Abstract
The predominant pathway for phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2) synthesis is thought to be phosphorylation of phosphatidylinositol 4-phosphate at the 5 position of the inositol ring by type I phosphatidylinositol phosphate kinases (PIPK): PIPKIα, PIPKIβ, and PIPKIγ. PIPKIγ has been shown to play a role in PI(4,5)P2 synthesis in brain, and the absence of PIPKIγ is incompatible with postnatal life. Conversely, mice lacking PIPKIα or PIPKIβ (isoforms are referred to according to the nomenclature of human PIPKIs) live to adulthood, although functional effects in specific cell types are observed. To determine the contribution of PIPKIα and PIPKIβ to PI(4,5)P2 synthesis in brain, we investigated the impact of disrupting multiple PIPKI genes. Our results show that a single allele of PIPKIγ, in the absence of both PIPKIα and PIPKIβ, can support life to adulthood. In addition, PIPKIα alone, but not PIPKIβ alone, can support prenatal development, indicating an essential and partially overlapping function of PIPKIα and PIPKIγ during embryogenesis. This is consistent with early embryonic expression of PIPKIα and PIPKIγ but not of PIPKIβ. PIPKIβ expression in brain correlates with neuronal differentiation. The absence of PIPKIβ does not impact embryonic development in the PIPKIγ knock-out (KO) background but worsens the early postnatal phenotype of the PIPKIγ KO (death occurs within minutes rather than hours). Analysis of PIP2 in brain reveals that only the absence of PIPKIγ significantly impacts its levels. Collectively, our results provide new evidence for the dominant importance of PIPKIγ in mammals and imply that PIPKIα and PIPKIβ function in the generation of specific PI(4,5)P2 pools that, at least in brain, do not have a major impact on overall PI(4,5)P2 levels.
Keywords: Gene Knockout; Neural Metabolism; Neurobiology; Phosphatidylinositol; Phosphatidylinositol Signaling; Phosphatidylinositol 4,5-Bisphosphate; Phosphatidylinositol 4-Phosphate 5-Kinase
Introduction
The role of inositol phospholipids as signaling molecules was first established more than 50 years ago with the finding that cholinergic-induced secretion in the pancreas and brain correlates with increased incorporation of 32P into these lipids (1). Since then, it has been shown that phosphorylation at the 3, 4, and 5 positions of the inositol ring to generate phosphoinositides along with further metabolism of such lipids by pyrophosphorylation, phospholipases, and phosphatases generate a broad spectrum of important signaling molecules (2). Key among them is PI(4,5)P2, a phosphoinositide concentrated in the plasma membrane and a defining feature of this membrane. PI(4,5)P22 is implicated either directly (though interactions of its cytosolically exposed headgroup) or via its metabolites (such as PI(3,4,5)P3, Ins(3,4,5)P3, and other inositol polyphosphates, diacylglycerol, and arachidonic acid) in the control of variety of physiological processes. These include receptor signaling and Ca2+ homeostasis, exo- and endocytosis, cytoskeletal dynamics, cytokinesis, cell migration, control of gene expression, and ion channel activity (for a recent review, see Refs. 2–6). Impaired metabolism of PI(4,5)P2 or of PI(4,5)P2-derived metabolites has also been implicated in human disorders including psychiatric and neurologic diseases such as mental retardation (7–10), bipolar disorder and schizophrenia (11–13), Alzheimer disease (14, 15), diabetes (16), cancer (17), and ciliopathies (18, 19). Furthermore, PI(4,5)P2 metabolism can be hijacked by pathogens to facilitate their invasion of cells and their proliferation (20, 21). Therefore, a precise understanding of PI(4,5)P2 synthesis and metabolism is a question of central importance in biology and medicine.
PI(4,5)P2 undergoes continuous turnover, with its levels being controlled by a tightly regulated balance between synthesis and either catabolism or metabolism to other lipid species. Additional regulatory mechanisms involve its segregation in metabolically segregated pools (22, 23). There exist two pathways leading to PI(4,5)P2 synthesis; from PI4P via type I PI4P 5-kinases (PIPKIs) or from PI5P via type II PI5P 4-kinases (PIPKIIs) (24). Both PIPKIs and PIPKIIs occur as three distinct isoforms (α, β, γ) (3, 6, 25, 26), encoded by distinct genes (a fourth PIPKI isoform, PIPKIδ, is catalytically inactive (27)). These enzymes share a conserved catalytic core domain (further conserved within the type I and the type II families) and have divergent N and C termini that have regulatory and targeting functions. Such flanking regions, which at least in some isoforms undergo alternative splicing and phosphorylation, define the specific role of each isoform by controlling its action in space and time (3, 6, 28–36).
The bulk of PI(4,5)P2 synthesis is thought to occur via PIPKIs given the abundance of PI4P. Furthermore, the yeast genome encodes only one PIPK, which has PI4P 5-kinase activity (37). However, the relative roles of each of the three mammalian PIPKIs (α, β, and γ) remain poorly characterized. Because of historical reasons that have created confusion in the literature, PIPKIα in mouse corresponds to PIPKIβ in humans and vice versa (25, 26). We will use the human nomenclature.
The function of individual mammalian PIPKI kinases, which have differential expression in different tissues (25, 26, 38, 39) (see also Fig. 1 of this study), has been explored in cultured cells by overexpression and knockdown studies as well as in KO mice. These studies led to the conclusion that each PIPKI controls a distinct PI(4,5)P2 pool within given cells (2, 3, 5, 6, 28–31, 40–42). We are particularly interested in PI(4,5)P2 synthesis in brain. Although we have established that the γ PIPKI isoform plays the dominant role in the generation of PI(4,5)P2 in neurons and at synapses in particular (32, 38), little is known about the contribution of the α and β PIPKI isoforms to the generation of this phosphoinositide in the nervous system.
FIGURE 1.
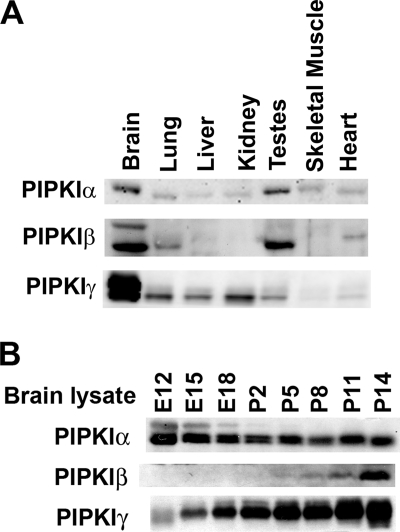
Tissue expression and developmental pattern of expression in brain of PIPKI isoforms. A, Western blots of α, β and γ PIPKI isoforms in total lysates from the tissues indicated are shown. Protein amounts were determined by BCA, and equal amounts of protein (50 μg) were loaded in each lane. This experiment was performed with two independent sets of protein samples. B, Western blots of the 3 PIPKI kinases in head (embryos) and brain (postnatal) lysates from the developmental stages indicated are shown.
Mice harboring a targeted disruption of the PIPKIγ gene, leading to complete lack of protein expression, show a major (∼30–50%) reduction of PI(4,5)P2 in the brain (7, 32). These mice have major synaptic defects and die by 12–24 h after birth, demonstrating a vital role for this isoform (32). Impaired nervous system function likely plays an important role in their death, although synaptic transmission can occur, albeit with lower efficiency, even in the absence of PIPKIγ. Surprisingly, an independently generated mouse line, PIPKIγ gene-trap KO mice, fail to develop beyond embryonic day 11.5 (43). The reason for the differences between the 2 mutant mice remains unclear and may reflect the different genetic backgrounds (43). Recently, a mutation in human PIPKIγ(D253N) that almost completely abolishes its enzymatic activity was shown to be responsible for a very severe condition leading to late prenatal or perinatal lethality, lethal congenital contractural syndrome (LCCS3). This syndrome is characterized by fetal akinesia, limb contractures, and muscle atrophy (44), a phenotype that could be explained by motor neuron failure. This finding supports the concept that lack of PIPKIγ activity becomes essential only late in embryonic development, when normal functioning of the nervous system becomes critical.
PIPKIα gene-trap mice, which express virtually no detectable PIPKIα, are viable with no obvious neurological defects, although they have some defects in fertility, decreased platelet levels (30%) of PI(4,5)P2 and impaired thrombin-induced IP3 production in platelets (45). PIPKIβ KO mice have defects in bone marrow-derived mast cell function that correlate with a 15% decrease in PI(4,5)P2 levels in these cells (46). However, overall, PIPKIβ KO mice are healthy, fertile, and show no obvious neurological defects.
Prompted by the observation that the nervous system can develop and at least partially function in the absence of PIPKIγ (32), we have now examined the contribution of PIPKIα and PIPKIβ to PI(4,5)P2 synthesis in the brain and the synergistic function of the 3 PIPKIs in supporting embryonic development and postnatal life.
EXPERIMENTAL PROCEDURES
Mice
Mice used for this study were PIPKIα gene trap (strain 129Ola;C57BL/6) (45), PIPKIβ KO mice (strain 129Ola;C57BL/6) (46), and PIPKIγ KO mice (strain 129SV/J; C57BL/6) (32). Unless otherwise indicated in the figure legends, all experiments were performed using E17-E19 brains.
Antibodies and Western Blotting
Primary antibodies were as follows. Antibodies directed against SHIP2 were raised in our laboratory (antigen raised against amino acids 887–1019 from human SHIP23). Antibodies directed against PIPKIγ (38), synaptojanin 1 (47), and OCRL (48) were described previously. Other antibodies include anti-human PIPKIβ from Santa Cruz Biotechnology (catalogue number sc-11778), anti-human PIPKIα from Santa Cruz Biotechnology (catalogue number sc-11774) or kindly provided by Dr. Martine Roussel (St. Jude Children's Research Hospital, Memphis TN (49)), and anti-PIPKIIα and anti-PIPKIIβ from Dr. Lew Cantley (Harvard Medical School). Western blotting of tissue extracts was carried out by standard procedures. Blots were visualized with enhanced chemiluminescence.
High Performance Liquid Chromatography (HPLC)
HPLC of deacylated lipids was performed essentially as described (7, 50). Tissues were homogenized using a motorized Dounce homogenizer in cold chloroform/methanol (1:2) (v/v), 50 mm HCl, and 2 mm AlCl3, which was shown to minimize PI(4,5)P2 breakdown. Resulting extracts were vortexed, and CHCl3 and HCl were added to break the phases. After centrifugation, the lower solvent phase was removed with a transfer pipette and washed with methanol and 2 mm oxalic acid. The solvent phase was again removed and dried under liquid nitrogen. The lipids were deacylated with 40% monomethylamine:water:butanol:methanol (36:8:9:47), dried in a SpeedVac, and resuspended in butanol:pertroleum ether:ethyl formate (20:40:1) and water. The aqueous phase was transferred and dried in a SpeedVac, and the resulting sample was resuspended in water. Phosphorylated glycerol headgroups were resolved using anion exchange HPLC with a KOH gradient and were detected by conductivity (ICS 3000 Dionex ion chromatography system; Ionpac analytical column AS11-HC 2 × 250 mm). Levels of each species were measured by quantifying the area under each peak and normalized to glycerophosphorylserine levels. Data were analyzed with analysis of variance and Dunnett's post-hoc test.
In Vitro Lipid Kinase Assays
Tissues were homogenized in buffer containing 50 mm Tris, pH 7.4, 10 mm MgCl2, 1 mm EGTA, and protease inhibitors. Homogenates were spun at 1000 × g, and this post-nuclear supernatant was further spun at 200,000 × g to obtain a membrane free supernatant (cytosol). To generate PI4P micelles, C16 PI4P (Echelon Biosciences Inc., Salt Lake City, UT) in chloroform:methanol was dried under N2 gas, suspended at 1 mg/ml in 50 mm Tris, pH 8.0, and bath-sonicated for 15 s. Fifty μg of cytosol was incubated for 15 min at 37 °C with 80 μm PI4P micelles with 10 μCi of [32γP]ATP, 50 μm ATP in 50 mm Tris, pH 7.4, 10 mm MgCl2, 1 mm EGTA at a final volume of 50 μl. Reactions were stopped with 700 μl of chloroform:methanol (2:1) containing 10 μg/ml brain phosphoinositides (Sigma, catalogue number P-6023) and 400 μl of 1 n HCl. After vortexing and centrifugation, the solvent phase was washed with 500 μl of methanol:HCl:water (20:20:1), and the solvent phase was transferred and dried under liquid nitrogen and resuspended in chloroform:methanol (2:1). Samples were spotted onto a silica plate (Fisher) and resolved by thin layer chromatography using water:acetic acid:methanol:acetone:chloroform (14:32:24:30:64) as a solvent. PI(4,5)P2 was quantified by either densitometry of autoradiography films using Image J software or by liquid scintillation spectroscopy of PI(4,5)P2 spots scraped from the TLC plate. Both methods of quantitation gave similar results.
Electrophysiology
Whole-cell patch clamp recordings of miniature excitatory post-synaptic currents (mEPSC) were performed on 13–16 days in vitro primary hippocampal neuronal cultures. During recordings, neurons were continuously perfused with an extracellular solution containing 140 mm NaCl, 3 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 10 mm HEPES, 20 mm glucose buffered to pH 7.3 with NaOH. The intracellular solution present in the pipette contained 115 mm CsMeSO4, 20 mm CsCl, 10 mm HEPES, 0.6 mm EGTA, 2.5 mm MgCl2, 0.4 mm Na3GTP, 4 mm Na2ATP, 10 mm sodium phosphocreatine. A concentration of 50 μm picrotoxin was used to block inhibitory synaptic transmission, and 500 nm tetrodotoxin was used to block the generation of action potentials. For readily releasable pool measurements, 4-s pulses of hypertonic (500 mm) sucrose were applied near neuronal perikarya in the presence of tetrodotoxin (500 nm) (51).
Recordings were acquired with a patch amplifier (EPC-9, HEK). Glass electrodes (Hilgenberg GmbH) were pulled with a Sutter P-97 micropipette puller (Sutter Instrument Co.) to a tip resistance of 2–3 megaohms. Data were filtered at 1 kHz and digitized at 2 kHz. mEPSCs were analyzed using the Mini Analysis Program (Synaptosoft, Leonia, NJ), and the threshold of mEPSC amplitude was set at 5 pA. The holding potential was −60 mV. Series resistance (RS) during whole-cell recordings was not compensated, and recordings with RS values greater than 20 megaohms were not included in the analysis.
RESULTS
Pattern of Expression of PIPKIs
To begin to characterize the contribution of the PIPKI isoforms to PI(4,5)P2 synthesis in the nervous system, we examined their pattern of expression in brains relative to other various tissues and during development. All three isoforms, referred to henceforth α, β, and γ (human nomenclature), had the highest level of expression in the brain and were also expressed at lower and variable levels in other tissues (Fig. 1A). The γ isoform, as shown by an antibody that recognizes all γ splice variants, was by far more concentrated in brain than in other tissues, as previously described (38, 39). α was the isoform with the lower variability of expression among tissues.
The three PIPKI isoforms also showed different patterns of expression in brain during development (Fig. 1B). Western blots of lysates from embryos heads and postnatal brains revealed that levels of α were highest early in embryogenesis and slightly decreased over time. In contrast, expression of γ increased with embryonic development and continued to steadily increase postnatally, coincident with synaptic development. Levels of β also greatly increased postnatally. Collectively, these data are consistent with a specialized role of β and γ in the nervous system and with a predominant house-keeping function of α.
PIPKIs, Embryonic Development, and Viability
Previously described mice with disrupting mutations in each of the three PIPKI genes were bred to each other to establish the impact of double and triple mutations at the organismal level. Two such mutations in the genes encoding β and γ, respectively, had been generated by conventional targeted mutagenesis; that is, deletion of exon 3 in the case of the β isoform (46) and of exons 2–6 in the case of the γ isoform (32). In both cases no protein product was detected in homozygous mutant mice. The mutation in the gene encoding α was generated by random insertion mutagenesis (gene trap). In the mutant gene, β-geo is inserted into intron 1 so that exon 1 splices with the β-geo creating a non-functional product (45). Mice homozygous for this mutation have virtually no expression of wild type product (45). α/β double KO mice survived to adulthood. Strikingly, this happened even in the presence of a single normal γ allele, demonstrating that a single copy of the γ gene is sufficient for life and, thus, the critical importance of the γ isoform.
β/γ double KO mice developed normally in utero and were born but died within minutes after birth (Fig. 2). For comparison, β KO mice live to adulthood without a major phenotype (46), and γ KO mice live for several hours (32). Immunoblots (Fig. 3A) confirmed the absence of β and γ isoforms in their brain. β/γ double KO mice that were born by natural delivery or by cesarean sections at E18 did initially respond to stimulation with slight movements and exhibited shallow respiratory movements. However, all movements soon ceased, and the mice turned cyanotic (Fig. 2) and died within minutes. All β/γ double KO mice delivered by C-section at E18 had beating hearts, and histological analysis of all major tissues did not reveal obvious defects.
FIGURE 2.
Perinatal lethality of β/γ double KO mice. Shown is a photograph of a P0 β/γ double KO pup and a β/γ double heterozygote littermate control.
FIGURE 3.
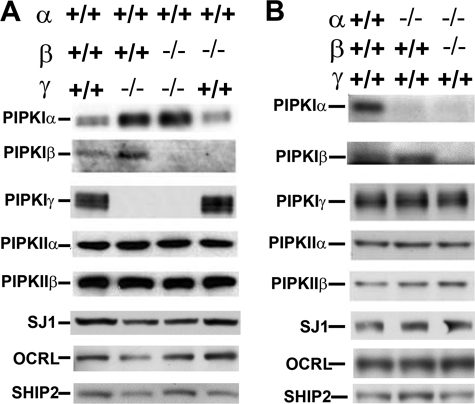
Levels of PI(4,5)P2-metabolizing enzymes in PIPKI mutant mice. Western blots for the enzymes indicated brain extracts from E18 mice with various genotypes. The only obvious adaptive change observed was the increase of α and β in mice that lack the γ isoform. A and B refer to different litters. Each blot is representative of results obtained with at least three independent experiments.
Thus, the lack of the β isoforms worsens the phenotype of γ KO mice leading to an even earlier postnatal death and indicating a synergistic function of the two PIPKIs. This severe phenotype is consistent with impaired nervous system function, as previously shown for γ KO mice. However, as in the single γ KO mice, nervous system dysfunction in the β/γ double KO mice did not seem to be catastrophic, because the brain underwent a grossly normal development, and some activity did occur. Furthermore, primary neuronal cultures obtained from β/γ KO mice had a normal appearance, and electrophysiological recordings of mEPSCs, i.e. of the electrical events that reflect exocytosis of single synaptic vesicles, revealed that the additional absence of β did not worsen the phenotype previously observed in γ KO neurons (supplemental Fig. 1). The frequency of mEPSCs and readily releasable pool of synaptic vesicles, as detected by recording mEPSCs in response to hyperosmolar sucrose application, were decreased to the same extent in γ and β/γ double KO neurons, with no change in mEPSC amplitude in both conditions (supplemental Fig. 1). Most likely, the more severe neurological phenotype of β/γ KO mice than of γ KO mice reflects defects at circuit levels resulting from minor additional impairments rather than a major synaptic failure. We conclude that the α PIPKI isoform is sufficient to support development, including development of the nervous system.
No α/γ double KO mice were born. Of 183 embryos from breeding that should have yielded α/γ double KOs, no such double KOs were found (Tables 1 and 2). Thus, although mice that lack the α isoform, like mice that lack the β isoform, survive to adulthood and are fertile, in the absence of the γ isoform, the additional absence of α produces a more severe phenotype than the additional absence of β. Consistent with the developmental pattern of expression of the three PIPKI isoforms during development, α plays a housekeeping function that overlaps with γ during development, whereas β synergizes with γ during postnatal life.
TABLE 1.
Mendelian ratios
| Crosses between β and γ mutants (all α wt) | Total embryos | β−/− |
γ−/− |
β−/−γ−/− |
|||
|---|---|---|---|---|---|---|---|
| Expected | Observed | Expected | Observed | Expected | Observed | ||
| β+/−; γ+/− × β+/−; γ+/− | 245 | 62 (25%) | 59 | 62 (25%) | 57 | 16 (6.25%) | 11 |
| β−/−; γ+/− × β−/−; γ+/− | 113 | 113 (100%) | 113 | 29 (25%) | 30 | 29 (25%) | 30 |
TABLE 2.
Mendelian ratios
| Crosses between α, β and γ mutants | Total embryos | α−/− |
β−/− |
γ−/− |
α−/−β−/− |
α−/−γ−/− |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Expected | Observed | Expected | Observed | Expected | Observed | Expected | Observed | Expected | Observed | ||
| α+/−; β+/−; γ+/− × α+/−; β+/−; γ+/− | 123 | 31 (25%) | 26 | 31 (25%) | 19 | 31 (25%) | 22 | 8 (6.25%) | 7 | 8 (6.25%) | 0 |
| α+/−; β+/−; γ+/− × α+/−; β−/−; γ+/− | 50 | 25 (50%) | 21 | 25 (50%) | 18 | 25 (50%) | 10 | 7 (12.5%) | 8 | 7 (12.5%) | 0 |
| α−/−; β+/−; γ+/− × α−/−; β+/−; γ+/− | 10 | 10 (100%) | 10 | 3 (25%) | 4 | 3 (25%) | 0 | 3 (25%) | 4 | 3 (25%) | 0 |
Adaptive Changes of PI(4,5)P2-metabolizing Enzymes in PIPKI KO Neurons
We next investigated whether a lack of PIPKI isoforms in brain resulted in adaptive increased levels of other PIPK isoforms or in decreased expression of PI(4,5)P2 phosphatases. Because γ KO mice do not survive beyond day 1, these comparisons were carried out in full-term embryos obtained by Cesarean section. A compensatory increase of the other two PIPKI isoforms was observed only in the absence of the γ isoform (see the increase of α and β isoforms in the γ KO brains in Fig. 3A) possibly because only the absence of this kinase results in a major reduction of PI(4,5)P2 levels in brain (Fig. 4 and 5). This result agrees with the previous observation that mRNAs for α and β isoforms increase in HeLa cells upon knockdown of γ by siRNA (52). No change was observed, not even in the γ KO, in the levels of the type II PIPKs for which antibodies were available (α and β isoforms, Fig. 3), speaking against a compensatory up-regulation of PI(4,5)P2 synthesis via the phosphorylation of PI5P at the 4 position of the inositol ring. There were no obvious changes in protein levels of the three major brain PI(4,5)P2 phosphatases tested, synaptojanin 1, OCRL, and SHIP2. Although it remains possible that levels of some of the other PI(4,5)P2 phosphatases encoded by the mammalian genome (53) are modified (for example INPP5e, which recent studies have implicated in brain function (18, 19)), clearly the absence of PIPKIs does not result in global compensatory change of levels of expression of the major PI(4,5)P2 phosphatases.
FIGURE 4.
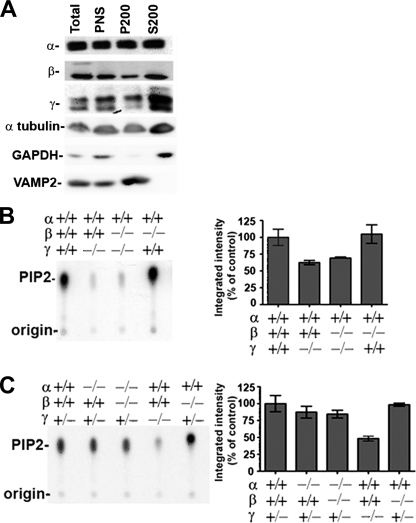
Impact of the lack of PIPKI isoforms on the PI4P 5-kinase activity present in mouse brain cytosol. A, Western blots are shown for the three PIPKI isoforms in total homogenate, postnuclear supernatant (PNS), and 200,000 × g pellet (P200 = membrane fraction) and supernatant (S200 = cytosol) generated from the post-nuclear supernatant. A large fraction of each kinase is recovered in the cytosol. Other proteins are shown as a control; note that VAMP2, a transmembrane protein, is absent from the cytosol, whereas GAPDH, a predominantly soluble protein, is enriched in this fraction. B and C, PIP2 was generated by brain cytosol from various genotypes in the presence of PI4P micelles and [γ-32P]ATP and then revealed by thin layer chromatography and autoradiography. Left panels, shown are representative examples of autoradiograms from PI4P 5-kinase assays involving mouse brain with the indicated genotypes. Right panels: shown is quantitation of the autoradiographic signal using data from independent experiments; bars in B represent averages (±S.E.) from at least three independent mice and in C represent averages from at least two independent mice.
FIGURE 5.
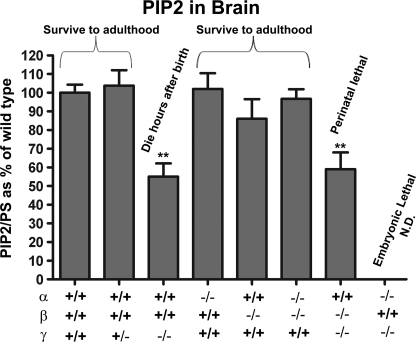
PIP2 levels in brains of PIPKI KO mice. All PIP2 values were normalized to glycerophosphorylserine. Data are expressed as percent of levels in WT and represent the mean (±S.E.). ** indicates p < 0.01. n = 16 for wild type, n = 7 for γ heterozygous, n = 8 for γ KO, n = 7 for α KO, n = 8 for β KO, n = 8 for α/β double KO, and n = 7 for β/γ double KO. N.D., not determined.
Contribution of PIPKI Isoforms to PI4P 5-Kinase Activity in Brain Cytosol
The availability of brain tissue lacking PIPKI isoforms allowed us to determine the contribution of each kinase to the PI4P 5-kinase activity present in the cytosol (200,000 × g supernatants). All three enzymes were present in the cytosol (Fig. 4A) (the contribution of the membrane-associated pools of these kinases to activity could not be determined because the PI4P 5-kinase activity was disrupted by detergents (54)). To analyze cytosolic PI4P 5-kinase activity, PI4P micelles were incubated with [32γP]ATP and cytosol from KO and control mice. Thin layer chromatography and autoradiography was then used to separate and detect PIP2 from PIP and PI.
As reported (32), there was a very strong decrease in the kinase activity of the γ KO cytosol relative to control (average 40% decrease in our assays) (Fig. 4, B and C) despite the up-regulation of the α and β isoforms in these mouse brains (see Fig. 3A). The combined absence of the β and γ isoforms showed a decreased activity similar to the one produced by the lack of the γ isoform alone, and lack of the β isoform alone did not produce an obvious decrease in kinase activity relative to control. Lack of the α isoform also produced only a minor decrease in the kinase activity (Fig. 4B) as shown by comparing mice with both or no alleles of the α gene in the γ heterozygous background.
Contributions of PIPKI Isoforms to Steady State Levels of Total PI(4,5)P2 in the Brain
Finally, we analyzed the impact of the absence of each PIPKI on the brain levels of PI(4,5)P2. To this aim, phospholipids were extracted from intact brains of E17-E19 mice (i.e. full term but before birth, given the lack of survival of some genotypes) and deacylated, and the resulting glycerophosphorylinositol headgroups were resolved from each other and from other deacylated lipids by an HPLC procedure based on anion exchange and conductivity detection (50). This method does not allow discrimination between the three different diphosphoinositides, but the bulk of PIP2 is known to be accounted for by PI(4,5)P2 (55). Hence, PIP2 values obtained by this method are thought to closely reflect PI(4,5)P2 levels.
Lack of both alleles of the γ isoform produced a significant decrease in PIP2 levels as reported (Fig. 5) (7, 32). In contrast, lack of the α and β isoforms alone or of both the α and β isoforms together produced no major reductions in PIP2 levels relative to control brains. Furthermore, the absence of both β and γ together did not enhance the decrease in PIP2 levels compared with mice lacking only the γ isoform. When a similar analysis was performed on liver extracts, no major differences in the PIP2 content was observed between the various genotypes, confirming the overlapping function of PIP kinases and the occurrence of major homeostatic mechanisms to control PIP2 levels (supplemental Fig. 2). The major impact of the absence of the γ isoform in brain but not in liver confirms that the unique and dominant function of this isoform in the nervous tissue.
DISCUSSION
It was shown previously that the γ PIPKI isoform is essential for postnatal life (32). Here we complement this finding by showing that a single allele of this kinase is sufficient to support life from conception to adulthood even when both the α and β isoforms are missing. Thus, PIPKIγ, which is expressed at very high levels in the nervous system (32, 38, 39) and is the predominant PIPKI in this tissue, is the most critical isoform for life. Most likely, the importance of γ for the function of the nervous system underlies the perinatal lethality resulting from its absence.
During embryonic development the function of the γ isoform overlaps in part with the function of α, as shown by the early embryonic lethality associated with the combined absence of these two kinases. Conversely, postnatally, β overlaps in function with γ, because the additional lack of β prevents even the short (hours) postnatal life that occurs in the absence of the γ isoform alone. These findings are in agreement with the developmental patterns of expression of the α and β isoforms in brain, which suggest a housekeeping function for α and a function in the developed nervous system for β. It may seem puzzling that the γ isoform alone can support life to adulthood given its presence in non-neuronal tissues at levels (as detected by immunoblotting) so much lower than in brain. However, this difference may be more a reflection of the extremely high concentration of γ in the nervous system than of a very low level expression outside the brain.
A variety of housekeeping function for α have been reported, including functions in gene expression and regulation (45, 56, 57). Clearly these functions must be nonessential or at least partially overlapping with (and compensated by in the absence of α) the functions of γ. Specialized functions of β in the nervous system may include the regulation of excitability and synaptic function via its interaction with PDZ domain-containing proteins such as EBP50/NHERF (41) that are expressed in neurons and localized at synapses (58). They may also include roles in axonal outgrowth and remodeling (59, 60), possibly through the effects of a PI(4,5)P2 pool controlled by β on the actin cytoskeleton (46). Given evidence for a role of specific interactions of γ with proteins with fundamental cellular functions such as cadherins (61), talin (33, 34, 62), and the clathrin adaptor AP-2 (28–31), it is surprising that mice can develop to full term in the absence of this kinase. Although gene-trap γ KO mice have an embryonic phenotype (43), no expression of either full-length or fragments of γ were identified upon an extensive search with a variety of antibodies in the brain of the previously described γ KO mouse used for this study (32). Thus, mice can clearly develop and live for few hours in the absence of γ, and the discrepancy between the two mice must be explained by a different genetic background.
Overall, our study reveals differences but also some overlap in the function of PIPKI isoforms. The absence of either α or β can be compensated by the other kinases. Hence, although it is clear that each kinase has a special role in the control of specific PI(4,5)P2 pools with important functional consequences on specific cellular functions (40, 42, 45, 46, 63), a significant level of mixing among these pools must also exist. The most critical role of γ may be explained in part by the selective concentration of this kinase at neuronal synapses (32, 38), whose distance from the cell body makes compensation by PI(4,5)P2 pools generated in other cell regions inefficient.
Supplementary Material
Acknowledgments
We thank Frank Wilson for outstanding support, Shawn Ferguson for discussion, Michelle Pirruccello for help, and Caroline Zeiss for histological analysis of control and β/γ double KO embryos.
This work was supported, in whole or in part, by National Institutes of Health Grants NS36251, DK45735, and DA018343 (to P. D. C.). This work was also supported by the Yale Center for Genomics and Proteomics and by a National Alliance for Research on Schizophrenia and Depression distinguished Neuroscience Award (to P. D. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
T. Itoh and P. De Camilli, unpublished information.
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PIPKI and PIPKII
- type I and type II phosphatidylinositol 4-phosphate 5-kinase, respectively
- mEPSC
- miniature excitatory post-synaptic currents.
REFERENCES
- 1.Hokin M. R., Hokin L. E. (1953) J. Biol. Chem. 203, 967–977 [PubMed] [Google Scholar]
- 2.Di Paolo G., De Camilli P. (2006) Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 3.Heck J. N., Mellman D. L., Ling K., Sun Y., Wagoner M. P., Schill N. J., Anderson R. A. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 15–39 [DOI] [PubMed] [Google Scholar]
- 4.Mao Y. S., Yin H. L. (2007) Pflugers Arch. 455, 5–18 [DOI] [PubMed] [Google Scholar]
- 5.Barlow C. A., Laishram R. S., Anderson R. A. (2010) Trends Cell Biol. 20, 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Bout I., Divecha N. (2009) J. Cell Sci. 122, 3837–3850 [DOI] [PubMed] [Google Scholar]
- 7.Voronov S. V., Frere S. G., Giovedi S., Pollina E. A., Borel C., Zhang H., Schmidt C., Akeson E. C., Wenk M. R., Cimasoni L., Arancio O., Davisson M. T., Antonarakis S. E., Gardiner K., De Camilli P., Di Paolo G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9415–9420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attree O., Olivos I. M., Okabe I., Bailey L. C., Nelson D. L., Lewis R. A., McInnes R. R., Nussbaum R. L. (1992) Nature 358, 239–242 [DOI] [PubMed] [Google Scholar]
- 9.Schurman S. J., Scheinman S. J. (2009) Nat. Rev. Nephrol. 5, 529–538 [DOI] [PubMed] [Google Scholar]
- 10.McCrea H. J., De Camilli P. (2009) Physiology 24, 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito T., Stopkova P., Diaz L., Papolos D. F., Boussemart L., Lachman H. M. (2003) Am. J. Med. Genet. B Neuropsychiatr Genet. 116B, 77–83 [DOI] [PubMed] [Google Scholar]
- 12.Stopkova P., Saito T., Fann C. S., Papolos D. F., Vevera J., Paclt I., Zukov I., Stryjer R., Strous R. D., Lachman H. M. (2003) Am. J. Med. Genet. B. Neuropsychiatr. Genet. 123B, 50–58 [DOI] [PubMed] [Google Scholar]
- 13.Soares J. C., Dippold C. S., Wells K. F., Frank E., Kupfer D. J., Mallinger A. G. (2001) Neurosci. Lett. 299, 150–152 [DOI] [PubMed] [Google Scholar]
- 14.Landman N., Jeong S. Y., Shin S. Y., Voronov S. V., Serban G., Kang M. S., Park M. K., Di Paolo G., Chung S., Kim T. W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19524–19529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berman D. E., Dall'Armi C., Voronov S. V., McIntire L. B., Zhang H., Moore A. Z., Staniszewski A., Arancio O., Kim T. W., Di Paolo G. (2008) Nat. Neurosci. 11, 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marion E., Kaisaki P. J., Pouillon V., Gueydan C., Levy J. C., Bodson A., Krzentowski G., Daubresse J. C., Mockel J., Behrends J., Servais G., Szpirer C., Kruys V., Gauguier D., Schurmans S. (2002) Diabetes 51, 2012–2017 [DOI] [PubMed] [Google Scholar]
- 17.Yuan T. L., Cantley L. C. (2008) Oncogene 27, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacoby M., Cox J. J., Gayral S., Hampshire D. J., Ayub M., Blockmans M., Pernot E., Kisseleva M. V., Compère P., Schiffmann S. N., Gergely F., Riley J. H., Pérez-Morga D., Woods C. G., Schurmans S. (2009) Nat. Genet. 41, 1027–1031 [DOI] [PubMed] [Google Scholar]
- 19.Bielas S. L., Silhavy J. L., Brancati F., Kisseleva M. V., Al-Gazali L., Sztriha L., Bayoumi R. A., Zaki M. S., Abdel-Aleem A., Rosti R. O., Kayserili H., Swistun D., Scott L. C., Bertini E., Boltshauser E., Fazzi E., Travaglini L., Field S. J., Gayral S., Jacoby M., Schurmans S., Dallapiccola B., Majerus P. W., Valente E. M., Gleeson J. G. (2009) Nat. Genet. 41, 1032–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norris F. A., Wilson M. P., Wallis T. S., Galyov E. E., Majerus P. W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14057–14059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terebiznik M. R., Vieira O. V., Marcus S. L., Slade A., Yip C. M., Trimble W. S., Meyer T., Finlay B. B., Grinstein S. (2002) Nat. Cell Biol. 4, 766–773 [DOI] [PubMed] [Google Scholar]
- 22.McLaughlin S., Murray D. (2005) Nature 438, 605–611 [DOI] [PubMed] [Google Scholar]
- 23.Krauss M., Haucke V. (2007) EMBO Rep. 8, 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rameh L. E., Tolias K. F., Duckworth B. C., Cantley L. C. (1997) Nature 390, 192–196 [DOI] [PubMed] [Google Scholar]
- 25.Ishihara H., Shibasaki Y., Kizuki N., Katagiri H., Yazaki Y., Asano T., Oka Y. (1996) J. Biol. Chem. 271, 23611–23614 [DOI] [PubMed] [Google Scholar]
- 26.Loijens J. C., Anderson R. A. (1996) J. Biol. Chem. 271, 32937–32943 [DOI] [PubMed] [Google Scholar]
- 27.Chang J. D., Field S. J., Rameh L. E., Carpenter C. L., Cantley L. C. (2004) J. Biol. Chem. 279, 11672–11679 [DOI] [PubMed] [Google Scholar]
- 28.Bairstow S. F., Ling K., Su X., Firestone A. J., Carbonara C., Anderson R. A. (2006) J. Biol. Chem. 281, 20632–20642 [DOI] [PubMed] [Google Scholar]
- 29.Nakano-Kobayashi A., Yamazaki M., Unoki T., Hongu T., Murata C., Taguchi R., Katada T., Frohman M. A., Yokozeki T., Kanaho Y. (2007) EMBO J. 26, 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krauss M., Kukhtina V., Pechstein A., Haucke V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thieman J. R., Mishra S. K., Ling K., Doray B., Anderson R. A., Traub L. M. (2009) J. Biol. Chem. 284, 13924–13939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Paolo G., Moskowitz H. S., Gipson K., Wenk M. R., Voronov S., Obayashi M., Flavell R., Fitzsimonds R. M., Ryan T. A., De Camilli P. (2004) Nature 431, 415–422 [DOI] [PubMed] [Google Scholar]
- 33.Ling K., Doughman R. L., Firestone A. J., Bunce M. W., Anderson R. A. (2002) Nature 420, 89–93 [DOI] [PubMed] [Google Scholar]
- 34.Di Paolo G., Pellegrini L., Letinic K., Cestra G., Zoncu R., Voronov S., Chang S., Guo J., Wenk M. R., De Camilli P. (2002) Nature 420, 85–89 [DOI] [PubMed] [Google Scholar]
- 35.Fairn G. D., Ogata K., Botelho R. J., Stahl P. D., Anderson R. A., De Camilli P., Meyer T., Wodak S., Grinstein S. (2009) J. Cell Biol. 187, 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min S. H., Abrams C. S. (2009) Biochem. J. 423, e5–8 [DOI] [PubMed] [Google Scholar]
- 37.Desrivières S., Cooke F. T., Parker P. J., Hall M. N. (1998) J. Biol. Chem. 273, 15787–15793 [DOI] [PubMed] [Google Scholar]
- 38.Wenk M. R., Pellegrini L., Klenchin V. A., Di Paolo G., Chang S., Daniell L., Arioka M., Martin T. F., De Camilli P. (2001) Neuron 32, 79–88 [DOI] [PubMed] [Google Scholar]
- 39.Ishihara H., Shibasaki Y., Kizuki N., Wada T., Yazaki Y., Asano T., Oka Y. (1998) J. Biol. Chem. 273, 8741–8748 [DOI] [PubMed] [Google Scholar]
- 40.Mao Y. S., Yamaga M., Zhu X., Wei Y., Sun H. Q., Wang J., Yun M., Wang Y., Di Paolo G., Bennett M., Mellman I., Abrams C. S., De Camilli P., Lu C. Y., Yin H. L. (2009) J. Cell Biol. 184, 281–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacalle R. A., Peregil R. M., Albar J. P., Merino E., Martínez-A. C., Mérida I., Mañes S. (2007) J. Cell Biol. 179, 1539–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasudevan L., Jeromin A., Volpicelli-Daley L., De Camilli P., Holowka D., Baird B. (2009) J. Cell Sci. 122, 2567–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Lian L., Golden J. A., Morrisey E. E., Abrams C. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11748–11753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narkis G., Ofir R., Landau D., Manor E., Volokita M., Hershkowitz R., Elbedour K., Birk O. S. (2007) Am. J. Hum. Genet. 81, 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Chen X., Lian L., Tang T., Stalker T. J., Sasaki T., Kanaho Y., Brass L. F., Choi J. K., Hartwig J. H., Abrams C. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14064–14069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasaki J., Sasaki T., Yamazaki M., Matsuoka K., Taya C., Shitara H., Takasuga S., Nishio M., Mizuno K., Wada T., Miyazaki H., Watanabe H., Iizuka R., Kubo S., Murata S., Chiba T., Maehama T., Hamada K., Kishimoto H., Frohman M. A., Tanaka K., Penninger J. M., Yonekawa H., Suzuki A., Kanaho Y. (2005) J. Exp. Med. 201, 859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cremona O., Di Paolo G., Wenk M. R., Lüthi A., Kim W. T., Takei K., Daniell L., Nemoto Y., Shears S. B., Flavell R. A., McCormick D. A., De Camilli P. (1999) Cell 99, 179–188 [DOI] [PubMed] [Google Scholar]
- 48.Erdmann K. S., Mao Y., McCrea H. J., Zoncu R., Lee S., Paradise S., Modregger J., Biemesderfer D., Toomre D., De Camilli P. (2007) Dev. Cell 13, 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis J. N., Rock C. O., Cheng M., Watson J. B., Ashmun R. A., Kirk H., Kay R. J., Roussel M. F. (1997) Mol. Cell. Biol. 17, 7398–7406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nasuhoglu C., Feng S., Mao J., Yamamoto M., Yin H. L., Earnest S., Barylko B., Albanesi J. P., Hilgemann D. W. (2002) Anal. Biochem. 301, 243–254 [DOI] [PubMed] [Google Scholar]
- 51.Stevens C. F., Tsujimoto T. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 846–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Padrón D., Wang Y. J., Yamamoto M., Yin H., Roth M. G. (2003) J. Cell Biol. 162, 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ooms L. M., Horan K. A., Rahman P., Seaton G., Gurung R., Kethesparan D. S., Mitchell C. A. (2009) Biochem. J. 419, 29–49 [DOI] [PubMed] [Google Scholar]
- 54.Parker G. J., Loijens J. C., Anderson R. A. (1998) Methods Mol. Biol. 105, 127–139 [DOI] [PubMed] [Google Scholar]
- 55.Wenk M. R., Lucast L., Di Paolo G., Romanelli A. J., Suchy S. F., Nussbaum R. L., Cline G. W., Shulman G. I., McMurray W., De Camilli P. (2003) Nat. Biotechnol. 21, 813–817 [DOI] [PubMed] [Google Scholar]
- 56.Pan W., Choi S. C., Wang H., Qin Y., Volpicelli-Daley L., Swan L., Lucast L., Khoo C., Zhang X., Li L., Abrams C. S., Sokol S. Y., Wu D. (2008) Science 321, 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mellman D. L., Gonzales M. L., Song C., Barlow C. A., Wang P., Kendziorski C., Anderson R. A. (2008) Nature 451, 1013–1017 [DOI] [PubMed] [Google Scholar]
- 58.Paquet M., Kuwajima M., Yun C. C., Smith Y., Hall R. A. (2006) J. Comp. Neurol. 494, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hernández-Deviez D. J., Roth M. G., Casanova J. E., Wilson J. M. (2004) Mol. Biol. Cell 15, 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamazaki M., Miyazaki H., Watanabe H., Sasaki T., Maehama T., Frohman M. A., Kanaho Y. (2002) J. Biol. Chem. 277, 17226–17230 [DOI] [PubMed] [Google Scholar]
- 61.Ling K., Bairstow S. F., Carbonara C., Turbin D. A., Huntsman D. G., Anderson R. A. (2007) J. Cell Biol. 176, 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee S. Y., Voronov S., Letinic K., Nairn A. C., Di Paolo G., De Camilli P. (2005) J. Cell Biol. 168, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y. J., Li W. H., Wang J., Xu K., Dong P., Luo X., Yin H. L. (2004) J. Cell Biol. 167, 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.