Abstract
GIPC1/synectin, a single PDZ domain-containing protein, binds to numerous proteins and is involved in multiple biological processes, including cell migration. We reported previously that MyoGEF, a guanine nucleotide exchange factor, plays a role in regulating breast cancer cell polarization and invasion. Here, we identify GIPC1 as an interacting partner of MyoGEF. Both in vitro and in vivo binding assays show that the GIPC1 PDZ domain binds to the PDZ-binding motif at the C terminus of MyoGEF. Immunofluorescence analysis shows that GIPC1 and MyoGEF colocalize to the cell leading edge. Depletion of GIPC1 by RNAi in MDA-MB-231 cells causes cells to shift from a polarized to a rounded morphology. Matrigel invasion assays show that RNAi-mediated depletion of GIPC1 dramatically decreases MDA-MB-231 cell invasion. Notably, an anti-MyoGEF peptide antibody, whose epitope is located at the C terminus of MyoGEF, interferes with GIPC1-MyoGEF complex formation. Treatment of MDA-MB-231 cells with the anti-MyoGEF peptide antibody disrupts cell polarization and invasion. Thus, our results suggest that GIPC1-MyoGEF complex formation plays an important role in regulating MDA-MB-231 breast cancer cell polarization and invasion.
Keywords: Cdc42, Cell Migration, Cell Motility, Protein-Protein Interactions, Rho, GIPC1, Guanine Nucleotide Exchange Factor, MyoGEF
Introduction
Rho GTPase signaling plays a central role in regulating cell migration (1). Evidence has accumulated indicating that small GTPase proteins, including Rac1, Cdc42, and RhoA, can be activated at the cell leading edge, where they regulate actin polymerization and membrane protrusion, thus contributing to the regulation of cell migration (2–5). Guanine nucleotide exchange factors (GEFs)2 activate the small GTPase proteins by catalyzing the exchange of bound GDP for GTP, whereas GTPase-activating proteins inactivate the small GTPase proteins by increasing their low intrinsic GTPase activity (6, 7). Therefore, localization of GEFs to specific subcellular locations is critically important for spatiotemporal activation of small GTPase proteins (3, 8). At least two different mechanisms have been implicated in regulating the localization of GEFs. First, protein-protein interactions involving the pleckstrin homology domain of GEFs can target GEFs such as Dbl and Trio to their destinations such as the actin cytoskeleton (9–11). Second, the PDZ-binding motif is found in ∼40% of GEFs, and binding of PDZ domain-containing proteins to the PDZ-binding motif of GEFs such as Syx1, kalirin-7, and bPIX can target GEFs to specific locations such as the plasma membrane (12–14).
GIPC1/synectin, a single PDZ domain-containing protein, acts as a scaffolding protein to function in multiple biological processes such as protein trafficking, endocytosis, and receptor clustering (15, 16). Accumulating evidence further indicates that GIPC1 plays a role in regulating cell polarity and motility. Murine primary arterial endothelial cells derived from GIPC1-ablated mice show decreased migration and impaired polarization (17). Syndecan-4, Syx1, and endoglin regulate cell migration through interactions with GIPC1/synectin (18–20). GIPC1 interacts with 5T4, a transmembrane glycoprotein that is involved in tumor metastasis (21–23). Moreover, a recent study shows that cancerous breast tissues express an increased level of GIPC1 compared with normal breast tissues (24). However, it is not clear whether GIPC1 plays a role in regulating breast cancer cell migration and/or invasion.
We reported previously that MyoGEF, a guanine nucleotide exchange factor, can activate RhoA/RhoC and is implicated in regulating breast cancer cell invasion (25). Using yeast two-hybrid screening, we identified GIPC1 as one of the MyoGEF-interacting proteins. In this study, we demonstrate that the GIPC1 PDZ domain can bind to the PDZ-binding motif at the C terminus of MyoGEF. Depletion of GIPC1 by RNAi inhibits the invasion activity of MDA-MB-231 breast cancer cells. Moreover, we show that an anti-MyoGEF peptide antibody can bind to the C terminus of MyoGEF and interfere with the in vitro interaction between GIPC1 and MyoGEF. Treatment of MDA-MB-231 cells with the anti-MyoGEF peptide antibody disrupts cell polarization and invasion. Thus, our results indicate that complex formation between GIPC1 and MyoGEF plays a role in regulating MDA-MB-231 cell polarization and invasion.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screening
Yeast two-hybrid screening was carried out as described previously (26). Briefly, full-length human MyoGEF was used as bait to screen a mouse 11-day embryo Matchmaker cDNA library (Clontech). Synthetic defined medium lacking leucine, tryptophan, and histidine was used to identify the positive yeast colonies. The filter lift assay for β-galactosidase activity was then carried out to confirm the positive colonies. The cDNA fragments encoding the potential MyoGEF-interacting partners were recovered from the positive yeast colonies and subjected to DNA sequencing. The mouse gipc1 cDNA was amplified using the following primer pair: 5′-GAATTCAATGCCACTGGGACTGGGG-3′ (forward primer; the underlined nucleotide sequence is the recognition site for EcoRI) and 5′-CTCGAGGTAGCGGCCAACCTTGGC-3′ (reverse primer; the underlined nucleotide sequence is the recognition site for XhoI).
Plasmids and Cell Culture
pEGFP-MyoGEF and pCS3-MyoGEF were described previously (27). GIPC1 and MyoGEF cDNA fragments were subcloned into pEGFP-C3 and pCS3+MT vectors to generate plasmids encoding GFP- or Myc-tagged polypeptides. All plasmids encoding GST-tagged MyoGEF or GIPC1 fragments were generated by subcloning the cDNA fragments into the pGEX-6p-1 vector. MDA-MB-231 breast cancer cells were purchased from American Type Culture Collection (Manassas, VA). MDA-MB-231 cells were grown in Leibovitz's L-15 medium supplemented with 10% fetal bovine serum. HeLa cells were purchased from Clontech and were grown in DMEM supplemented with 10% fetal bovine serum. Transfection was done with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNAs specific for human GIPC1 were purchased from Invitrogen (siRNA1, GCU ACG CCU UCA UCA AGC GCA UCA A; siRNA2, CCA ACG UCA AGG AGC UGU AUG GCA A; and siRNA3, UGU GGA GCC UGU UAC CUC CGC AUU U).
Protein Expression and in Vitro Translation
GST-fused polypeptides were expressed in a bacterial expression system. BL21 bacterial cells expressing GST-fused polypeptides were homogenized by sonication and lysed in PBS containing 1% Triton X-100 for 1 h at 4 °C. The GST fusion proteins were purified using glutathione-conjugated agarose beads, eluted with 100 mm Tris-HCl (pH 7.5) and 5 mm glutathione, and dialyzed against 50 mm Tris-HCl (pH 7.5) and 50 mm NaCl. In vitro translated Myc-tagged proteins were synthesized using the TnT SP6 quick coupled transcription/translation system (Promega) according to the manufacturer's instructions.
Immunoprecipitation and GST Pulldown Assays
Immunoprecipitation and GST pulldown assays were carried out as described previously (27, 28). Briefly, transfected cells were lysed in radioimmune precipitation assay lysis buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.25% deoxycholate, 1% Nonidet P-40, 1 mm EDTA, 1 mm PMSF, 1 mm Na3VO4, and 1 mm NaF with protease inhibitor mixture) for 10 min on ice. Cell extracts were collected and precleared with protein A/G-agarose beads. The precleared lysate was incubated with agarose-conjugated anti-Myc antibody overnight at 4 °C. After washing four times with radioimmune precipitation assay lysis buffer, the bound proteins were eluted with SDS loading buffer. For GST pulldown experiments, the immobilized GST-fused polypeptides were incubated with in vitro translated Myc-tagged proteins or with cell lysates from transfected cells overnight at 4 °C. After washing four times with binding buffer (50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 0.05% Triton X-100, 10% glycerol, 0.2 mm EDTA, and 1 mm DTT), the beads were resuspended in SDS loading buffer to elute the bound proteins.
Immunoblotting
Cell lysates and immunoprecipitated and GST pulldown proteins were separated on 7 or 4–12% SDS-polyacrylamide gel, transferred to an Immobilon-P transfer membrane (Millipore), blocked in 5% nonfat milk, and incubated with primary antibodies as indicated. The following primary antibodies were used: mouse anti-Myc (9E10, 1:1000), rabbit anti-GFP (1:1000), and rabbit anti-β-tubulin (1:2000) (Santa Cruz Biotechnology); goat anti-GIPC1 (1:250; Novus Biologicals, Littleton, CO); and rabbit anti-MyoGEF (1:100) (25, 27). The blots were washed and incubated with horseradish peroxide-conjugated secondary antibodies (1:5000; Santa Cruz Biotechnology) for 1 h at 23 °C. The blots were visualized by SuperSignal West Pico luminol/enhancer solution (Pierce).
Immunofluorescence
Immunofluorescence was carried out as described previously (25, 26). MDA-MB-231 cells transfected with plasmids or siRNA were trypsinized; cultured on fibronectin-coated coverslips for an additional 1, 3, or 5 h; and then fixed with 4% paraformaldehyde. For RhoA staining, cells were fixed with 10% TCA for 10 min on ice. The primary antibodies used for immunofluorescence were mouse monoclonal anti-Myc (9E10, 1:1000), rabbit polyclonal anti-MyoGEF (1:50), and goat polyclonal anti-GIPC1 (1:100; Abcam). The following secondary antibodies were purchased from Invitrogen: Alexa Fluor 594-labeled donkey anti-mouse IgG (1:500), Alexa Fluor 350-labeled donkey anti-mouse IgG (1:500), Alexa Fluor 594-labeled donkey anti-goat IgG (1:500), Alexa Fluor 488-labeled donkey anti-goat IgG (1:500), Alexa Fluor 594-labeled donkey anti-rabbit IgG (1:500), and Alexa Fluor 488-labeled donkey anti-rabbit IgG (1:500). Actin filaments were stained with rhodamine- or FITC-phalloidin (Invitrogen). Images were taken using a Leica DMI 6000 B microscope and processed by blind deconvolution. To determine the cell polarity, long (L) and short (S) axes of individual cells were measured using the NIH ImageJ program. Cells were counted as polarized (L/S ratio > 2.0) or nonpolarized (L/S ratio < 2.0).
Matrigel Invasion Assays
Transfected or antibody-treated MDA-MB-231 cells were trypsinized, and ∼1 × 105 cells (in Leibovitz's L-15 medium containing 3% BSA) were seeded on the upper wells of BioCoat Matrigel chambers (BD Biosciences). The lower wells were filled with Leibovitz's L-15 medium containing 10% FBS. The transfected cells then underwent chemoattraction across the Matrigel and filter (pore size of 8 μm) to the lower surface of the transwells for 22 h. The nonmigrating cells on the upper chambers were removed with a cotton swab. The migrated cells on the lower surface of the membrane were fixed in 4% paraformaldehyde, stained with 1% crystal violet, and then photographed at five different and random fields with a 20× objective. Data were collected from three independent experiments, each done in triplicate. Migrated cells were counted, and mean differences (±S.E.) between groups were analyzed using Student's t test.
In Vitro Antibody Delivery
MDA-MB-231 cells were treated with normal IgG or anti-MyoGEF antibody using in vitro PULSin protein/antibody and peptide delivery reagent (Genesee Scientific, San Diego, CA) according to the manufacturer's instructions. Cells were grown in a 24-well tissue culture plate. 1 μg of normal rabbit IgG (Sigma) or anti-MyoGEF antibody was used for each well of a 24-well tissue culture plate. 4 h after antibody treatment, the treated cells were subjected to Matrigel invasion assays as described above or processed for immunofluorescence staining with phalloidin and Alexa Fluor 594-labeled goat anti-rabbit IgG (Invitrogen).
RhoA/RhoC Activation Assays
RhoA/RhoC activation assays were preformed as described previously (25, 29).
RESULTS
GIPC1 PDZ Domain Is Required for Interactions with MyoGEF
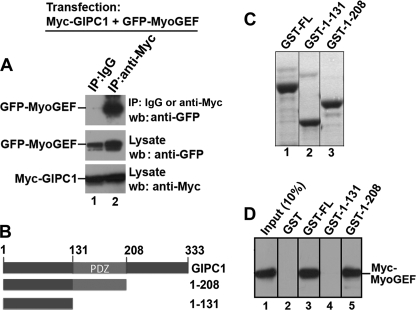
We reported previously that MyoGEF is involved in the regulation of cytokinesis and breast cancer cell invasion (25–27). Yeast two-hybrid screening led us to identify GIPC1 as an interacting partner of MyoGEF. GIPC1 plays a role in regulating cell migration (17–20). Furthermore, GIPC1 levels increase in breast cancerous cells and tissues (24). These findings prompted us to further characterize the interactions between MyoGEF and GIPC1 as well as to examine whether the GIPC1-MyoGEF interaction has a role in regulating breast cancer cell invasion. To confirm the interaction between MyoGEF and GIPC1 in mammalian cells, HeLa cells were transfected with plasmids encoding Myc-GIPC1 and GFP-MyoGEF. The transfected cells were subjected to immunoprecipitation with anti-Myc antibody, followed by immunoblotting with anti-GFP antibody. As shown in Fig. 1A, Myc-GIPC1 was co-immunoprecipitated with GFP-MyoGEF from transfected HeLa cell lysates, suggesting that GIPC1 and MyoGEF can interact in vivo. GST pulldown assays showed that GST-GIPC1 could also pull down in vitro translated Myc-MyoGEF (Fig. 1D, lane 3), suggesting that GIPC1 can physically interact with MyoGEF. In addition, GST-tagged GIPC1 fragment 1–208 (containing the PDZ domain), but not 1–131 (without the PDZ domain), could pull down in vitro translated Myc-MyoGEF (Fig. 1D, compare lanes 4 and 5), suggesting that the PDZ domain of GIPC1 is required for interactions with MyoGEF.
FIGURE 1.
GIPC1 PDZ domain is required for interaction with MyoGEF. A, in vivo interactions between Myc-GIPC1 and GFP-MyoGEF. IP, immunoprecipitation; wb, Western blotting. B, schematic diagram of GIPC1 and its truncated fragments. The numbers represent the positions of the amino acid residues. C, Coomassie Blue-stained gel of GST-tagged full-length GIPC1 (GST-FL) and truncated fragments (GST-1–131 and GST-1–208). D, in vitro interactions between Myc-MyoGEF and GST-tagged full-length GIPC1 and truncated fragments.
GIPC1 Binds to the C-terminal PDZ-binding motif of MyoGEF
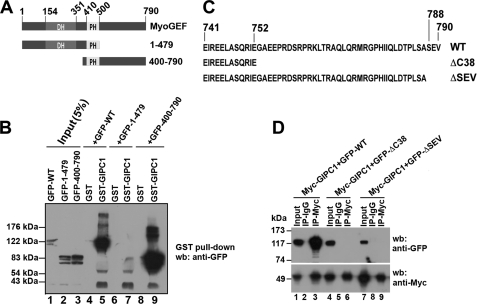
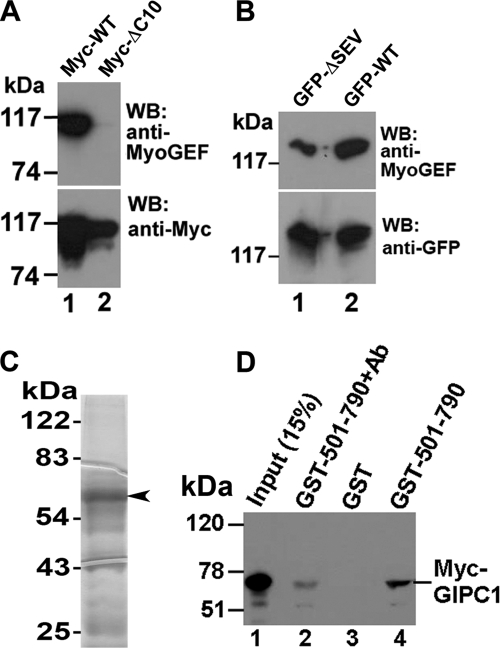
We then asked which regions of MyoGEF are required for interactions with GIPC1. Plasmids encoding GFP-tagged full-length MyoGEF (GFP-WT MyoGEF) and truncated fragments (N-terminal half (GFP-MyoGEF-(1–479)), and C-terminal half (GFP-MyoGEF-(400–790)) were transfected into HeLa cells. 24 h following transfection, the transfected cell lysates were subjected to GST pulldown assays to determine which fragments could bind to GST-GIPC1. As shown in Fig. 2B, GFP-WT MyoGEF and GFP-MyoGEF-(400–790), but not GFP-MyoGEF-(1–479), could be pulled down by GST-GIPC1 (compare lanes 5 and 9 with lane 7), suggesting that the C-terminal region of MyoGEF is required for interactions with GIPC1. The consensus sequence of type I PDZ-binding motifs is (S/T)X(V/A) (30). Analysis of the MyoGEF amino acid sequence revealed that three amino acid residues (SEV) at the C terminus of MyoGEF appear to be a type I PDZ-binding motif. Therefore, we generated several truncated or mutated versions of MyoGEF (Fig. 2C): ΔC38 (lacking the C-terminal 38 amino acid residues) and ΔSEV (lacking the putative type I PDZ-binding motif). Plasmids encoding GFP-MyoGEFΔC38, GFP-MyoGEFΔSEV, or GFP-WT MyoGEF were cotransfected with a plasmid encoding Myc-GIPC1 into HeLa cells. The transfected cell lysates were then subjected to immunoprecipitation with anti-Myc antibody, followed by immunoblotting with anti-GFP antibody. As shown in Fig. 2D, Myc-GIPC1 could be co-immunoprecipitated with GFP-WT MyoGEF, but not with GFP-MyoGEFΔC38 and GFP-MyoGEFΔSEV (compare lane 3 with lanes 6 and 9), suggesting that three amino acid residues (SEV) at the C terminus of MyoGEF are critical for interactions with GIPC1.
FIGURE 2.
GIPC1 binds to the C-terminal PDZ-binding motif of MyoGEF. A, schematic diagram of full-length MyoGEF and truncated fragments. The numbers represent the positions of amino acid residues. DH, Dbl homology domain; PH, pleckstrin homology domain. B, in vitro interactions between GST-GIPC1 and GFP-WT MyoGEF (GFP-WT), GFP-MyoGEF-(1–479) (GFP-1–479), or GFP-MyoGEF-(400–790) (GFP-400–790). C, schematic diagram of the C terminus of MyoGEF mutants. The numbers represent the positions of amino acid residues. D, in vivo interactions between Myc-GIPC1 and GFP-tagged full-length MyoGEF and fragments. The membrane was stripped and reblotted with anti-Myc antibody. GFP-ΔC38, GFP-MyoGEFΔC38; GFP-ΔSEV, GFP-MyoGEFΔSEV; IP, immunoprecipitation; wb, Western blotting.
Colocalization of MyoGEF and GIPC1 at the Cell Periphery and Cell Leading Edge
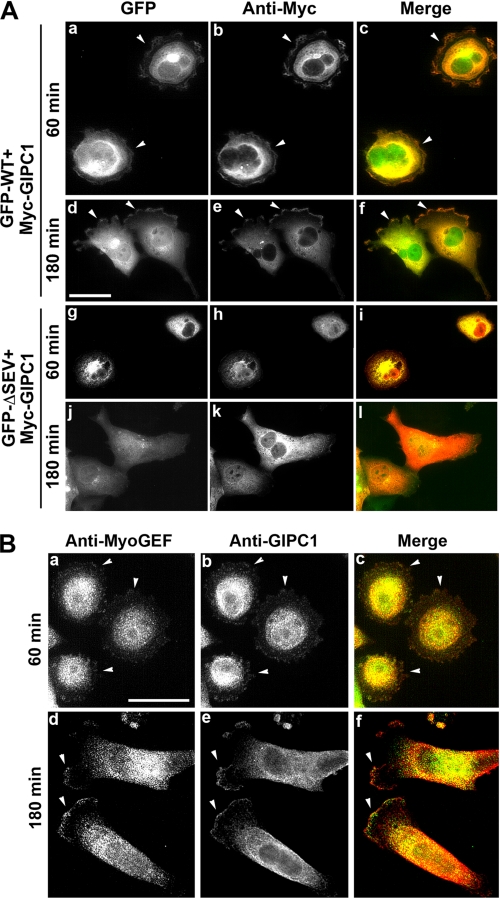
To further confirm the interaction between MyoGEF and GIPC1, we transfected plasmids encoding GFP-MyoGEF and Myc-GIPC1 into MDA-MB-231 cells to examine whether both proteins colocalize in transfected cells. 24 h after transfection, the transfected cells were trypsinized and replated on fibronectin-coated coverslips. After incubation for an additional 60 or 180 min, the transfected cells were fixed and processed for immunofluorescence analysis. Before cells became polarized (after a 60-min incubation), GFP-MyoGEF and Myc-GIPC1 colocalized to the cell periphery (Fig. 3A, arrowheads in panels a–c). After cells became polarized (after a 180-min incubation), GFP-MyoGEF and Myc-GIPC1 were concentrated to the cell leading edge (arrowheads in panels d–f). Because three amino acid residues (SEV) at the C terminus of MyoGEF were required for interactions with GIPC1 (Fig. 2D), we asked whether a MyoGEF mutant lacking these three C-terminal amino acids (ΔSEV) could still colocalize with GIPC1 in transfected cells. MDA-MB-231 cells were transfected with plasmids encoding Myc-GIPC1 and GFP-MyoGEFΔSEV. GFP-MyoGEFΔSEV showed diffuse distributions in the cytoplasm and did not colocalize with Myc-GIPC1 to the cell periphery or to the cell leading edge (Fig. 3A, panels g–l). These results further confirm that GIPC1 interacts with MyoGEF through binding to the PDZ-binding motif at the C terminus of MyoGEF.
FIGURE 3.
Colocalization of MyoGEF and GIPC1 in MDA-MB-231 cells. A, colocalization of Myc-GIPC1 and GFP-WT MyoGEF (GFP-WT) or GFP-MyoGEFΔSEV (GFP-ΔSEV). Scale bar = 25 μm. B, colocalization of endogenous MyoGEF and GIPC1. Scale bar = 20 μm.
To determine whether endogenous MyoGEF colocalizes with endogenous GIPC1, untransfected MDA-MB-231 cells were grown on fibronectin-coated coverslips for 60 or 180 min and then subjected to immunofluorescence staining for MyoGEF and GIPC1. As shown in Fig. 3B, MyoGEF and GIPC1 were colocalized to the cell periphery (arrowheads in panels a–c) or to the cell leading edge (arrowheads in panels d–f).
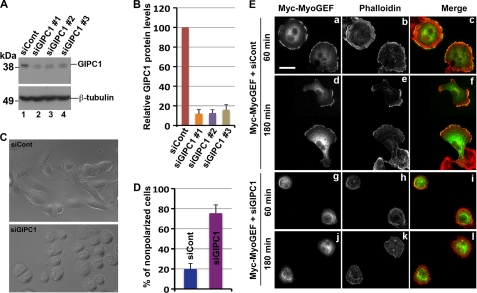
Depletion of GIPC1 Disrupts Cell Polarity and Localization of MyoGEF
We reported previously that depletion of MyoGEF by RNAi disrupts MDA-MB-231 cell polarity (25). The interaction between GIPC1 and MyoGEF led us to ask whether depletion of GIPC1 also affects MDA-MB-231 cell polarization. MDA-MB-231 cells were transfected with control or GIPC1 siRNAs. 48 h after transfection, MDA-MB-231 cells treated with GIPC1 siRNA exhibited a rounded morphology (Fig. 4C). We then examined whether depletion of GIPC1 has an impact on the localization of MyoGEF. A plasmid encoding Myc- MyoGEF was cotransfected with control or GIPC1 siRNAs into MDA-MB-231 cells. 48 h after transfection, the transfected cells were trypsinized, replated on fibronectin-coated coverslips, and incubated for an additional 60 min or 180 min. The transfected cells were fixed and stained with anti-Myc antibody and phalloidin. In MDA-MB-231 cells transfected with control siRNA and the Myc-MyoGEF-expressing plasmid, MyoGEF localized to the cell periphery before cells became polarized (Fig. 4E, panels a–c) or to the cell leading edge after cells became polarized (panels d–f). Cells transfected with GIPC1 siRNA and the Myc-MyoGEF-expressing plasmid did not become polarized (Fig. 4E, compare panels d–f with panels j–l). In addition, MyoGEF did not localize to the cell periphery (Fig. 4E, panels g–l). Our results suggest that GIPC1 is required for the localization of MyoGEF during cell spreading and cell polarization.
FIGURE 4.
Depletion of GIPC1 disrupts cell polarity and the localization of MyoGEF. A, immunoblot analysis of MDA-MB-231 cells transfected with siRNA against GIPC1 (siGIPC1). B, the image in A was quantitated using the NIH ImageJ program. C, phase-contrast images of MDA-MB-231 cells transfected with control (siCont) or GIPC1 siRNAs. D, quantitation of nonpolarized MDA-MB-231 cells treated with control or GIPC1 siRNAs. E, localization of Myc-MyoGEF in MDA-MB-231 cells transfected with control or GIPC1 siRNAs. Scale bar = 20 μm.
Binding of an Anti-MyoGEF Peptide Antibody to the C Terminus of MyoGEF Interferes with the GIPC1-MyoGEF Interaction
Because GIPC1 could bind to the C-terminal PDZ-binding motif of MyoGEF (Fig. 2D), we reasoned that a peptide antibody against the C terminus of MyoGEF might bind to the C terminus of MyoGEF, thus interfering with the binding of GIPC1 to the PDZ-binding motif at the C terminus of MyoGEF. As described previously (27), a peptide corresponding to the C-terminal 18 amino acid residues (773MRGPHIIQLDTPLSASEV790) was used to raise an anti-MyoGEF antibody. This peptide contains the PDZ-binding motif (SEV). To identify the epitope of the anti-MyoGEF peptide antibody, HeLa cells exogenously expressing different truncated versions of GFP- or Myc-tagged MyoGEF were subjected to immunoblot analysis with the anti-MyoGEF peptide antibody as well as with the anti-GFP or anti-Myc antibody. As expected, the anti-MyoGEF peptide antibody recognized the C-terminal half of MyoGEF (amino acids 501–790), but not the N-terminal half (amino acids 1–500) or a MyoGEF mutant lacking the C-terminal 38 amino acids (data not shown). Furthermore, the anti-MyoGEF peptide antibody did not recognize a MyoGEF mutant lacking the C-terminal 10 amino acids (Fig. 5A). Deletion of the PDZ-binding motif (SEV) from the C terminus of MyoGEF also decreased the ability to bind the anti-MyoGEF peptide antibody (Fig. 5B), suggesting that the PDZ-binding motif (SEV) contributes directly to the epitope.
FIGURE 5.
Binding of the anti-MyoGEF peptide antibody to the C terminus of MyoGEF interferes with the GIPC1-MyoGEF interaction. A, immunoblot analysis of MDA-MB-231 cells transfected with plasmids encoding Myc-tagged wild-type MyoGEF (Myc-WT) or mutant MyoGEF lacking the C-terminal 10 amino acid residues (Myc-ΔC10). WB, Western blot. B, immunoblot analysis of MDA-MB-231 cells transfected with plasmids encoding GFP-WT MyoGEF (GFP-WT) or GFP-MyoGEFΔSEV (GFP-ΔSEV). C, 10 μg of purified GST-MyoGEF-(501–790) was subjected to SDS-PAGE, followed by Coomassie Blue staining. D, in vitro interactions between GST-MyoGEF-(501–790) (GST-501–790) and Myc-GIPC1 in the presence or absence of the anti-MyoGEF peptide antibody (Ab).
We then asked whether binding of the anti-MyoGEF peptide antibody to the C terminus of MyoGEF interferes with the interaction between GIPC1 and MyoGEF. GST-tagged MyoGEF fragment 501–790 (GST-MyoGEF-(501–790)) (Fig. 5C) was incubated with the anti-MyoGEF peptide antibody. Antibody-treated GST-MyoGEF-(501–790) was then used in GST pulldown assays to examine its interaction with in vitro translated Myc-GIPC1. As shown in Fig. 5D, pretreatment with the anti-MyoGEF peptide antibody decreased the binding of GST-MyoGEF-(501–790) to Myc-GIPC1 (compare lanes 2 and 4). These results suggest that the anti-MyoGEF peptide antibody can compete with GIPC1 for binding to the C terminus of MyoGEF.
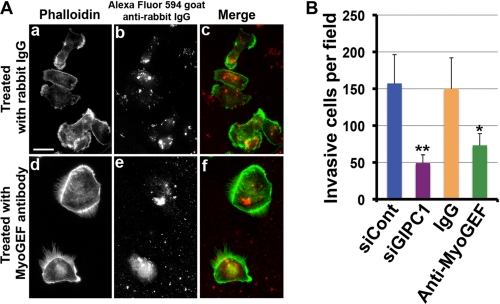
Treatment of MDA-MB-231 Cells with the Anti-MyoGEF Peptide Antibody Interferes with Cell Polarization and Invasion
We reported previously that depletion of MyoGEF decreases the invasion activity of MDA-MB-231 cells (25). In vitro binding assays showed that the anti-MyoGEF peptide antibody could compete with GIPC1 for binding to the C terminus of MyoGEF (Fig. 5D). Thus, we reasoned that treatment of MDA-MB-231 cells with the anti-MyoGEF peptide antibody might interfere with the GIPC1-MyoGEF interaction and disrupt cell polarization and invasion. To test this possibility, we examined the effect of antibody treatment on MDA-MB-231 cell polarization and invasion. 4 h after treatment with normal rabbit IgG or anti-MyoGEF antibody, the treated cells were subjected to immunofluorescence staining with phalloidin and Alexa Fluor 594-labeled goat anti-rabbit IgG. MDA-MB-231 cells treated with the anti-MyoGEF peptide antibody did not polarize (Fig. 6A, compare panels a–c and d–f). In addition, MDA-MB-231 cells treated with normal rabbit IgG or anti-MyoGEF antibody were subjected to Matrigel invasion assays. Fig. 6B shows that treatment with anti-MyoGEF antibody decreased the invasion activity of MDA-MB-231 cells. Consistent with these findings, depletion of GIPC1 by RNAi in MDA-MB-231 cells also decreased cell invasion (Fig. 6B). These results indicate that GIPC1-MyoGEF complex formation may play a role in regulating the polarization and invasion activity of MDA-MB-231 cells.
FIGURE 6.
Treatment with the anti-MyoGEF peptide antibody interferes with MDA-MB-231 cell polarization and invasion. A, shown is the immunofluorescence staining of MDA-MB-231 cells treated with normal IgG or anti-MyoGEF antibody. Scale bar = 20 μm. B, MDA-MB-231 cells treated with anti-MyoGEF antibody or GIPC1 siRNA (siGIPC1) were subjected to Matrigel invasion assays. The results were quantitated as described under “Experimental Procedures.” *, p < 0.05; **, p < 0.01.
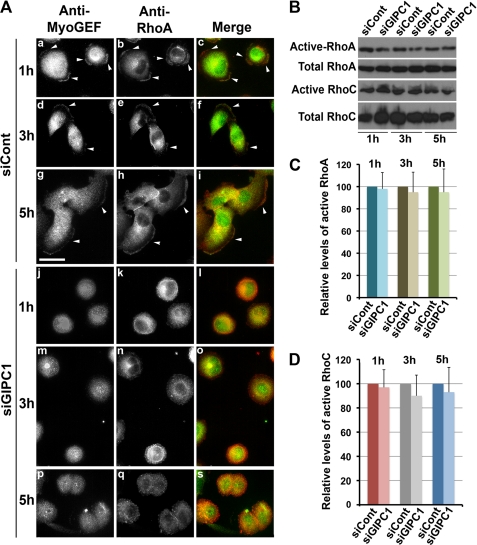
Depletion of GIPC1 Interferes with RhoA Localization but Not with Activation of RhoA and RhoC
Disruption of GIPC1 function interfered with the localization of MyoGEF (Fig. 4). We have also shown previously that depletion of MyoGEF interferes with the activation of RhoA/RhoC and decreases the invasion activity of MDA-MB-231 cells (25). Thus, we asked whether depletion of GIPC1 has an impact on the activation and/or localization of RhoA/RhoC. Immunofluorescence staining of TCA-fixed cells with anti-RhoA antibody has been used to monitor the localization of active RhoA (31, 32). To examine the effect of GIPC1 depletion on the localization of active RhoA, MDA-MB-231 cells were transfected with control or GIPC1 siRNAs. 48 h after transfection, the transfected cells were trypsinized and replated on fibronectin-coated coverslips. After incubation for an additional 1, 3, or 5 h, the transfected cells were fixed with TCA and then subjected to immunofluorescence staining with antibodies specific for MyoGEF and RhoA. In cells transfected with control siRNA, both MyoGEF and RhoA localized to the cell periphery (Fig. 7A, arrowheads in panels a–c) or to the cell leading edge (arrowheads in panels d–i). Conversely, cells transfected with GIPC1 siRNA did not polarize, and both MyoGEF and RhoC did not localize to the cell periphery (Fig. 7A, panels j–s). These findings suggest that depletion of GIPC1 interferes with the localization of MyoGEF and active RhoA. We then asked whether depletion of GIPC1 has an impact on the activation of RhoA/RhoC. MDA-MB-231 cells transfected with control or GIPC1 siRNAs for 72 h were subjected to rhotekin pulldown assays for RhoA activation (25, 27). As shown in Fig. 7 (B–D), depletion of GIPC1 did not interfere with the activation of RhoA and RhoC.
FIGURE 7.
Effect of GIPC1 depletion on the activation and localization of RhoA/RhoC. A, shown is the immunofluorescence staining of MyoGEF and RhoA in MDA-MB-231 cells transfected with control (siCont) or GIPC1 (siGIPC1) siRNAs. B, depletion of GIPC1 did not decrease the amount of active RhoA and RhoC in MDA-MB-231 cells. C and D, the image in B was quantitated using the NIH ImageJ program.
Expression of GIPC1 in Breast Cancer Cell Lines
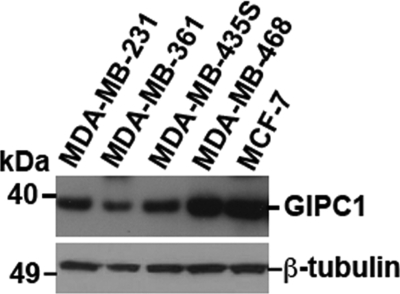
We showed previously that MyoGEF is expressed in invasive breast cancer cells (MDA-MB-231 and MDA-MB-435S) but is not detectable in noninvasive (MDA-MB-361 and MCF-7) or poorly invasive (MDA-MB-468) breast cancer cells (25). As shown in Fig. 8, however, GIPC1 is expressed in invasive, noninvasive, and poorly invasive breast cancer cells, suggesting that the interplay between GIPC1 and other specific factors such as MyoGEF may be critical for breast cancer cell invasion.
FIGURE 8.
Expression of GIPC1 in breast cancer cell lines. Breast cancer cell lysates were subjected to immunoblot analysis with antibodies specific for GIPC1 (upper panel) or β-tubulin (lower panel).
DISCUSSION
In this study, we have demonstrated that the GIPC1 PDZ domain can bind to the PDZ-binding motif at the C terminus of MyoGEF. Depletion of GIPC1 in MDA-MB-231 breast cancer cells by RNAi disrupts cell polarization and decreases cell invasion. Treatment of MDA-MB-231 breast cancer cells with an anti-MyoGEF peptide antibody that can interfere with the in vitro interaction between GIPC1 and MyoGEF also leads to impaired cell polarity and decreased cell invasion. Our results suggest that GIPC1-MyoGEF complex formation plays an important role in regulating the polarization and invasion activity of MDA-MB-231 breast cancer cells.
We have shown previously that MyoGEF can activate RhoA and RhoC in MDA-MB-231 cells (25). However, we found that depletion of GIPC1 by RNAi did not affect RhoA and RhoC activation in MDA-MB-231 cells (Fig. 7). Thus, binding of GIPC1 to MyoGEF may not have a role in controlling MyoGEF activity toward RhoA and RhoC. Instead, our results suggest that the GIPC1-MyoGEF interaction may be important for the recruitment of MyoGEF to the cell leading edge (Figs. 3 and 4). Consistently, depletion of GIPC1 interferes with the localization of active RhoA (Fig. 7). It has also been shown that GIPC1/synectin can bind to a Rho GEF called Syx1 and that the GIPC1-Syx1 interaction plays a role in regulating endothelial cell migration and tube formation (19). In addition, binding of GIPC1 to Syx1 is responsible for targeting Syx1 to the cell membrane (19). Therefore, GIPC1 likely acts as a scaffolding protein to recruit Rho GEFs such as MyoGEF and Syx1 to specific subcellular locations, thus leading to localized activation of Rho GTPase proteins. Activation and/or localization of Rho GTPase proteins has been implicated in the regulation of cell migration and/or invasion (2–5).
Membrane vesicle trafficking is also implicated in regulating cell polarization and migration (16, 33–37). A line of evidence indicates that GIPC1 is implicated in endocytosis (15, 16, 38, 39). Furthermore, depletion of PLEKHG6/MyoGEF represses dextran uptake in EGF-stimulated A431 cells (40), suggesting that MyoGEF may also play a role in regulating endocytosis. However, it is not clear at present whether the GIPC1-MyoGEF interaction is implicated in regulating membrane vesicle trafficking.
GIPC1/synectin can bind to a number of proteins, including syndecan-4 (41), the GTPase-activating protein RGS-GAIP (42), the transmembrane protein M-SemF (43), receptor tyrosine kinases TrkA and TrkB (44, 45), integrins α5 and α6 (46), neuropilin-1 (47), the insulin-like growth factor type 1 receptor (48), the myeloid cell-surface marker CD93 (49), the melanosomal membrane protein gp75 (50), the human T-cell lymphotrophic virus type 1 Tax oncoprotein (51), megalin (LDL receptor) (52, 53), 5T4 (21), the TGFβIII receptor (54), the β-adrenergic receptor (55), the human lutropin receptor (56), dopamine D2 and D3 receptors (57), and GLUT1 and myosin VI (38, 58). It is likely that the list of GIPC1-interacting partners will continue to increase. Thus, it appears to be important to dissect out the roles of specific binding partners and the causes of the various cell phenotypes resulting from depletion of GIPC1. Our results show that the anti-MyoGEF peptide antibody can interfere with GIPC1-MyoGEF complex formation and that treatment with the anti-MyoGEF peptide antibody impairs cell polarity and decreases cell invasion (Fig. 6). These findings suggest that binding of GIPC1 to MyoGEF plays an important role in regulating MDA-MB-231 breast cancer cell polarization and invasion. However, it remains to be determined whether binding of GIPC1 to other interacting partners also contributes to the regulation of breast cancer cell polarization and invasion.
Acknowledgments
We thank Drs. Robert S. Adelstein and Mary Anne Conti for critical reading and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant P20 RR015563 from the National Center for Research Resources. This work was also supported by the Terry C. Johnson Center for Basic Cancer Research. This is Contribution 10-211-J from the Kansas Agricultural Experiment Station (Manhattan, KS).
- GEF
- guanine nucleotide exchange factor.
REFERENCES
- 1.Raftopoulou M., Hall A. (2004) Dev. Biol. 265, 23–32 [DOI] [PubMed] [Google Scholar]
- 2.Burridge K., Wennerberg K. (2004) Cell 116, 167–179 [DOI] [PubMed] [Google Scholar]
- 3.Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 4.Pertz O., Hodgson L., Klemke R. L., Hahn K. M. (2006) Nature 440, 1069–1072 [DOI] [PubMed] [Google Scholar]
- 5.Kurokawa K., Matsuda M. (2005) Mol. Biol. Cell 16, 4294–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackay D. J., Hall A. (1998) J. Biol. Chem. 273, 20685–20688 [DOI] [PubMed] [Google Scholar]
- 7.Zheng Y. (2001) Trends Biochem. Sci. 26, 724–732 [DOI] [PubMed] [Google Scholar]
- 8.Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 9.Bellanger J. M., Astier C., Sardet C., Ohta Y., Stossel T. P., Debant A. (2000) Nat. Cell Biol. 2, 888–892 [DOI] [PubMed] [Google Scholar]
- 10.Seipel K., O'Brien S. P., Iannotti E., Medley Q. G., Streuli M. (2001) J. Cell Sci. 114, 389–399 [DOI] [PubMed] [Google Scholar]
- 11.Vanni C., Parodi A., Mancini P., Visco V., Ottaviano C., Torrisi M. R., Eva A. (2004) Oncogene 23, 4098–4106 [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Mata R., Burridge K. (2007) Trends Cell Biol. 17, 36–43 [DOI] [PubMed] [Google Scholar]
- 13.Penzes P., Johnson R. C., Sattler R., Zhang X., Huganir R. L., Kambampati V., Mains R. E., Eipper B. A. (2001) Neuron 29, 229–242 [DOI] [PubMed] [Google Scholar]
- 14.Park E., Na M., Choi J., Kim S., Lee J. R., Yoon J., Park D., Sheng M., Kim E. (2003) J. Biol. Chem. 278, 19220–19229 [DOI] [PubMed] [Google Scholar]
- 15.Hasson T. (2003) J. Cell Sci. 116, 3453–3461 [DOI] [PubMed] [Google Scholar]
- 16.Buss F., Luzio J. P., Kendrick-Jones J. (2002) Traffic 3, 851–858 [DOI] [PubMed] [Google Scholar]
- 17.Chittenden T. W., Claes F., Lanahan A. A., Autiero M., Palac R. T., Tkachenko E. V., Elfenbein A., Ruiz de Almodovar C., Dedkov E., Tomanek R., Li W., Westmore M., Singh J. P., Horowitz A., Mulligan-Kehoe M. J., Moodie K. L., Zhuang Z. W., Carmeliet P., Simons M. (2006) Dev. Cell 10, 783–795 [DOI] [PubMed] [Google Scholar]
- 18.Tkachenko E., Elfenbein A., Tirziu D., Simons M. (2006) Circ. Res. 98, 1398–1404 [DOI] [PubMed] [Google Scholar]
- 19.Liu M., Horowitz A. (2006) Mol. Biol. Cell 17, 1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee N. Y., Ray B., How T., Blobe G. C. (2008) J. Biol. Chem. 283, 32527–32533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Awan A., Lucic M. R., Shaw D. M., Sheppard F., Westwater C., Lyons S. A., Stern P. L. (2002) Biochem. Biophys. Res. Commun. 290, 1030–1036 [DOI] [PubMed] [Google Scholar]
- 22.Myers K. A., Rahi-Saund V., Davison M. D., Young J. A., Cheater A. J., Stern P. L. (1994) J. Biol. Chem. 269, 9319–9324 [PubMed] [Google Scholar]
- 23.Carsberg C. J., Myers K. A., Evans G. S., Allen T. D., Stern P. L. (1995) J. Cell Sci. 108 (Pt 8), 2905–2916 [DOI] [PubMed] [Google Scholar]
- 24.Rudchenko S., Scanlan M., Kalantarov G., Yavelsky V., Levy C., Estabrook A., Old L., Chan G. L., Lobel L., Trakht I. (2008) BMC Cancer 8, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu D., Asiedu M., Wei Q. (2009) Oncogene 28, 2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asiedu M., Wu D., Matsumura F., Wei Q. (2009) Mol. Biol. Cell 20, 1428–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D., Asiedu M., Adelstein R. S., Wei Q. (2006) Cell Cycle 5, 1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei Q. (2005) J. Biol. Chem. 280, 37790–37797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B. P., Burridge K. (2000) Mol. Cell. Biol. 20, 7160–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., Cantley L. C. (1997) Science 275, 73–77 [DOI] [PubMed] [Google Scholar]
- 31.Charras G. T., Hu C. K., Coughlin M., Mitchison T. J. (2006) J. Cell Biol. 175, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yonemura S., Hirao-Minakuchi K., Nishimura Y. (2004) Exp. Cell Res. 295, 300–314 [DOI] [PubMed] [Google Scholar]
- 33.Mellor H. (2004) Curr. Biol. 14, R434–435 [DOI] [PubMed] [Google Scholar]
- 34.Ulrich F., Heisenberg C. P. (2009) Traffic 10, 811–818 [DOI] [PubMed] [Google Scholar]
- 35.Caswell P., Norman J. (2008) Trends Cell Biol. 18, 257–263 [DOI] [PubMed] [Google Scholar]
- 36.Jones M. C., Caswell P. T., Norman J. C. (2006) Curr. Opin. Cell Biol. 18, 549–557 [DOI] [PubMed] [Google Scholar]
- 37.Le Roy C., Wrana J. L. (2005) Dev. Cell 9, 167–168 [DOI] [PubMed] [Google Scholar]
- 38.Reed B. C., Cefalu C., Bellaire B. H., Cardelli J. A., Louis T., Salamon J., Bloecher M. A., Bunn R. C. (2005) Mol. Biol. Cell 16, 4183–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naccache S. N., Hasson T., Horowitz A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12735–12740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Angelo R., Aresta S., Blangy A., Del Maestro L., Louvard D., Arpin M. (2007) Mol. Biol. Cell 18, 4780–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y., Li M., Chen W., Simons M. (2000) J. Cell. Physiol. 184, 373–379 [DOI] [PubMed] [Google Scholar]
- 42.De Vries L., Lou X., Zhao G., Zheng B., Farquhar M. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12340–12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L. H., Kalb R. G., Strittmatter S. M. (1999) J. Biol. Chem. 274, 14137–14146 [DOI] [PubMed] [Google Scholar]
- 44.Lou X., Yano H., Lee F., Chao M. V., Farquhar M. G. (2001) Mol. Biol. Cell 12, 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato H., Ohno K., Hashimoto K., Sato K. (2004) FEBS Lett. 572, 123–128 [DOI] [PubMed] [Google Scholar]
- 46.El Mourabit H., Poinat P., Koster J., Sondermann H., Wixler V., Wegener E., Laplantine E., Geerts D., Georges-Labouesse E., Sonnenberg A., Aumailley M. (2002) Matrix Biol 21, 207–214 [DOI] [PubMed] [Google Scholar]
- 47.Cai H., Reed R. R. (1999) J. Neurosci. 19, 6519–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J., O'Donnell M., Gitler A. D., Klein P. S. (2006) Development 133, 3651–3660 [DOI] [PubMed] [Google Scholar]
- 49.Bohlson S. S., Zhang M., Ortiz C. E., Tenner A. J. (2005) J. Leukoc. Biol. 77, 80–89 [DOI] [PubMed] [Google Scholar]
- 50.Liu T. F., Kandala G., Setaluri V. (2001) J. Biol. Chem. 276, 35768–35777 [DOI] [PubMed] [Google Scholar]
- 51.Rousset R., Fabre S., Desbois C., Bantignies F., Jalinot P. (1998) Oncogene 16, 643–654 [DOI] [PubMed] [Google Scholar]
- 52.Gotthardt M., Trommsdorff M., Nevitt M. F., Shelton J., Richardson J. A., Stockinger W., Nimpf J., Herz J. (2000) J. Biol. Chem. 275, 25616–25624 [DOI] [PubMed] [Google Scholar]
- 53.Lou X., McQuistan T., Orlando R. A., Farquhar M. G. (2002) J. Am. Soc. Nephrol. 13, 918–927 [DOI] [PubMed] [Google Scholar]
- 54.Blobe G. C., Liu X., Fang S. J., How T., Lodish H. F. (2001) J. Biol. Chem. 276, 39608–39617 [DOI] [PubMed] [Google Scholar]
- 55.Hu L. A., Chen W., Martin N. P., Whalen E. J., Premont R. T., Lefkowitz R. J. (2003) J. Biol. Chem. 278, 26295–26301 [DOI] [PubMed] [Google Scholar]
- 56.Hirakawa T., Galet C., Kishi M., Ascoli M. (2003) J. Biol. Chem. 278, 49348–49357 [DOI] [PubMed] [Google Scholar]
- 57.Jeanneteau F., Diaz J., Sokoloff P., Griffon N. (2004) Mol. Biol. Cell 15, 696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bunn R. C., Jensen M. A., Reed B. C. (1999) Mol. Biol. Cell 10, 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]