Abstract
Recent advances in our understanding of translational dynamics indicate that codon usage and mRNA secondary structure influence translation and protein folding. The most frequent cause of cystic fibrosis (CF) is the deletion of three nucleotides (CTT) from the cystic fibrosis transmembrane conductance regulator (CFTR) gene that includes the last cytosine (C) of isoleucine 507 (Ile507ATC) and the two thymidines (T) of phenylalanine 508 (Phe508TTT) codons. The consequences of the deletion are the loss of phenylalanine at the 508 position of the CFTR protein (ΔF508), a synonymous codon change for isoleucine 507 (Ile507ATT), and protein misfolding. Here we demonstrate that the ΔF508 mutation alters the secondary structure of the CFTR mRNA. Molecular modeling predicts and RNase assays support the presence of two enlarged single stranded loops in the ΔF508 CFTR mRNA in the vicinity of the mutation. The consequence of ΔF508 CFTR mRNA “misfolding” is decreased translational rate. A synonymous single nucleotide variant of the ΔF508 CFTR (Ile507ATC), that could exist naturally if Phe-508 was encoded by TTC, has wild type-like mRNA structure, and enhanced expression levels when compared with native ΔF508 CFTR. Because CFTR folding is predominantly cotranslational, changes in translational dynamics may promote ΔF508 CFTR misfolding. Therefore, we propose that mRNA “misfolding” contributes to ΔF508 CFTR protein misfolding and consequently to the severity of the human ΔF508 phenotype. Our studies suggest that in addition to modifier genes, SNPs may also contribute to the differences observed in the symptoms of various ΔF508 homozygous CF patients.
Keywords: ABC Transporter, Cystic Fibrosis, DNA, MRNA, Translation, F508del CFTR, Codon Usage, mRNA Structure, Synonymous SNP
Introduction
Changes in mRNA secondary structures have major biological consequences (1–4). Identification of disease causing mutations that alter the regulatory regions of mRNAs (5) and single nucleotide polymorphisms (SNPs) in the coding regions that control gene expression (6) support this idea. Studies on the dynamics of translation also establish that the secondary structure of the mRNA determines the length of the pause cycles and the rate of translation (7). Membrane integration and folding of multi-spanning membrane proteins is mainly cotranslational (8, 9), and therefore mRNA structure-related changes in translational dynamics are likely to influence membrane protein folding.
Cystic fibrosis (CF)2 results from mutations in the CFTR gene (10–12). CFTR, a multi-spanning membrane glycoprotein in the ABC transporter superfamily, functions as a chloride channel in epithelial cells and regulates complex cellular functions (13, 14). The most common disease-causing mutation in CFTR is the deletion of three nucleotides (CTT) that results in the loss of Phe-508 (ΔF508) and a synonymous SNP (ATT) for Ile-507 (11, 15). Because ΔF508 CFTR is misfolded and subject to rapid endoplasmic reticulum-associated degradation (ERAD) (16–18), CF has become an excellent model for protein folding disorders (19–21). Previous studies, however, have focused on the CFTR protein only, without considering the possible effects of the mutation on CFTR mRNA structure, translation, and cotranslational protein folding.
The concept that mutations may alter mRNA structure and protein folding led to the experiments presented herein. We compared the structures of WT and ΔF508 CFTR mRNAs via molecular modeling (22, 23) and circular dichroism (CD) spectroscopy (24). To confirm the differences in mRNA structures, we developed an mRNA folding assay based on published methods (25–27). We also analyzed the stability and compared the translational rates of WT, ΔF508 (Ile507ATT), and an SNP variant (Ile507ATC) of ΔF508 CFTR mRNAs. Based on the observed differences between the translational rates of two variants of ΔF508 CFTR mRNAs, we developed cell lines and compared the expression of the native (Ile507ATT) and variant (Ile507ATC) ΔF508 CFTR. The results indicate that mRNA “misfolding” may contribute to the severity of the ΔF508 phenotype.
EXPERIMENTAL PROCEDURES
Molecular Modeling
mRNA structural models were created using the MFOLD algorithm (22, 28).
Plasmids
pcDNA3.1 constructs containing WT (pcDNA3.1-WT) or ΔF508 CFTR (pcDNA3.1- ΔF508CFTR-Ile507ATT) were used for in vitro transcription and translation experiments as described previously (29, 30).
Construction of pcDNA3.1 ΔF508CFTR-Ile507ATC
The Stratagene QuikChange Primer Design Program was used to create mutagenic primers. The construct was generated using QuickChange Lightning Site-Directed Mutagenesis kit (Stratagene 210518) with pcDNA3.1ΔF508 (Ile507ATT)) as a template. All constructs used in these studies were sequenced prior to experiments at the UAB Department of Genetics Sequencing Core Facility.
In Vitro Transcription of CFTR mRNA for Circular Dichroism (CD) Studies
CFTR transcripts were synthesized from Xhol-cut pcDNA-WT or pcDNA-ΔF plasmids using MEGAscript T7 (Ambion, AM133) and purified using MEGAclear (Ambion AM1908) according to the manufacturer's recommendations.
Circular Dichroism Spectroscopy
CD spectra of refolded WT and ΔF508 CFTR mRNA (0.35 mg/ml) samples were acquired with a JASCO J-815 spectrophotometer (31, 32) at 4.6 °C in a 0.1-cm cell with a data pitch of 0.2 nm and 8 s averaging time. Three individually synthesized and purified mRNA samples from each construct were tested. Each sample was run six times to obtain the CD spectra. The results represent the average and standard deviation of 18 samples. The results are expressed in molar ellipticity [θ] as function of the wavelength (λ) (28–30).
mRNA Folding Assays
The mRNA folding assays were designed based on the principles of the selective 2′-hydroxyl acylation reaction analyzed by primer extension (SHAPE) assay (25–27) with the following modifications. We inserted ∼150-nucleotide fragments from the mutation region of the CFTR gene (WT, ΔF508 (Ile507ATT) or variant ΔF508 (Ile507ATC)) into a cassette containing T7 promoter and linker sequences that allow the synthesis of the mRNA fragments. The purified products were placed in RNA folding buffer (27) and subjected to digestion with RNase T1 (Ambion AM2283) that cleaves the 3′-end of single-stranded G residues. Digestion times and RNase T1 concentrations were first optimized, and experiments were performed using 0.01 unit or 0.05 unit of T1 for 15 min at 25 °C. RNase T1 activity was stopped with the addition of 3 m ammonium acetate and ethanol. Following a 70% ethanol wash and air-drying, samples containing the RNA digests were solubilized in H2O and used in one-step semi-quantitative RT-PCR (Applied Biosystems 4309169) to amplify WT and ΔF508 CFTR-specific fragments. Primers were designed based on the differences in the predicted secondary structures of the WT and ΔF508 CFTR mRNA fragments and the expected digests following RNase T1 digestion. WT CFTR fragment specific primers: WT Primer 1: 5′-CCT GGC ACC ATT AAA GAA; WT Primer 2 (complementary): 5′-TTC TGT ATC TAT ATT CAT-3′; ΔF508 fragment-specific primers: ΔF508 primer 1 (complementary): 5′-TTT AAT GGT GCC AGG C-3′; ΔF508 primer 2: 5′-TGG AAG AAT TTC ATT CTG TTC TCA G-3′. PCR products were analyzed on 5% agarose gels and visualized with ethidium bromide.
In Vitro Transcription/Translation in the Presence of Microsomes
In vitro translation reactions in the presence of microsomes were performed as described previously (29) with specific modifications. Full-length WT, ΔF508 (Ile507ATT), and ΔF508 (Ile507ATC) CFTR proteins were synthesized from pCDNA3.1 WT, pCDNA3.1 ΔF508 (Ile507ATT), and pCDNA 3.1 ΔF508 (Ile507ATC) plasmids. Six parallel translation reactions were performed for each construct. Ten minutes after addition of canine pancreatic microsomal membranes (Promega Y4041), translation initiation was stopped with aurintricarboxylic acid (ATA, 100 μm final concentration, Sigma A1895). Translation rates were determined from the reaction mixture based on the incorporation of [35S]methionine via TCA precipitation at different time points following addition of ATA. Results are presented as the means ± S.D. of six samples.
WT and ΔF508 CFTR mRNA Half-life Measurements
CFTR mRNA half-lives were measured in HeLaWT or HeLaΔF cells (33) cultured under standard conditions as described (57). The mRNA half-lives were calculated from the exponential decay based on: C/C0 = e−kdt (C and C0 are mRNA amounts after time t and at t0, respectively, and kd is the mRNA decay constant). CFTR mRNA levels were measured by real time RT-PCR (32).
Development of Cell Lines Expressing Native (Ile507ATT) or Variant (Ile507ATC) ΔF508 CFTR
293F cells (Invitrogen) were used to express native and variant ΔF508 CFTR under the control of the tetracycline-responsive element (TRE) as described previously (34–40). See supplemental materials for details.
Western Blotting
Cellular proteins were isolated and separated using standard protocols as described (35, 56). Following Western transfer, CFTR was detected using the MM13–4 anti-CFTR N-terminal monoclonal antibody and ECL (Millipore).
Statistical Analysis
Results are expressed as means ± S.D. Statistical significance among means was determined using the Student's t test. A “p” value of <0.05 was considered significant.
RESULTS
Molecular Models of Human WT and ΔF508 CFTR mRNAs Predict Secondary Structural Differences
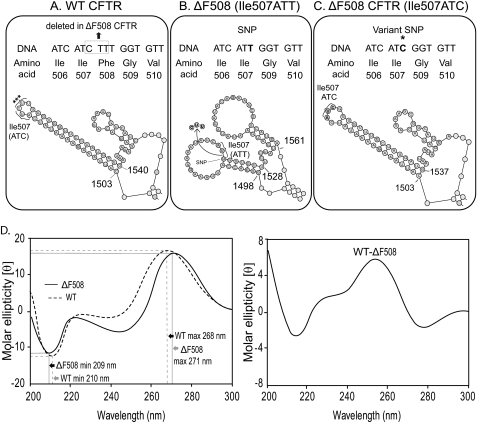
We utilized the full-length coding sequences of human WT and ΔF508 CFTR (NCBI Reference Sequence: NM_000492.3) (11) to obtain theoretical models of the resulting mRNA transcripts. Using the MFOLD algorithm (23), fifty theoretical structure models were generated with different energy values. The calculated minimal free energy values for WT and ΔF508 CFTR were Ewt = [−1148.49 to −1136.05] and EΔF = [−1144.04 to −1127.82], respectively. Within most of the models, and more importantly, in all models generated with the lowest energy values, the ΔF508 CFTR mRNA formed two enlarged loops with long single-stranded regions close to the mutation site (Fig. 1, A and B). We did not observe significant changes in the predicted secondary structures outside of the region containing the mutation (supplemental Fig. S1, A and B). Although there were only slight differences in the minimal energy values for WT and ΔF508 CFTR mRNA, the WT CFTR mRNA had a lower energy minimum than the mutant.
FIGURE 1.
Alterations in ΔF508 CFTR mRNA secondary structure as shown by theoretical modeling and circular dichoism spectroscopy. CFTR gene sequences for WT (A), ΔF508 (Ile507ATT) (B), and ΔF508 (Ile507ATC) (C) and corresponding amino acid sequences are shown (top panels). Theoretical models of the mRNA secondary structures in this region (bottom panel) are shown. The synonymous SNP (T) introduced by the mutation (*) and the exchanged nucleotide in the variant (Ile507ATC) ΔF508 CFTR are illustrated in the top panel. Nucleotides 1503 and 1540 (WT), 1498, 1528, and 1561 (ΔF508 CFTR Ile508ATT), and 1503 and 1537 (ΔF508 CFTR Ile507ATC) are labeled for easy comparison of the models (A in the ATG start codon = 1; the CTT deletion corresponds to nucleotides 1521–23); Ile-507 is labeled by brackets; ***: the ΔF508 mutation site. Full-length mRNA secondary structures (MFOLD) are shown in supplemental Fig. S1, A–C). D, CD spectra and differential CD spectrum of the WT and ΔF508 (Ile507ATT) CFTR mRNAs. The results are expressed in molar ellipticity [θ] as a function of the wavelength (λ); representative spectra of three individual experiments. Spectra were obtained from six individual runs of the same mRNA preparation. Maximum and minimum values and shifts in the wavelengths (± S.D. (n = 18)) are indicated by black and gray arrows, respectively.
Restoration of the Native Codon for Ile-507 (ATC) Eliminates the Predicted mRNA Structural Differences between the WT and ΔF508 CFTR mRNA in the Vicinity of the Mutation
Deletion of CTT from CFTR involves the last nucleotide (C) of Ile507 (ATC) and the first two nucleotides (TT) of Phe-508 (TTT). Therefore, the mutation introduces a synonymous codon, ATT for Ile-507 (Fig. 1, A and B). Based on this observation, we restored the native codon for Ile-507 (ATC) in the mutant and obtained theoretical models (Fig. 1C and supplemental Fig. S1C). Importantly, there were no enlarged single-stranded loops in the models of Ile507ATC ΔF508 CFTR mRNA (compare Fig. 1, A–C). Furthermore, the minimal energy value of the Ile507ATC ΔF508 CFTR variant (EΔFIle507ATC = [−1147.94 −1134]) was closer in value to the WT CFTR mRNA. Analysis of all synonyms in the vicinity of the mutation also revealed that CTT deletion from a CFTR with a Phe508TTC codon would result in the restoration of the native codon for Ile507 (ATC) in ΔF508 CFTR. This implies that the Ile507ATC ΔF508 CFTR variant may exist naturally. In summary, the models suggest that the Ile507ATT SNP is responsible for the formation of the enlarged single-stranded loops in the ΔF508 CFTR mRNA.
CD Spectroscopy Reveals Structural Differences between WT and ΔF508 CFTR mRNA
To confirm the structural alterations of the ΔF508 CFTR mRNA, we analyzed in vitro transcribed WT and ΔF508 CFTR mRNA by CD spectroscopy as described (24, 32). The CD spectra of the ΔF508 CFTR mRNAs had a decrease in maximum intensity (16.68; S.D. = 0.05; n = 18) at 271 nm, with a 3-nm shift of the maximum wavelength position compared with WT maximum (16.8; S.D. = 0.01; n = 18, p = <0.002) at 268 nm. The ΔF508 CFTR spectrum shows an increase in the intensity of the minimum (11.42; S.D. = 0.01; n = 18) at 209 nm with a 1-nm shift toward longer wavelengths compared with the WT CFTR mRNA spectrum minimum (12.4; S.D. = 0.09; n = 18; p = <0.001) (Fig. 1D, left panel). The differences between WT and ΔF508 CFTR mRNA CD spectra are shown as difference spectra in Fig. 1D, right panel. Because these types of spectral alterations result from unstacked bases in single-stranded regions or from disruption of helical base paired loops (31, 41, 42), the CD spectra support the predicted structural differences between the WT and the ΔF508 CFTR mRNA.
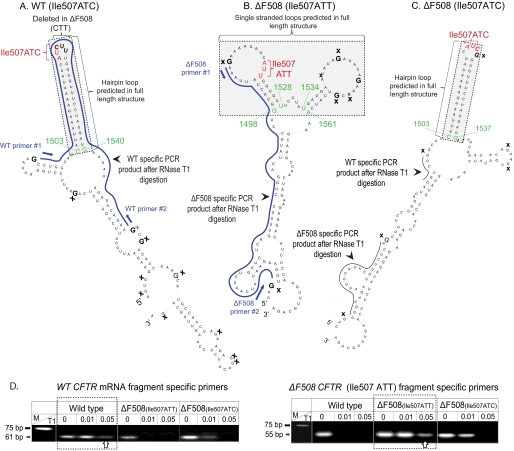
RNA Folding Assays Confirm the Predicted mRNA Structural Differences within the Mutated Region
Because the CD spectra do not provide information about the location of the structural differences, we developed a biochemical RNA folding assay to confirm the presence of the hairpin loop (WT and ΔF508 Ile507ATC; Fig. 1, A and C) or the two enlarged single-stranded loops (ΔF508 Ile507ATT; Fig. 1B) in the CFTR mRNAs. Because the full-length CFTR mRNA is not suitable for secondary structural analysis using presently available techniques, we tested whether the predicted structural differences exist in excised segments of the CFTR mRNAs using a modification of a previously developed assay (27). For these studies, we selected the region of interest, a ∼150-base long sequence in the WT, ΔF508 (Ile507ATT), and variant ΔF508 (Ile507ATC) CFTR, with the mutation located in the center of the sequence. We chose the 150 base length fragments of the CFTR mRNA for our analysis based on the original protocol by Wilkinson et al. (27) indicating that the maximum length of an RNA fragment that allowed the proper function of the attached folding elements was ∼150 bases. Then, we obtained theoretical models for the resulting CFTR mRNA segments. Importantly, the theoretical models predicted identical structural elements for all three fragments as seen in the full-length mRNAs (nucleotides corresponding to 1503–1540 (WT), 1503–1537 (ΔF508 Ile507ATC), and 1498–1561 (ΔF508 Ile507ATT) (Fig. 2, A–C)). We also analyzed the models of several different length mRNA fragments and found identical elements at the vicinity of the mutation as in the full-length mRNAs (data not shown).
FIGURE 2.
Theoretical models of CFTR mRNA fragments and RNA folding assay results. A, WT CFTR fragment model. B, ΔF508 (Ile507ATT) CFTR fragment model. C, variant (Ile507ATC) ΔF508 CFTR fragment model. Ile-507 (red); the CTT deletion site (CUU) is indicated by brackets; the structures that are identical to the full-length mRNA models are highlighted in gray. Nucleotides corresponding to the full-length sequences are labeled green; blue lines represent the projected PCR products predicted following T1 RNase digestion; single-stranded Gs (T1 site) are indicated by X; primers 1 and 2: complementary RT-PCR primer sequences used to amplify WT or ΔF508 (Ile507ATT) CFTR-specific products. D, RT-PCR amplification of WT and ΔF508 (Ile507ATT) CFTR-specific mRNA fragments following RNase T1 digestion. Arrows indicate WT (61 bp) ΔF508 (55 bp) fragment structure specific PCR products in samples digested with 0.05 units of RNase T1. M: molecular weight marker. A representative gel of four individual experiments is shown.
To confirm the presence of these mRNA secondary structural elements, we cloned the regions of interest of the WT, ΔF508 (Ile507ATT) and variant ΔF508 (Ile507ATC) CFTR cDNAs into a cassette that allowed the synthesis of the RNAs (27). Following in vitro transcription and purification of the CFTR mRNA fragments, we placed them into a folding buffer that promotes the formation of secondary structures (27). To map the predicted loop regions, we digested the folded RNAs with single-strand specific RNase T1. If the theoretical models are correct, the T1 digestion site mapping should generate different fragments for the WT, ΔF508 (ATT) and variant ΔF508 (ATC) constructs (Fig. 2, A–C, solid lines). Based on the proposed RNase T1 digest fragment sizes and sequences, we designed primers for the WT and the ΔF508 CFTR constructs. Following optimization of the T1 RNase reactions, we digested the folded RNA samples with RNase T1 (0.01 and 0.05 units for 15 min at 25 °C). Then, we purified the digested fragments, performed RT-PCR and analyzed the results on agarose gels (Fig. 2, bottom panels). The predicted WT CFTR construct specific fragment (61 bp) was amplified from only the WT samples following digestion with T1 (Fig. 2, lower left panel, white arrow). No 61-bp fragment was amplified from the Ile507ATT ΔF508 CFTR samples using the WT-specific primers. Whereas, in the variant ΔF508 (Ile507ATC) samples, we detected a WT CFTR-specific PCR product following digestion with 0.01 U T1 (Fig. 2, lower left panel, black arrow) but not after digestion with 0.05 units of T1. When we added ΔF508 CFTR fragment-specific primers to the reaction mixture, the PCR product of the predicted size (55 bp) was only present following digestion with the higher activity T1 (0.05 unit) in the ΔF508 (Ile507ATT) samples (Fig. 2, lower right panel, white arrow). Because of the slightly different structure of the variant ΔF508 (Ile507ATC) CFTR compared with the WT, neither WT nor ΔF508 specific products were seen in the Ile507ATC variant samples following complete digestion (0.05 unit of T1) (Fig. 2, right panel). The presence of both WT and ΔF508 specific fragments in the variant ΔF508 samples after digestion with 0.01 unit of T1 probably result from less efficient digestion caused by the more WT-like structure, or the presence of mixed structures in the Ile507ATC variant (Fig. 2). These results further support the predicted secondary structural differences between WT and ΔF508 CFTR mRNA in the region of the mutation and confirm the formation of the predicted structures in the fragments. Because of a lack of biochemical assays to confirm the secondary structural differences in the full-length CFTR mRNAs, it is notable that prediction of identical structural elements in the full-length and fragment CFTR mRNAs implies that a single nucleotide change in the third position of the Ile507 codon (C to T) carries a strong mRNA secondary structure determination potential.
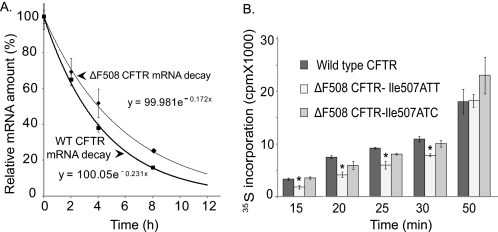
Misfolding of ΔF508 CFTR mRNA Does Not Affect mRNA Half-life
To investigate the consequences of mRNA “misfolding” on ΔF508 CFTR mRNA stability, we measured the half-lives of WT and ΔF508 CFTR mRNA in HeLaWT and HelaΔF cells (43, 44). The half-life of WT CFTR mRNA was 3 h, 4 min, or about 50 min shorter than the ΔF508 CFTR mRNA half-life. Based on a number of CFTR mRNA stability assays performed in different cell lines (data not shown), this difference between the half-lives of WT and mutant CFTR is not significant (Fig. 3A). This observation supports the concept that misfolding of the mutant mRNA does not decrease mRNA stability.
FIGURE 3.
CFTR mRNA half-life and translational rate measurements. A, WT and ΔF508 CFTR mRNA half-life measurements. Cells expressing HeLaWT and HeLaΔF cells were treated with actinomycin D to inhibit transcription. RNA was isolated at the time points specified and relative CFTR mRNA levels were measured by real time RT-PCR. CFTR mRNA levels at the start of the experiment (T0) were set as 100%. Results are plotted as % of CFTR mRNA/t0. CFTR mRNA half-lives were calculated from the exponential decay based on trend line equation C/C0 = e−kdt (C and C0 are mRNA amounts after time t and t0, respectively, and kd is the mRNA decay constant; R2 = 0.99). Results were validated in three independent experiments. B, in vitro CFTR translation rates in the presence of microsomes and ATA. ATA was added 10 min after initiation of translation. Incorporation of [35S]methionine was measured 15, 20, 25, 30, 50, and 60 min following addition of ATA. *, significant differences were found in 35S incorporation levels between WT and ΔF508 (Ile507ATT) CFTR translation at 15, 20, 25, and 30 min; n = 6; p < 0.01. No significant differences were found in the translation rates of WT and variant ΔF508 (Ile507ATC) CFTR.
Misfolding of ΔF508 CFTR mRNA Reduces the Translational Rate
We began these studies by comparing the transcription of WT and ΔF508 CFTR. Although we did not expect differences in transcription efficiency, it was important to rule out this possibility. The results indicated that WT and ΔF508 CFTR mRNA levels were identical following in vitro transcription.
To test whether the enlarged single stranded loops in the ΔF508 CFTR mRNA altered the translation of the mutant protein, we performed in vitro translation reactions using equal amounts of WT, ΔF508 (Ile507ATT), and variant ΔF508 (Ile507ATC) CFTR mRNAs as templates in the presence of microsomes that lacked proteasomes. We eliminated the mRNA relaxation step that would abolish the secondary structures and placed the mRNAs into folding buffer prior to translation. Translation initiation was blocked 10 min after the reaction start and [35S]methionine incorporation was monitored at different time points following the block. The results indicate a higher [35S]methionine incorporation into WT than ΔF508 (Ile507ATT) CFTR at earlier time points (p < 0.01 at 15, 20, 25, and 30 min; Fig. 3B). The rate of radioactivity incorporation into the variant ΔF508 (Ile507ATC) CFTR was similar to that observed for the WT (Fig. 3B). By 50 min, the rate of [35S]methionine incorporation was equal for all three constructs. These results support the idea that the enlarged single-stranded regions in the vicinity of the mutation slow down translation.
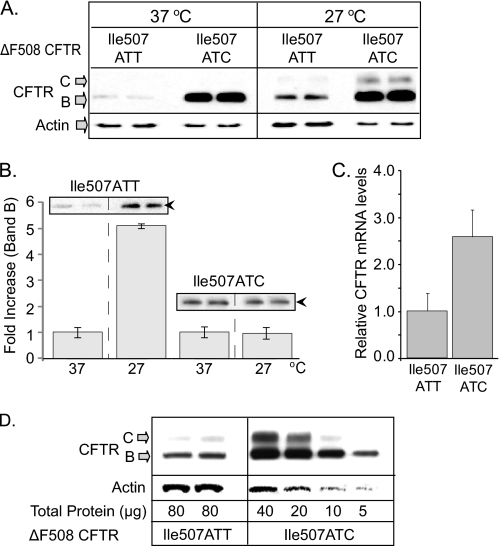
Higher Protein Expression of Ile507ATC ΔF508 CFTR in 293F Cells Compared with the Native Ile507ATT ΔF508 CFTR
To investigate the effect of the synonymous (Ile507ATC) SNP on ΔF508 CFTR protein expression, we developed 293F cells with inducible native (Ile507ATT) or variant (Ile507ATC) ΔF508 CFTR expression and selected clonal lines. After standardizing CFTR mRNA levels, we compared native and variant ΔF508 CFTR protein levels by Western blotting (Fig. 4). Because the selected native Ile507ATT ΔF508 CFTR clone expressed ∼35% of the CFTR mRNA expressed in the variant (Ile507ATC) clone, we loaded one-third of the protein lysate from ΔF508 variant cells than from the native Ile507ATT ΔF508 CFTR-expressing cells (Fig. 4A). As demonstrated in Fig. 4A (left panel), the expression of the variant (Ile507ATC) was significantly higher than native ΔF508 CFTR. Because ΔF508 CFTR is rapidly degraded from the ER (16–18, 38), increased expression of the Ile507ATC variant (Band B) may be the result of enhanced translation or reduced ERAD. Therefore, based on our previous studies indicating that ERAD is inhibited at low temperature (46), and ΔF508 CFTR can be rescued from ERAD when cells are cultured at 27 °C (47, 48), we tested the effect of low temperature culture on the two ΔF508 CFTR variants using Western blotting. As shown in Fig. 4A (right panel), a 4-h culture at low temperature resulted in significant increase in native ΔF508 CFTR Band B levels. In contrast, variant (Ile507ATC ΔF508 CFTR) Band B levels did not increase significantly at 27 °C. Interestingly, we did observe rescue (Band C expression) from the Ile507ATC variant. The minimal change in variant (Ile507ATC) Band B levels following low temperature culture suggested that the protein produced from the Ile507ATC variant is more resistant to ERAD. To confirm this, we compared the effect of low temperature on Band B levels of native and variant ΔF508 CFTR Fig. 4B. Whereas we observed ∼5-fold increase in native (Ile507ATT) ΔF508 CFTR Band B levels at low temperature (4 h), there was no significant change in variant (Ile507ATC) ΔF508 CFTR Band B, supporting the hypothesis that the protein expressed from the Ile507ATC CFTR variant is less accessible to ERAD.
FIGURE 4.
Expression of native (Ile507ATT) and variant (Ile507ATC) ΔF508 CFTR in 293F cells. A, CFTR expression levels at 37 and 27 °C. 293F cells expressing native or variant ΔF508 CFTR were induced with 4 ng/ml doxacyclin and cultured at 37 or 27 °C for 4 h. Cells were lysed in RIPA, and proteins were separated by PAGE (6%). CFTR was detected with MM13–4 monoclonal antibody and ECL. Higher expression of Band B CFTR in the Ile507ATC ΔF508 CFTR-expressing cells at 37 °C and significant levels of Band C CFTR following a 4-h culture at 27 °C are shown. B, native (Ile507ATT) ΔF508 CFTR Band B levels increase at 27 °C. Changes in Band B levels of native (Ile507ATT) and variant (Ile507ATC) ΔF508 CFTR were measured following a 4 h culture at 27 °C by Western blotting and densitometry. Results are expressed as fold increase in Band B density at 27 °C compared with 37 °C controls; n = 3. Inlay, representative gels; black arrowheads: Band B CFTR. C, CFTR mRNA levels following doxacyclin induction and 4 h at 27 °C. Ile507ATT and Ile507ATC ΔF508 CFTR mRNA levels were compared in real time RT-PCR. mRNA levels in the variant (Ile507ATC) cells are plotted as fold increase over native (Ile507ATT) ΔF508 CFTR, (n = 6). D, CFTR protein levels in serial dilutions of variant, Ile507ATC ΔF508 CFTR expressing cell lysates. Cellular proteins were isolated following doxacyclin induction and 4-h culture at 27 °C. Western blots were performed as described in A. Enhanced Band B and significant Band C formation in the Ile507ATC samples compared with native (Ile507ATT) ΔF508 CFTR.
As additional support for enhanced Band B expression from the Ile508ATC variant and because the efficiency of induction may vary between experiments, we measured CFTR mRNA levels (Fig. 4C) from the same samples tested by Western blotting. Based on the measured mRNA levels, we performed a serial dilution of cell lysates to compare protein expression levels to mRNA expression (Fig. 4D). The results indicate that there was more immature protein (Band B) and mature protein (Band C) of the variant (Ile507ATC) ΔF508 CFTR even when only one-eighth the amount of native (Ile507ATT) ΔF508 CFTR cell lysate samples were loaded.
The results obtained from cell lines expressing the native (Ile507ATT) or the variant (Ile507ATC) ΔF508 CFTR strongly support the idea that the effects of mRNA misfolding and Phe deletion on the biogenesis of ΔF508 CFTR are closely connected. Although the majority of the ΔF508 CFTR mutants (native and variant) are degraded from the ER, the variant appears to be more stable and available for rescue. Importantly, any improvement in “escape” from ERAD could reduce the severity of the ΔF508 phenotype. Therefore, these results provide a direct connection to the possible in vivo effects of SNPs and a rationale for further studies to identify the detailed trafficking and potential functional differences between the native and variant ΔF508 CFTR.
DISCUSSION
The experiments presented herein draw attention to the effects of nucleotide deletions and synonymous SNPs on mRNA secondary structure and the consequences of the mRNA structural alterations on translation and protein folding. Our results also demonstrate that high fidelity computational methods (49, 50) and biochemical mRNA folding assays (27, 51) can provide background for studies investigating the dynamics of translation and protein folding.
Importantly, while several studies proposed that alterations in the secondary structures of mRNAs in the coding region may alter translation and protein folding (6, 7, 40, 41), there is limited experimental data in the literature to support this hypothesis. The results presented herein offer significant experimental proof regarding this important prediction.
In an elegant study, Tinoco and co-workers (7) emphasized the importance of mRNA secondary structural elements on translational dynamics. Measuring the length of the pauses and translocation steps during translation, they noted that the mRNA hairpin loops influenced the length of pause cycles during translation without affecting translocation between codons. Therefore, these and our results have a direct correlation to what may occur during ΔF08 CFTR biogenesis. The elongated translational pauses during the synthesis of the ΔF508 CFTR NBD1 domain may delay cotranslational folding and signal the quality control (ERAD) machinery. If the elongated pauses initiate, or allow ubiquitination, this could result in early ERAD of ΔF508 CFTR, as demonstrated by previous studies (52, 53). The observation that ΔF508 CFTR is cotranslationally ubiquitinated and quickly degraded through ERAD (45) supports this idea. Because strict rules govern the ERAD-associated processes of ubiquitin activation, ubiquitin linkage to proteins and retro-translocation (54), it is likely that translational rate modifications affect this process. Therefore, it is tempting to speculate that the slower translation of ΔF508 CFTR (Ile507ATT) facilitates ERAD. The results presented in Fig. 4 support this hypothesis because the expression of the variant, Ile507ATC ΔF508 CFTR resulted in higher levels of band B CFTR than the native (Ile507ATT ΔF508 CFTR), likely because it was less accessible to ERAD (Fig. 4B). However, the details how mRNA structural changes affect protein stability remains to be determined.
Although our studies concentrated on the mRNA secondary structural alterations caused by the deletion of CTT in the human CFTR and introduction of the synonymous codon (ATT) for Ile-507, we also analyzed the codon frequency for Ile in the human genome and in CFTR. We did this based on previous studies suggesting that clusters of rare codons may cause translational pauses (55–58). We found that that while ATT is a less frequent codon than ATC, ATA is the rarest codon for Ile in the human genome. In CFTR, however, ATT is the most frequently used codon for Ile. Therefore, it is unlikely that the slight reduction of the codon frequency in a single locus (47% (ATC) to 35% (ATT)) with relative synonymous codon usage values changing from 20.9 to 15.8 would significantly affect tRNA availability. A study investigating the role of SNPs on CAT synthesis supports this idea (57). Because excess amounts of Escherichia coli tRNAs did not alter the pause cycles, the authors suggested that mRNA structural alterations, rather than the limited availability of rare tRNAs, were responsible for translational pausing.
Codon usage, however, has been shown to be important for the folding and function of the MDR1 gene product, P-glycoprotein (P-gp). Studies by Gottesman and co-workers (59) demonstrated that SNPs that introduced rare codons into MDR1 altered both the structure and ligand specificity of P-gp. Considering that P-gp is an ABC transporter with structural similarities to CFTR (60), we asked the question whether the mRNA structure, the codon frequency, or the combination of the two were responsible for decreased ΔF508 CFTR translation rates. Our studies on the variant, (Ile507ATC) ΔF508 CFTR, helped to address this question. We showed that replacement of the Ile507ATT with ATC in the mutant resulted in a more WT CFTR-like mRNA structure. Most importantly, both the translational rate and cellular expression levels of the variant (Ile507ATC) ΔF508 CFTR improved compared with the native (Ile507ATT), ΔF508 CFTR. Therefore, these results are consistent with the idea that the reduced translational rates resulted from the mRNA secondary structural differences rather than the rarity of the ATT codon.
An earlier study provides further support for our hypothesis. It has been proposed that synonymous mutations that alter the cytosine content result in base pairing shifts that have significant effects on mRNA secondary structure (61). Indeed, the ΔF508 mutation in CFTR removes a cytosine from the third position of Ile507 by changing the codon from ATC to ATT. In contrast, if Phe-508 was encoded by the synonym TTC in the WT CFTR, deletion of CTT would not change the cytosine content of the region. Therefore, as suggested by Chamary and Hurst (61) and supported by our results from studies on the Ile507ATC ΔF508 CFTR variant, the mRNA structural instability caused by the removal of cytosine, i.e. a longer single-stranded region in the mutation region, could be responsible for the reduced translational rate.
Furthermore, the poor correlation between the ΔF508 homozygous genotype and the severity of symptoms implies the involvement of environmental factors and modifier genes in the development of CF (62). Our results presented herein suggest that in addition to CF modifier genes (63), SNPs that result in mRNA structural changes may also contribute to the differences observed in the severity of symptoms between CF patients homozygous for ΔF508.
Although we used the CFTR mRNA as our model, genetic mutations are likely to affect the structure of other mRNAs as well. Because, as in a number protein folding diseases involving early degradation of a partially functional protein, the exact amount of functional ΔF508 CFTR sufficient to ameliorate the serious symptoms of CF is not known, it is critical that we understand all of the mutation-associated defects. Therefore, the results presented here provide the foundation for future studies to test the role of mRNA structure on protein folding and ERAD. This type of analysis will improve our understanding of the complex dynamics associated with membrane protein translation, membrane integration and folding.
Supplementary Material
Acknowledgments
We thank Drs. David Bedwell, Kevin Kirk, Casey Morrow, and Lianwu Fu for critical reading of the manuscript and for helpful suggestions.
This work was supported, in whole or in part, by Grants R01HL076587 (to Z. B.) and DK060065 (to J. F. C.), CFF, and the Genetically Defined Microbe and Expression Core of the UAB Mucosal HIV and Immunobiology Center Grant R24DK64400 (to J. K.) from the National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1, A–C.
- CF
- cystic fibrosis
- P-gp
- P-glycoprotein
- ERAD
- endoplasmic reticulum-associated degradation
- CFTR
- cystic fibrosis transmembrane conductance regulator.
REFERENCES
- 1.Kozak M. (2005) Gene 361, 13–37 [DOI] [PubMed] [Google Scholar]
- 2.Svoboda P., Di Cara A. (2006) Cell Mol. Life Sci. 63, 901–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batey R. T. (2006) Curr. Opin. Struct. Biol. 16, 299–306 [DOI] [PubMed] [Google Scholar]
- 4.Leontis N. B., Lescoute A., Westhof E. (2006) Curr. Opin. Struct. Biol. 16, 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cazzola M., Skoda R. C. (2000) Blood 95, 3280–3288 [PubMed] [Google Scholar]
- 6.Kudla G., Murray A. W., Tollervey D., Plotkin J. B. (2009) Science 324, 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen J. D., Lancaster L., Hodges C., Zeri A. C., Yoshimura S. H., Noller H. F., Bustamante C., Tinoco I. (2008) Nature 452, 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alder N. N., Johnson A. E. (2004) J. Biol. Chem. 279, 22787–22790 [DOI] [PubMed] [Google Scholar]
- 9.Kleizen B., van Vlijmen T., de Jonge H. R., Braakman I. (2005) Mol. Cell 20, 277–287 [DOI] [PubMed] [Google Scholar]
- 10.Kerem B., Rommens J. M., Buchanan J. A., Markiewicz D., Cox T. K., Chakravarti A., Buchwald M., Tsui L. C. (1989) Science 245, 1073–1080 [DOI] [PubMed] [Google Scholar]
- 11.Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L. (1989) Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 12.Rommens J. M., Iannuzzi M. C., Kerem B., Drumm M. L., Melmer G., Dean M., Rozmahel R., Cole J. L., Kennedy D., Hidaka N. (1989) Science 245, 1059–1065 [DOI] [PubMed] [Google Scholar]
- 13.Collins F. S. (1992) Science 256, 774–779 [DOI] [PubMed] [Google Scholar]
- 14.Sheppard D. N., Welsh M. J. (1999) Physiol. Rev. 79, S23–45 [DOI] [PubMed] [Google Scholar]
- 15.Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. (1990) Cell 63, 827–834 [DOI] [PubMed] [Google Scholar]
- 16.Ward C. L., Kopito R. R. (1994) J. Biol. Chem. 269, 25710–25718 [PubMed] [Google Scholar]
- 17.Ward C. L., Omura S., Kopito R. R. (1995) Cell 83, 121–127 [DOI] [PubMed] [Google Scholar]
- 18.Bebök Z., Mazzochi C., King S. A., Hong J. S., Sorscher E. J. (1998) J. Biol. Chem. 273, 29873–29878 [DOI] [PubMed] [Google Scholar]
- 19.Fadiel A., Eichenbaum K. D., Hamza A., Tan O., Lee H. H., Naftolin F. (2007) Curr. Protein Pept. Sci. 8, 29–37 [DOI] [PubMed] [Google Scholar]
- 20.Gregersen N. (2006) J. Inherit. Metab. Dis. 29, 456–470 [DOI] [PubMed] [Google Scholar]
- 21.Cohen F. E., Kelly J. W. (2003) Nature 426, 905–909 [DOI] [PubMed] [Google Scholar]
- 22.Zuker M. (2000) Curr. Opin. Struct. Biol. 10, 303–310 [DOI] [PubMed] [Google Scholar]
- 23.Zuker M., Mathews D. H., Turner D. H. (1999) Algorithms and Thermodynamics for RNA Secondary Structure Prediction: A Practical Guide, Kluwer Academic Publishers [Google Scholar]
- 24.Woody R. W. (1995) Methods Enzymol. 246, 34–71 [DOI] [PubMed] [Google Scholar]
- 25.Merino E. J., Wilkinson K. A., Coughlan J. L., Weeks K. M. (2005) J. Am. Chem. Soc. 127, 4223–4231 [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson K. A., Merino E. J., Weeks K. M. (2005) J. Am. Chem. Soc. 127, 4659–4667 [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson K. A., Merino E. J., Weeks K. M. (2006) Nat. Protoc. 1, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 28.Markham N. R., Zuker M. (2005) Nucleic Acids Res. 33, W577–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oberdorf J., Skach W. R. (2002) Methods Mol. Med. 70, 295–310 [DOI] [PubMed] [Google Scholar]
- 30.Merrick W. C., Barth-Baus D. (2007) Methods Enzymol. 429, 1–21 [DOI] [PubMed] [Google Scholar]
- 31.Nuzzaci M., Bochicchio I., De Stradis A., Vitti A., Natilla A., Piazzolla P., Tamburro A. M. (2009) J. Virol. Methods 155, 118–121 [DOI] [PubMed] [Google Scholar]
- 32.Sosnick T. R., Fang X., Shelton V. M. (2000) Methods Enzymol. 317, 393–409 [DOI] [PubMed] [Google Scholar]
- 33.Bebok Z., Collawn J. F., Wakefield J., Parker W., Li Y., Varga K., Sorscher E. J., Clancy J. P. (2005) J. Physiol. 569, 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen W., Wu X., Levasseur D. N., Liu H., Lai L., Kappes J. C., Townes T. M. (2000) Stem Cells 18, 352–359 [DOI] [PubMed] [Google Scholar]
- 35.Doerfler W., Hoeveler A., Weisshaar B., Dobrzanski P., Knebel D., Langner K. D., Achten S., Müller U. (1989) Cell Biophys. 15, 21–27 [DOI] [PubMed] [Google Scholar]
- 36.Naldini L., Blömer U., Gage F. H., Trono D., Verma I. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11382–11388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zufferey R., Donello J. E., Trono D., Hope T. J. (1999) J. Virol. 73, 2886–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. (1997) Nat. Biotechnol. 15, 871–875 [DOI] [PubMed] [Google Scholar]
- 39.Szymczak A. L., Workman C. J., Wang Y., Vignali K. M., Dilioglou S., Vanin E. F., Vignali D. A. (2004) Nat. Biotechnol. 22, 589–594 [DOI] [PubMed] [Google Scholar]
- 40.Urlinger S., Baron U., Thellmann M., Hasan M. T., Bujard H., Hillen W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7963–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piazzolla P., Crescenzi A., De Biasi M., Tamburro A. M. (1998) Arch. Virol. 143, 2305–2312 [DOI] [PubMed] [Google Scholar]
- 42.Wells B. D., Yang J. T. (1974) Biochemistry 13, 1311–1316 [DOI] [PubMed] [Google Scholar]
- 43.Rab A., Bartoszewski R., Jurkuvenaite A., Wakefield J., Collawn J. F., Bebok Z. (2007) Am. J. Physiol. Cell Physiol. 292, C756–C766 [DOI] [PubMed] [Google Scholar]
- 44.Varga K., Jurkuvenaite A., Wakefield J., Hong J. S., Guimbellot J. S., Venglarik C. J., Niraj A., Mazur M., Sorscher E. J., Collawn J. F., Bebök Z. (2004) J. Biol. Chem. 279, 22578–22584 [DOI] [PubMed] [Google Scholar]
- 45.Sato S., Ward C. L., Kopito R. R. (1998) J. Biol. Chem. 273, 7189–7192 [DOI] [PubMed] [Google Scholar]
- 46.Jurkuvenaite A., Chen L., Bartoszewski R., Goldstein R., Bebok Z., Matalon S., Collawn J. F. (2010) Am. J. Respir. Cell Mol. Biol. 42, 363–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denning G. M., Anderson M. P., Amara J. F., Marshall J., Smith A. E., Welsh M. J. (1992) Nature 358, 761–764 [DOI] [PubMed] [Google Scholar]
- 48.Varga K., Goldstein R. F., Jurkuvenaite A., Chen L., Matalon S., Sorscher E. J., Bebok Z., Collawn J. F. (2008) Biochem. J. 410, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doshi K. J., Cannone J. J., Cobaugh C. W., Gutell R. R. (2004) BMC Bioinformatics 5, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dowell R. D., Eddy S. R. (2004) BMC Bioinformatics 5, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deigan K. E., Li T. W., Mathews D. H., Weeks K. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang F., Kartner N., Lukacs G. L. (1998) Nat. Struct. Biol. 5, 180–183 [DOI] [PubMed] [Google Scholar]
- 53.Farinha C. M., Amaral M. D. (2005) Mol. Cell Biol. 25, 5242–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta S. K., Majumdar S., Bhattacharya T. K., Ghosh T. C. (2000) Biochem. Biophys. Res. Commun. 269, 692–696 [DOI] [PubMed] [Google Scholar]
- 56.Komar A. A., Lesnik T., Reiss C. (1999) FEBS Lett. 462, 387–391 [DOI] [PubMed] [Google Scholar]
- 57.Ramachandiran V., Kramer G., Horowitz P. M., Hardesty B. (2002) FEBS Lett. 512, 209–212 [DOI] [PubMed] [Google Scholar]
- 58.Marin M. (2008) Biotechnol. J. 3, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 59.Kimchi-Sarfaty C., Oh J. M., Kim I. W., Sauna Z. E., Calcagno A. M., Ambudkar S. V., Gottesman M. M. (2007) Science 315, 525–528 [DOI] [PubMed] [Google Scholar]
- 60.Hoof T., Demmer A., Hadam M. R., Riordan J. R., Tümmler B. (1994) J. Biol. Chem. 269, 20575–20583 [PubMed] [Google Scholar]
- 61.Chamary J. V., Hurst L. D. (2005) Genome Biol. 6, R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zielenski J. (2000) Respiration 67, 117–133 [DOI] [PubMed] [Google Scholar]
- 63.Knowles M. R. (2006) Curr. Opin. Pulm. Med. 12, 416–421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.