Abstract
We searched for proteins whose synthesis is enhanced by polyamines at the stationary phase of cell growth using an Escherichia coli polyamine-requiring mutant in which cell viability is greatly decreased by polyamine deficiency. The synthesis of ribosome modulation factor (RMF) was strongly enhanced by polyamines at the level of translation at the stationary phase of cell growth. In rmf mRNA, a Shine-Dalgarno (SD) sequence is located 11 nucleotides upstream of the initiation codon AUG. When the SD sequence was moved to the more common position 8 nucleotides upstream of the initiation codon, the degree of polyamine stimulation was reduced, although the level of RMF synthesis was markedly increased. Polyamine stimulation of RMF synthesis was found to be caused by a selective structural change of the bulged-out region of the initiation site of rmf mRNA. The decrease in cell viability caused by polyamine deficiency was prevented by the addition of a modified rmf gene whose synthesis is not influenced by polyamines. The results indicate that polyamines enhance cell viability of E. coli at least in part by enhancing RMF synthesis.
Keywords: Bacteria, Polyamines, RNA Structure, Translation, Translation Regulation, Bulged-out Nucleotides of Double-stranded RNA, Cell Viability, Polyamine Modulon, Polyamine Stimulation
Introduction
Polyamines (putrescine, spermidine, and spermine) are necessary for normal cell growth (1, 2). Because polyamines interact with nucleic acids and exist mostly as polyamine-RNA complexes in cells (3, 4), their proliferative effects are presumed to be caused by changes in RNA function. It was found that polyamines enhanced the formation of active 30 S ribosomal subunits for stimulation of general protein synthesis (5–8). Furthermore, we found that synthesis of several kinds of proteins was stimulated by polyamines and proposed that a set of genes whose expression is enhanced by polyamines at the level of translation can be classified as a “polyamine modulon” (2, 9). It was also shown that stimulation of specific types of protein synthesis by polyamines was based on a selective structural change in the bulged-out region of double-stranded RNA caused by spermidine, a polycation, and cannot be fulfilled by K+ and Mg2+ ions (10, 11). The complex of spermidine and bulged-out ribonucleotides mimics double-stranded RNA, and such a structural change in the initiation region of oppA- and rpoE-mRNAs enhanced the synthesis of OppA, an oligopeptide binding protein in its transport system, and RpoE (σ24), one of the sigma factors for transcription of a set of heat shock response genes (10, 11).
In Escherichia coli, we have thus far identified 11 kinds of genes as components of the polyamine modulon at the logarithmic phase of cell growth using a polyamine requiring mutant MA261. Expression of oppA (12); cya, encoding adenylate cyclase (13); rpoS, encoding σ38 sigma factor of RNA polymerase, which is involved in expression of genes at stationary phase (14); fecI, encoding σ18 sigma factor for the iron transport operon (9); fis, a global regulator of transcription of some growth-related genes, including genes for rRNA and some tRNAs (9); and prfB, encoding polypeptide release factor 2 (15) was enhanced by polyamines at the level of translation in cells cultured with glucose as an energy source. The rpoN, a σ54 sigma factor for nitrogen metabolism genes, hns, a positive regulator of expression of genes involved in flagellin synthesis and ribosomal protein synthesis, and cra, a global transcription factor for a large number of genes for glycolysis and glyconeogenesis were found to be members of the polyamine modulon in cells cultured with glucose and glutamate (16). Recently, we have reported that rpoE encoding a σ24 sigma factor and stpA which are involved in the transcription of a number of heat shock response genes are new members of the polyamine modulon under heat shock conditions (11).
In this study, we looked for new members of polyamine modulon involved in cell viability at the stationary phase of cell growth in E. coli, and found that rmf, a gene encoding ribosome modulation factor (RMF)2 is a member of the polyamine modulon at the stationary phase. RMF comprises 55 amino acid residues and is known to facilitate the formation of 100 S dimers from 70 S ribosomes and inhibit translation for preserving translational machinery at stationary phase (17, 18). Disruption of the rmf gene caused loss of 100 S dimers and reduction of cell viability at stationary phase (19). Polyamines caused a structural change of the bulged-out region of the initiation site of rmf mRNA, and enhanced RMF synthesis. Accordingly, polyamines enhanced cell viability at least partially through stimulation of synthesis of RMF at the level of translation. This is the first report of the existence of a polyamine modulon at the stationary phase, in which cell growth is limited.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
A polyamine-requiring mutant of E. coli MA261 (speB speC gly leu thr thi) (20) and MA261 lacZ::Em (21) were cultured in medium A (0.4% glucose (22.4 mm), 40.2 mm K2HPO4, 22.1 mm KH2PO4, 1.7 mm sodium citrate, 7.6 mm (NH4)2SO4, 0.41 mm MgSO4, 6 μm thiamine, 40 μm biotin, 0.8 mm leucine, 0.8 mm threonine, 0.7 mm methionine, 1 mm serine, 1 mm glycine, 0.6 mm ornithine, pH 6.8) in the presence and absence of 100 μg/ml (0.6 mm) putrescine dihydrochloride. MA261 rmf::Cm was prepared from C600 rmf::Cm by P1 transduction (22) and cultured as described above. Cell growth was monitored by measuring the absorbance at 540 nm. Cell viability was determined by counting colony numbers grown on Luria-Bertani (LB)-containing 1.5% agar plate at 37 °C for 24 h (23).
Plasmids
Total chromosomal DNA from E. coli W3110 was prepared according to the method of Wilson et al. (24). To make pMW-lacRMF(WT), PCR was performed using total chromosomal DNA as template and 5′-TAACTGGGATCCCACATTTAGTAATCACTG-3′ (P1) and 5′-CAGACAGGAATTCAAAAGGCGAAACCTCCG-3′ (P2) as primers. After cutting with BamHI and EcoRI, the 282-bp PCR fragment was inserted into the same restriction site of a low copy number vector pMW119 (Nippon Gene). Site-directed mutagenesis by overlap extension using PCR (25) was performed to prepare pMW-lacRMF(SD) and pMW-lacRMF(SD*). To make pMW-lacRMF(SD), the first PCR was performed using P1 and P2(SD) (5′-GTCTCTTCATGCCTCGCCTCCGTGATACTG-3′) and P1(SD) (5′-GAAACCAGTATCACGGAGGCGAGGCATGAA-3′) and P2 as primers, and pMW-lacRMF(WT) as a template. To make pMW-lacRMF(SD*), the first PCR was performed using P1 and P2(SD*) (5′-TGCTTTGTCTCCGTGATACTGGTTTCTGGT-3′) and P1(SD*) (5′-CACGGAGACAAAGCATGAAGAGACAAAAAC-3′) and P2 as primers, and pMW-lacRMF(WT) as a template. The second PCR was performed using the first PCR products as templates and P1 and P2 as primers. After cutting with BamHI and EcoRI, the 282-bp PCR fragment was inserted into the same restriction site of pMW119. The plasmids pMW-lacRMF −6(C → G) and pMW-lacRMF −35(ΔU) were also prepared as described above with site-directed mutagenesis by overlap extension using PCR (25).
To make the rmf-lacZ fusion gene, PCR was performed using total chromosomal DNA as template and 5′-CGTCCCCGGGATGTTGCCTG-3′ (P3) and 5′-AGCCCGGGTGACCTTTGATT-3′ (P4) as primers. The amplified rmf gene (a 227-nucleotide 5′-upstream region and a 115-nucleotide open reading frame) was digested with XmaI and inserted into the same restriction site of pMC1871 (26) to make the pMCrmf-lacZ fusion plasmid. For construction of pMWrmf-lacZ, the SalI fragment containing the rmf-lacZ gene of pMCrmf-lacZ was inserted into the same restriction site of pMW119. Site-directed mutagenesis for the construction of the plasmids encoding a mutated fusion gene with modified initiation region (pMWrmf [−6(C→G)]−lacZ, pMWrmf [−35(ΔU)]−lacZ, pMWrmf [−36(ΔC)]−lacZ, and pMWrmf [+56(C→G)]−lacZ) was performed by overlap extension using PCR (25). A list of oligonucleotide primers for mutagenesis is available from the authors upon request.
To prepare pMW-lacRMF(SD*)−RpoS(33CAG), PCR was performed using pMW33CAG encoding RpoS(33CAG), which has CAG codon at 33rd amino acid position of rpoS mRNA (14), as template and 5′-CAACGAATTCCGTGACCTTGCTCAGCGCA-3′ (P5) and 5′-GATGGGAATTCGACCTTTTATTGTGCACAG-3′ (P6) as primers. The EcoRI fragment containing rpoS(33CAG) gene was inserted into the same site of pMW-lacRMF(SD*). Plasmid pMW-lacRpoS(33CAG) was constructed by inserting the EcoRI fragment containing rpoS(33CAG) gene into the same site of pMW119. The nucleotide sequence of the plasmids was confirmed by the 3130 Genetic Analyzer (Applied Biosystems).
Dot Blot Analysis
E. coli MA261 cells were cultured at A540 = 0.05 in the presence and absence of putrescine and harvested at A540 = 0.2 and 24 h. Total RNA was prepared from these cells by the method of Emory and Belasco (27). Dot blot analysis was performed according to the standard method (28) using the ECL direct nucleic acid labeling and detection systems (GE Healthcare Bio-Sciences). Total RNA used for dot blotting was 0.2 to 3 μg. The probe was made by PCR, and the size of the probe used for rmf mRNA was 300 nucleotides. Chemical luminescence was detected by a LAS-3000 luminescent image analyzer (Fuji Film).
Western Blot Analysis
Western blot analysis was performed by the method of Nielsen et al. (29), using ECL Western blotting reagents (GE Healthcare Bio-Sciences). Antibodies against RMF and RpoS were prepared as described previously (22, 30). Antibody against OmpC was kindly supplied by H. Mori, Nara Institute of Science and Technology. Antibody against β-galactosidase was obtained from Sigma-Aldrich. Although the level of proteins was 1.3–1.6-fold higher in polyamine-containing cells than in polyamine-deficient cells, the same amount of proteins were used for Western blot analysis. The level of protein was quantified with a LAS-3000 luminescent image analyzer (Fuji Film).
Measurement of RMF Synthesis by Immunoprecipitation of Proteins Labeled with [35S]Methionine
E. coli MA261 cells were cultured in medium A containing 0.2 mm methionine instead of 0.7 mm in the absence of putrescine. At A540 = 0.6 and 24 h, the culture was divided into 5-ml aliquots, and continued to grow in the presence (100 μg/ml) and absence of putrescine for 20 min. Then, [35S]methionine (1 MBq) was added to each 5-ml aliquot, and the cells were allowed to grow for additional 5 min. After the addition of unlabeled methionine at a final concentration of 20 mm, the cells were harvested, resuspended in 1 ml of buffer A (10 mm sodium phosphate, at pH 7.4, 100 mm NaCl, 1% Triton X-100, and 0.1% SDS), and disrupted by grinding for 30 s at 2,500 rpm six times with a Multi-Beads Shocker (Yasui Kikai). Immunoprecipitation of [35S]methionine-labeled RMF was carried out using whole cell lysate containing 500,000 cpm of [35S]methionine-labeled proteins and antiserum against RMF according to the method of Philipson et al. (31). After SDS-PAGE, the radioactivity associated with RMF was quantified using a BAS-1800II imaging analyzer (Fuji Film).
Sucrose Density Gradient Centrifugation of Ribosomes
The cells cultured until A540 = 0.2 or for 24 h in 40 ml were harvested, resuspended in buffer containing 20 mm Tris-HCl, 10 mm magnesium acetate, and 100 mm ammonium acetate (pH 7.6) and disrupted by grinding for 30 s at 1,500 rpm three times with a Multi-Beads Shocker. The suspension was centrifuged at 10,000 rpm for 15 min at 4 °C. The cell lysate (7 A260 units) was loaded on a 12-ml linear sucrose gradient (5–20%). The tubes were centrifuged for 4 h at 25,000 rpm in a Hitachi P28S2. The sucrose gradients were fractionated into 53–54 tubes. Absorbance at 260 nm was measured after 2-fold dilution of each fraction.
Measurement of Polyamines in Whole Cells
Polyamines in whole cells were extracted by treatment of the cells with 10% trichloroacetic acid at 70 °C for 15 min with occasional shaking. Polyamine content was determined by high pressure liquid chromatography as described previously (32). Protein content was determined by the method of Bradford (33).
Prediction of the Secondary Structure of RNA
Optimal computer folding of mRNAs was performed by the method of Zuker (34). Free energy (ΔG) for the formation of the secondary structure was calculated on the basis of the data of Turner et al. (35).
Circular Dichroism (CD) Measurement of RNA
RMF WT RNA (5′-UGUUUUCUUUUCCACCAGGAAACGAGGCA-3′) constituting 17 nucleotides (−42th to −26th nucleotides upstream from the initiation codon AUG) and 12 nucleotides (−11th to +1st nucleotides of open reading frame) of rmf mRNA, RMF −6(C → G) RNA (5′-UGUUUUCUUUUCCACCAGGAAAGGAGGCA-3′), which substitutes cytidine with guanosine nucleotide at −6th of RMF WT RNA, and RMF −35(ΔU) RNA (5′-UGUUUUCUUUCCACCAGGAAACGAGGCA-3′), which lacks one uridine nucleotide at −35th position of RMF WT RNA were obtained from Hokkaido System Science. CD spectra were recorded over 200–320 nm on a Jasco J-820 spectropolarimeter (Jasco International Co.) using a 0.1 cm path-length cuvette at 37 °C (36). Scan speed was 100 nm/min, and CD samples contained 10 mm Tris-HCl (pH 7.5), 50 mm KCl, and 50 μm RNA. Where indicated, magnesium acetate, putrescine or spermidine was added to the CD samples. Typical spectra at 37 °C corresponded to the average of three scans.
RESULTS
Decrease in Cell Viability by Polyamine Deficiency
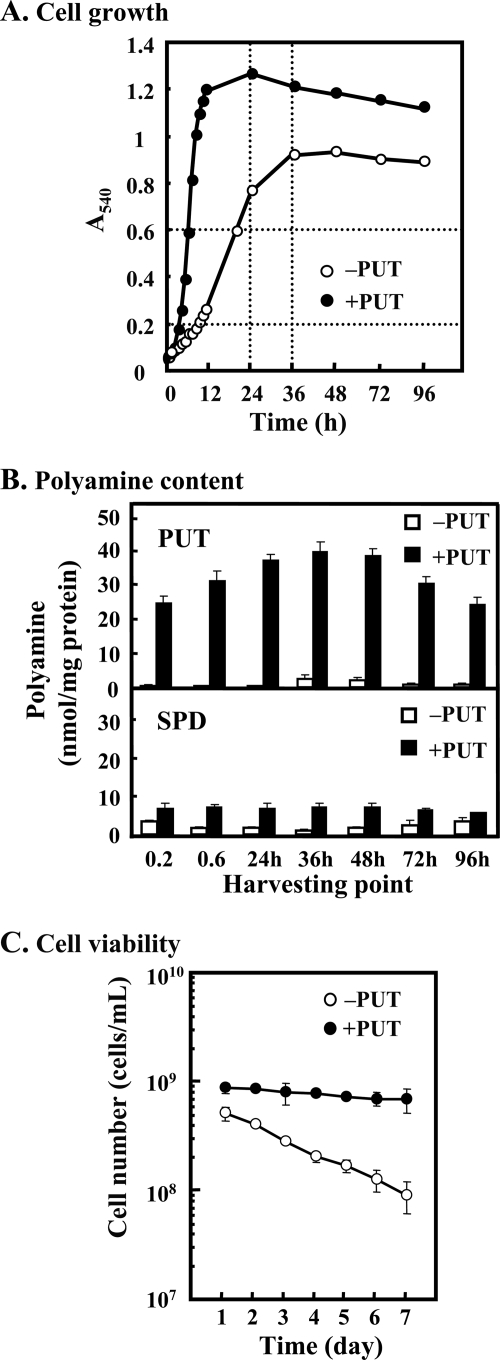
Polyamines are known to be involved in normal cell growth. Such a polyamine requirement for cell growth was confirmed using a polyamine-requiring mutant E. coli MA261. In this strain, putrescine added to the medium is taken up into cells and can be converted to spermidine. When 100 μg/ml putrescine was added to the medium, levels of spermidine and putrescine in cells increased, and cell growth was enhanced 3–5-fold (Fig. 1, A and B). It was determined whether polyamines enhance viability of MA261 cells. For cells cultured in the presence of putrescine, viability did not decrease significantly over 7 days, whereas cells cultured in the absence of putrescine showed a marked decrease in viability. After 7 days in culture, the increase in viability in the presence of putrescine was ∼8-fold (Fig. 1C).
FIGURE 1.
Growth, polyamine content, and cell viability of E. coli MA261. A, culture of E. coli MA261 cells with (●) or without (○) 100 μg/ml putrescine. Polyamine effect was examined at A540 = 0.2, 0.6, and 24 and 36 h after the onset of cell culture. Time points are shown as dotted lines. B, polyamine content in cells was measured at the indicated time. Contents of putrescine (upper) and spermidine (lower) of cells cultured in the absence (open column) and presence (closed column) of 100 μg/ml putrescine were shown. C, cell viability was determined as described under “Experimental Procedures.” Values are means ± S.E. of triplicate determinations.
Polyamine Stimulation of RMF Synthesis and Its Mechanism
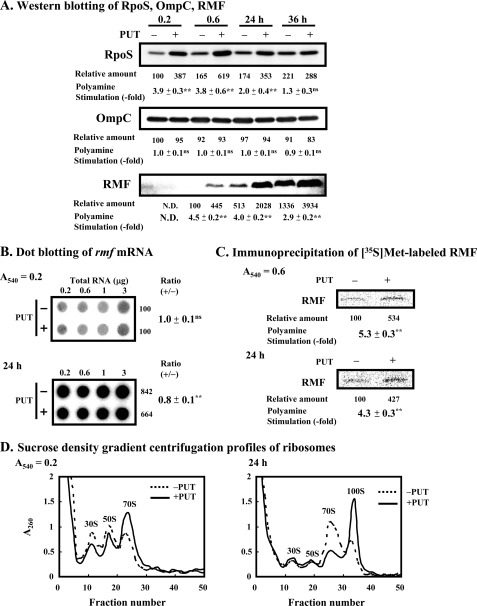
Protein synthesis is involved in cell viability. Thus, it was determined what kinds of protein synthesis are enhanced by polyamines. We previously reported a decrease in cell viability due to deficiency of the σ38 subunit (RpoS) of RNA polymerase, the outer membrane protein C (OmpC), and/or the RMF (37). RMF is synthesized in the stationary phase and is uniquely associated with 100 S dimers (17–19). Among them, we reported that rpoS gene, encoding σ38 subunit of RNA polymerase, is a polyamine modulon (14). Polyamines enhance the synthesis of RpoS through stimulation of read-through of amber codon UAG-dependent Gln-tRNASupE on ribosome-associated rpoS mRNA (14). The levels of RpoS, OmpC, and RMF were measured at the early logarithmic (A540 = 0.2), the late logarithmic (A540 = 0.6), and the stationary phase (24 and 36 h after the onset of cell culture) (see Fig. 1A). It was confirmed that synthesis of RpoS at the early to late logarithmic phase was enhanced by polyamines, judging from the level of RpoS protein (Fig. 2A). Synthesis of OmpC was not influenced by polyamines. Synthesis of RMF at early logarithmic phase was not observed, as reported previously (19). Synthesis of RMF at late logarithmic to the stationary phase (24 h) was strongly enhanced by polyamines (Fig. 2A). Because the level of rmf mRNA was nearly equal in cells cultured with or without 100 μg/ml putrescine (Fig. 2B), enhancement of RMF synthesis was at the level of translation. To confirm that increase in RMF is at the level of translation, synthesized RMF was pulse-labeled with [35S]methionine and immunoprecipitated with the antibody against RMF. As shown in Fig. 2C, RMF synthesis was significantly stimulated by polyamines at both late logarithmic and stationary phases. At the stationary phase (24 h), most of ribosomes in cells cultured in the presence polyamines were 100 S dimers, but those were mostly 70 S monomers in the absence of polyamines (Fig. 2D), confirming that RMF enhanced 100 S dimer formation (17–19).
FIGURE 2.
Levels of RpoS (σ38), OmpC, and RMF in E. coli MA261 cultured in the presence and absence of putrescine. A, Western blot analysis was performed using 5 μg of protein for RpoS (σ38) and OmpC, and 25 μg of protein for RMF. Percentage of acrylamide used in SDS-polyacrylamide gel electrophoresis was 10.5% for RpoS (σ38) and OmpC, and 15% for RMF. B, dot blot analysis of rmf mRNA in E. coli MA261 harvested at A540 = 0.2 or 24 h after the onset of cell culture was performed using various amounts of RNA shown in the figure. C, immunoprecipitation of [35S]methionine-labeled RMF was performed using E. coli MA261 cells incubated with [35S]methionine for 5 min as described under “Experimental Procedures.” Values shown in A–C are means ± S.E. of triplicate determinations. D, ribosomal profiles were analyzed by sucrose density gradient centrifugation using the cell lysate of E. coli MA261 cells harvested at A540 = 0.2 or 24 h after the onset of cell culture. Student's t test was performed for the value obtained in the presence of putrescine versus in the absence of putrescine. ns, p > 0.05; **, p ≤ 0.01.
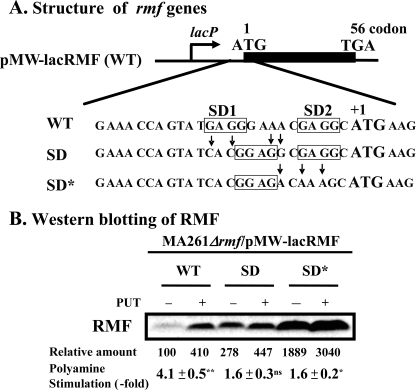
The mechanism of polyamine stimulation of RMF synthesis was studied using modified rmf mRNAs. When the nucleotide sequence of rmf mRNA was determined, two candidate SD sequences were observed (Fig. 3A). Thus, two modified rmf mRNAs were constructed. In one construct, pMW-lacRMF(SD), the first SD sequence (SD1) located at 11 nucleotides upstream of the initiation codon AUG was shifted to the more common position located at 8 nucleotides upstream of the initiation codon. In the other construct, pMW-lacRMF(SD*), the second SD sequence (SD2) was modified together with first SD1 sequence. When the rmf (SD) mRNA was used instead of rmf (WT) mRNA, the degree of polyamine stimulation was reduced from 4.1 to 1.6-fold, although the level of RMF protein in cells cultured without polyamines was increased. When the rmf (SD*) mRNA was used, the degree of polyamine stimulation was also reduced from 4.1 to 1.6-fold, and the level of RMF was markedly increased about 19-fold in cells cultured without polyamines because of the elimination of the inhibitory SD sequence located at 2 nucleotides upstream of the initiation codon (Fig. 3B). The results indicate that synthesis of RMF is enhanced by polyamines because of the presence of an unusual SD sequence in rmf mRNA, as seen in other mRNAs including oppA-, fecI-, fis-, rpoN-, hns-, rpoE-, and stpA (9, 11, 12, 16).
FIGURE 3.
Effect of the position of SD sequence on polyamine stimulation of RMF synthesis. A, nucleotide sequence of 5′-untranslated region of rmf mRNA was shown. Site-directed mutagenesis to change the position of SD sequence was performed as described under “Experimental Procedures.” B, E. coli MA261 rmf::Cm (MA261 Δrmf) carrying pMW-lacRMF(WT), pMW-lacRMF(SD), or pMW-lacRMF(SD*) was cultured in medium A with 0.5 mm isopropyl-β-d-thiogalactopyranoside in the presence and absence of 100 μg/ml putrescine for 24 h. Western blotting of RMF synthesized from wild type and SD sequence-modified rmf mRNAs was performed using 5 μg of protein. Values are means ± S.E. of triplicate determinations. ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01.
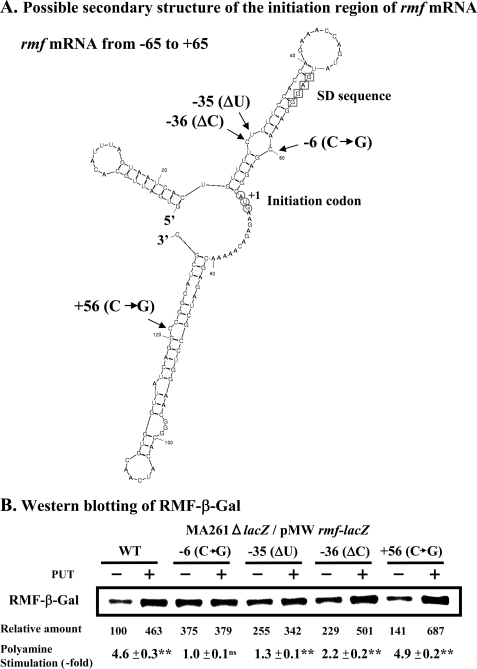
We have previously reported that polyamine stimulation of OppA synthesis and rat liver Ile-tRNA formation likely involves a structural change of the bulged-out region of double-stranded RNA (12, 38) and have recently shown that a selective structural change of the bulged-out region of oppA mRNA and rat liver tRNAIle is important for polyamine stimulation (10). Thus, a possible secondary structure of the initiation region of rmf mRNA (−65 to +65) was constructed by the method of Zuker (34), and it was found that a bulged-out region existed, in which selective structural change by spermidine would occur (Fig. 4A). The nucleotide sequence of the bulged-out region of double-stranded RNA, near the initiation codon AUG and the SD sequence of rmf mRNA, was modified to make three different forms of the double-stranded RNA. Those mRNAs are rmf [−6(C→G)] mRNA, rmf [−35(ΔU)] mRNA, and rmf [−36(ΔC)] mRNA. As a control, rmf [+56(C→G)] mRNA was constructed to remove the bulged-out region of another double-stranded region that is located distantly from the initiation site of rmf mRNA. The predicted secondary structure of the initiation region (positions −65 to +65) of four mutants of rmf mRNA was essentially the same as that of wild-type rmf mRNA (data not shown).
FIGURE 4.
Effect of polyamines on the synthesis of RMF-β-Gal fusion proteins derived from wild and mutated rmf-lacZ mRNAs in the 5′-untranslated region. A, possible secondary structure of the initiation region of rmf mRNA was constructed by the method of Zuker (34). The positions in which the sequence was mutated are shown. Symbols of ▵ and → in parenthesis represent removal and replacement of nucleotide, respectively. B, effect of polyamines on RMF-β-Gal fusion protein synthesis was evaluated by Western blot analysis using 5 μg of protein of cell lysate and antibody against β-galactosidase. Values are means ± S.E. of triplicate determinations. ns, p > 0.05; **, p ≤ 0.01.
The effects of polyamines on the synthesis of an RMF-β-Gal fusion protein were studied using these various rmf-lacZ fusion mRNAs. As shown in Fig. 4B, polyamines stimulated the synthesis of RMF-β-Gal from wild-type rmf-lacZ mRNA by 4.6-fold. Polyamine stimulation of the synthesis of RMF- β-Gal fusion protein from rmf [−6(C→G)]−lacZ, rmf [−35(ΔU)]−lacZ, and rmf [−36(ΔC)]−lacZ mRNAs was reduced to 1.1–2.2-fold, although the basal level of protein synthesis in the absence of polyamines was enhanced. When polyamine stimulation of the synthesis of the RMF-β-Gal fusion protein from rmf [+56(C→G)]−lacZ mRNA was examined as a control, the degree of polyamine stimulation was nearly equal to that of synthesis from wild-type rmf-lacZ mRNA. The results support the idea that a structural change of the bulged-out region of double-stranded RNA close to the initiation codon AUG and the SD sequence of rmf mRNA is involved in polyamine stimulation of RMF synthesis.
Selective Structural Change of the Bulged-out Region in Double-stranded RNA of rmf mRNA by Spermidine
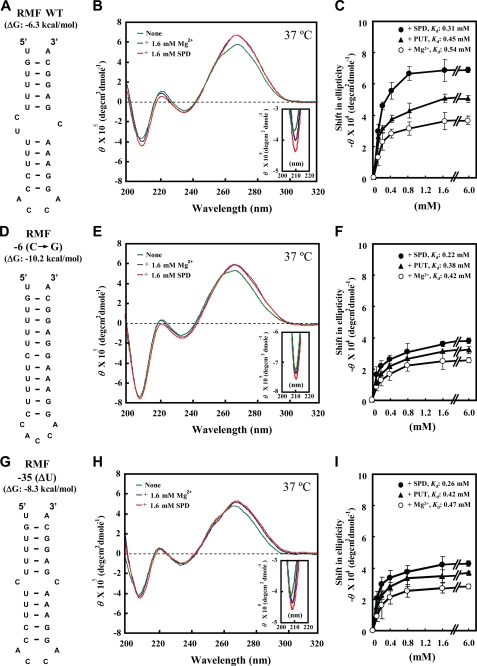
Structural changes of the bulged-out region induced by spermidine were studied using synthetic RNAs containing the wild type or the mutated initiation region of the rmf mRNA. Structural changes induced by spermidine of RMF WT RNA, which corresponds to the double-stranded region of −42nd to −26th and −11th to +1st of rmf mRNA (Fig. 5A), RMF −6(C→G) RNA, which substitutes cytidine with guanosine nucleotide at the −6th position of RMF WT RNA (Fig. 5D), and RMF −35(ΔU) RNA, which lacks one uridine nucleotide at −35th position of RMF WT RNA (Fig. 5G), were analyzed in the presence of 10 mm Tris-HCl (pH 7.5) and 50 mm KCl at 37 °C. A substantial increase in the relative intensity of the negative band at 208 nm in CD reflects stabilization (or an increase) of the A-form double-stranded RNA (36). There was a marked increase in the relative intensity of the negative band at 208 nm induced by 1.6 mm spermidine in RMF WT RNA including the bulged-out region in double-stranded RNA, and this was greater than the increase in RMF −6(C→G) RNA and RMF −35(ΔU) RNA, which do not have the marked bulged-out region in the double-stranded RNA (Fig. 5, B, E, and H). In contrast, 1.6 mm Mg2+ or putrescine had a much smaller effect than spermidine on RMF WT RNA, RMF −6(C→G) RNA and RMF −35(ΔU) RNA. A marked increase in the relative intensity of the negative band at 208 nm in RMF WT RNA by 1.6 mm spermidine was also observed in the presence of 1.6 mm Mg2+ (data not shown). Concentration-dependent shifts of RMF WT RNA, RMF −6(C→G) RNA and RMF −35(ΔU) RNA induced by spermidine, putrescine, or Mg2+ at 208 nm were then measured (Fig. 5, C, F, and I). It was confirmed that the structural change of RMF WT RNA produced by spermidine was much greater than that produced by putrescine or Mg2+, but that spermidine had little effect on the RMF −6(C→G) RNA and RMF −35(ΔU) RNA. The structural changes of three RMF RNAs induced by putrescine were slightly greater than those induced by Mg2+. The apparent dissociation constants (Kd) for spermidine, putrescine, and Mg2+ are shown on Fig. 5, C, F, and I. These results clearly show that spermidine caused a selective structural change of the bulged-out region in double-stranded RNA at the initiation site of rmf mRNA.
FIGURE 5.
CD spectra of RMF WT RNA, RMF −6(C→G) RNA, and RMF −35(ΔU) RNA. A, D, and G, structure of RMF WT RNA, RMF −6(C→G) RNA, and RMF −35(ΔU) RNA. B, E, and H, CD spectra were recorded as described under “Experimental Procedures.” Green line, no addition; blue line, 1.6 mm Mg2+; red line, 1.6 mm spermidine. C, F, and I. Concentration-dependent shifts induced by Mg2+ (○), putrescine (▴), and spermidine (●) at 37 °C in magnitude at 208 nm are shown. Values are means ± S.E. of triplicate determinations. The Kd values of spermidine, putrescine, and Mg2+ for RMF WT RNA, RMF −6(C→G) RNA, and RMF −35(ΔU) RNA at 37 °C were determined according to the double-reciprocal equation plot.
Increase in Cell Viability by RMF
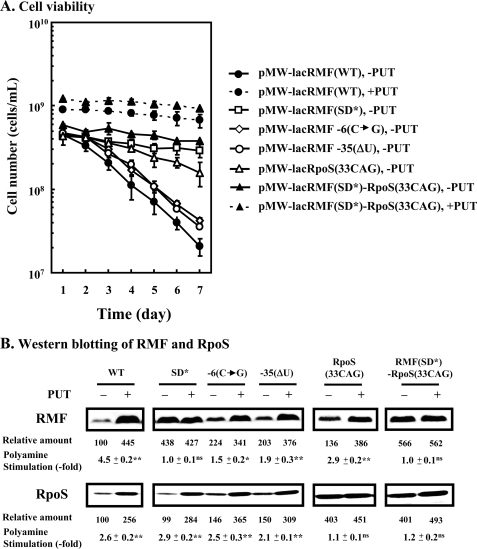
It was shown that polyamine deficiency greatly reduces cell viability (see Fig. 1C). It was also shown that RMF is strongly involved in cell viability and RpoS assists the function of RMF in cell viability (19, 22, 39, 40), and synthesis of both proteins are enhanced by polyamines (see Fig. 2A). Thus, we examined whether reductions in RMF and RpoS caused by polyamine deficiency are involved in reduced cell viability when cells are cultured without polyamines. For these experiments, plasmids encoding modified rmf and rpoS genes, in which RMF and RpoS synthesis is not influenced by polyamines, were used. These constructs were pMW-lacRMF(SD*) (see Fig. 3B) pMW-lacRMF −6(C→G), pMW-lacRMF −35(ΔU) (see Fig. 4), pMW-lacRpoS (33CAG) (14), and pMW-lacRMF(SD*)-RpoS(33CAG). With these constructs, RMF and RpoS were synthesized effectively even in the absence of polyamines (Fig. 6B). In the absence of polyamines, cell viability was enhanced in parallel with the increase in the level of RMF and RpoS (Fig. 6A). Effect of RMF on cell viability was greater than that of RpoS. Under these conditions, cell viability of E. coli MA261 rmf::Cm cultured in the presence of polyamines on day 7 decreased to 10% of that of E. coli MA261 cultured in the presence of polyamines (data not shown). In the presence of polyamines, RMF and RpoS did not enhance cell viability significantly (Fig. 6A). The results indicate that polyamines contribute to cell viability of E. coli at least in part by enhancing RMF synthesis.
FIGURE 6.
Recovery of the decrease in cell viability under polyamine deficiency by RMF and/or RpoS. A, E. coli MA261 cells carrying various plasmids were cultured in medium A with 0.5 mm isopropyl-β-d-thiogalactopyranoside in the presence and absence of 100 μg/ml putrescine. Cell viability was determined as described under “Experimental Procedures.” ●---●, MA261/pMW-lacRMF(WT) cultured with putrescine; ▴---▴, MA261/pMW-lacRMF(SD*)-RpoS(33CAG) cultured with putrescine; ●—●, MA261/pMW-lacRMF(WT) cultured without putrescine; □—□, MA261/pMW-lacRMF(SD*) cultured without putrescine; ◇—◇, MA261/pMW-lacRMF −6(C→G) cultured without putrescine; ○—○, MA261/ pMW-lacRMF −35(ΔU) cultured without putrescine; ▵—▵, MA261/pMW-lacRpoS(33CAG) cultured without putrescine; ▴—▴, MA261/pMW-lacRMF(SD*)-RpoS(33CAG) cultured without putrescine. B, level of RMF and RpoS in cells cultured with or without putrescine for 36 h was measured by Western blotting using 5 μg of protein. Values are means ± S.E. of triplicate determinations. ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01.
DISCUSSION
We have shown thus far that polyamines enhance cell growth through an increase in general protein synthesis by activating 30 S ribosomal subunits (2, 7, 8) and increase in specific kinds of protein synthesis, which is important for cell growth (2, 9, 10). We proposed that a set of genes whose expression is enhanced by polyamines at the level of translation can be classified as a “polyamine modulon.”
In this study, we found that polyamine deficiency caused a decrease in cell viability at the stationary phase. Therefore, we looked for members of the polyamine modulon that are involved in cell viability. It was found that RMF synthesis is strongly enhanced by polyamines at the level of translation. So, the rmf gene is the first member of the polyamine modulon identified at the stationary phase. Transformation of a plasmid encoding a modified rmf gene, in which RMF synthesis is not influenced by polyamines, reversed the effects on cell viability caused by polyamine deficiency, confirming that RMF is strongly involved in cell viability (19). A decrease in RpoS was also involved in the decrease in cell viability caused by polyamine deficiency, because polyamines enhanced the synthesis of RpoS about 2.0-fold at the stationary phase. We previously reported that overaccumulation of spermidine in a spermidine acetyltransferase-deficient E. coli CAG2242 also caused a decrease in cell viability (40). When 2 mm spermidine was added to medium during the culture of this strain, spermidine content became about 200 nmol/mg protein although putrescine content became negligible. The level of spermidine increased more than 20-fold compared with that in polyamine-containing MA261 cells (see Fig. 1B). Under these conditions, RMF synthesis was greatly decreased and RpoS synthesis was slightly decreased, confirming that RMF is strongly involved in cell viability, and RpoS assisted the function of RMF in cell viability (40). Thus, maintenance of the optimal polyamine concentration, which stimulates the synthesis of RMF and RpoS, is important for cell viability.
Polyamines enhance specific kinds of protein synthesis by three different mechanisms in E. coli. First, polyamine stimulation of protein synthesis takes place when the SD sequence is distant from the initiation codon AUG or obscure as in case of oppA-, fecI-, fis-, rpoN-, hns-, rpoE- stpA-, and rmf-mRNAs (9, 11, 12, 16, and this study). Second, polyamines enhance translation initiation from the inefficient initiation codon UUG or GUG in cya- and cra-mRNAs (13, 16). Third, polyamines stimulate read-through of termination codon or stimulate a +1 frameshift at the termination codon existing in the open reading frame in rpoS- and prfB-mRNAs (14, 15). Although these conditions are essential for polyamine stimulation of protein synthesis, it also requires the existence of a bulged-out structure in double-stranded RNA. Such structures are selectively changed by spermidine, but not by Mg2+ (2, 10, 11). In the case of RMF synthesis, SD sequence of rmf mRNA was distant from the initiation codon AUG, and a bulged-out region in double-stranded RNA existed at the initiation site of rmf mRNA. Thus, the initiation of RMF synthesis was fulfilled effectively in the presence of polyamines through the selective structural change of the bulged-out region in double-stranded RNA at the initiation site of rmf mRNA.
Excess spermidine also inhibited RMF synthesis (40). In this case, the structure of the bulged-out region in double-stranded RNA may become too tight, so that the SD sequence of rmf mRNA may not be able to come close to the initiation codon AUG. Thus, it is important to clarify how different concentrations of each polyamine, that is, putrescine and spermidine, cause the structural change of RNA. From our present data, putrescine can cause smaller structural changes than spermidine even at high concentrations. Because more than 10 mm spermidine precipitated RNA during CD measurement of RNA, another strategy is necessary to evaluate the structure of RNA in the presence of high concentrations of spermidine.
With regard to the function of RMF on cell viability, it is thought that inhibition of wasteful protein synthesis through formation of 100 S dimers of ribosomes is strongly related to cell viability (18). It has been also shown that the stringent response factor, ppGpp, which is required for long-term survival (41), positively regulates rmf transcription (42), and synthesis of ppGpp is enhanced by polyamines (43). Thus, polyamines contribute to the increase in cell viability by regulating synthesis of both ppGpp and RMF, which inhibit RNA and protein synthesis. It should be clarified whether RMF inhibits protein synthesis in general or specific kinds of proteins.
It has been reported that decarboxylated S-adenosyl-l-methionine (dcAdoMet) accumulates at concentrations of 300 to 500 μm as a consequence of the inhibition of putrescine biosynthesis in mammalian cells (44, 45). However, it was not clear whether dcAdoMet is involved in the inhibition of cell growth during polyamine deficiency. During the growth of E. coli MA261 in the absence of polyamines, ∼500 μm dcAdoMet accumulated at the logarithmic phase, but the level of dcAdoMet at the stationary phase was negligible (data not shown). Thus, it is clear that dcAdoMet is not involved in the decrease in cell viability and RMF synthesis by polyamine deficiency. In a cell-free protein synthetic system, 1 mm spermidine greatly enhanced RMF synthesis (data not shown), confirming that polyamine stimulation of RMF synthesis is directly involved in the increase in cell viability.
Acknowledgments
We thank Dr. K. Williams for critical reading of manuscript. Thanks are due to Dr. H. Mori for providing an antibody against OmpC. We also thank Dr. Y. Ikeguchi, Faculty of Pharmaceutical Sciences, Josai University, for providing decarboxylated S-adenosyl-l-methionine.
This work was supported by a grant-in aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan.
- RMF
- ribosome modulation factor
- SD
- Shine-Dalgarno
- PUT
- putrescine
- SPD
- spermidine.
REFERENCES
- 1.Cohen S. S. (1998) A Guide to the Polyamines, Oxford University Press, New York, NY [Google Scholar]
- 2.Igarashi K., Kashiwagi K. (2010) Int. J. Biochem. Cell Biol. 42, 39–51 [DOI] [PubMed] [Google Scholar]
- 3.Watanabe S., Kusama-Eguchi K., Kobayashi H., Igarashi K. (1991) J. Biol. Chem. 266, 20803–20809 [PubMed] [Google Scholar]
- 4.Miyamoto S., Kashiwagi K., Ito K., Watanabe S., Igarashi K. (1993) Arch. Biochem. Biophys. 300, 63–68 [DOI] [PubMed] [Google Scholar]
- 5.Echandi G., Algranati I. D. (1975) Biochem. Biophys. Res. Commun. 62, 313–319 [DOI] [PubMed] [Google Scholar]
- 6.Echandi G., Algranati I. D. (1975) Biochem. Biophys. Res. Commun. 67, 1185–1191 [DOI] [PubMed] [Google Scholar]
- 7.Igarashi K., Kashiwagi K., Kishida K., Watanabe Y., Kogo A., Hirose S. (1979) Eur. J. Biochem. 93, 345–353 [DOI] [PubMed] [Google Scholar]
- 8.Igarashi K., Kashiwagi K., Kishida K., Kakegawa T., Hirose S. (1981) Eur. J. Biochem. 114, 127–131 [DOI] [PubMed] [Google Scholar]
- 9.Yoshida M., Kashiwagi K., Shigemasa A., Taniguchi S., Yamamoto K., Makinoshima H., Ishihama A., Igarashi K. (2004) J. Biol. Chem. 279, 46008–46013 [DOI] [PubMed] [Google Scholar]
- 10.Higashi K., Terui Y., Suganami A., Tamura Y., Nishimura K., Kashiwagi K., Igarashi K. (2008) J. Biol. Chem. 283, 32989–32994 [DOI] [PubMed] [Google Scholar]
- 11.Terui Y., Higashi K., Tabei Y., Tomitori H., Yamamoto K., Ishihama A., Igarashi K., Kashiwagi K. (2009) J. Bacteriol. 191, 5348–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida M., Meksuriyen D., Kashiwagi K., Kawai G., Igarashi K. (1999) J. Biol. Chem. 274, 22723–22728 [DOI] [PubMed] [Google Scholar]
- 13.Yoshida M., Kashiwagi K., Kawai G., Ishihama A., Igarashi K. (2001) (2001) J. Biol. Chem. 276, 16289–16295 [DOI] [PubMed] [Google Scholar]
- 14.Yoshida M., Kashiwagi K., Kawai G., Ishihama A., Igarashi K. (2002) J. Biol. Chem. 277, 37139–37146 [DOI] [PubMed] [Google Scholar]
- 15.Higashi K., Kashiwagi K., Taniguchi S., Terui Y., Yamamoto K., Ishihama A., Igarashi K. (2006) J. Biol. Chem. 281, 9527–9537 [DOI] [PubMed] [Google Scholar]
- 16.Terui Y., Higashi K., Taniguchi S., Shigemasa A., Nishimura K., Yamamoto K., Kashiwagi K., Ishihama A., Igarashi K. (2007) J. Bacteriol. 189, 2359–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wada A., Yamazaki Y., Fujita N., Ishihama A. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2657–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada A., Igarashi K., Yoshimura S., Aimoto S., Ishihama A. (1995) Biochem. Biophys. Res. Commun. 214, 410–417 [DOI] [PubMed] [Google Scholar]
- 19.Yamagishi M., Matsushima H., Wada A., Sakagami M., Fujita N., Ishihama A. (1993) EMBO J. 12, 625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham-Rundles S., Maas W. K. (1975) J. Bacteriol. 124, 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashiwagi K., Watanabe R., Igarashi K. (1994) Biochem. Biophys. Res. Commun. 200, 591–597 [DOI] [PubMed] [Google Scholar]
- 22.Apirakaramwong A., Fukuchi J., Kashiwagi K., Kakinuma Y., Ito E., Ishihama A., Igarashi K. (1998) Biochem. Biophys. Res. Commun. 251, 482–487 [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edition, pp. A1–A4, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Wilson K., Ausubel F. M., Brent R., Kingston R. E. (1987) Current Protocols in Molecular Biology, pp. 2.4.1–2.4.2, John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 25.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 26.Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. (1983) Gene 25, 71–82 [DOI] [PubMed] [Google Scholar]
- 27.Emory S. A., Belasco J. G. (1990) J. Bacteriol. 172, 4472–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edition, pp. 7.46–7.50, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Nielsen P. J., Manchester K. L., Towbin H., Gordon J., Thomas G. (1982) J. Biol. Chem. 257, 12316–12321 [PubMed] [Google Scholar]
- 30.Jishage M., Ishihama A. (1995) J. Bacteriol. 177, 6832–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philipson L., Andersson P., Olshevsky U., Weinberg R., Baltimore D., Gesteland R. (1978) Cell 13, 189–199 [DOI] [PubMed] [Google Scholar]
- 32.Igarashi K., Kashiwagi K., Hamasaki H., Miura A., Kakegawa T., Hirose S., Matsuzaki S. (1986) J. Bacteriol. 166, 128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 34.Zuker M. (2003) Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner D. H., Sugimoto N., Freier S. M. (1988) Annu. Rev. Biophys. Chem. 17, 167–192 [DOI] [PubMed] [Google Scholar]
- 36.Nakano S., Kanzaki T., Sugimoto N. (2004) J. Am. Chem. Soc. 126, 1088–1095 [DOI] [PubMed] [Google Scholar]
- 37.Raj V. S., Füll C., Yoshida M., Sakata K., Kashiwagi K., Ishihama A., Igarashi K. (2002) Biochem. Biophys. Res. Commun. 299, 252–257 [DOI] [PubMed] [Google Scholar]
- 38.Kusama-Eguchi K., Watanabe S., Irisawa M., Watanabe K., Igarashi K. (1991) Biochem. Biophys. Res. Commun. 177, 745–750 [DOI] [PubMed] [Google Scholar]
- 39.Hengge-Aronis R., Lange R., Henneberg N., Fischer D. (1993) J. Bacteriol. 175, 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apirakaramwong A., Kashiwagi K., Raj V. S., Sakata K., Kakinuma Y., Ishihama A., Igarashi K. (1999) Biochem. Biophys. Res. Commun. 264, 643–647 [DOI] [PubMed] [Google Scholar]
- 41.Primm T. P., Andersen S. J., Mizrahi V., Avarbock D., Rubin H., Barry C. E., 3rd (2000) J. Bacteriol. 182, 4889–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izutsu K., Wada A., Wada C. (2001) Genes Cells 6, 665–676 [DOI] [PubMed] [Google Scholar]
- 43.Igarashi K., Mitsui K., Kubota M., Shirakuma M., Ohnishi R., Hirose S. (1983) Biochim. Biophys. Acta 755, 326–331 [DOI] [PubMed] [Google Scholar]
- 44.Pegg A. E., Pösö H., Shuttleworth K., Bennett R. A. (1982) Biochem. J. 202, 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mamont P. S., Danzin C., Wagner J., Siat M., Joder-Ohlenbusch A. M., Claverie N. (1982) Eur. J. Biochem. 123, 499–504 [DOI] [PubMed] [Google Scholar]