the tested sites, the interaction between the 3′1 and 5′1 sites was most frequently observed, indicating their CTCF binding property and potential CTCF-dependent higher-order chromosome configuration (Fig. 4A). In accordance with estrogen-independent CTCF binding at the 5′1 and 3′1 sites, their interaction was also not dependent on estrogen, in contrast to the estrogen-stimulated interaction between the TFF1 promoter and 3′1 site. Considering a weak ERα binding at 3′1 site (Fig. 1B), the TFF1 promoter-3′1 interaction might be attributed to estrogen-stimulated ERα binding, consistent with a previous investigation (10, 38). In addition, a weak and estrogen-independent interaction between the TFF2 promoter and the 3′1 boundary was also detected. However, limited by the short distance between the TFF2 promoter and the 5′1 site in a linear DNA context, it is not clear whether this interaction is a natural configuration or a side effect of the strong interaction between the 5′1 and 3′1 sites. On the other hand, we did not observe any interaction of the 3′1 site with other elements outside the 5′1–3′1 block including the TFF3 promoter, the 5′2 site, and the 3′2 site (data not shown). The distant spatial interaction between the 5′1 and 3′1 sites and the expression profile of the genes in their confined region (Fig. 1B) support the proposition that CTCF-mediated higher-order chromatin configuration predisposes the estrogen response program.
FIGURE 4.
CTCF-dependent and transcription-independent spatial interaction of the 5′1 and 3′1 boundary elements. A, estrogen-independent spatial interaction of the 5′1 and 3′1 boundaries. MCF-7 cells were deprived of estrogen for 3 days and treated with E2 for 15, 45, or 90 min. DNAs were extracted from the cells for 3C assays with 3′1 primer as the anchoring primer and the 5′1 and TFF1/2 promoter primers (pro) as the interacting primers. Fragments covering two adjacent DpnII sites were amplified as a loading control. Arrowheads indicate the expected PCR products. B, transcription-independent spatial interaction of the 5′1 and 3′1 boundaries. MCF-7 cells were treated with tamoxifen (TAM) for 12 h or α-amanitin for 2 h. Genomic DNAs were extracted from the cells for 3C analysis with the 3′1-5′1 PCR primers. C, CTCF-dependent spatial interaction of the 5′1 and 3′1 boundaries. MCF-7 cells were transfected with control siRNA (siNS), or specific siRNA molecules for ERα, FOXA1, or CTCF, deprived of estrogen for 3 days, and then subjected to the 3C assay with the 3′1-5′1 primers (upper panel). The expression of ERα, FOXA1, and CTCF in these cells was examined by Western blotting. Each bar represents the mean ± S.D. for triplicate experiments. D, cell type-specific spatial interaction of the 5′1 and 3′1 boundaries. The spatial interaction of the 5′1 and 3′1 boundaries was examined in MCF-7 and MDA-MB-231 cells by 3C assays. The DNA looping of the GAPDH locus was analyzed as a control. siNS, control siRNA; siCTCF, CTCF-specific siRNA.
The estrogen-independent CTCF binding and long range interaction of the 5′1 and 3′1 boundaries suggest that the higher-order chromatin structure is configured independent of active transcription. To investigate this hypothesis, we treated MCF-7 cells with either tamoxifen, an antagonist of estrogen that competes for ERα binding (28, 39), or α-amanitin, which generates a synchronized cell population in which the genome is devoid of transactivating factors (7–9). Then the long range interaction between the 5′1 and 3′1 boundaries was determined using 3C assays. As shown in Fig. 4B, the interaction of the 5′1-3′1 was not affected by either tamoxifen or α-amanitin. We then investigated the dependence of the 5′1-3′1 interaction on CTCF and other estrogen signaling components. In these experiments, 3C assays were performed in MCF-7 cells with knockdown of the expression of ERα, FOXA1, or CTCF. The results indicated that depletion of either ERα or FOXA1 had no effect or limited effect on the 5′1-3′1 interaction. However, in contrast, knockdown of CTCF impaired the 5′1-3′1 interaction (Fig. 4C). Collectively these results suggest that the 5′1-3′1 interaction and the higher order chromatin configuration occurs independent of an active transcription event.
Next, we investigated whether the higher-order chromatin structure also exhibited a cell-specific pattern. For this purpose, 3C assays were performed in estrogen-responsive MCF-7 cells and estrogen non-responsive MDA-MB-231 cells. But first the DNA looping of the GAPDH locus was evaluated to exclude a possible variation resulting from potentially differential enzymatic activity on different cellular DNAs (40). As evident in Fig. 4D, a comparable frequency of DNA looping of the GAPDH locus was observed in these two cell lines. However, association of the 5′1-3′1 elements was lost in estrogen non-responsive MDA-MB-231 cells (Fig. 4D). Taken together, these results suggest that the 3′1 boundary clusters with a neighboring 5′1 boundary, dependent on the insulator-binding protein CTCF but independent of active transcription and estrogen stimulation, forming a “pioneer” platform to facilitate the binding of ERα and the genomic response to estrogen.
Loss of CTCF Binding at the 3′1 Boundary Renders PRC2-mediated Spread of Heterochromatin in the TFF1 Locus
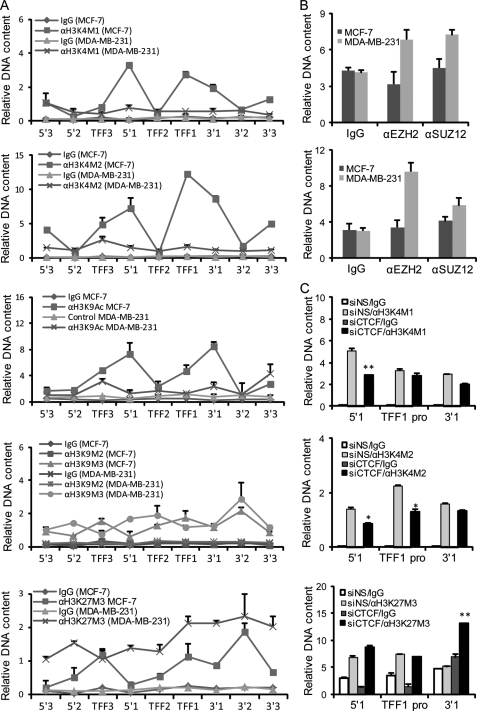
To further gain a mechanistic insight into the role of the boundary elements in the estrogen-triggered transcription program, we compared active and repressive histone markers across the TFF1 locus in estrogen-responsive MCF-7 cells and non-responsive MDA-MB-231 cells. H3K4M1/2 were surveyed as active markers, because they were found not only to be enriched in active enhancers genomewide (41), but also to display a cell type-specific occurrence pattern correlating with transcription activation by nuclear receptors (12). As shown in Fig. 5A, H3K4M1/2 was detected in both the 5′1 and 3′1 boundaries as well as in the TFF1 promoter in MCF-7 cells, whereas in MDA-MB-231 cells, none of the three sites exhibited detectable H3K4M1/2 (Fig. 5A, first and second panels). Corroborating this, the active marker H3K9 acetylation was detected in MCF-7 cells but not in MDA-MB-231 cells in the TFF1 promoter (Fig. 5A, third panel). Comparison of repressive markers H3K9M2/3 and H3K27M3 on the 5′1 and 3′1 boundaries and the TFF1 promoter in these two cell lines revealed a similar H3K9M2/3 modification pattern (Fig. 5A, fourth panel). However, in estrogen non-responsive MDA-MB-231 cells, the 3′1 boundary and TFF1 promoter showed a 2–3-fold elevation and the 5′1 boundary displayed a 5-fold enrichment of H3K27M3 modification (Fig. 5A, fifth panel). The trimethylation reaction of H3K27 is catalyzed by the polycomb repressive complex (PRC2) consisting of EZH2, SUZ12, and EED (42, 43). Examination of the binding profile of PRC2 components indicated that, compatible with the enrichment of H3K27M3 in the 5′1 and 3′1 boundaries in MDA-MB-231 cells, EZH2 and SUZ12 occupied these elements (Fig. 5B). Thus the change in epigenetic markers to repressive histone modifications within the 5′1-3′1-delimited region in MDA-MB-231 cells suggests that the enclosed region was invaded by facultative heterochromatin.
FIGURE 5.
Loss of CTCF binding at the 3′1 boundary is associated with a PRC2-mediated spread of heterochromatin in the TFF1 locus. A, comparison of the active and repressive histone makers across the TFF locus between estrogen responsive and non-responsive cells. Soluble chromatins were prepared from MCF-7 and MDA-MB-231 cells, and qChIP assays were performed with primers covering the indicated boundary elements and the gene promoters and with antibodies against the indicated histone modifications. Each point represents the mean ± S.D. for triplicate experiments. B, the binding of PRC2 proteins at the 5′1 and 3′1 boundaries. Soluble chromatins were prepared from MCF-7 or MDA-MB-231 cells and qChIP assays were performed with primers covering the 5′1 (upper) and 3′1 (lower) boundaries and with antibodies against the indicated proteins. Each bar represents the mean ± S.D. for triplicate experiments. C, the effect of CTCF on the establishment of the epigenetic marks in the TFF1 locus in MCF-7 cells. MCF-7 cells were transfected with control siRNA (siNS) or CTCF-specific siRNA (siCTCF). Soluble chromatins were prepared from the cells and qChIP assays were performed with antibodies against H3K4M1 (top), H3K4M2 (middle), and H3K27M3 (bottom) and with primers covering the 5′1 and 3′1 boundaries and the TFF1 promoter (TFF1 Pro). Each bar represents the mean ± S.D. for triplicate experiments. *, p < 0.05; **, p < 0.01 (compared with siNS transfected cells).
To further support the pioneering role of CTCF/boundary elements and the higher order chromatin structure in the genomic response to estrogen, we investigated the effect of CTCF on establishment of epigenetic marks in the TFF1 locus in MCF-7 cells. Knockdown of CTCF led to a differential alteration of the epigenetic marks at the regulatory elements of the TFF1 locus, with the 3′1 boundary, to which CTCF directly binds, exhibiting an increase in H3K27M3, and the 5′1 boundary, to which CTCF also directly binds, displaying a significant reduction in both H3K4M1 and H3K4M2. Of further importance, the interior TFF1 promoter, to which CTCF does not bind, also showed a significant decrease (t test, p < 0.05) in H3K4M2 (Fig. 5C). These results suggest a pattern of heterochromatin spreading in the interior of the 3′1-5′1-defined region upon the loss of CTCF and support a pioneering role of CTCF/boundary elements in establishing the genomic responsiveness to estrogen in this locus.
CTCF Acts Upstream of FOXA1
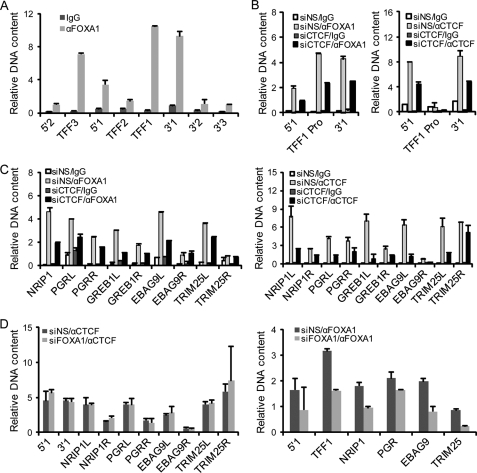
As stated before, FOXA1 has been defined as a pioneer factor for its ability to translate the H3K4M1/2 code and consequently instruct a poised nuclear receptor-directed transcriptional program (10, 12). In light of our observations of transcription-independent but CTCF-dependent genomic clustering between 5′1 and 3′1 and the histone modification changes upon CTCF depletion, it is possible that the CTCF/boundary elements may also contribute to pre-defining an estrogen-triggered transcription program via the higher order chromatin configuration favoring the maintenance of the H3K4M1/2 modifications. It was therefore of interest to investigate the functional hierarchy between CTCF and FOXA1. To this end, we first determined the FOXA1 binding pattern in the TFF1 locus. Using the genomewide FOXA1 binding profile in MCF-7 cells (12), we located its binding sites in the TFF1 promoter and also in the 5′1 and 3′1 boundaries, which also contain CTCF-binding sites. qChIP assays confirmed the binding of FOXA1 in the 5′1 and 3′1 boundaries and in the TFF1 promoter (Fig. 6A). The coincident existence of both FOXA1- and CTCF-binding sites at the 5′1 and 3′1 sites suggests that CTCF might cooperate with its nearby FOXA1 on the linear DNA template to establish a functional coordination. However, extensive analysis of genomewide ChIP-on-chip data (12, 33) indicated that the FOXA1- and CTCF-binding sites are located within 1 kb of each other in only a small portion of the genome (about 2%). This argues against the possibility that a physical interaction between FOXA1 and CTCF underlies their functional coordination. Nevertheless, the heterochromatin spreading exemplified at the interior TFF1 promoter supports a model in which CTCF might influence locus-wide FOXA1 binding by directing euchromatin/heterochromatin distribution.
FIGURE 6.
CTCF acts upstream of the pioneer factor FOXA1. A, FOXA1 binding pattern in the TFF locus. Soluble chromatins were prepared from MCF-7 cells, and qChIP assays were performed with primers covering the indicated boundary elements and gene promoters and antibodies against FOXA1. Each bar represents the mean ± S.D. for triplicate experiments. B, the binding of FOXA1 at the TFF1 locus is dependent on CTCF. MCF-7 cells were transfected with control siRNA (siNS) or CTCF-specific siRNA (siCTCF). Soluble chromatins were prepared from the cells and qChIP assays were performed with primers covering the 5′1 boundary, 3′1 boundary, and the TFF1 promoter (TFF1 Pro) and with antibodies against FOXA1 (left) or CTCF (right). Each bar represents the mean ± S.D. for triplicate experiments. C, the dependence of FOXA1 recruitment on CTCF in estrogen responsive gene loci. MCF-7 cells were transfected with control siRNA or CTCF-specific siRNA. Soluble chromatins were prepared from the cells, and qChIP assays were performed with primers covering the FOXA1-binding sites and CTCF-binding sites of the indicated genes and antibodies against FOXA1 (left) or CTCF (right). The L or R following gene symbols represents the primer set used to amplify FOXA1- or CTCF-binding sites located to the left or right (from the 5′ to 3′ direction) within the CTCF-confined regions in these genes. Each bar represents the mean ± S.D. for triplicate experiments. D, the binding of CTCF does not rely on FOXA1. MCF-7 cells were transfected with control siRNA (siNS) or FOXA1-specific siRNA (siFOXA1). Soluble chromatins were prepared from the cells, and qChIP assays were performed with antibodies against CTCF (left) or FOXA1 (right) and with primers described in C. Each bar represents the mean ± S.D. for triplicate experiments.
Next, we knocked down the expression of either CTCF or FOXA1 in MCF-7 cells and performed qChIP assays to investigate their potential interdependence in DNA binding. The results of these experiments revealed that although depletion of FOXA1 had no effect on CTCF binding at the 5′1 and 3′1 boundaries (Fig. 6D), depletion of CTCF resulted in impaired FOXA1 binding at both the 5′1 and 3′1 boundaries (Fig. 6B). In accordance with the spreading of heterochromatic markers and the reduced H3K4M2 in the TFF1 promoter, the binding of FOXA1 in this region was also significantly decreased with CTCF depletion (Fig. 6B). These results suggest a hierarchical recruitment and functional connection of CTCF and FOXA1 in the TFF1 locus in which the recruitment and the action of CTCF precedes that of FOXA1. To generalize this upstream functionality of higher order chromosomal structure established by CTCF, we determined the dependence of FOXA1 recruitment on CTCF in other well defined estrogen responsive gene loci. Under CTCF depletion, all the five selected genes, NRIP1, PGR, GREB1, EBAG9, and TRIM25, which scatter in different chromosomes and have at least one FOXA1-binding site, showed lost FOXA1 recruitment within the CTCF-confined region in MCF-7 cells (Fig. 6C), whereas depletion of FOXA1 did not compromise CTCF binding in these loci (Fig. 6D).
DISCUSSION
Despite the fact that ERα has been successfully used as a model system in delineating several aspects of transcriptional regulation, information on the role of CTCF and the involvement of higher order chromatin structures in ERα-mediated gene transcription is scarce. On the other hand, long-range interactions of genomic regulatory sites (10) and even interchromosomal interactions (38) have been reported for ERα-mediated gene transcription. We showed in the current study that CTCF binding boundaries at the TFF locus possess an intrinsic insulator function and demarcate estrogen responsiveness. We demonstrated that these boundaries cluster in the nuclear space and may represent a loop domain to facilitate ERα recognition of the target. In support of the notion that the higher order chromatin configuration wires a domain and represents another pioneer factor for ERα target recognition, we found that CTCF binding and the clustering of these elements are independent of estrogen stimulation and active transcription. Indeed, CTCF binding and clustering are lost in estrogen non-responsive cells, and, in addition, ablation of CTCF in estrogen responsive cells resulted in loss of the higher order chromatin configuration in this region and estrogen responsiveness in this locus.
Genomewide analyses revealed the presence of the FOXA1 protein in close proximity to ERα-binding sites, leading to the proposal that FOXA1 acts as a pioneer factor in ERα-mediated gene transcription (10–12). We showed in the current study that depletion of FOXA1 did not affect the configuration of the higher order chromatin structure in the TFF1 locus, whereas ablation of CTCF impeded FOXA1 binding in this region, placing CTCF and the insulators upstream of FOXA1 in defining the region for estrogen responsiveness. In addition, it has been reported that FOXA1 binds to chromatin with H3K4M1/2 and functions in translating this epigenetic language in ERα-mediated gene transcription (12). Considering the general belief that H3K4 methylation marks active gene transcription, it is logical to speculate that additional protein factor(s)/chromatin modifier(s) act between CTCF and FOXA1 in a hierarchical event to add the H3K4M1/2 marks in predisposing the region to ERα recognition. If that is the case, the loop domain resulting from the higher order chromatin configuration may facilitate the addition of the H3K4M1/2 marks prior to binding of FOXA1 and ERα. Meanwhile, it is plausible that this epigenetic mark, in addition to its role in signaling the recruitment of FOXA1 and ERα, in turn aids in the stabilization/organization of the loop domain. Future investigations are warranted to connect these “dots” in the event.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 30830032 and 30921062 (to Y. S.) and 973 Program Grants 2005CB522404 and 2007CB914503 (to Y. S.) from the Ministry of Science and Technology of China.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- ERα
- estrogen receptor α
- CTCF
- CCCTC-binding factor
- FOXA1
- Forkhead box A1
- PRC2
- polycomb repressive complex 2
- 3C
- chromosome conformation capture
- E2
- estradiol
- qChIP
- quantitative ChIP.
REFERENCES
- 1.Shang Y. (2006) Nat. Rev. Cancer 6, 360–368 [DOI] [PubMed] [Google Scholar]
- 2.Doisneau-Sixou S. F., Sergio C. M., Carroll J. S., Hui R., Musgrove E. A., Sutherland R. L. (2003) Endocr. Relat. Cancer 10, 179–186 [DOI] [PubMed] [Google Scholar]
- 3.Musgrove E. A., Sutherland R. L. (2009) Nat. Rev. Cancer 9, 631–643 [DOI] [PubMed] [Google Scholar]
- 4.McDonnell D. P., Norris J. D. (2002) Science 296, 1642–1644 [DOI] [PubMed] [Google Scholar]
- 5.Chambon P. (2004) Nat. Med. 10, 1027–1031 [DOI] [PubMed] [Google Scholar]
- 6.O'Malley B. W. (2005) Mol. Endocrinol. 19, 1402–1411 [DOI] [PubMed] [Google Scholar]
- 7.Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Cell 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 8.Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. (2000) Cell 103, 843–852 [DOI] [PubMed] [Google Scholar]
- 9.Reid G., Hübner M. R., Métivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F. (2003) Mol. Cell 11, 695–707 [DOI] [PubMed] [Google Scholar]
- 10.Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 11.Laganière J., Deblois G., Lefebvre C., Bataille A. R., Robert F., Giguère V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11651–11656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupien M., Eeckhoute J., Meyer C. A., Wang Q., Zhang Y., Li W., Carroll J. S., Liu X. S., Brown M. (2008) Cell 132, 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips J. E., Corces V. G. (2009) Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaszner M., Felsenfeld G. (2006) Nat. Rev. Genet. 7, 703–713 [DOI] [PubMed] [Google Scholar]
- 15.Wallace J. A., Felsenfeld G. (2007) Curr. Opin. Genet. Dev. 17, 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell A. C., West A. G., Felsenfeld G. (1999) Cell 98, 387–396 [DOI] [PubMed] [Google Scholar]
- 17.Ohlsson R., Renkawitz R., Lobanenkov V. (2001) Trends Genet. 17, 520–527 [DOI] [PubMed] [Google Scholar]
- 18.Filippova G. N. (2008) Curr. Top. Dev. Biol. 80, 337–360 [DOI] [PubMed] [Google Scholar]
- 19.Splinter E., Heath H., Kooren J., Palstra R. J., Klous P., Grosveld F., Galjart N., de Laat W. (2006) Genes Dev. 20, 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell A. C., Felsenfeld G. (2000) Nature 405, 482–485 [DOI] [PubMed] [Google Scholar]
- 21.Hark A. T., Schoenherr C. J., Katz D. J., Ingram R. S., Levorse J. M., Tilghman S. M. (2000) Nature 405, 486–489 [DOI] [PubMed] [Google Scholar]
- 22.Li R., Zhang H., Yu W., Chen Y., Gui B., Liang J., Wang Y., Sun L., Yang X., Zhang Y., Shi L., Li Y., Shang Y. (2009) EMBO J. 28, 2763–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L., Shi L., Li W., Yu W., Liang J., Zhang H., Yang X., Wang Y., Li R., Yao X., Yi X., Shang Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10195–10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., Li R., Li Y., Zhang Y., Li Q., Yi X., Shang Y. (2009) Cell 138, 660–672 [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., Sun L., Liang J., Yu W., Zhang Y., Wang Y., Chen Y., Li R., Sun X., Shang Y. (2006) EMBO J. 25, 4223–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Yi X., Sun X., Yin N., Shi B., Wu H., Wang D., Wu G., Shang Y. (2004) Genes Dev. 18, 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Zhang H., Liang J., Yu W., Shang Y. (2007) EMBO J. 26, 2645–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H., Chen Y., Liang J., Shi B., Wu G., Zhang Y., Wang D., Li R., Yi X., Zhang H., Sun L., Shang Y. (2005) Nature 438, 981–987 [DOI] [PubMed] [Google Scholar]
- 29.Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W. (2002) Mol. Cell 10, 1453–1465 [DOI] [PubMed] [Google Scholar]
- 30.Giamarchi C., Solanas M., Chailleux C., Augereau P., Vignon F., Rochefort H., Richard-Foy H. (1999) Oncogene 18, 533–541 [DOI] [PubMed] [Google Scholar]
- 31.Jeltsch J. M., Roberts M., Schatz C., Garnier J. M., Brown A. M., Chambon P. (1987) Nucleic Acids Res. 15, 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 33.Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I., Green R. D., Zhang M. Q., Lobanenkov V. V., Ren B. (2007) Cell 128, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao L., Zhou M., Cui Y. (2008) Nucleic Acids Res. 36, D83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C. F., Wolffe A., Ohlsson R., Lobanenkov V. V. (2000) Curr. Biol. 10, 853–856 [DOI] [PubMed] [Google Scholar]
- 36.Dekker J., Rippe K., Dekker M., Kleckner N. (2002) Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 37.Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forné T. (2007) Nat. Protoc. 2, 1722–1733 [DOI] [PubMed] [Google Scholar]
- 38.Hu Q., Kwon Y. S., Nunez E., Cardamone M. D., Hutt K. R., Ohgi K. A., Garcia-Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G., Fu X. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19199–19204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shang Y., Brown M. (2002) Science 295, 2465–2468 [DOI] [PubMed] [Google Scholar]
- 40.Tan-Wong S. M., French J. D., Proudfoot N. J., Brown M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5160–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 42.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 43.Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O'Connor M. B., Kingston R. E., Simon J. A. (2002) Cell 111, 197–208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.