Abstract
The critical and multiple roles of thrombin in blood coagulation are regulated by ligands and cofactors. Zymogen activation imparts proteolytic activity to thrombin and also affects the binding of ligands to its two principal exosites. We have used the activation peptide fragment 1.2 (F12), a ligand for anion-binding exosite 2, to probe the zymogenicity of thrombin by isothermal titration calorimetry. We show that F12 binding is sensitive to subtle aspects of proteinase formation beyond simply reporting on zymogen cleavage. Large thermodynamic differences in F12 binding distinguish between a series of thrombin species poised along the transition of zymogen to proteinase. Active-site ligands transitioned a zymogen-like state to a proteinase-like state. Conversely, removal of Na+ converted proteinase-like thrombin to a more zymogen-like form. Thrombin mutants, with deformed x-ray structures, previously considered to be emblematic of specific regulated states of the enzyme, are instead shown to be variously zymogen-like and can be made proteinase-like by active-site ligation. Thermodynamic linkage between anion-binding exosite 2, the Na+-binding site, and the active site arises from interconversions of thrombin between a continuum of zymogen- and proteinase-like states. These interconversions, reciprocally regulated by different ligands, cast new light on the problem of thrombin allostery and provide a thermodynamic framework to explain the regulation of thrombin by different ligands.
Keywords: Allosteric Regulation, Blood Coagulation Factors, Serine Protease, Thermodynamics, Thrombin
Introduction
Thrombin is a pivotal serine proteinase product of the blood coagulation cascade. In addition to catalyzing fibrinogen cleavage and platelet activation to form the blood clot, it also plays essential regulatory roles (1, 2). It catalyzes proteolytic activation steps that amplify flux toward thrombin formation following initiation of the coagulation cascade. Thrombin also functions as a negative regulator by activating protein C in the anticoagulant pathway (3). These opposing roles of thrombin in coagulation derive from its ability to act with specificity on a range of protein substrates, with differing sequences flanking the cleavage sites, in reactions that are regulated by cofactors and two anion-binding exosites (ABE1 and ABE2)2 found on opposite faces of the proteinase (4, 5). Allosteric control, arising from binding of ligands to different sites, is considered to underlie the regulation of thrombin specificity and its dual role as procoagulant and anticoagulant proteinase (4, 5).
Thrombin and the other serine proteinases of blood coagulation contain catalytic domains bearing the chymotrypsin fold (6). In this family, the zymogen precursor is converted to proteinase by cleavage following Arg15 (7).3 The nascent N terminus inserts in a sequence-specific manner into an N-terminal binding pocket to form a salt bridge with Asp194 (7). Salt bridge formation is associated with changes in activation domains, optimization of the primary specificity pocket to facilitate substrate binding, and a flip in the Gly193 amide bond to form the oxyanion hole (7). These transitions imbue the product with proteolytic activity.
Early studies with chymotrypsin suggested the persistence of zymogen-like species at near-neutral pH, related to the partial deprotonation of the new N terminus responsible for salt bridge formation (8). It is nevertheless implicitly assumed that irreversible cleavage at Arg15 and the concerted transitions that ensue highly favor proteinase formation. This is despite the likely presence of intervening equilibria between substates that lie on the pathway for conversion of zymogen to fully stabilized proteinase. A notable exception is factor VIIa which possesses poor catalytic activity and has been proposed to be zymogen-like when free and stabilized in the proteinase state when bound to tissue factor (9).
Na+ is considered an essential player in thrombin allostery (10). The Na+-binding site, absent in the zymogen but present in the proteinase, is proposed to control transitions of the enzyme between a putatively inactive E* form, a Na+-free slow form, and a Na+-bound fast species (10–12). These enzymic species are considered to exhibit varying specificity and contribute differentially to the functions of thrombin as anticoagulant versus procoagulant enzyme (10). The explosive growth in mutagenesis and x-ray crystallography investigating these phenomena has yielded a myriad of structures of variously distorted mutant forms of the enzyme interpreted to represent specific regulated states (10, 13). The alternative suggestion that they instead represent zymogen-like species of thrombin forms the basis for this investigation (14).
Zymogen activation to yield thrombin also produces large increases in the affinity of ligands for ABE1 (15). Based on differences in affinity for the zymogen relative to proteinase, substrates, inhibitors, Na+, and ABE1 ligands are all expected to energetically favor the proteinase. These ideas are supported by recent findings in NMR studies (16). The conversion of prothrombin to thrombin releases the N-terminal fragment 1.2 region (F12) as a propiece that reversibly associates with the proteinase as well as a precursor zymogen species through ABE2 (17). F12 represents the authentic protein ligand for ABE2, with particular relevance to the process of thrombin formation. The recent observation that F12 binds to a zymogen precursor with higher affinity than to thrombin unexpectedly suggests that the conversion of zymogen to proteinase is accompanied by reciprocal changes in ligand affinity for the two exosites in thrombin (18). Whereas ligand binding to ABE1 is expected to favor the proteinase, ligation at ABE2 would instead favor the zymogen. If zymogen-like substates of thrombin can be significantly populated following zymogen cleavage, such reciprocal effects of liganding at ABE1 and ABE2 would provide a previously unanticipated mechanism for the regulation of its function.
We now employ F12 as a tool in thermodynamic measurements to assess whether thrombin can exhibit zymogen-like behavior. Our finding that thrombin can interconvert between zymogen- and proteinase-like states in a ligand-dependent manner provides a new framework for understanding how its multiple functions are regulated.
EXPERIMENTAL PROCEDURES
Materials
The thrombin inhibitor dansyl-l-arginine-N-(3-ethyl-1,5-pentanediyl)amide (DAPA; Hematologic Technologies), the peptidyl substrate l-pyroglutamyl-l-prolyl-l-arginyl-p-nitroanilide (S2366; Chromogenix), d-Phe-l-Pro-l-Arg chloromethyl ketone (FPRck; Calbiochem), and BisTris propane (Sigma) were from the indicated suppliers. Stock solutions of S2236 and DAPA were prepared in water, and concentrations were determined using E342 = 8270 m−1 cm−1 (S2236) and E340 = 4105 m−1 cm−1 (DAPA) (19, 20). Choline chloride (ICN Biomedicals) was recrystallized from boiling ethanol and dried extensively in vacuo to yield a white, odorless, crystalline powder. Alternatively, high purity choline chloride (catalogue no. 219770500) was obtained from Acros Organics. Both types of choline chloride yielded equivalent results that were different from those obtained with lower grades of the salt.
Proteins
Prothrombin variants bearing the indicated mutations were expressed in HEK293 cells and purified from conditioned medium as described previously (21). The variants were preparatively converted to thrombin (IIa) or to prethrombin 2 (P2) and purified as described (21). The variants of P2 or IIa are subscripted to indicate the substitutions in the catalytic domain: IIaTAT contains Thr-Ala-Thr in place of Ile16-Val17-Glu18; P2S195A and IIaS195A contain Ala in place of Ser195; IIaS195A,W215A,E217A contains Ala substitutions at Ser195, Trp215, Glu217; and IIaD102N,S195A contains Asn in place of Asp102 and Ala in place of Ser195. The variant IIaE217K,S195A (with Lys substituted for Glu217 and Ala for Ser195) was prepared as described previously (22). F12 was prepared and purified from prothrombin isolated from plasma using established procedures (21). IIa was prepared by preparative activation of prothrombin from plasma, followed by purification (23). IIai was prepared by inactivating plasma-derived thrombin with FPRck, followed by purification as described (21). Protein concentrations were determined using the following molecular weights and extinction coefficients (E280, mg−1 cm2): F12, 34,800 and 1.12; and all P2 and thrombin variants, 37,500 and 1.94 (24).
Isothermal Titration Calorimetry
Measurements were performed using an iTC200 system (MicroCal) at 25 °C. All protein species were dialyzed for 18 h versus 10 liters of 20 mm BisTris propane, 0.15 m NaCl, and 5 mm Ca2+ (pH 7.4); concentrated by centrifugal ultrafiltration using an Ultracel-10K device (Amicon), if necessary; and centrifuged to remove particulates before use. The cell (204 μl) contained either P2S195A or the indicated thrombin variant (5.5–23.9 μm), and the injection syringe (45 μl) contained F12 (97.9–436.4 μm). Heat flow was measured with continuous stirring, following injections of F12 spaced at 180-s intervals. In a typical experiment, the first injection was 0.5 μl, followed by 20 injections of 2 μl each. Trivial heat flow due to ligand dilution and buffer mismatch was determined from an identical series of injections of F12 into dialysate. In some cases, the additional control of injecting dialysate into the cell containing P2S195A or a thrombin variant was also performed. For studies with DAPA, the indicated concentration of inhibitor was present in the syringe and the cell. For measurements with increasing NaCl concentrations, proteins were dialyzed into buffer containing either 150 mm NaCl or choline chloride. The desired protein and NaCl concentrations were obtained by mixing the two dialyzed samples. Small differences in thermodynamic constants reported here in comparison with a previous study done using Hepes (18) at least partly reflect the use of BisTris propane as a buffer to permit studies with varying NaCl concentrations. Proteolysis during the experiment was limited by pursuing studies with S195A derivatives of IIa or P2 whenever possible. Lack of proteolysis during the experiment was verified by SDS-PAGE analysis of samples following dialysis and of expended reaction mixtures.
Initial Velocity Studies
Reaction mixtures (200 μl) in wells of a 96-well plate containing the indicated concentrations of S2366 were initiated with either 1 or 2 nm thrombin. Initial rates of substrate hydrolysis were determined in a SpectraMax kinetic plate reader (Molecular Devices) by measuring the increase in absorbance at 405 nm. When necessary, initial rates were expressed in concentration terms using E405 = 9887 m−1 cm−1 (19) and an effective path length of 0.59 cm for a 200-μl reaction volume. Studies were performed in 20 mm BisTris propane, 0.1% (w/v) PEG-8000, 5 mm Ca2+, and the indicated concentrations of NaCl with ionic strength maintained at 0.15 m using choline chloride. Buffers were adjusted to pH 7.4 or 8.0 with HCl following equilibration of the solution to either 25 or 37 °C. Initial velocities at 37 °C were determined by performing experiments in a room thermostatted at 37 °C.
Data Analysis
Origin scripts provided with the instrument were used to process heat flow traces, compute concentrations in the cell, integrate peaks, and subtract areas for the buffer control. Data were then analyzed using SedPhat (25). The A + B hetero-association model was used to obtain fitted values of Kd, ΔH, and base line and the calculated values of ΔG and ΔS, assuming a stoichiometry of 1. Slight deviations from the assumed stoichiometry were accommodated by fitting a value for the non-binding and heat silent fraction of incompetent protein in the cell. For global fits in the three-component experiments, analysis was according to the A + B + C hetero-association model. Errors in fitted parameters were computed by the Monte Carlo method to yield mean 67% confidence limits. Errors were propagated into the calculated terms (26).
Initial velocities were globally analyzed according to Scheme 3 employing the rapid equilibrium assumption using Dynafit (27). Errors in the fitted parameters reflect linear approximations of 95% confidence limits.
SCHEME 3.
Nonessential activation by Na+ of substrate (S) hydrolysis by thrombin (E).
RESULTS
Distinguishing Zymogen-like and Proteinase States of Thrombin
Thermodynamic studies have previously established that F12 binds with ∼20-fold higher affinity to the zymogen P2 than it does to thrombin (18). We therefore investigated the binding of F12 to a thrombin variant (IIaTAT) bearing Thr-Ala-Thr in place of the N-terminal Ile16-Val17-Glu18 sequence, intended to impair salt bridge formation and the conformational transition to proteinase. In agreement with this intent, IIaTAT exhibits undetectable activity (<0.02%) toward peptidyl substrates (28). Isothermal titration calorimetry of F12 binding to IIaTAT yielded heat traces and an isotherm (Fig. 1) intermediate to those expected from previous measurements of F12 binding to P2 or thrombin (18). Thus, the thermodynamic constants for F12 binding are apparently sensitive to subtle aspects of proteinase formation beyond simply reporting on the cleavage, or lack thereof, at the Arg15–Ile16 bond.
FIGURE 1.
Binding of F12 to IIaTAT assessed by isothermal titration calorimetry. Upper panel, heat flow tracings were obtained upon successive injections (2 μl) of 293 μm F12 into 20.1 μm IIaTAT in 20 mm BisTris propane, 0.15 m NaCl, and 5 mm Ca2+ (pH 7.4) at 25 °C. The initial injection (0.5 μl) was eliminated from the analysis. The heat flow profile from an identical injection sequence of F12 into dialysate is shown offset for reference. Lower panel, integrated heats were analyzed, and the line is drawn according to the fitted parameters listed in Table 1.
To place the thermodynamic parameters obtained with cleaved but zymogen-like IIaTAT in context, we employed additional reference states capable of binding F12 but differentially poised along the pathway for the conversion of zymogen to proteinase: P2S195A is the uncleaved zymogen; IIaS195A (catalytic Ser195 substituted with Ala) was employed as the proteinase with an unoccupied active site; and IIai is wild-type thrombin covalently inactivated with FPRck to lock the enzyme into its most stable “proteinase-like” configuration by irreversibly occupying the active site.
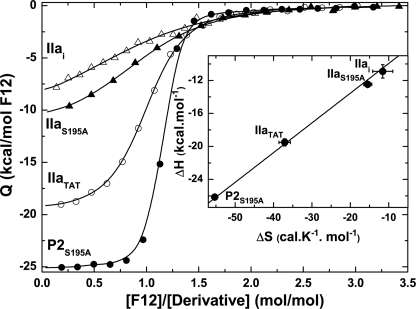
Isotherms for F12 binding yielded a family of curves wherein both shape and amplitude changed with the increasingly proteinase-like character of the reference species (Fig. 2). Fitted thermodynamic constants revealed that F12 binding to the zymogen P2S195A exhibited the highest affinity with large negative enthalpic and entropic contributions (Table 1). The equilibrium dissociation constant for F12 binding increased overall by ∼40-fold, progressing from zymogen, to cleaved but zymogen-like (IIaTAT), to proteinase with an unoccupied active site (IIaS195A), and to the ultimate stabilized proteinase (IIai) (Table 1). In comparison with the modest increase in ΔG, progression through this series yielded very large increases in both ΔH and TΔS (Table 1).
FIGURE 2.
Binding of F12 to reference states in the pathway for conversion of zymogen to proteinase. Integrated heats from F12 titrations with the species listed on the left are illustrated. The lines were drawn according to the fitted parameters listed in Table 1. Inset, dependence of ΔH on ΔS for these reference states. The line was drawn following weighted linear regression analysis.
TABLE 1.
Thermodynamic constants for the binding of F12 to reference states
Parameters were derived from titration of F12 into the indicated species at 298 K in 20 mm BisTris propane, 0.15 m NaCl, and 5 mm Ca2+ (pH 7.4). Thermodynamic parameters are listed ±67% confidence limits.
| Species | Kd | ΔG | ΔH | ΔS | Fia |
|---|---|---|---|---|---|
| μm | kcal mol−1 | kcal mol−1 | cal mol−1K−1 | ||
| P2S195A | 0.082 ± 0.005 | −9.66 ± 0.08 | −26.15 ± 0.20 | −55.33 ± 0.60 | 0.075 ± 0.001 |
| IIaTAT | 0.63 ± 0.04 | −8.45 ± 0.05 | −19.51 ± 0.44 | −37.11 ± 1.48 | 0.044 ± 0.004 |
| IIaS195A | 1.67 ± 0.18 | −7.87 ± 0.05 | −12.49 ± 0.32 | −15.48 ± 1.10 | 0.041 ± 0.002 |
| IIai | 3.40 ± 1.20 | −7.46 ± 0.21 | −10.90 ± 0.81 | −11.56 ± 2.64 | 0 ± 0.03 |
a Binding stoichiometry was assumed to be 1. Uncertainty in stoichiometry was accommodated by fitting the fraction of incompetent species in the cell (Fi).
Large and compensating changes in the thermodynamic constants were evident as a linear relationship between ΔH and ΔS with the reference states positioned along this plot in order of their increasing proteinase-like character (Fig. 2, inset). The extra thermodynamic information content of such enthalpy-entropy compensation has been debated (29). However, a quasi-linear relationship between ΔH and ΔS, determined by calorimetry, is expected for the binding of a ligand to related states of a protein in reversible equilibrium with each other (29, 30). The linear relationship between ΔH and ΔS indicates that a common source of additivity distinguishes F12 binding to the different reference states with increasing proteinase-like character (29, 31). We surmise that this relationship defines the thermodynamic trajectory for the conversion of zymogen to proteinase. The ability of the species to interconvert along this trajectory represents an essential feature of this interpretation.
Driving Zymogen-like Thrombin to a Proteinase-like State
DAPA binds with high affinity to the active site of thrombin (20). Titrations of IIaTAT with F12 in the presence of different fixed concentrations of DAPA yielded isotherms with decreasing amplitudes (Fig. 3). At high concentrations of DAPA, the isotherms resembled those obtained with the ultimate proteinase-like state (IIai) (Fig. 3). The data could be globally analyzed to yield thermodynamic constants for the stepwise binding interactions of DAPA and F12 with IIaTAT to form a ternary complex (Scheme 1). In contrast to the nanomolar affinity of DAPA for thrombin, the inferred binding of DAPA to IIaTAT was ∼1000-fold weaker (Scheme 1), consistent with the zymogen-like character of this species (28). The inferred constants for F12 binding to IIaTAT saturated with DAPA were similar to those obtained for the binding of F12 to IIai (Scheme 1 and Table 1). The results indicate that binding of DAPA to the zymogen-like variant IIaTAT stabilizes it in a proteinase-like state.
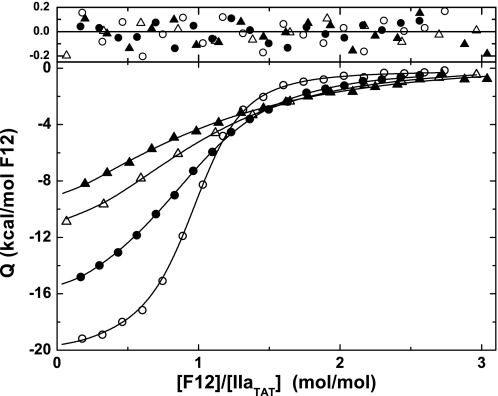
FIGURE 3.
DAPA modulates F12 binding to IIaTAT. Shown are integrated heats obtained from titrations of F12 into IIaTAT in the presence of 0 (○), 15 (●), 60 (▵), and 250 (▴) μm DAPA. The lines were drawn following global analysis using the fitted parameters listed in Scheme 1. Residuals to the fitted lines are shown in the upper panel.
SCHEME 1.
Linked thermodynamics for F12 and DAPA binding to IIaTAT.
To provide a mechanism-independent test of this conclusion, ΔH and ΔS pairs were derived from individual fits of the traces. Values obtained for F12 binding to IIaTAT with increasing concentrations of DAPA were found to extend along the linear trajectory toward the IIai reference state (Fig. 4).
FIGURE 4.
Ligand-dependent transitions of IIaTAT and IIaS195A. Isotherms for F12 binding to IIaTAT at increasing concentrations of DAPA (0–250 μm; red circles) or for IIaS195A in the presence of decreasing concentrations of Na+ (150–0 mm; blue circles) were individually fitted to derive ΔH and ΔS. The values for the indicated reference states are shown (black circles) along the line as presented in Fig. 2 (inset).
Converting Thrombin to a Zymogen-like Form
The foregoing studies were conducted at 150 mm NaCl to replicate the concentration of Na+ in blood. We examined the binding of F12 to IIaS195A in the presence of decreasing concentrations of Na+ with ionic strength maintained at 0.15 m with choline chloride. Integrated heats increased with decreasing concentrations of Na+, yielding isotherms that increasingly resembled those obtained with the zymogen-like thrombin variant (Fig. 5). Global analysis of these data yielded thermodynamic constants for the stepwise binding interactions of Na+ and F12 with IIaS195A to form a ternary complex (Fig. 5 and Scheme 2). Minor differences were evident in isotherms for F12 binding to P2S195A or to IIai in the presence or absence of 150 mm Na+ (Fig. 5, inset). The zymogen is not expected to contain a Na+-binding site. Furthermore, IIai is irreversibly locked into the proteinase configuration and minimally affected by bound Na+ (16, 32). The observations suggest minor contributions from heat effects arising from Na+ binding to F12 and point to the validity of the global analysis according to Scheme 2. The principal findings are that IIaS195A is saturated with Na+ at 150 mm NaCl and that F12 binding to IIaS195A in the absence of Na+ closely mirrors its binding to zymogen-like IIaTAT at 150 mm NaCl (Scheme 2). The logarithmic relationship between affinity and energetic terms accounts for the close similarity in thermodynamic constants despite an ∼4-fold difference in inferred Kd for F12 binding to IIaTAT versus IIaS195A in the absence of Na+ (Schemes 1 and 2). Thus, bound Na+ is part of the stabilized structure of the proteinase state, and its dissociation converts IIaS195A to a more zymogen-like form. Accordingly, ΔH and ΔS pairs obtained from individual fits of the traces were found distributed along the linear trajectory extending toward IIaTAT with decreasing Na+ (Fig. 4).
FIGURE 5.
Na+-dependent binding of F12 to IIaS195A. Integrated heats for the binding of F12 to IIaS195A were determined from titrations conducted in the presence of 0 (○), 4.4 (●), 12 (▵), 50 (▴), and 150 (▿) mm Na+. The lines were drawn following global analysis using the fitted parameters listed in Scheme 2. Residuals to the fitted lines are shown in the upper panel. The inset illustrates isotherms obtained with IIai using 0 (◁) and 150 (◀) mm Na+ or with P2S195A using 0 (□) and 150 mm (■) Na+.
SCHEME 2.
Linked thermodynamics for F12 and Na+ binding to IIaS195A.
Na+ Binding to Thrombin
There is a discrepancy between Kd = 5.8 ± 0.5 mm for Na+ binding to thrombin obtained from our linkage analysis (Scheme 2) and published values of ∼35 mm at 25 °C and ∼120 mm at 35 °C (10). To assess whether this discrepancy arose from a peculiar property of IIaS195A, we pursued initial velocity studies of peptidyl substrate hydrolysis to infer the binding of Na+ to wild-type thrombin (33). Increasing concentrations of Na+ yielded a modest increase in the rate of S2366 hydrolysis (Fig. 6). Global analysis according to the kinetic model for nonessential activation (Scheme 3) provided an adequate description of the data (Fig. 6). The inferred binding constant for Na+ (KE,N) was in agreement with that determined from the thermodynamic linkage studies (Table 2). This further validates the conclusions derived from studies of the Na+-dependent binding of F12 to IIaS195A.
FIGURE 6.
Kinetics of peptidyl substrate hydrolysis. Initial velocities for S2366 hydrolysis were measured at 25 °C in 20 mm BisTris propane and 0.1% (w/v) PEG-8000 (pH 7.4) with 2 nm IIa and increasing concentrations of Na+ corresponding to 0, 1, 3, 5, 10, 15, 20, 75, and 150 mm (bottom curve to top curve). Ionic strength was maintained at 0.15 m using choline chloride. The lines were drawn following analysis according to Scheme 3 using the fitted constants in Table 2.
TABLE 2.
Kinetic constants for the Na+-dependent modulation of peptidyl substrate cleavage by thrombin
Parameters were derived from initial velocity studies as illustrated in Fig. 6 and analyzed according to Scheme 3 employing the rapid equilibrium assumption. Fitted parameters are listed ±95% confidence limits.
| T | pH | KE,S | KEN,S | kcat(E) | kcat(EN) | KE,N | KES,N |
|---|---|---|---|---|---|---|---|
| μm | μm | s−1 | s−1 | mm | mm | ||
| 25 °C | 7.4 | 90 ± 5 | 150 ± 5 | 60 ± 2 | 130 ± 1.6 | 4.4 ± 0.34 | 7.3 ± 0.5 |
| 25 °C | 8.0 | 80 ± 4 | 96 ± 5 | 71 ± 1 | 121 ± 1.4 | 5.7 ± 0.55 | 6.8 ± 0.7 |
| 37 °C | 7.4 | 330 ± 80 | 170 ± 35 | 159 ± 20 | 229 ± 19 | 33.7 ± 13.6 | 16.9 ± 6.5 |
| 37 °C | 8.0 | 280 ± 14 | 70 ± 8 | 164 ± 4 | 252 ± 6 | 159 ± 23 | 39.8 ± 7.1 |
Recognizing the importance attached to the significance of Na+ affinity measured at physiologic temperature (10), additional kinetic studies were undertaken at 37 °C. At pH 7.4 and at 37 °C, KE,N increased to ∼34 mm (Table 2), still well below the published value and the physiologic concentration of Na+. However, further investigation revealed that the prior studies were apparently conducted at pH 8.0 (33). At pH 8.0 and at 37 °C, we could reproduce the previously published KE,N ∼ 160 mm (Table 2).
Distorted or “Anticoagulant” Thrombins Are Zymogen-like
The relationship between Na+ binding to thrombin and its interconversion between zymogen- and proteinase-like states motivated studies with the thrombin variants IIaW215A,E217A, IIaE217K, and IIaD102N, that exhibit distorted structures and are considered emblematic of the slow or E* forms of the enzyme (34–36). Binding studies with F12 using these variants also containing the S195A substitution yielded ΔH and ΔS values distributed along the linear relationship between these parameters and variously shifted toward the zymogen-like IIaTAT (Fig. 7). For IIaS195A,E217K or IIaS195A,W215A,E217A, the addition of DAPA yielded data consistent with their stabilization in the proteinase-like state (Fig. 7). Rather than representing particular states of the enzyme with special properties, our findings are more consistent with the interpretation that these destabilizing mutations favor zymogen-like forms, which can be variably rectified by ligands that bind to the active site.
FIGURE 7.
Enthalpy-entropy compensation observed with thrombin variants. Closed black symbols denote thermodynamic parameters measured for the four reference states as well as three mutants measured at 150 mm Na+. Closed red circles represent findings with the zymogen-like variant IIaTAT with increasing concentrations of DAPA. Closed blue circles show the results with IIaS195A with decreasing concentrations of Na+. Open blue circles show the results at 0 mm Na+ obtained with P2S195A or with IIai. The results of rescue of IIaS195A,E217K with 33 μm DAPA or of IIaS195A,W215A,E217A (IIaS195A,WE) with 60 μm DAPA are shown by closed green symbols. Error bars denote 67% confidence limits. The line was drawn following weighted linear regression with a slope of 321 ± 10 K.
Continuum between Zymogen- and Proteinase-like States
The idea that any one state of thrombin lies on the trajectory for the conversion of zymogen to proteinase is illustrated by the agreement of all ΔH and ΔS values determined in this study with the linear relationship initially established with the reference states (Fig. 7). We conclude that thrombin can exist in a range of substates that fall on a continuum bounded by zymogen and fully stabilized proteinase. Interconversions between these states are reciprocally linked to binding interactions at the active site and the Na+-binding site, as well as the binding of F12 to ABE2. This relationship also satisfactorily accounts for the properties of a series of mutants previously proposed to represent the slow or E* forms of thrombin.
DISCUSSION
We have provided a unifying thermodynamic framework for understanding how linked binding interactions with different ligands regulate thrombin. Linkage arises from the ability of thrombin to interconvert between zymogen- and proteinase-like forms that differ in energetics for their interactions with active-site ligands, Na+, and the activation peptide F12, which engages ABE2. Shuttling of thrombin between these states in a ligand-dependent manner has wide-ranging implications for understanding how different ligands regulate thrombin in opposing ways and could affect its multiple roles.
Major differences in ΔH and ΔS with modest differences in ΔG underlie the binding of F12 to zymogen-like versus proteinase-like species. F12 is a surprisingly useful probe for the zymogenicity of thrombin, uniquely amenable to analysis by isothermal titration calorimetry. The reliable inferences permitted by this approach would be inaccessible from measurements of just ligand binding affinity or from thermodynamic constants derived from the temperature dependence of kinetic or binding studies, expected to yield artificially correlated parameters. An elliptical relationship is expected between ΔH and ΔS for ligand binding to related states in reversible equilibrium (29, 30). We presume that the species that we have studied reflect energetic states that lie on a portion of this ellipse. The zymogen-to-proteinase transition provides the source of additivity that distinguishes F12 binding to the reference states and the ligand-dependent transitions along this trajectory (31).
Na+ enhances the binding of hirugen or a thrombomodulin fragment to thrombin (37, 38). These findings can readily be accommodated by the idea that both ABE1 ligands and Na+ energetically favor proteinase-like forms as documented by recent NMR studies (16). Binding of F12 to ABE2, which energetically favors the zymogen-like state, will oppose binding of hirugen or thrombomodulin to ABE1. Consequently, binding of F12 to ABE2 is expected to be deleterious for ligand binding at ABE1 and vice versa. Previous studies documenting linkage in the binding of ligands to ABE1 and ABE2 have been based on small differences in affinity and remain controversial (39, 40). It is likely that appreciation of the true magnitude of the extent of reciprocally regulated binding to these exosites will require calorimetric studies of the type presented here.
Interpretation of thrombin allostery in terms of ligand-dependent shuttling between zymogen- and proteinase-like states has particular impact on the voluminous structure-function work in the field. Zymogen-like thrombin forms are expected to predominate in the absence of Na+ and would therefore exhibit variably impaired activity toward different substrates, particularly when judged by measurements of kcat/Km. Binding of thrombomodulin to ABE1 favors the proteinase-like state and also inhibits the procoagulant functions of thrombin (3). Thus, measurements of protein C activation done in Na+-free buffer but at saturating concentrations of the cofactor would yield the appearance of minimally perturbed anticoagulant function. This provides an alternative explanation, supported by structural evidence, for the contention that Na+-free “slow” thrombin is anticoagulant, whereas Na+-bound “fast” thrombin has both anticoagulant and procoagulant functions (10, 41). Mutations in thrombin that destabilize the proteinase would adversely affect Na+ binding and similarly affect function, appearing as anticoagulant thrombins (11, 12). This likely accounts for the diverse range of mutations in thrombin, including removal of several residues in the autolysis loop or a highly conserved disulfide bond, that have been shown to yield varyingly anticoagulant-specific or E* proteinase forms (11, 42). In line with this reasoning, distorted structures reported for various thrombin mutants are rectified to proteinase-like structures following reaction with FPRck or complexation with ABE1 ligands (32, 34). Equivalently distorted structures in a diverse series of mutants have been taken as evidence for the unambiguous identification of the E* state (10). Instead, we suggest that ligand- and mutation-dependent redistribution of thrombin between zymogen- and proteinase-like states provides a more complete and parsimonious explanation.
We have shown that at the pH and Na+ concentration in blood, thrombin is essentially saturated with Na+, which is part of the structure of the stabilized proteinase. The fractional saturation is less at 37 °C but nevertheless exceeds 80%. These ideas cast doubt on the preeminence ascribed to the fact that ∼50% of thrombin is bound to Na+ in the physiologic milieu, thereby yielding equally populated Na+-bound and Na+-free states that differ in their spectra of activities (10). Although this may apply in an indirect way at pH 8.0, it is difficult to see its relevance to normal mammalian physiology.
The previous suggestion that Na+-free or mutant forms of thrombin are zymogen-like has been dismissed because the N terminus appears correctly inserted for salt bridge formation (13). It has also been argued that Na+-free or mutant thrombins must represent species with distinct properties because they exhibit different Vmax values for peptidyl substrates (43). These arguments are seemingly based on considering the zymogen-to-proteinase transition as a concerted change following the formation of the nascent N terminus. Instead, our observations are more consistent with a series of coupled but stepwise changes that follow Arg15 cleavage that account for thermodynamic additivity. This idea is supported by the findings of NMR studies of thrombin variously ligated to Na+, hirugen, and FPRck (16). In this scenario, more zymogen-like species represent forms that have failed to undergo one or more of the changes associated with proteinase formation. Extent of rescue to the proteinase state and the measured kinetic properties would be dependent on the nature of the impaired transition(s), as well as the substrate or active-site ligand used.
The large thermodynamic differences in the binding of F12 to zymogen relative to proteinase are surprisingly not observed with fragment 2 produced by proteolysis of F12 (18). Studies with EDTA did not reduce F12 binding to that observed with fragment 2 (data not shown). It appears unlikely that effects seen with F12 arise from the Ca2+-stabilized structure of the fragment 1 region. Rather, it seems probable that proteolysis of F12 significantly impacts the interactions between the fragment 2 region and ABE2. Our observations suggest caution in equating the behavior of any one ABE2 ligand with another.
The literature on thrombin allostery has been dominated by the lexicon of Na+ binding, fast and slow thrombins, anticoagulant thrombins, as well as an E* form supposedly inactive but not zymogen-like (13). Our findings instead provide a more general framework for the interpretation of thrombin function based on its ready ability to interconvert between a continuum of states bounded by zymogen and the ultimate stabilized proteinase. Because of thermodynamic differences in the binding of different ligands to these states, these equilibria are modulated in opposing ways by ligands relevant to the function of thrombin in vivo. The various substrates and ligands of thrombin ligate ABE1 and ABE2 to varying extents (1, 5). Zymogen-like forms favored by ligation at ABE2 would bind ABE1 ligands less favorably and possibly exhibit diminished active-site function with respect to certain substrates. Therefore, we propose that ligand-dependent shuttling of thrombin between zymogen- and proteinase-like states plays an important role in modulating its multiple regulatory roles in blood coagulation and cell biology.
Acknowledgments
We are grateful to colleagues Rodney Camire, Pete Lollar, George Vlasuk, and Bill Church for critical review of the manuscript.
The work was supported, in whole or in part, by National Institutes of Health Grant HL-74124 (to S. K.).
Residues in IIa and P2 are numbered according to the homologous residues in chymotrypsinogen (4).
- ABE
- anion-binding exosite
- F12
- fragment 1.2
- DAPA
- dansyl-l-arginine-N-(3-ethyl-1,5-pentanediyl)amide
- FPRck
- d-Phe-l-Pro-l-Arg chloromethyl ketone
- BisTris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- IIa
- thrombin
- P2
- prethrombin 2
- IIai
- FPRck-inactivated IIa.
REFERENCES
- 1.Davie E. W., Kulman J. D. (2006) Semin. Thromb. Hemostasis 32, Suppl. 1, 3–15 [DOI] [PubMed] [Google Scholar]
- 2.Mann K. G. (2003) Chest 124, 4S–10S [DOI] [PubMed] [Google Scholar]
- 3.Esmon C. T. (2006) Semin. Thromb. Hemostasis 32, Suppl. 1, 49–60 [DOI] [PubMed] [Google Scholar]
- 4.Bode W. (2006) Semin. Thromb. Hemostasis 32, Suppl. 1, 16–31 [DOI] [PubMed] [Google Scholar]
- 5.Lane D. A., Philippou H., Huntington J. A. (2005) Blood 106, 2605–2612 [DOI] [PubMed] [Google Scholar]
- 6.Rawlings N. D., Barrett A. J. (1993) Biochem. J. 290, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan A. R., James M. N. (1998) Protein Sci. 7, 815–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fersht A. R., Requena Y. (1971) J. Mol. Biol. 60, 279–290 [DOI] [PubMed] [Google Scholar]
- 9.Higashi S., Matsumoto N., Iwanaga S. (1996) J. Biol. Chem. 271, 26569–26574 [DOI] [PubMed] [Google Scholar]
- 10.Di Cera E. (2008) Mol. Aspects Med. 29, 203–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bah A., Carrell C. J., Chen Z., Gandhi P. S., Di Cera E. (2009) J. Biol. Chem. 284, 20034–20040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gandhi P. S., Page M. J., Chen Z., Bush-Pelc L., Di Cera E. (2009) J. Biol. Chem. 284, 24098–24105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Cera E. (2009) IUBMB Life 61, 510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huntington J. A. (2009) J. Thromb. Haemost. 7, Suppl. 1, 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bock P. E., Panizzi P., Verhamme I. M. (2007) J. Thromb. Haemost. 5, Suppl. 1, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lechtenberg B., Johnson D. J., Freund S., Huntington J. A. (2010) Proc. Natl. Acad. Sci. U.S.A., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arni R. K., Padmanabhan K., Padmanabhan K. P., Wu T. P., Tulinsky A. (1993) Biochemistry 32, 4727–4737 [DOI] [PubMed] [Google Scholar]
- 18.Kamath P., Krishnaswamy S. (2008) J. Biol. Chem. 283, 30164–30173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lottenberg R., Jackson C. M. (1983) Biochim. Biophys. Acta 742, 558–564 [DOI] [PubMed] [Google Scholar]
- 20.Nesheim M. E., Prendergast F. G., Mann K. G. (1979) Biochemistry 18, 996–1003 [DOI] [PubMed] [Google Scholar]
- 21.Orcutt S. J., Krishnaswamy S. (2004) J. Biol. Chem. 279, 54927–54936 [DOI] [PubMed] [Google Scholar]
- 22.Johnson D. J., Adams T. E., Li W., Huntington J. A. (2005) Biochem. J. 392, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundblad R. L., Kingdon H. S., Mann K. G. (1976) Methods Enzymol. 45, 156–176 [DOI] [PubMed] [Google Scholar]
- 24.Mann K. G., Elion J., Butkowski R. J., Downing M., Nesheim M. E. (1981) Methods Enzymol. 80, 286–302 [DOI] [PubMed] [Google Scholar]
- 25.Houtman J. C., Brown P. H., Bowden B., Yamaguchi H., Appella E., Samelson L. E., Schuck P. (2007) Protein Sci. 16, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bevington P. R., Robinson K. D. (1992) Data Reduction and Error Analysis for the Physical Sciences, 2nm Ed., McGraw-Hill Book Co., New York [Google Scholar]
- 27.Kuzmic P. (1996) Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 28.Bianchini E. P., Orcutt S. J., Panizzi P., Bock P. E., Krishnaswamy S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10099–10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp K. (2001) Protein Sci. 10, 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eftink M. R., Anusiem A. C., Biltonen R. L. (1983) Biochemistry 22, 3884–3896 [DOI] [PubMed] [Google Scholar]
- 31.Lumry R. (1995) Methods Enzymol. 259, 628–720 [DOI] [PubMed] [Google Scholar]
- 32.Pineda A. O., Carrell C. J., Bush L. A., Prasad S., Caccia S., Chen Z. W., Mathews F. S., Di Cera E. (2004) J. Biol. Chem. 279, 31842–31853 [DOI] [PubMed] [Google Scholar]
- 33.Wells C. M., Di Cera E. (1992) Biochemistry 31, 11721–11730 [DOI] [PubMed] [Google Scholar]
- 34.Pineda A. O., Chen Z. W., Caccia S., Cantwell A. M., Savvides S. N., Waksman G., Mathews F. S., Di Cera E. (2004) J. Biol. Chem. 279, 39824–39828 [DOI] [PubMed] [Google Scholar]
- 35.Carter W. J., Myles T., Gibbs C. S., Leung L. L., Huntington J. A. (2004) J. Biol. Chem. 279, 26387–26394 [DOI] [PubMed] [Google Scholar]
- 36.Pineda A. O., Chen Z. W., Bah A., Garvey L. C., Mathews F. S., Di Cera E. (2006) J. Biol. Chem. 281, 32922–32928 [DOI] [PubMed] [Google Scholar]
- 37.Kroh H. K., Tans G., Nicolaes G. A., Rosing J., Bock P. E. (2007) J. Biol. Chem. 282, 16095–16104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vindigni A., White C. E., Komives E. A., Di Cera E. (1997) Biochemistry 36, 6674–6681 [DOI] [PubMed] [Google Scholar]
- 39.Verhamme I. M., Olson S. T., Tollefsen D. M., Bock P. E. (2002) J. Biol. Chem. 277, 6788–6798 [DOI] [PubMed] [Google Scholar]
- 40.Petrera N. S., Stafford A. R., Leslie B. A., Kretz C. A., Fredenburgh J. C., Weitz J. I. (2009) J. Biol. Chem. 284, 25620–25629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams T. E., Li W., Huntington J. A. (2009) J. Thromb. Haemost. 7, 1688–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bush-Pelc L. A., Marino F., Chen Z., Pineda A. O., Mathews F. S., Di Cera E. (2007) J. Biol. Chem. 282, 27165–27170 [DOI] [PubMed] [Google Scholar]
- 43.Page M. J., Di Cera E. (2006) Physiol. Rev. 86, 1049–1092 [DOI] [PubMed] [Google Scholar]