Abstract
The ErbB2 and ErbB3 receptor tyrosine kinases act synergistically to promote cellular properties associated with tumor development. Previous studies indicate that endogenous ErbB3 protein is markedly elevated in mouse mammary tumors induced by transgenic ErbB2 overexpression. However, this occurs in the absence of elevated ErbB3 transcript, indicating that post-transcriptional regulatory mechanisms play crucial roles in suppressing ErbB3 protein in normal tissue. Our previous studies also demonstrate that protein levels of Nrdp1, an E3 ubiquitin ligase that targets ErbB3 for degradation, are markedly suppressed in tumors from ErbB2 transgenic animals relative to normal tissue. Here we demonstrate that transgenic expression of Nrdp1 cDNA in the mouse mammary gland is not sufficient to suppress elevated ErbB3 levels or tumor initiation and growth in ErbB2 transgenic mice. Unexpectedly, Nrdp1 protein is absent in tumors from Nrdp1/ErbB2 bigenic mice, and real time PCR analysis indicates that Nrdp1 protein levels are suppressed post-transcriptionally. Nrdp1 protein is more resistant to proteasome-dependent degradation when exogenously expressed in cultured MCF10A nontransformed human breast epithelial cells than in breast tumor cells. These observations indicate that mammary tumors use potent post-transcriptional mechanisms to suppress Nrdp1 protein levels and that protein destabilization may play a central role in Nrdp1 loss in tumors.
Keywords: Mammary Gland, Protein Degradation, Receptor Tyrosine Kinase, Transgenic, Ubiquitin Ligase
Introduction
Overexpression and aberrant activation of members of the ErbB family of receptor tyrosine kinases are thought to contribute to the development and progression of a variety of tumor types (1). Notably, amplification of the erbB2 gene is observed in 25–30% of breast cancer patients, and overexpression of the ErbB2 protein correlates with earlier relapse and poor prognosis (2, 3). Numerous studies with cultured cells suggest that overexpression of the ErbB2 protein is sufficient to activate its protein-tyrosine kinase activity, which is necessary for ErbB2-induced cellular transformation. ErbB2 overexpression in the mouse mammary gland is sufficient to induce the formation of metastatic tumors (4), underscoring the functional importance of ErbB2 in tumor development. These observations have pointed to a central role for ErbB2 in breast tumor initiation and progression, and ErbB2-directed antibody and small molecule inhibitors are currently employed clinically in the treatment of breast cancer (5, 6).
Members of the ErbB family take part in a complex array of combinatorial interactions through the formation homo- and heterodimers between the different family members, which in turn activate distinct signaling pathways (7–10). It has been suggested that signaling by the ErbB2-ErbB3 heterodimer is the most potent of the 10 ErbB receptor dimeric complexes (11), and the ErbB2-ErbB3 complex is a proposed oncogenic unit (12). Recent evidence supports a central role for ErbB3 in ErbB2-amplified breast cancer (13), and several reports point to roles for ErbB3 activation in mediating resistance to cancer therapeutics (14, 15). ErbB3 overexpression has been reported in up to 63% of human breast tumors (16) and is positively associated with lymph node metastases, histological grade, recurrence, and worsened prognosis (17–21). Together these observations strongly suggest that ErbB3 function is an important aspect of ErbB2-mediated breast cancer, prompting the need for a more thorough understanding of ErbB3 expression and dysregulation in breast cancer.
Transgenic mouse models have demonstrated that ErbB2 overexpression in the mammary gland using the MMTV4 promoter/enhancer is sufficient to induce to metastatic cancer (4, 22). The prolonged latency of tumor development in these animals relative to mammary tumors driven by more potent oncogenes suggests that other rate-limiting processes also contribute to tumor onset. For example, it has been demonstrated that the acquisition of deletions in ErbB2 that facilitate its homodimerization through extracellular disulfide bonding is required for ErbB2-induced tumorigenesis (22, 23). However, transgenic expression of these mutants in the mammary gland still exhibit prolonged latencies (24), suggesting the involvement of other processes.
Interestingly, the vast majority of tumors from transgenic animals that overexpress ErbB2 also overexpress endogenous ErbB3 protein by well over 10-fold relative to normal tissue (16, 24), again pointing to a central role for ErbB3 in ErbB2-driven breast tumors. Remarkably, elevated ErbB3 protein is observed in tumors, even though transcript levels are similar to uninvolved (normal) tissue from the same animals (16, 24, 25). These observations indicate that normal mammary tissue utilizes potent post-transcriptional mechanisms to keep ErbB3 protein levels in check and that tumors disrupt these mechanisms to create a permissive environment for receptor overexpression (26, 27).
Nrdp1 is a RING finger ubiquitin ligase that has been demonstrated to mediate the ubiquitination and degradation of ErbB3 to regulate steady-state (ligand independent) receptor levels (28, 29). Overexpression of Nrdp1 in cultured cells suppresses ErbB3 levels, whereas overexpression of a dominant-negative form or knockdown of endogenous Nrdp1 elevates ErbB3 (16). Interestingly, Nrdp1 protein is consistently lost in mammary tumors from ErbB2-overexpressing transgenic mice, and we have observed a strong inverse correlation in the levels of Nrdp1 and ErbB3 proteins in breast cancer patient primary tumors (16). These observations raise the possibility that the suppression of Nrdp1 protein by breast tumors contributes to malignancy by allowing ErbB3 overexpression.
In this study we set out to test the prediction that restoration of Nrdp1 to mouse mammary tumors suppresses ErbB3 levels, increases tumor latency, and lowers tumor burden in transgenic mice with ErbB2-induced mammary tumors. Unexpectedly, we find that transgenic expression is not a viable approach to the restoration of Nrdp1 to ErbB2 overexpression-induced mouse mammary tumors because tumors employ potent post-transcriptional mechanisms to suppress both endogenous and transgene-derived Nrdp1 protein. Our observations suggest that tumors dysregulate mechanisms involved in maintaining Nrdp1 protein stability.
EXPERIMENTAL PROCEDURES
Transgenic Mice
All of the mouse procedures were approved by the Institutional Animal Care and Use Committee of the University of California, Davis. The cDNA encoding human Nrdp1 was subcloned into the HindIII and EcoRI restriction sites of MMTV-LTR vector, and immunofluorescence microscopy of transiently transfected MMTV-Nrdp1 or MMTV-GFP plasmids was used to confirm MMTV-LTR-driven expression in dexamethasone-treated cells as previously described (30, 31). ScaI-linearized plasmid was injected into fertilized FvB mouse eggs, which were transplanted into pseudopregnant females. Potential founder animals were screened by PCR analysis of tail DNA using primers GCTGTCCTGCTTCTATTGT (forward) and GCAGTAGCCTCATCATCAC (reverse), and founders were confirmed by Southern blotting. Three independent transgenic lines were established by mating founder animals with wild type mice. Both FvB MMTV-Nrdp1 and MMTV-NDL mice overexpressing the rat ErbB2 transgene (NDL2–5; 24) were bred and maintained at the animal facilities at the University of California, Davis. For aging studies, the mice were palpated to identify mammary tumors every other week from 15 to 19 weeks of age and then weekly from 20 to 60 weeks of age. Following tumor development, mammary fat pads were collected for whole mount and immunohistochemical analysis. In addition, dissected tumor and adjacent normal fat pad tissues were collected following tumor development in these mice and snap frozen in liquid nitrogen for RNA and protein analysis.
Cell Culture Experiments
MCF10A, BT474, and MDA-MB-231 cells were obtained from ATCC and grown in the recommended media. The derivation and growth of the NDL cell line were described previously (25). For proteasome inhibitor experiments, cells stably transduced with pMX-pie or pMX-pie-Nrdp1 retroviruses (16) were treated for 6 h without and with 10 μm MG132. Whole cell lysates were resolved by SDS-PAGE and immunoblotted as described below. For half-life experiments, transduced cells were treated with 50 μm cycloheximide for various times, and whole cell lysates were immunoblotted with antibodies to FLAG and actin.
Western Blot Analysis
Mammary fat pads from transgenic mice were homogenized as described (25). 20 μg of total protein was then collected in sample buffer (62.5 mm Tris-HCl, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, 0.05% bromphenol blue), boiled for 5 min at 95 °C, and resolved by 10% SDS-PAGE. Following transfer to nitrocellulose (Pall Life Science), the proteins were immunoblotted with various primary antibodies, detected using horseradish peroxidase-conjugated secondary antibodies (Invitrogen), and developed using SuperSignal West chemicals (Pierce). An Alpha Innotech imaging station with FluorChem FC2 software was used to capture and quantify immunoblotting images. The antibodies used in these studies include: FLRF/Nrdp1 (Bethyl, Inc.), affinity-purified rabbit Nrdp1 previously described (28), ErbB3 C-17 (Santa Cruz Biotechnology), GAPDH (Sigma), FLAG (Sigma), and actin AC15 (Sigma).
Immunofluorescence Analysis
MCF10A and MDA-MB-231 cells on coverslips were transiently transduced with pMXpie-Nrdp1 retroviral particles (16), resulting in nearly 100% transduction efficiency as assessed by viral-mediated GFP expression. Three days following transduction, the cells were fixed and processed for immunofluorescence as described previously (30, 31) using anti-FLAG antibodies and DAPI nuclear stain. For direct comparisons among cell lines, camera shutter times and imaging parameters were kept constant for all samples.
Real Time PCR
Tail genomic DNA was collected by NaOH extraction. Extraction of RNA from mouse tissues, cDNA synthesis, and real time PCR analysis were all carried out as previously described (25). Exogenous and endogenous Nrdp1 transcript in mouse tissues was quantified using forward primer TGAACCGACGCTACTATGAGAACT, reverse primer CTGGTTCTCACAGGCCATCAC, and probe 6FAM-TGGCCAAGCGCATCCCTGG-MGBNFQ. Mouse and human ErbB3 and actin real time primers and probes were purchased from ABI. Nrdp1 and ErbB3 transcript content in tissue samples were normalized to actin. T cell receptor delta gene reference was quantified using forward primer CAGACTGGTTATCTGCAAAGCAA, reverse primer TCTATGCCAGTTCCAAAAAACATC, and probe 6FAM-ATTATAACGTGCTCCTGGGACACCC-MGBNFQ.
Immunohistochemistry and Mammary Whole Mounts
All of the tissues were fixed in 10% formalin and processed by the Pathology Laboratory of the University of California, Davis. Four-micrometer-thick paraffin sections were stained with Mayer's hematoxilin and eosin. Images of slides were captured using 4×, 20×, and 40× objectives on a Zeiss Axioskop microscope with an Axiocam camera and Axiovision 3.1 software. Mammary whole mounts were prepared as previously described (32), and the images were collected using an Epson scanner and processed using Adobe Photoshop 7.0 software.
RESULTS
Construction of MMTV-Nrdp1 Transgenic Mice
To test the hypothesis that Nrdp1 restoration to ErbB2-induced mouse mammary tumors will suppress tumor onset by lowering ErbB3 protein levels, we sought to examine tumor development in ErbB2/Nrdp1 bigenic mice. We first created a vector to express human Nrdp1 cDNA in the mouse mammary gland using the MMTV-LTR promoter/enhancer (Fig. 1A) and demonstrated that this construct could drive Nrdp1 expression in cultured mammary epithelial cells (not shown). Three founder FvB strain transgenic mouse lines (called lines 12, 15, and 19) were then generated using a linearized MMTV-Nrdp1 plasmid.
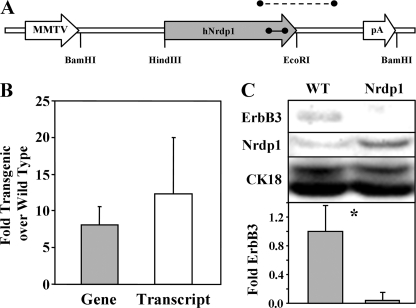
FIGURE 1.
MMTV-Nrdp1 expression in transgenic mice. A, the MMTV-Nrdp1 vector employed to generate transgenic mice is illustrated, depicting the segment amplified for transgene screening (dashed line) and the segment amplified in real time PCR experiments (solid line). B, genomic DNA from tails and RNA from mammary glands were collected from four to six wild type or line 19 MMTV-Nrdp1 nulliparous female mice and used in real time PCR analysis. The ratio of signal corresponding to Nrdp1 gene or transcript in transgenic relative to wild type animals is plotted. C, lysates from mammary gland tissue derived from WT and MMTV-Nrdp1 transgenic mice were blotted with antibodies to ErbB3, Nrdp1, or cytokeratin-18. Blots from four independent determinations were quantified, and relative ErbB3 levels (normalized to CK18) were plotted. *, p < 0.05.
To estimate gene copy number and transcript levels, PCR primers that amplify an 82-bp region of human Nrdp1 exon 7 that is 100% identical to mouse were employed in real time PCR strategies. Real time PCR analysis of genomic DNA from nulliparous female mice revealed that founder lines 12, 15, and 19 contained ∼6, 2, and 14 copies of the Nrdp1 transgene, respectively, using the chromosome 14 T cell receptor delta gene as a reference. Real time analysis of genomic and mammary gland cDNA from several wild type and line 19-derived animals revealed that the transgenic mice contained on average 8-fold higher Nrdp1-containing genomic DNA than wild type animals, and ∼12-fold higher message levels (Fig. 1B). The lack of suitable antibodies precluded immunohistochemical analysis of ErbB3 and Nrdp1 in a mouse mammary gland. However, immunoblotting lysates of mammary glands taken from nulliparous adult wild type and MMTV-Nrdp1 female mice revealed that Nrdp1 protein levels were elevated severalfold in transgenic animals (Fig. 1C). This relatively modest elevation in Nrdp1 protein in transgenic versus wild type animals might suggest that normal mammary tissue establishes an upper limit to Nrdp1 protein expression that cannot be exceeded, even with significantly higher transcript levels. Although ErbB3 protein was close to the limit of detection by immunoblotting, it appeared that its levels were suppressed in the mammary glands of MMTV-Nrdp1 mice, consistent with the role of Nrdp1 in promoting ErbB3 degradation (16, 28, 29).
The impact of Nrdp1 overexpression on mammary gland development and morphology in transgenic mice appeared modest. Using whole mount analysis we observed that mammary gland development during puberty was delayed in some MMTV-Nrdp1 animals but eventually caught up to development observed in wild type mice (not shown). No deficiency in mammary development during pregnancy was noted, and MMTV-Nrdp1 dams were able to nurse their pups normally. Inspection of nulliparous MMTV-Nrdp1 transgenic female mice at very advanced ages of 52–74 weeks revealed a lobuloalveolar hyperplasic phenotype, sometimes coupled with a high degree of vacuolation and casein staining, in three of six animals examined. This phenotype was observed in only one of six wild type mice. However, the characteristics of the hyperplastic lesions were very similar to those described previously for elder FvB strain mice (33), where hyperplastic growths in the mammary gland were suggested to be secondary to spontaneously arising prolactin-secreting lesions of the pituitary gland independent of transgene expression.
Transgenic Nrdp1 Overexpression Does Not Alter ErbB2-induced Tumors
To assess the effect of Nrdp1 restoration on ErbB2-driven mammary tumors, we crossed our MMTV-Nrdp1 line 19 mice with the NDL 2–5 line of ErbB2 transgenic mice (24). We commonly refer to the resulting bigenic line as NxN. As expected, normal mammary tissue from NxN animals exhibited ∼12-fold elevated levels of Nrdp1 transcript relative to NDL mice (Fig. 2A), but normal tissue from both strains contained similar levels of ErbB3 transcript (Fig. 2B) as assessed by real time RT-PCR. These observations indicate that the Nrdp1 transgene is efficiently transcribed in bigenic mice.
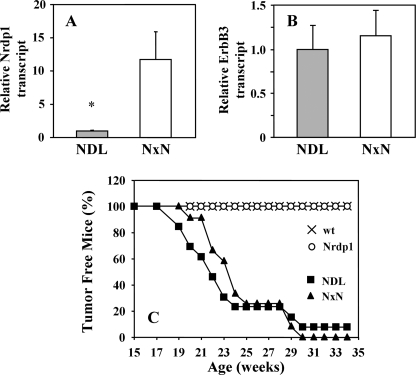
FIGURE 2.
Nrdp1 overexpression does not significantly alter tumor onset in MMTV-NDL mice. Real time RT-PCR was used to measure the relative transcript levels of Nrdp1 (A) and ErbB3 (B) in normal mammary tissue of 4–6 each MMTV-NDL and MMTV-Nrdp1 × MMTV-NDL (NxN) mice. The data are plotted as the fold transcript level of each gene in bigenic animals relative to MMTV-NDL mice. C, 10–12 each wild type, NDL, Nrdp1, and NxN mice were aged, and the time to the first palpable tumor was recorded. The percentage of tumor-free mice is plotted as a function of age. *, p < 0.05.
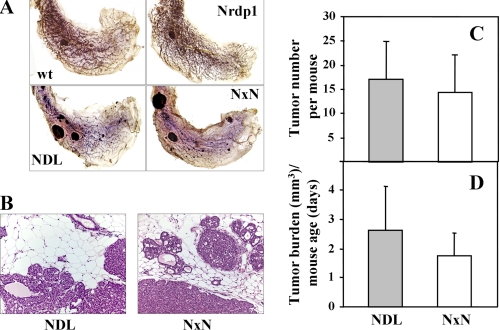
Surprisingly, we found that the latency of tumor formation in bigenic mice was statistically indistinguishable from the latency in NDL animals (Fig. 2C). None of the wild type or Nrdp1 transgenic mice employed in the study developed tumors over 35 weeks, whereas both NDL and NxN mice exhibited average tumor latencies of 21–23 weeks. A similar latency was observed when MMTV-Nrdp1 transgenic line 12 was crossed into the NDL tumor model (data not shown). Tumors derived from NDL and NxN strains were histologically similar (Fig. 3, A and B). The average numbers of tumors present throughout the mammary glands (Fig. 3C) as well as the overall tumor burden at the time of sacrifice (Fig. 3D) were also indistinguishable between the two groups. These observations indicate that reintroduction of the Nrdp1 transcript under an exogenous promoter has little discernable impact on ErbB2-induced tumor development.
FIGURE 3.
Nrdp1 overexpression does not significantly alter tumor phenotype in MMTV-NDL mice. A, mammary gland whole mounts from wild type, NDL, Nrdp1, and NxN mice were stained with hematoxalin/eosin. B, tumor sections from NDL and NxN mice were stained with hematoxilin and eosin. The images from A and B are representative of tissue obtained from at least seven animals of the indicated genotypes. C, the number of mammary gland tumors was determined for each mouse at sacrifice, and the averages were plotted for NDL and NxN mice. The error bars represent standard errors of seven to nine mice. D, the total volume of all mammary tumors for each mouse was determined at sacrifice, and that number was divided by the age at sacrifice to control for tumor growth with time. The averages of this ratio are plotted for NDL transgenics and NxN bigenics.
Transgene-derived Nrdp1 Protein Is Post-transcriptionally Suppressed during the Normal-to-Tumor Transition
The observation that Nrdp1 overexpression in the mammary gland does not impact tumor formation and growth contrasts with in vitro results, where stable overexpression suppressed the proliferation and survival of cultured breast cancer cells (16). One possible explanation is that, in contrast to cultured cells, overexpressed Nrdp1 may not efficiently remove ErbB3 from the tumors of bigenic mice. To test this possibility, we immunoblotted normal and tumor tissue lysates from NDL and NxN mice with antibodies to ErbB3. Consistent with previous reports (16, 24), we observed that ErbB3 protein levels are 10–100-fold higher in tumors from NDL mice than in normal tissue (Fig. 4A, top panel), from virtually undetectable signal in normal tissue to a very robust band in tumors. This difference in protein levels did not result from elevated ErbB3 transcript in tumors because real time RT-PCR analysis indicated a difference of less than 2-fold (Fig. 4B). Interestingly, very similar differences were observed in ErbB3 protein and message levels in normal and tumor tissue from NxN animals. These observations indicate that, in contrast with in vitro studies, overexpression of Nrdp1 cDNA in the mammary gland has little impact on ErbB3 protein levels in tumors. Thus, the inability of overexpressed Nrdp1 to suppress ErbB3 protein levels in mouse tumors may underlie its inability to impact tumor development.
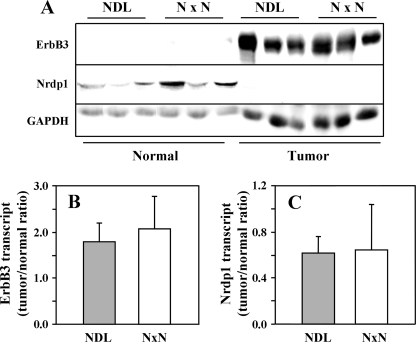
FIGURE 4.
Nrdp1 protein is post-transcriptionally suppressed by mouse mammary tumors. A, lysates of normal or tumor mammary tissue from three each NDL transgenic or NxN bigenic mice were blotted with antibodies to ErbB3, Nrdp1, or GAPDH control. B and C, transcript levels of ErbB3 (B) and Nrdp1 (C) were determined by real time PCR analysis of cDNA from mammary glands of NDL and NxN mice and plotted as the tumor to normal ratio. The data in each panel are representative of two or three independent determinations.
The discrepancy between the in vitro and in vivo results raises the question as to why transgenically overexpressed Nrdp1 does not function to suppress ErbB3 in tumors. One possibility is that despite the elevated transcript, transgene-derived Nrdp1 protein in tumors is nonfunctional. To address this possibility, we compared Nrdp1 protein and transcript expression levels in normal and tumor tissue derived from NDL and NxN mice. Consistent with the comparison of MMTV-Nrdp1 and wild type animals (Fig. 1C), Nrdp1 protein levels in normal mammary tissue were on average severalfold higher in NxN mice than in NDL mice (Fig. 4A, middle panel). More importantly, Nrdp1 protein was completely absent in tumors from both strains, even though Nrdp1 transcript levels were only suppressed by ∼35% during the transition from normal mammary tissue to tumor (Fig. 4C). These observations indicate that endogenous Nrdp1 protein is efficiently eliminated during the normal-to-tumor transition, raising the possibility that the suppression of Nrdp1 protein levels may be an important aspect of mammary ErbB2-mediated tumor initiation or growth. Critically, overexpression of Nrdp1 cDNA by better than 10-fold via an exogenous promoter cannot overcome Nrdp1 suppression by tumors because protein levels are potently suppressed via post-transcriptional mechanisms.
Nrdp1 Protein Is More Labile in Breast Tumor Cells than in Nontransformed Cells
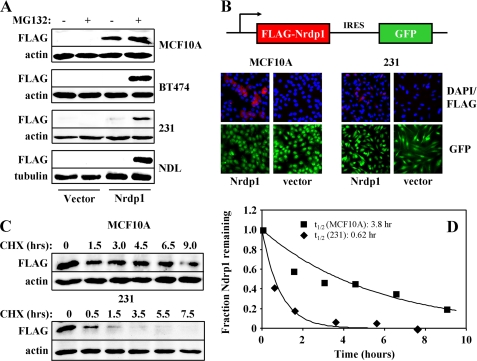
To test the hypothesis that Nrdp1 protein is inherently more labile in tumor cells than in nontransformed cells, we developed an in vitro system for comparing the stability of exogenously expressed Nrdp1 in various cell lines. Immunoblotting of lysates from transiently transduced cells revealed that exogenous FLAG-tagged Nrdp1 protein expression is readily detected in non-transformed MCF10A breast epithelial cells, and its levels are only modestly increased (2–3-fold) upon treatment of cells with the proteasome inhibitor MG132 (Fig. 5A). However, Nrdp1 protein is not detected in ErbB2/3-positive BT474 breast tumor cells unless treated with MG132, where very robust expression is observed. Likewise, cells cultured from NDL tumors (25) are resistant to Nrdp1 expression, suggesting that tumors arise from a population of cells that have lost the ability to maintain the Nrdp1 protein levels characteristic of normal tissue. MDA-MB-231 breast tumor cells supported a low level of Nrdp1 expression, which was markedly increased upon MG132 treatment (Fig. 5A). These observations indicate that breast tumor cell lines efficiently dispose of Nrdp1 via a proteasome-dependent mechanism.
FIGURE 5.
Nrdp1 protein is more labile in human breast tumor cells than in nontransformed breast epithelial cells. A, MCF10A, BT474, NDL, or MDA-MB-231 cells were transduced with control virus or virus expressing FLAG-tagged Nrdp1 and treated for 6 h without or with 10 μm MG132, and lysates were immunoblotted with antibodies to FLAG and either actin or tubulin. B, MCF10A and MDA-MB-231 cells were transiently transduced with pMX-pie retrovirus encoding either nothing (vector) or FLAG-Nrdp1 (Nrdp1) followed by an IRES sequence and GFP cDNA (16). 72 h following transduction, the cells were fixed and imaged for GFP expression (green), FLAG-Nrdp1 expression (red), and nuclei (blue). C, MCF10A or MDA-MB-231 cells expressing FLAG-Nrdp1 were treated with 50 μm cycloheximide (CHX) for the indicated times, and proteins in lysates were immunoblotted with antibodies to FLAG or actin. D, blots from C were quantified, and the fraction of remaining Nrdp1 in MCF10A cells (squares) and MDA-MB-231 cells (diamonds) was plotted against the time of cycloheximide treatment. FLAG-Nrdp1 half-lives were estimated by fitting the data to a single exponential. The data from each panel are representative of two to five independent experiments.
To examine the nature of Nrdp1 suppression by tumor cells, we compared Nrdp1 staining in MCF10A and MDA-MB-231 cells by immunofluorescence. In these experiments, the cells were transiently transduced with retrovirus encoding either nothing or FLAG-Nrdp1 immediately preceding an IRES-GFP cassette and examined for the presence of GFP, FLAG epitope, and nuclei. Comparison of GFP expression with DAPI nuclear staining revealed that transduction efficiency was essentially 100% for all conditions, verifying the presence of GFP- and Nrdp1-encoding message in all cells. However, only about half of the GFP-positive MCF10A cells exhibited detectable staining for FLAG-Nrdp1 protein (Fig. 5B), suggesting that post-transcriptional mechanisms suppressed Nrdp1 protein production in some cells. More importantly, less than 10% of the MDA-MB-231 cells were capable of supporting detectable FLAG-Nrdp1 protein, and Nrdp1 protein levels were significantly lower in these cells than in the MCF10As. These observations indicate that only a subpopulation of cells within a given culture is capable of supporting the efficient expression of Nrdp1 protein and that this subpopulation is far less prevalent in tumor cells than in nontransformed cells.
To directly compare Nrdp1 lability in nontransformed and tumor cells, we compared its half-life when expressed in MCF10A and MDA-MB-231 cells by monitoring levels of exogenously expressed protein after treatment with cycloheximide (Fig. 5, C and D). We found that the half-life of FLAG-Nrdp1 is at least 6-fold longer in the nontransformed line than in the tumor line. Taken together, our observations strongly suggest that enhanced protein degradation contributes to the post-transcriptional loss of Nrdp1 protein observed during the breast normal-to-tumor transition.
DISCUSSION
Although overexpression of ErbB2 protein in patient breast tumors very strongly correlates with gene amplification, the mechanisms by which ErbB3 protein is overexpressed have not been elucidated. Amplification of the erbB3 gene has been reported, but PCR and FISH results suggest that this is not as prevalent as erbB2 amplification (34, 35) and does not account for the frequency of elevated ErbB3 protein observed in tumors (17). Tumors from transgenic mice overexpressing ErbB2 in the mammary gland exhibit elevated and tyrosine-phosphorylated endogenous ErbB3 (24), pointing to a selective advantage for the co-expression and activation of this receptor in ErbB2-overexpressing tumors. Interestingly, we (16, 25) and others (24) have observed that despite a 10–100-fold elevation in ErbB3 protein levels during the normal-to-tumor transition, ErbB3 message levels remain virtually constant. These observations indicate that very potent post-transcriptional regulatory mechanisms suppress endogenous ErbB3 protein levels in normal mammary tissue. Such mechanisms may be important in tightly regulating receptor levels in cells, ensuring sufficient signaling for tissue development and maintenance but preventing oversignaling leading to aberrant development and neoplasia. Importantly, it appears that tumors inactivate these post-transcriptional mechanisms to augment receptor signaling and promote their growth and malignancy (27).
Because Nrdp1 regulates ErbB3 protein levels in cultured breast tumor cells by targeting receptors for degradation, we postulated that restoration of Nrdp1 to ErbB2-induced mouse mammary tumors would suppress tumor onset or progression by suppressing ErbB3 levels. Our hope was that the creation of the MMTV-Nrdp1 mouse would allow us to test this possibility. We initially found that Nrdp1 overexpression in the mammary gland suppresses ErbB3 protein levels in adult mice and elicits a minor delay in mammary gland development during puberty. It has previously been observed that ErbB3 deletion partially disrupts mammary gland morphogenesis (36), pointing to a role for this receptor in mammary development. Although the disruption of mammary development in MMTV-Nrdp1 mice was not nearly as striking, it is possible that Nrdp1 protein levels are post-transcriptionally suppressed during mammary gland development to permit ErbB3 function as they are in tumors. The necessity of Nrdp1 in mammary gland development and ErbB2-induced mammary tumor formation will be the focus of future studies. A hyperplastic phenotype observed in a subset of older wild type and transgenic mice was consistent with lesions associated with lactogenic hormone-producing pituitary tumors common to FvB strain animals independent of transgene (33).
Crossing MMTV-Nrdp1 mice into the ErbB2 overexpression model did not affect the latency or extent of mammary tumor formation nor the expression of ErbB3 protein. These results were initially surprising because there is a strong inverse relationship between Nrdp1 and ErbB3 protein levels when the analogous experiment is carried out with cultured cells (16, 28). Comparison of Nrdp1 transcript and protein levels in normal and tumor tissue revealed that transcript was significantly elevated in bigenic mice, as expected, but protein was undetectable. These observations indicate that like ErbB3, Nrdp1 is post-transcriptionally regulated by tumors. These post-transcriptional regulatory mechanisms must be sufficiently powerful to rid tumors of Nrdp1 protein despite a 12-fold elevation in its message.
Although other mechanisms such as translation and microRNA-mediated suppression may play a role in the post-transcriptional loss of Nrdp1, our observations point to a key role for protein degradation. We have previously observed that the Nrdp1 protein can be markedly unstable when transiently expressed in selected cell lines, and mutation of the RING finger domain significantly elevates its stability (37). These observations suggest that Nrdp1 autoubiquitination can promote is own degradation, in turn raising the possibility that tumor cells inactivate mechanisms that thwart the efficiency of Nrdp1 degradation. The resulting loss of Nrdp1 allows ErbB3 accumulation, which then contributes to tumor initiation and progression. In support of this hypothesis, we observed that Nrdp1 protein is more stable in nontransformed MCF10A breast epithelial cells than in several breast cancer cell lines and that proteasome-dependent pathways mediate the efficient degradation of Nrdp1 in tumor cells. Immunofluorescence studies revealed that differences in Nrdp1 expression between cell lines were related to both the percentage of cells capable of supporting Nrdp1 expression and the amount of Nrdp1 protein produced in each cell. Our previous ability to stably express Nrdp1 in breast tumor cells (16) was likely due to the clonal selection of expressing subpopulations.
It should be noted that Nrdp1 is likely not the only protein dysregulated in either mouse or patient mammary tumors to augment ErbB2/ErbB3 signaling (25, 27, 38). Numerous proteins have been described that contribute to the degradation of ErbB receptors by either mediating their trafficking to lysosomes or suppressing the signaling activity of receptors. The loss or suppression of many of these has been reported in various tumors. For example, we have observed that like Nrdp1, the ErbB negative regulatory protein LRIG1 is also suppressed during the mammary normal-to-tumor transition in NDL mice (25). Thus, it is possible that tumors effectively suppress or inactivate combinations of genes encoding ErbB negative regulators to elevate ErbB receptor levels and/or augment receptor signaling activity. Future studies will be aimed at characterizing combinatorial contributions of ErbB negative regulatory mechanisms that are suppressed by mammary tumors.
More broadly, our observations underscore the importance of post-transcriptional mechanisms in regulating key events in breast tumor progression. Both ErbB3 and Nrdp1 are dysregulated post-transcriptionally in tumors, and it is known that other oncogenic and tumor suppressor proteins are similarly targeted by tumor cells at the level of protein synthesis or stability. Because such events are overlooked by gene expression screens, strategies for accurately comparing the levels of a given oncogenic or anti-oncogenic protein in normal and tumor tissue are essential to provide a complete picture of oncogenic signaling.
This work was supported, in whole or in part, by National Institutes of Health Grants CA123541 and GM068994 (to K. L. C.), CA118384 (to C. S.), and T32 RR07038 (to H. C. W.).
- MMTV
- murine mammary tumor.
REFERENCES
- 1.Holbro T., Civenni G., Hynes N. E. (2003) Exp. Cell Res. 284, 99–110 [DOI] [PubMed] [Google Scholar]
- 2.Slamon D. J., Clark G. M., Wong S. G., Levin W. J., Ullrich A., McGuire W. L. (1987) Science 235, 177–182 [DOI] [PubMed] [Google Scholar]
- 3.Slamon D. J., Godolphin W., Jones L. A., Holt J. A., Wong S. G., Keith D. E., Levin W. J., Stuart S. G., Udove J., Ullrich A., et al. (1989) Science 244, 707–712 [DOI] [PubMed] [Google Scholar]
- 4.Guy C. T., Webster M. A., Schaller M., Parsons T. J., Cardiff R. D., Muller W. J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10578–10582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pal S. K., Pegram M. (2007) Rev. Endocr. Metab. Disord. 8, 269–277 [DOI] [PubMed] [Google Scholar]
- 6.Di Cosimo S., Baselga J. (2008) Eur. J. Cancer 44, 2781–2790 [DOI] [PubMed] [Google Scholar]
- 7.Carraway K. L., 3rd, Cantley L. C. (1994) Cell 78, 5–8 [DOI] [PubMed] [Google Scholar]
- 8.Alroy I., Yarden Y. (1997) FEBS Lett. 410, 83–86 [DOI] [PubMed] [Google Scholar]
- 9.Riese D. J., 2nd, Stern D. F. (1998) Bioessays 20, 41–48 [DOI] [PubMed] [Google Scholar]
- 10.Yarden Y., Sliwkowski M. X. (2001) Nat. Rev. Mol. Cell Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 11.Citri A., Skaria K. B., Yarden Y. (2003) Exp. Cell Res. 284, 54–65 [DOI] [PubMed] [Google Scholar]
- 12.Holbro T., Beerli R. R., Maurer F., Koziczak M., Barbas C. F., 3rd, Hynes N. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee-Hoeflich S. T., Crocker L., Yao E., Pham T., Munroe X., Hoeflich K. P., Sliwkowski M. X., Stern H. M. (2008) Cancer Res. 68, 5878–5887 [DOI] [PubMed] [Google Scholar]
- 14.Stern D. F. (2008) J. Mammary Gland Biol. Neoplasia 13, 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell M. R., Amin D., Moasser M. M. (2010) Clin. Cancer Res. 16, 1373–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yen L., Cao Z., Wu X., Ingalla E. R., Baron C., Young L. J., Gregg J. P., Cardiff R. D., Borowsky A. D., Sweeney C., Carraway K. L., 3rd (2006) Cancer Res. 66, 11279–11286 [DOI] [PubMed] [Google Scholar]
- 17.Lemoine N. R., Barnes D. M., Hollywood D. P., Hughes C. M., Smith P., Dublin E., Prigent S. A., Gullick W. J., Hurst H. C. (1992) Br. J. Cancer 66, 1116–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Travis A., Pinder S. E., Robertson J. F., Bell J. A., Wencyk P., Gullick W. J., Nicholson R. I., Poller D. N., Blamey R. W., Elston C. W., Ellis I. O. (1996) Br. J. Cancer 74, 229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naidu R., Yadav M., Nair S., Kutty M. K. (1998) Br. J. Cancer 78, 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witton C. J., Reeves J. R., Going J. J., Cooke T. G., Bartlett J. M. (2003) J. Pathol. 200, 290–297 [DOI] [PubMed] [Google Scholar]
- 21.Hamburger A. W. (2008) J. Mammary Gland Biol. Neoplasia 13, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel P. M., Dankort D. L., Hardy W. R., Muller W. J. (1994) Mol. Cell Biol. 14, 7068–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen K., Angelini P. D., Laos S., Bach-Faig A., Cunningham M. P., Ferrer-Ramón C., Luque-García A., García-Castillo J., Parra-Palau J. L., Scaltriti M., Ramón y Cajal S., Baselga J., Arribas J. (2009) Mol. Cell Biol. 29, 3319–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegel P. M., Ryan E. D., Cardiff R. D., Muller W. J. (1999) EMBO J. 18, 2149–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J. K., Shattuck D. L., Ingalla E. Q., Yen L., Borowsky A. D., Young L. J., Cardiff R. D., Carraway K. L., 3rd, Sweeney C. (2008) Cancer Res. 68, 8286–8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carraway K. L., 3rd, Yen Y., Ingalla E., Sweeney C. (2008) in Cancer Drug Discovery and Development: EGFR Signaling Networks in Cancer Therapy (Haley J. D., Gullick W. J. eds) pp. 169–186, Humana Press, Totowa, NJ [Google Scholar]
- 27.Fry W. H., Kotelawala L., Sweeney C., Carraway K. L., 3rd (2009) Exp. Cell Res. 315, 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamonti A. J., Guy P. M., Ivanof C., Wong K., Sweeney C., Carraway K. L., 3rd (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2866–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu X. B., Goldberg A. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funes M., Miller J. K., Lai C., Carraway K. L., 3rd, Sweeney C. (2006) J. Biol. Chem. 281, 19310–19319 [DOI] [PubMed] [Google Scholar]
- 31.Cao Z., Wu X., Yen L., Sweeney C., Carraway K. L., 3rd (2007) Mol. Cell Biol. 27, 2180–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen S. B., Young L. J., Smith G. H. (2000) in Methods in Mammary Gland Biology and Breast Cancer Research (Ip M. M., Asch B. eds) pp. 75–85, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 33.Wakefield L. M., Thordarson G., Nieto A. I., Shyamala G., Galvez J. J., Anver M. R., Cardiff R. D. (2003) Comp. Med. 53, 424–432 [PubMed] [Google Scholar]
- 34.Zaczek A., Welnicka-Jaśkiewicz M., Bielawski K. P., Jaśkiewicz J., Badzio A., Olszewski W., Rhone P., Jassem J. (2008) J. Cancer Res. Clin. Oncol. 134, 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sassen A., Rochon J., Wild P., Hartmann A., Hofstaedter F., Schwarz S., Brockhoff G. (2008) Breast Cancer Res. 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson-Fisher A. J., Bellinger G., Breindel J. L., Tavassoli F. A., Booth C. J., Duong J. K., Stern D. F. (2008) Breast Cancer Res. 10, R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu X., Yen L., Irwin L., Sweeney C., Carraway K. L., 3rd (2004) Mol. Cell Biol. 24, 7748–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sweeney C., Miller J. K., Shattuck D. L., Carraway K. L., 3rd (2006) J. Mammary Gland Biol. Neoplasia 11, 89–99 [DOI] [PubMed] [Google Scholar]