Abstract
The interaction of Bcl-2 family proteins at the mitochondrial outer membrane controls membrane permeability and thereby the apoptotic program. The anti-apoptotic protein Bcl-2 binds to the pro-apoptotic protein Bax to prevent Bax homo-oligomerization required for membrane permeabilization. Here, we used site-specific photocross-linking to map the surfaces of Bax and Bcl-2 that interact in the hetero-complex formed in a Triton X-100 micelle as a membrane surrogate. Heterodimer-specific photoadducts were detected from multiple sites in Bax and Bcl-2. Many of the interaction sites are located in the Bcl-2 homology 3 (BH3) region of Bax and the BH1–3 groove of Bcl-2 that likely form the BH3-BH1–3 groove interface. However, other interaction sites form a second interface that includes helix 6 of Bax and the BH4 region of Bcl-2. Loss-of-function mutations in the BH3 region of Bax and the BH1 region of Bcl-2 disrupted the BH3-BH1–3 interface, as expected. Surprisingly the second interface was also disrupted by these mutations. Similarly, a loss-of-function mutation in the BH4 region of Bcl-2 that forms part of the second interface also disrupted both interfaces. As expected, both kinds of mutation abolished Bcl-2-mediated inhibition of Bax oligomerization in detergent micelles. Therefore, Bcl-2 binds Bax through two interdependent interfaces to inhibit the pro-apoptotic oligomerization of Bax.
Keywords: Membrane Proteins, Mitochondrial Apoptosis, Protein Conformation, Protein Cross-linking, Protein/Protein Interactions, BH3-only Protein, Bax, Bcl-2, Oligomerization, Pore
Introduction
Bcl-2 and Bax function oppositely during apoptosis initiation (1, 2). Both proteins contain multiple Bcl-2 homology (BH)4 motifs (3) and have similar monomeric structures (4, 5). In healthy cells, Bcl-2 is anchored at the mitochondrial outer membrane (MOM) through its C-terminal transmembrane (TM) α-helix 9, whereas Bax is a monomeric protein in the cytosol with its C-terminal hydrophobic α-helix 9 in a hydrophobic groove formed by its BH1–3 sequences (Fig. 1A) (6–10). A similar hydrophobic groove in Bcl-2 is empty according to the NMR structure of a Bcl-2 protein lacking α-helix 9 (Fig. 1A) (4). During apoptosis, truncated Bid (tBid) and other Bcl-2 family proteins that only contain a single BH3 motif (hence called BH3-only proteins) interact with both Bcl-2 and Bax at the surface of MOM changing them to a multispanning conformation with helices 5, 6, and 9 embedded in the membrane (11–14). Cytosolic Bax can also be converted to the membrane-bound form through interaction with the Bax that is already bound to the membrane, a mechanism that was termed auto-activation (15, 16). The MOM-bound multispanning Bax proteins form oligomers that permeabilize the membrane to release cytochrome c and other intermembrane space proteins that activate caspases and nucleases to kill the cell (9, 17–19). Bcl-2 inhibits the Bax-mediated MOM permeabilization as follows: (i) by competing with Bax for binding with tBid thereby inhibiting Bax targeting to mitochondria, (ii) by binding Bax at the membrane to inhibit the conformational change from the soluble to the multispanning state, and (iii) by binding the multispanning Bax in the membrane to inhibit Bax oligomerization (11, 20–23). Because the structures of Bcl-2 and Bax are very similar (Fig. 1A) and both proteins change to similar conformation in response to tBid, we proposed that Bcl-2 can function as a dead-end binding partner for Bax to inhibit Bax activation and oligomerization (1, 14). However, it has been difficult to discern the molecular mechanism, particularly the binding interface, that allows Bcl-2 to block the oligomerization of Bax. The problem is exacerbated by the fact that the proteins interact with each other in membranes, an environment that hampers using conventional methods, such as crystallography and NMR, to obtain structural information (1).
FIGURE 1.
Structures and interactions of Bax and Bcl-2. A, monomeric structures of Bax (left) and Bcl-2 (right) were drawn using PyMOL (DeLano Scientific) based on the coordinates 1F16 and 1G5M in the Protein Data Bank (4, 5). All α-helices and the loop between α1 and α2 are indicated. The front surface that includes the BH1–3 region is colored in medium gray. The rear surface formed by α1, the loop between α1 and α2, and α6 are colored in dark gray. The schematic below under each structure has the front and rear surfaces indicated and will be used in other figures to represent Bax and Bcl-2. B, model for Bcl-2 homo-complex formation that is proposed based on previous studies (36–39). Neighboring molecules in the homo-oligomers are shaded differently for clarity, although they have the same conformation and interaction. C, Bax and Bcl-2 can potentially bind through their front surfaces (left) or rear surfaces (right) to form heterodimers.
Previous studies have revealed certain regions in Bcl-2 and Bax that are important for their interaction. Mutagenesis studies show that the BH1–3 regions of Bcl-2 and the BH3 region of Bax are indispensable for their interaction (14, 24–27). Structures of Bcl-2-like proteins in complex with the BH3 peptide from Bak or BH3-only proteins provide explanations for the mutagenesis results (28–32). In all of the complex structures determined so far, the BH3 peptide forms an amphipathic α-helix whose hydrophobic surface interacts with a hydrophobic groove on the Bcl-2-like protein formed by the BH1–3 regions. Because the hydrophobic surface of the BH3 helix is buried in the structure of monomeric Bax, binding of Bax to Bcl-2 would require a conformational change in Bax (5). This conformational change can be induced by binding with BH3-only proteins. The binding site on Bax for BH3-only proteins is likely a pocket formed by helices 1 and 6 as well as the loop between helices 1 and 2 located on the opposite side of the protein from the BH1–3 groove (Fig. 1A) (33–35). On the other hand, our previous study has shown that Bcl-2 forms a homodimer in which the BH1–3 regions form part of the interface (Fig. 1B, step 1) (36). Thus, binding of Bcl-2 to Bax would require dissociation of the Bcl-2 homodimer. However, the same study also revealed that the BH4 region located on the rear side of the Bcl-2 molecule opposite the BH1–3 groove is part of the Bcl-2 homodimer interface, suggesting there may be another type of homodimer in which the two molecules bind to each other through the rear surface, including the BH4 region (Fig. 1B, step 2). This type of Bcl-2 homodimer would have an exposed BH1–3 groove that can bind the BH3 region of Bax. Indeed, a Bcl-2 homodimer that is cross-linked via a cysteine located in the rear surface can interact with Bax to form a heterotrimer (36). Consistent with our Bcl-2 homodimer model, crystallographic and NMR structures of Bcl-xL homodimer show an opened BH1–3 groove on both protomers, and the Bcl-xL homodimer binds BH3 peptides (37, 38). In these structural studies, the Bcl-xL homodimer was formed either at basic pH or high temperature. When Bcl-xL protein was studied in Triton X-100, two different kinds of homodimers were found (39). One formed at acidic pH can bind BH3 peptides, whereas the other formed at basic pH cannot. Taken together, these studies suggest that Bcl-2-like proteins may exist in a monomer-dimer equilibrium with two forms of homodimers that are formed via the BH1–3 (front) interface or the rear interface as shown in Fig. 1B.
Recently, we systematically mapped the surface of Bax that mediates Bax homo-oligomerization in Triton X-100 micelles using a site-specific photocross-linking technique (40). The results suggest that there are two separate interaction surfaces as follows: the front surface consisting of helices 2 and 3, and the rear surface including helices 1 and 6 as well as the loop between helices 1 and 2 (Fig. 1A, left panel). Because helices 2 and 3 of Bax overlap with the BH3 motif that likely binds to the BH1–3 groove of Bcl-2 in the heterodimer (Fig. 1C, left panel), binding to Bcl-2 would prevent Bax homodimerization through the front surface. Because the formation of Bax oligomers larger than a dimer requires interactions through both the front and rear surfaces, binding of Bcl-2 to the front surface may be sufficient to block Bax oligomerization. However, the alternative mechanism cannot be ruled out. For example, Bcl-2 may bind to the rear surface that is exposed on the Bax dimer, which is formed through interaction via the front surface, a binding that may dissociate the Bax dimer. Because the BH1–3 groove of Bcl-2 would be used to bind the BH3 motif or helix 2 in the front surface of Bax, another surface of Bcl-2 may be used to bind the rear surface of Bax. A candidate for this Bcl-2 surface would be the rear surface that is located at the opposite side of the molecule from the BH1–3 groove (Fig. 1, A, right panel, and C, right panel), based on the structural homology between Bcl-2 and Bax.
To identify the interactions between Bcl-2 and Bax that inhibit the oligomerization of Bax, we studied Bax interaction with Bcl-2 using the detergent system and the site-specific photocross-linking methods that were used in our previous study of Bax homo-interaction (40). The results show that the surfaces of Bax and Bcl-2 involved in the hetero-interaction overlap with the surfaces used for the homo-interaction of each protein. Bcl-2 binds to both the front and rear surfaces of Bax. Surprisingly, the formation of the two interfaces is interdependent. Therefore, our current and previous studies support the notion that Bcl-2 inhibits Bax by forming heterodimers and trimers that are structurally similar to the Bax homo-complexes but that are not competent for further oligomerization.
EXPERIMENTAL PROCEDURES
Plasmids and Proteins
Construction of the pSPUTK plasmids containing coding sequences of wild type and mutant human Bax and Bcl-2 for in vitro transcription and translation was described previously (36, 40). For retroviral infection of Rat-1ERTAM cells, the BamHI/SalI fragment of pBABEhygro plasmid (Cell Biolabs, San Diego) was replaced by the BglII/XhoI fragment of the pSPUTK plasmid containing the coding sequence of wild type (WT) or mutant Bcl-2 resulting in the pBABEhygro-wild type or mutant Bcl-2 plasmid.
Expression and purification of His6-tagged Bcl-2 lacking the TM sequence (His6-Bcl-2ΔTM) and the mutant protein with Gly145 or Val15 replaced by Ala or Glu (His6-Bcl-2ΔTM-G145A or V15E, respectively) were carried out as described previously (36). Expression and purification of His6-tagged Bax (His6-Bax) and the mutant protein with Ile66 and Asp68 replaced by Glu and Arg (His6-Bax-I66E/D68R) were done as described previously (40, 41). The peptide, HHHHHHGGDPELIRTIMGWTLDFLRER, containing α-helix 6 (residues 130–147, in boldface) of human Bax with an N-terminal His6 tag and three linker residues (underlined) was synthesized by Abgent (La Jolla, CA).
Anti-apoptotic Activity of Bcl-2 Mutants in Rat-1ERTAM Cells
pBABEhygro plasmids encoding the various Bcl-2 mutants were transfected into Rat-1ERTAM cells as described previously (42). Stable clones were isolated by selection in hygromycin, and inhibition of apoptosis by the mutants was assayed in cells treated with etoposide. We have demonstrated previously that monitoring cleavage of the caspase 3/7 substrate poly(ADP-ribose) polymerase (PARP) in these cells by immunoblotting provides a quantitative estimation of apoptotic cell death (42).
Photocross-linking and Characterization of Photoadducts
Preparation of [35S]Met- and/or Nϵ-(5-azido-2-nitrobenzoyl) (ϵANB)-Lys-labeled Bax or Bcl-2 protein was carried out in a rabbit reticulocyte lysate or wheat germ extract-based in vitro translation system, respectively, as described previously (36, 43). Purified His6-tagged Bax, Bcl-2ΔTM, and/or the mutant protein was added to 10 μl of the in vitro synthesized protein to a final concentration of 2.2 μm, and if indicated, 0.25% (v/v) of Triton X-100 was added to induce oligomerization. Photocross-linking, purification of the photoadduct using Ni2+-nitrolotriacetic acid-agarose, and characterization of the photoadduct by reducing SDS-PAGE and phosphorimaging were as described previously (36, 40).
Gel Filtration Chromatography to Assay Bcl-2 Inhibition of Bax Oligomerization
Purified His6-tagged Bax (1.5 μm), Bax-I66E/D68R (1.5 μm), Bcl-2ΔTM (9.0 μm), and/or Bcl-2ΔTM-V15E (9.0 μm) proteins were incubated in 0.25% (v/v) Triton X-100. The samples were bound to Ni2+-nitrilotriacetic acid-agarose to exchange Triton X-100 with 2% CHAPS. The eluted proteins were subjected to gel filtration chromatography using Superdex 200 HR10/30 column (Amersham Biosciences), and the eluted fractions were analyzed by SDS-PAGE and immunoblotting with Bax or Bcl-2-specific antibody. Details of this assay were described previously (40).
RESULTS
Bax/Bcl-2 Heterodimer Detected by Photocross-linking
To determine which sites on Bax and Bcl-2 are located in the complex interface, we employed the same site-specific photocross-linking approach we used before to map the interface of Bcl-2-Bcl-2 and Bax-Bax complexes (36, 40). The initial interaction between Bax and Bcl-2 occurs between the full-length cytosolic Bax and the cytosolic domain of Bcl-2 anchored in the MOM through its C-terminal TM sequence. To mimic this interaction in the cross-linking study, we used full-length Bax and Bcl-2 lacking the TM sequence (Bcl-2ΔTM). The RNA encoding Bax or Bcl-2ΔTM was translated in vitro in the presence of ϵANB-Lys-tRNALys, a functional lysyl-tRNA analog that recognizes the lysine codon in RNA and incorporates a lysine analog with the photocross-linker 5-azido-2-nitrobenzoyl (ANB) covalently attached to the ϵ-amino group of the lysine side chain into the polypeptide (44). Because the RNA of Bax or Bcl-2ΔTM contains nine and two lysine codons, respectively, the resulting Bax and Bcl-2ΔTM proteins would have the ANB-Lys incorporated at multiple locations. [35S]Met was also included in the in vitro translation to incorporate a radioactive tracer into the proteins. The resulting ANB-Lys/[35S]Met-labeled Bax or Bcl-2ΔTM protein was mixed with purified recombinant His6-tagged Bcl-2ΔTM or Bax protein, respectively, in Triton X-100 micelles, a membrane surrogate that can activate Bax/Bcl-2 interaction (45). Upon exposure to UV light, a nitrene was generated from the ANB moiety, which is a powerful electrophile that rapidly reacts with heteroatoms, double or even single bonds in the bound His6-tagged proteins that are near the ANB-Lys residue. The photolysis thus generates a photoadduct that covalently links [35S]Met-Bax to His6-Bcl-2ΔTM or [35S]Met-Bcl-2ΔTM to His6-Bax. The resulting photoadduct was enriched by binding to Ni2+-chelating resin, separated from other proteins by reducing SDS-PAGE and detected by phosphorimaging. Because the photoadduct covalently links the two proteins, the cross-linked species displays an apparent molecular weight similar to the combined molecular weight of the two proteins on SDS-PAGE.
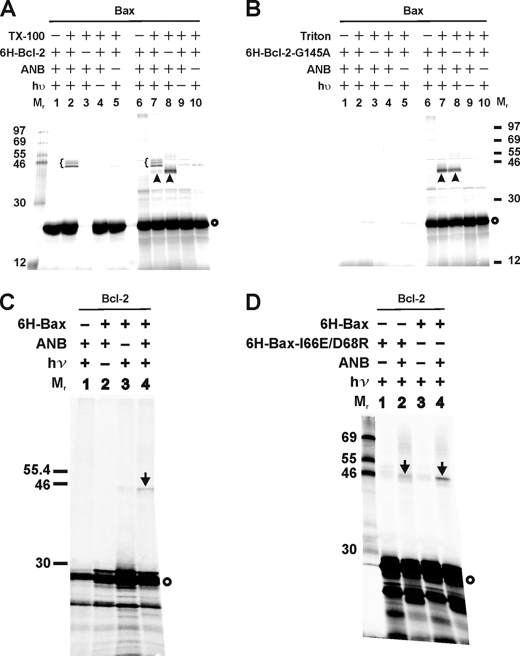
As shown in Fig. 2A (lane 2, indicated by a brace), specific photoadducts were formed between [35S]Met-Bax and His6-Bcl-2ΔTM. In control experiments, the adducts were not detected when His6-Bcl-2ΔTM (Fig. 2A, 6H-Bcl-2; lane 3), UV irradiation (hυ; lane 4), or ϵANB-Lys-tRNALys (ANB; lane 5) was omitted. The adducts were preferentially bound to Ni2+ resins compared with other products seen in the total photocross-linking sample (Fig. 2A, lanes 6–10), demonstrating that the adducts contain His6-Bcl-2ΔTM. Consistently, as shown in Fig. 2A, the in vitro synthesized [35S]Met-Bax monomer (indicated by a circle on the right of the phosphorimage) was detected in lanes 1, 2, 4, and 5 when the His6-tagged Bcl-2ΔTM was present in the reaction but not in lane 3 when the His6-tagged Bcl-2ΔTM was absent. Thus, [35S]Met-Bax bound to Ni2+ resin only when bound to His6-tagged Bcl-2. The molecular weight of the major adduct between Bax and His6-Bcl-2ΔTM is close to 46, the predicted molecular weight for a Bax/His6-Bcl-2ΔTM dimer. The difference in the apparent molecular weight among the various adducts most likely resulted from the ANB probes at different lysine residues of Bax reacting with the Bcl-2 because the in vitro synthesized [35S]Met-Bax protein is nearly homogeneous, and the purity of His6-tagged Bcl-2 protein is >90% as assessed by reducing SDS-PAGE and silver staining (data not shown). Among the other radioactive products detected, the most abundant one is the in vitro synthesized Bax (Fig. 2A, indicated by circle). The other one is a photoadduct between Bax and protein that is likely from the reticulocyte lysate used for translation in vitro as it formed in the absence of His6-Bcl-2ΔTM (Fig. 2A, lane 8, indicated by arrowhead). Because this adduct did not bind to Ni2+ resin, we conclude that it did not have a His6 tag. This photoadduct is unlikely formed between two in vitro synthesized Bax proteins because it was not detected when the ANB probe was attached to most of the interacting sites in Bax homo-complex as shown by our recent study (40).
FIGURE 2.
Photocross-linking detected Bax/Bcl-2 heterodimers. A, photocross-linking of in vitro synthesized ANB-Lys/[35S]Met-labeled Bax to His6-tagged Bcl-2ΔTM in the absence and presence of Triton X-100 (TX-100). Phosphorimaging data shown are from the total samples (lanes 6–10), and their corresponding Ni2+ resin-bound fractions (lanes 1–5). The heterodimer-specific photoadducts were detected in lanes 2–7 and are indicated by braces. A photoadduct formed by Bax and a protein in the reticulocyte lysate were detected in lanes 7 and 8 and indicated by arrowheads. In vitro synthesized [35S]Met-Bax monomer is indicated by a circle on the right of the phosphorimage. The molecular weight (Mr) of protein standards is indicated on the left. B, photocross-linking of the ANB-Lys/[35S]Met-labeled Bax to His6-tagged Bcl-2ΔTM-G145A (6H-Bcl-2-G145A). Both total (lanes 6–10) and the Ni2+ resin-bound (lanes 1–5) samples are shown. The symbols used to indicate the photoadduct of Bax and reticulocyte protein and the [35S]Met-Bax monomer are defined in A. C and D, photocross-linking of in vitro synthesized ANB-Lys/[35S]Met-labeled Bcl-2ΔTM to His6-tagged Bax or Bax-I66E/D68R. Data shown were from the Ni2+ resin-bound fractions of the samples. The heterodimer-specific photoadduct is indicated by an arrow, and the [35S]Met-Bcl-2ΔTM monomer by a circle.
Consistent with the finding that Triton X-100 is required for activating the Bax/Bcl-2 interaction (45), Bax/Bcl-2 photoadducts were not detected in the absence of Triton X-100 (Fig. 2A, lane 1). Furthermore, when purified His6-Bcl-2ΔTM-G145A protein was added to the cross-linking reaction with the ANB-Lys/[35S]Met-Bax, no photoadduct was detected from the fraction bound to Ni2+ resin (Fig. 2B, lane 2) as expected, because the G145A mutation prevents Bcl-2 binding to Bax in cells (14, 24, 25). The photoadduct detected in the total reaction (Fig. 2B, lane 7) is not the photoadduct of ANB-Lys-Bax and His6-tagged Bcl-2 mutant, because it was also formed in the absence of the Bcl-2 mutant (Fig. 2B, lane 8, indicated by arrowhead) and did not bind to Ni2+ resin (Fig. 2B, lane 2). As discussed above, this photoadduct is likely formed by the ANB-Lys-Bax and a protein in reticulocyte lysate. Interestingly, the formation of this photoadduct was inhibited by wild type His6-Bcl-2 but not the G145A mutant (compare the arrowhead-indicated bands in lanes 7 and 8 of Fig. 2, A and B), suggesting that binding by wild type Bcl-2 blocks the interaction of Bax with the reticulocyte protein. The mutant Bcl-2 does not bind to Bax, and therefore it does not have the inhibitory effect. Other evidence for the lack of interaction between the mutant Bcl-2 and Bax is that no [35S]Met-Bax produced in the total reaction was recovered from the Ni2+ resin-bound fraction (Fig. 2B, compare lanes 1, 2, 4, and 5 with 6, 7, 9, and 10, indicated by circle), suggesting that the [35S]Met-Bax does not bind to the His6-tagged mutant Bcl-2 that is bound to the Ni2+ resin. In contrast, [35S]Met-Bax was recovered from the Ni2+ resin-bound fraction in the presence of His6-tagged WT Bcl-2 (Fig. 2A, compare lanes 1, 2, 4, and 5 with 6, 7, 9, and 10, indicated by circle). Taken together, these results suggest that the photocross-linking strategy used here captures a biologically relevant Bax/Bcl-2 interaction.
The Bax/Bcl-2 interaction was also captured when the ANB probe was incorporated into the Bcl-2ΔTM protein (Fig. 2C). A Bcl-2ΔTM/His6-Bax-specific photoadduct was only detected when all of the necessary components, including His6-Bax, ϵANB-Lys-tRNALys, and UV irradiation, were present (Fig. 2C, compare lane 4, band indicated by arrow with the controls in lanes 1–3). The photoadduct bound to Ni2+ resin and its molecular weight is close to the predicted molecular weight (∼47) of Bcl-2ΔTM/His6-Bax dimer. Furthermore, formation of the photoadduct was inhibited by mutation of two conserved residues (I66E/D68R) in the BH3 region of Bax (Fig. 2D) that are critical for binding to Bcl-2 in cells (26, 27, 46). Therefore, the photocross-linking via ANB probes on Bcl-2 also captured biologically relevant Bcl-2/Bax heterodimers.
Generation of Functional Bax and Bcl-2 Mutants with a Single Lysine for Site-specific Photocross-linking
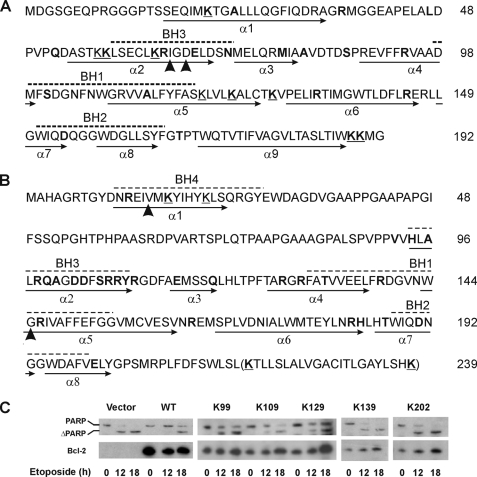
The above cross-linking data show that one or more ANB-Lys residues are close to or within the interface of Bax-Bcl-2 complex thereby resulting in heterodimer-specific photoadducts. However, which Lys residues are in or near the interface would be difficult to determine, and residues other than the Lys residues that are located in or near the interface would not be seen. To use the photocross-linking approach to map the binding surfaces, we generated a series of Bax and Bcl-2 mutants, each with a single Lys located at a specific location where the photoreactive probe can be incorporated. Each single Lys mutant has a Lys substituting for a residue in a Lys-null Bax or Bcl-2ΔTM (Bax K0 or Bcl-2ΔTM K0, respectively) that was constructed by replacing all of the Lys residues in WT Bax and Bcl-2ΔTM with Arg (Fig. 3, A and B). As a simple naming convention for these mutants, we refer to them as Bax or Bcl-2ΔTM followed by K and the location of the residue in the amino acid sequence of the protein. Thus, Bax K21 is a mutant Bax with a single lysine at amino acid position 21.
FIGURE 3.
Sequences of Bax and Bcl-2 mutants and anti-apoptotic activity of Bcl-2 mutants. Bax (A) and Bcl-2 (B) sequences are shown with BH motifs highlighted by dashed lines above and α-helices identified by arrows below. All nine lysines (underlined Ks) were changed to Arg to create Bax K0. Single Lys Bax mutants were created by replacing each of the residues highlighted in boldface with a Lys. Arrowheads indicate Ile66 and Asp68 of Bax that were changed to Glu and Arg, respectively, in the I66E/D68R mutant; and Val15 and Gly145 of Bcl-2 that were changed to Glu and Ala in the V15E and G145A mutants, respectively. The last 22 residues (in parentheses) were deleted in all Bcl-2ΔTM mutants. C, activity of wild type human Bcl-2 (WT) and the single Lys mutant proteins in Rat-1ERTAM cells. Cleavage of PARP to ΔPARP and expression of the Bcl-2 proteins in the etoposide-treated cells were followed by SDS-PAGE and immunoblotting of the cell extracts with PARP- (top panels) and Bcl-2 (bottom panels)-specific antibody, respectively. The Bcl-2 protein expressed stably in the cells is indicated above each panel. Cells were treated with etoposide for the time indicated below each panel prior to analysis. In the controls (vector and WT Bcl-2), the blot probed for human Bcl-2 was deliberately overexposed to show lack of background in the vector-transfected cells.
The following criteria were employed to design these single Lys mutants. 1) Only residues on the surface of the soluble protein were replaced by Lys to avoid disrupting the protein core. 2) Conserved substitutions were preferred, wherever suitable, to minimize the impact on the protein surface. Some of the lysines in WT Bax and Bcl-2ΔTM were kept as single Lys mutants. 3) Representative Bax and Bcl-2 mutants with a single Lys in some of the structurally important motifs were assayed alongside WT Bax and Bcl-2 and the K0 mutants in mammalian cells for pro- and anti-apoptotic activity, respectively. The results for Bax K0, K21, K58, K73, K87, K123, and K128, and Bcl-2 K0 were published previously (36, 40). Examples of additional activity data for Bcl-2 mutants expressed in Rat-1ERTAM cells that did (K99, K109, and K129) and did not (K139 and K202) inhibit etoposide-induced apoptosis as measured by cleavage of the caspase 3/7 substrate PARP are shown in Fig. 3C. Functionally inactive mutants such as K139 and K202 were not used in the cross-linking experiments.
In this study, detergent micelles were used as membrane surrogates. Although the structure of protein in detergent micelles may more closely resemble the structure in membranes than the structure determined in solution, it is unlikely to be identical to the structure in membranes. Moreover, the structures adopted in detergent will most certainly be different from the solution structures. However, throughout this study, we refer to the locations of the cross-linking sites by mapping them onto the solution structures of the proteins. Therefore, the use of the descriptors such as front and rear is arbitrary and unlikely to reflect the surfaces as they exist in the membrane. The terms are used only to permit readers to more easily orient the diagrams in Fig. 1 and relate the text to the schematics used throughout as a guide to interpreting the cross-linking data.
Bax/Bcl-2 Heterodimer Detected by Site-specific Photocross-linking
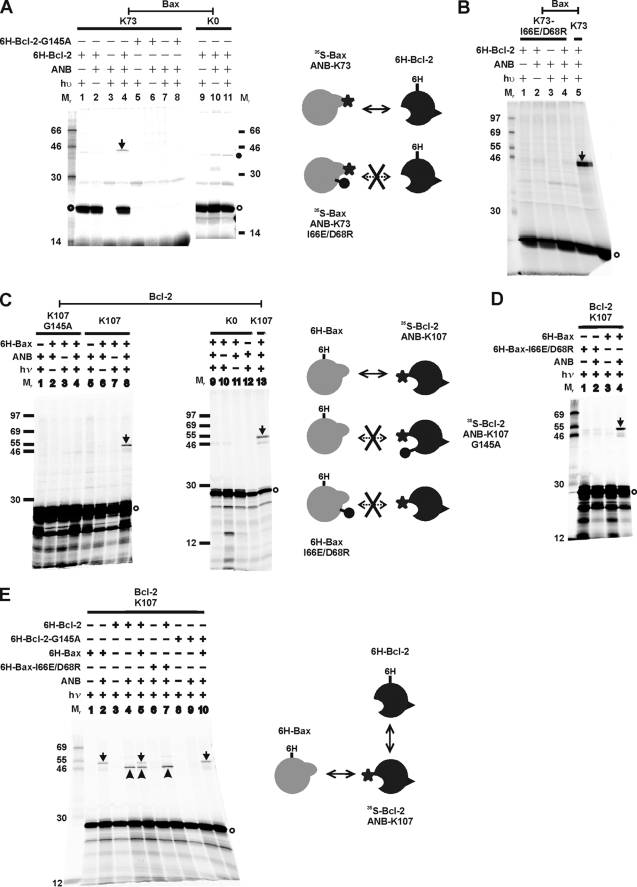
To determine whether site-specific photocross-linking can detect the hetero-interactions of Bax and Bcl-2, we used Bax K73 and Bcl-2 K107, two mutants with a single Lys replacing a residue in the BH3 regions of these proteins. Cross-linking through the photoreactive probe attached to these sites was expected to occur as the BH3 region is known to play an important role in the Bax/Bcl-2 interaction (4, 26, 27, 47, 48). The Bax or Bcl-2 mutant was synthesized in vitro to incorporate ANB-Lys and [35S]Met and incubated with His6-tagged Bcl-2ΔTM or Bax, respectively, in the presence of Triton X-100. Specific photoadducts were detected in the photolyzed samples (Fig. 4, A, lane 4, and C, lane 8; indicated by arrow), because these products were not detected in the control experiment when ϵANB-Lys-tRNALys or UV irradiation was omitted (Fig. 4, A, lanes 1 or 2, and C, lanes 7 or 6, respectively). We concluded that the photoadducts were formed between the [35S]Met-labeled proteins and their corresponding His6-tagged binding partners because the photoadducts were radioactive, bound to Ni2+ resin, and were not detected in the absence of the His6-tagged proteins (Fig. 4, A, lane 3, and C, lane 5). The apparent molecular weight of the photoadducts is close to the predicted molecular weight for the corresponding heterodimer (46 or 47 for [35S]Met-Bax/His6-Bcl-2ΔTM or [35S]Met-Bcl-2ΔTM/His6-Bax, respectively).
FIGURE 4.
Site-specific photocross-linking detected Bax/Bcl-2 heterodimer. A, photocross-linking of in vitro synthesized [35S]Met-labeled Bax with a single ANB probe attached to K73 to His6-tagged Bcl-2ΔTM (lanes 1–4) or Bcl-2ΔTM-G145A (lanes 5–8). The control reactions with in vitro synthesized Bax K0 are shown in lanes 9–11. The photoadduct between the [35S]Met-Bax K73 and His6-Bcl-2ΔTM is indicated by an arrow. The adduct was absent when either His6-Bcl-2ΔTM-G145A (lane 8) or in vitro synthesized Bax K0 (lane 11) was used. The filled circle adjacent to lane 11 indicates a nonphotoadduct because it was also detected in the absence of UV irradiation (lane 10). The open circle indicates the [35S]Met-Bax K73 or K0 monomer. B, photocross-linking of the ANB/[35S]Met-labeled Bax K73-I66E/D68R to His6-tagged Bcl-2ΔTM. The control reaction with ANB/[35S]Met-labeled Bax K73 is shown in lane 5. C, photocross-linking of His6-tagged Bax to in vitro synthesized [35S]Met-labeled Bcl-2ΔTM with a single ANB probe attached to K107 (lanes 5–8). The heterodimer-specific photoadduct was detected in lane 8 and indicated by an arrow. The adduct was not detected when either Bcl-2ΔTM-G145A-K107 (lane 4) or Bcl-2ΔTM K0 (lane 9) was synthesized in vitro and used in the cross-linking reaction. The open circle indicates the [35S]Met-Bcl-2ΔTM monomer. D, photocross-linking of His6-tagged Bax-I66E/D68R with in vitro synthesized ANB/[35S]Met-labeled Bcl-2ΔTM K107. The control reaction with His6-Bax is shown in lane 4. E, competitive photocross-linking of His6-Bax and His6-Bcl-2ΔTM with the ANB/[35S]Met-labeled Bcl-2ΔTM K107. The photoadduct was formed between the ANB/[35S]Met-Bcl-2 K107 and His6-Bax or His6-Bcl-2ΔTM and indicated by an arrow or arrowhead, respectively. In all panels, the phosphorimaging data shown were from the Ni2+ resin-bound fractions of the samples. The schematics on the side of each panel illustrate the proteins (Bax, gray; Bcl-2, black) in the corresponding cross-linking reaction and their interaction (indicated by the double-headed arrow). The star indicates the ANB photocross-linker. The lollipop indicates a mutation that abolished the interaction (indicated by the double-headed arrow with cross).
When RNA encoding Bax K0 or Bcl-2ΔTM K0 was used to program the in vitro translation, ANB-Lys should not be incorporated into the corresponding polypeptide because there is no lysine codon in the RNA. As expected, no photoadduct was detected when these polypeptides were used (Fig. 4, A, lane 11, and C, lane 9). Therefore, the photoadduct formed by [35S]Met-Bax K73 and His6-Bcl-2ΔTM or [35S]Met-Bcl-2ΔTM K107 and His6-Bax was via the ANB probe attached to Lys73 of the [35S]Met-Bax or Lys107 of the [35S]Met-Bcl-2ΔTM, respectively. The formation of these photoadducts is due to the position 73 in Bax and 107 in Bcl-2 being close to the interface of the Bax/Bcl-2 heterodimer because of the following: (i) the ANB-Lys in the [35S]Met-labeled Bax or Bcl-2 molecule was able to react specifically with micromolar concentrations of the binding partner, His6-tagged Bcl-2ΔTM, or Bax molecule, respectively, in reactions containing high concentrations of reticulocyte lysate proteins, including ∼150 mm globin; and (ii) the cross-linking occurred via the ANB-derived reactive nitrene that has a relatively short lifetime of nanoseconds.
To determine whether the site-specific photocross-linking captured heterodimer is relevant to the heterodimer form in biological systems, we first tested the mutant Bax and Bcl-2 that do not form heterodimer in cells. The results show that G145A mutation in His6- or [35S]Met-Bcl-2ΔTM abolished the photoadduct formation with [35S]Met- or His6-Bax (Fig. 4, A, lane 8, or C, lane 4), whereas I66E/D68R mutation in His6- or [35S]Met-Bax abolished the photoadduct formation with [35S]Met- or His6-Bcl-2ΔTM (Fig. 4, D, lane 2, or B, lane 4), respectively. Next we tested whether His6-tagged Bax and Bcl-2ΔTM can compete for cross-linking to [35S]Met/ANB-Lys107-Bcl-2ΔTM that can cross-link with each His6-tagged protein when they were presented individually (Fig. 4E, lanes 2 and 4, indicated by arrow and arrowhead, respectively). The amount of the [35S]Met-Bcl-2ΔTM/His6-Bax photoadduct formed in the reaction was reduced in the presence of His6-Bcl-2ΔTM (Fig. 4E, compare the arrow-indicated band in lanes 2 and 5). The presence of His6-Bax also reduced the amount of Bcl-2 homo-photoadduct (Fig. 4E, compare the arrowhead-indicated band in lanes 4 and 5). As predicted, this competition disappeared when the competitor was the mutant protein, His6-tagged Bcl-2ΔTM-G145A or Bax-I66E/D68R, that does not interact with the wild type [35S]Met-labeled Bax or Bcl-2ΔTM, respectively (Fig. 4E, compare the arrow-indicated bands in lanes 2 and 10 or the arrowhead-indicated bands in lanes 4 and 7). Taken together, these data suggest that the Bax/Bcl-2 heterodimer formed in the detergent system and detected by the site-specific photocross-linking is biologically relevant.
Interface of Bax/Bcl-2 Heterodimer Revealed by Site-specific Photocross-linking
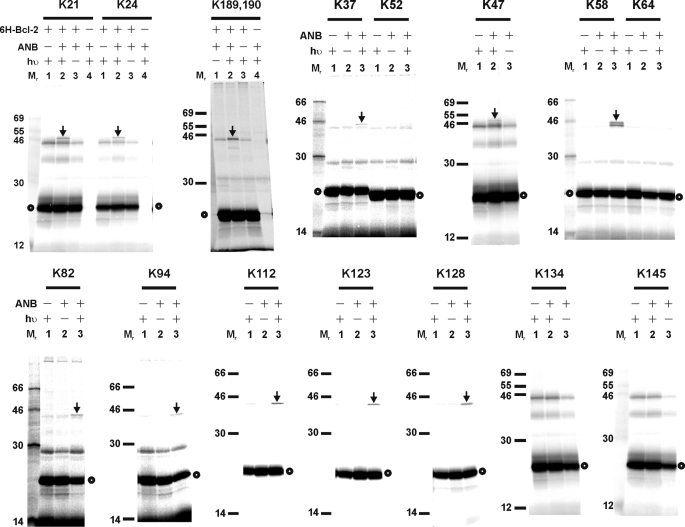
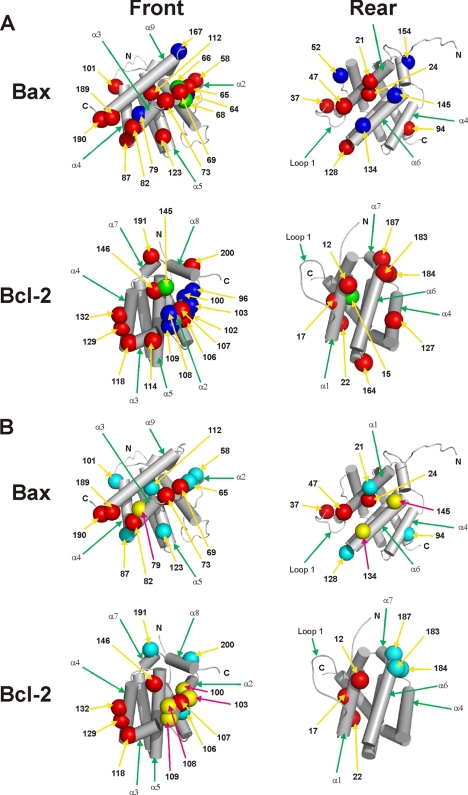
To map the heterodimer interface, we performed the photocross-linking reaction with 23 and 26 additional single ANB-Lys-labeled Bax and Bcl-2ΔTM mutants to His6-tagged Bcl-2ΔTM and Bax protein, respectively. The results shown in Figs. 5 and 6 indicate that the following residues in Bax are close to the interface with Bcl-2ΔTM: 21 and 24 in α1; 37 and 47 in the loop between α1 and α2 (termed loop 1 hereafter; loops between other α-helices are termed similarly); 58, 65 (data not shown), and 69 (data not shown) in α2; 73 in loop 2; 82 and 87 (data not shown) in loop 3; 94 in α4; 101 in loop 4 (data not shown); 112 and 123 in α5; 128 in loop 5; 189 and/or 190 near the C terminus (Table 1); and the following residues in Bcl-2ΔTM are close to the interface with Bax: 12, 17, and perhaps 22 (data shown in Fig. 2C) in α1; 106 and 107 in α2; 114 and 118 in α3; 127, 129, and 132 in α4; 146 in α5; 164 in loop 5; 183 and 184 in α6; 187 and 191 in α7; and 200 in α8 (Table 2).
FIGURE 5.
Photocross-linking of His6-tagged Bcl-2ΔTM protein to [35S]Met-Bax mutant proteins, each with a single ANB-labeled lysine residue. The phosphorimaging data shown were from the Ni2+ resin-bound fractions of the samples. The position of the ANB-lysine in each [35S]Met-Bax mutant is indicated at the top of each image. The heterodimer-specific photoadduct and the [35S]Met-Bax mutant monomer are indicated by an arrow and circle, respectively.
FIGURE 6.
Photocross-linking of His6-tagged Bax protein to [35S]Met-Bcl-2ΔTM mutant proteins, each with a single ANB-labeled lysine residue. The phosphorimaging data shown were from the Ni2+ resin-bound fractions of the samples. The position of the ANB-lysine in each [35S]Met-Bcl-2ΔTM mutant is indicated at the top of each image. The heterodimer-specific photoadduct and the [35S]Met-Bcl-2ΔTM mutant monomer are indicated by an arrow and circle, respectively.
TABLE 1.
Photoreactive probe locations in Bax structure that result in heterodimer-specific adducts with Bcl-2
| Residue | 21 | 24 | 37 | 47 | 58 | 65 | 69 | 73 | 82 | 87 | 94 | 101 | 112 | 123 | 128 | 189, 190 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | α1 | α1 | Loop 1 | α2 | α2 | α2 | Loop 2 | Loop 3 | α4 | Loop 4 | α5 | α5 | Loop 5 | Near COOH | ||
TABLE 2.
Photoreactive probe locations in Bcl-2 structure that result in heterodimer-specific adducts with Bax
| Residue | 12 | 17 | 22 | 106 | 107 | 114 | 118 | 127 | 129 | 132 | 139 | 146 | 164 | 183 | 184 | 187 | 191 | 200 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | α1 | α1 | α1 | α2 | α2 | α3 | α3 | α4 | α4 | α4 | Loop 4 | α5 | Loop 5 | α6 | α6 | α7 | α7 | α8 |
A model for the surfaces of Bax and Bcl-2 that are involved in hetero-interaction was built based on the photocross-linking data (Fig. 7). Not surprisingly, residues 58, 65, 68, 69, and 73 in the BH3 region of Bax and residues in the hydrophobic groove of Bcl-2, including residues 106, 107, 132, 146, 191, and 200 in BH1–3 regions as well as residues 114, 118, and 129 in α3-α4 regions, are located in the hetero-complex interface, consistent with the previous data that the BH3 region of Bax is critical for binding with the hydrophobic groove of Bcl-2 (26, 27, 47, 48). However, residues outside this canonical interface are also indicated by our photocross-linking data. For example, Bax residues 21 and 24 in BH4/α1; 37 and 47 in loop 1; 128 in loop 5; and Bcl-2 residues 12, 17, and perhaps 22 in BH4/α1; 164 in loop 5; and 183 and 184 in α6 are in the hetero-complex interface. Therefore, we proposed a two-interface model to account for most of the cross-linking data, in which the front-front interface is formed by the BH1–3 regions of Bax and Bcl-2, and the rear-rear interface by the BH4/α1, loop 1 and α6 regions located on the opposite side from the BH1–3 regions of the proteins (Fig. 1, A and C).
FIGURE 7.
Predicted interfaces of Bax-Bcl-2 hetero-complexes. A, based on the site-specific photocross-linking data shown above, the surfaces of Bax and Bcl-2 that may form the interface in the hetero-complex were modeled on the previously determined Bax and Bcl-2 structures (4, 5). The residues that, when replaced by ANB-Lys, generated a heterodimer-specific photoadduct and hence close to or within the interface are shown in red, and the residues that did not generate the photoadduct and hence likely far from the interface are shown in blue. The radii of these residues are increased to 6 Å to reflect the uncertainty of the photocross-linking-based mapping technique (36). The position of these residues in the sequence of corresponding protein is indicated by the number. The mutations, I66E/D68R in Bax and G145A and V15E in Bcl-2, are shown in green. B, surfaces of Bax and Bcl-2 that may form the interface in hetero-complex are overlaid with the surfaces that may form the interface in homo-complex according to our previous studies (36, 40). The red residues are involved in both homo- and hetero-interfaces, yellow residues in homo-interface only, and cyan residues in hetero-interface only. In all panels, the view on the left or right shows the front or rear surface, respectively. The residues assigned to each surface are those appeared in the corresponding surface in a space-filled structure that is oriented as the skeleton structure shown.
Two-interface Heterocomplex Model Is Supported by Cross-linking of Site-specific Photoreactive Probe-labeled Bcl-2 Proteins with a Surface-specific Bax Peptide
The front-front interface has been well supported by structural studies using peptides homologous to the BH3 region of Bax and proteins homologous to Bcl-2. For example, the BH3 peptide of Bak forms an amphipathic α-helix that binds to the BH1–3 hydrophobic groove of Bcl-xL (28). In addition, mutations in the BH3 region of Bax and the BH1–3 groove of Bcl-2 inhibit the Bax/Bcl-2 interaction in cell-free systems and cells (e.g. Fig. 2) (24, 26, 49, 50). On the other hand, the rear surface of Bax has been shown to bind a stabilized helical peptide from the BH3 region of Bim (34), and the rear/rear interaction is involved for Bax as well as Bak to form homo-oligomer (40, 51). Thus, the predicated rear-rear interface in Bax-Bcl-2 complex is logical but nevertheless needs further evidence.
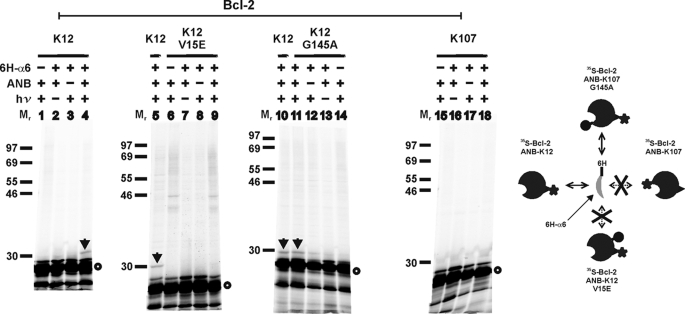
To test this interface more directly, we used a peptide containing the α6 region of Bax with a His6 tag at the N terminus in the photocross-linking experiments with [35S]Met-Bcl-2ΔTM with an ANB probe attached to K12 (in α1) on the rear or K107 (in α2) on the front surface. As shown in Fig. 8, photocross-linking of [35S]Met-Bcl-2ΔTM-K12 protein/His6-Bax α6 peptide was detected (lane 4, indicated by arrow) in the presence of the peptide, photoreactive probe, and UV irradiation but not in the absence of any of these components (lanes 1–3). The photoadduct is radioactive and bound to Ni2+-chelating resin, further demonstrating the specificity. Moreover, a loss-of-function mutation in α1 of Bcl-2 (V15E) abolished the photocross-linking (Fig. 8, lane 9) (52). The photocross-linking data support a rear-rear binding model because α6 of Bax and K12/α1 of Bcl-2 are part of the rear interface of the hetero-complex. As expected, when the photoreactive probe was attached to K107 in the front surface of Bcl-2ΔTM, no photoadduct was detected with the Bax α6 peptide (Fig. 8, lane 18), suggesting that the rear surface of Bax does not interact with the front surface of Bcl-2. Consistent with this result, the G145A mutation located in the front surface of Bcl-2 did not prevent the Bax α6 peptide cross-linking to Bcl-2 via the photoreactive probe attached to K12 in the rear surface (Fig. 8, compare lane 10 with 11). Therefore, the cross-linking results and previous structural and mutagenesis data provide strong support to the two-interface model for the Bax-Bcl-2 hetero-complex.
FIGURE 8.
Photocross-linking of His6-tagged Bax peptide to [35S]Met/ANB-Lys-labeled Bcl-2ΔTM protein. The phosphorimaging data shown were from the Ni2+ resin-bound samples from cross-linking of [35S]Met-Bcl-2ΔTM protein with or without the indicated mutation via the ANB probe attached to K12 or K107 in the rear or front surface, respectively, with His6-tagged Bax helix 6 peptide (6H-α6) that is part of the rear surface of Bax. The position of the single lysine is indicated at the top. The photoadduct containing [35S]Met-Bcl-2ΔTM K12 and 6H-α6 is indicated by arrow. [35S]Met-Bcl-2ΔTM monomer is indicated by an open circle. The schematic on the right illustrates the cross-linking of the Bax peptide with corresponding Bcl-2 proteins that is dependent on the location of photoreactive probe and sensitive to one but not the other mutation.
Two Interfaces in the Heterocomplex Are Interdependent
To test whether the two interfaces in the Bax-Bcl-2 hetero-complex can form independently, we used proteins with mutations in either the front or rear surface in cross-linking experiments with the photoreactive probe at the same or the opposite surface. As expected, no heterodimer-specific cross-linking was observed in control experiments in which the mutation and the photoreactive probe were in the same interface. Thus, the mutations in the front surface, I66E/D68R in Bax and G145A in Bcl-2ΔTM, inhibited the cross-linking from the ANB probe attached to the front surface, K73 in Bax, and K107 in Bcl-2ΔTM (Fig. 4, A–D, and Tables 3 and 4), whereas the V15E mutation in the rear surface of Bcl-2ΔTM inhibited the cross-linking from the ANB probe attached to the rear surface, K24 in Bax and K12 in Bcl-2ΔTM (Fig. 9A, compare lane 2 with 8, and C, compare lane 15 with 14; refer to the corresponding schematic in Fig. 9D and Tables 3 and 4). Surprisingly, the mutation in the front or rear surface also inhibited the heterodimer-specific cross-linking from the photoreactive probe attached to the opposite surface. Thus, the mutations in the front interface, I66E/D68R in Bax and G145A in Bcl-2ΔTM, inhibited the cross-linking from the ANB probe attached to the rear surface, K24 in Bax and K12 in Bcl-2ΔTM (Fig. 9, A, compare lane 2 with 4 and 6, and C, compare lane 1 with 2, and 10 with 9; refer to the corresponding schematic in Fig. 9D and Tables 3 and 4), whereas the V15E mutation in the rear surface of Bcl-2ΔTM inhibited the cross-linking from the ANB probe attached to the front surface, K73 in Bax and K107 in Bcl-2ΔTM (Fig. 9, A, compare lane 10 with 12, and B, compare lane 1 with 2; refer to the corresponding schematic in Fig. 9D and Tables 3 and 4). Together these data indicate that the formation of the two interfaces is most likely coupled such that the formation of the two interfaces is either all or none.
TABLE 3.
Effect of mutations on site-specific photocross-linking of [35S]Met-Bax with His6-Bcl-2ΔTM
| Photocross-linking | [35S]Met-Bax |
||||
|---|---|---|---|---|---|
| ANB-K73-Front |
ANB-K24-Rear |
||||
| WT | I66E/D68R-Front | WT | I66E/D68R-Front | ||
| His6-Bcl-2ΔTM | WT | + | − | + | − |
| G145A-Front | − | − | |||
| V15E-Rear | − | − | |||
TABLE 4.
Effect of mutations on site-specific photocross-linking of [35S]Met-Bcl-2ΔTM with His6-Bax
| Photocross-linking | [35S]Met-Bcl-2ΔTM |
||||||
|---|---|---|---|---|---|---|---|
| ANB-K107-Front |
ANB-K12-Rear |
||||||
| WT | G145A-Front | V15E-Rear | WT | G145A-Front | V15E-Rear | ||
| His6-Bax | WT | + | − | − | + | − | − |
| I66E/D68R-Front | − | − | |||||
FIGURE 9.
Effect of mutation in the front or rear surface of Bax or Bcl-2ΔTM on heterodimer-specific cross-linking via photoreactive probe attached to the front or rear surface. Phosphorimaging data shown are from the Ni2+ resin-bound samples from cross-linking of [35S]Met-Bax with ANB attached to the indicated Lys to His6-Bcl-2ΔTM (A) or from cross-linking of [35S]Met-Bcl-2ΔTM with ANB attached to the indicated Lys to His6-Bax (B and C). In these panels, the arrow indicates the corresponding photoadduct, and the open circle indicates the monomer of 35S-labeled Bax or Bcl-2ΔTM protein. Mutant Bax and Bcl-2 were used if indicated. The arrowhead-indicated band in lane 14 of C is not a photoadduct with His6-Bax because it was also detected in the absence of His6-Bax (lane 11). D, schematic illustration of the corresponding proteins used in the cross-linking reactions here and in Fig. 4 with the locations of photoreactive probe and mutation, and the effect of mutation is indicated. The information is also summarized in Tables 3 and 4.
We used the gel filtration assay to assess the effect of the mutations that inhibited the hetero-cross-linking on Bcl-2-mediated inhibition of Bax oligomerization in detergent, which was observed before (40). Bax-I66E/D68R oligomerized in Triton X-100 micelles similarly to WT Bax as detected by gel filtration chromatography (Fig. 10, compare A with B), suggesting that homo- oligomerization is not abolished by the mutations. The oligomerization of the His6-Bax-I66E/D68R was not inhibited by Bcl-2ΔTM (Fig. 10, compare B with C), consistent with our observation that this mutant Bax did not interact with Bcl-2 in the cross-linking experiments (Figs. 2, 4, and 9 and Tables 3 and 4). Initially, we were surprised that the I66E/D68R mutation did not inhibit Bax homo-oligomerization because in our previous study a G67R mutation inhibited Bax homo-cross-linking (40) and homo-oligomerization in the same gel filtration assay (data not shown). However, in the soluble Bax structure, these three residues are in helix 2 and hence the side chains are pointed in different directions (separated by ∼100°). Furthermore, unlike the G67R mutant, the I66E/D68R mutant retained partial pro-apoptotic activity when assayed in bax/bak double knock-out cells (data not shown), a result consistent with the oligomerization seen here.
FIGURE 10.
Effect of the interface mutations on Bcl-2 inhibition of Bax oligomerization in Triton X-100. Oligomerization of Triton X-100-treated His6-Bax or the I66E/D68R mutant in the absence or presence of His6-tagged Bcl-2ΔTM or the V15E mutant examined by gel filtration chromatography. The chromatographic fractions 21–32 were analyzed by SDS-PAGE and immunoblotting with a Bax- or Bcl-2-specific antibody to detect His6-tagged Bax (left panels) or Bcl-2 (right panels). The molar ratio of Bax versus Bcl-2 in the Bcl-2 containing sample is 1:6. The elution positions for protein standards are indicated at the top with the molecular weight. The predicted molecular weight for Bax or Bcl-2ΔTM monomer in a CHAPS micelle is 27 or 31, respectively. Thus, the WT and mutant Bax proteins are mainly in dimeric and oligomeric forms under all of the conditions, whereas the WT and mutant Bcl-2ΔTM proteins are in monomeric and dimeric forms.
Similarly, Bcl-2ΔTM-G145A and V15E mutants that did not cross-link to Bax (Figs. 2, 4, and 9 and Tables 3 and 4) did not inhibit Bax oligomerization (Fig. 10, compare A with D) (40). Therefore, mutation in one of the two interfaces in the hetero-complex was sufficient to disrupt interactions at both interfaces and abolished the inhibitory activity of Bcl-2 on Bax oligomerization.
DISCUSSION
We have used site-specific photocross-linking to systematically map the interface of Bax/Bcl-2 hetero-oligomers. By comparing this interface map with those of Bcl-2 and Bax homo-oligomers revealed in our previous studies (36, 40), it is possible to picture how Bcl-2 can inhibit Bax activation and oligomerization.
Accumulating data suggest that Bcl-2 can bind to the BH3-only protein such as tBid and Bim to prevent their interaction with soluble/peripheral Bax thereby inhibiting the initial activation of Bax (14, 34, 35, 53). The cross-linking data from our current study demonstrate a physical interaction between the hydrophobic BH1–3 groove of Bcl-2 and the BH3 region of Bax that resulted in the formation of front-front interface. We speculate that this interaction may occur when Bax is still in the activation complex with a BH3-only protein, because previous studies suggest that BH3-only activators engage Bax from the rear surface, which alters the conformation of Bax expelling the C-terminal α9 helix from the BH1–3 groove on the front and exposing the BH3 motif (33, 34, 54). The BH1–3 groove of Bcl-2 monomers may bind the exposed BH3 motif of Bax. The Bcl-2 homodimer formed by interactions between the rear-rear interface may also be able to bind the Bax BH3 motif because this Bcl-2 homodimer has the BH1–3 groove available (Fig. 1B, step 2). The consequence of this Bcl-2/Bax interaction would be 2-fold as follows: preventing the active Bax from using the exposed BH3 motif to activate another soluble/peripheral Bax (15) and inhibiting the homo-interaction between two active Bax proteins via their front surfaces (40).
Additional physical contacts between Bax and Bcl-2 were revealed by our cross-linking study. These contacts occur at the rear surface of the proteins, opposite to the BH1–3 front surface. Because BH3-only activator proteins have been shown to bind to the rear surface of Bax (33–35), binding of Bcl-2 to this surface may occur after the BH3-only protein dissociates. The dissociation of BH3-only protein from the rear surface may be facilitated by binding of Bcl-2 with the front surface of Bax. Accordingly, Bcl-2 may only bind to the rear surface of a Bax protein that already has a Bcl-2 bound to its front surface. Consistent with this possibility, mutations in the front surface that disrupted the binding of Bcl-2 to the front surface of Bax also ablated the binding to the rear surface. However, a mutation in the rear surface that disrupted the binding of Bcl-2 to the rear surface of Bax also abolished the binding to the front surface. Therefore, the minimal stable form of the hetero-complex seems to be one Bax sandwiched by two Bcl-2. Alternatively, the surface-specific mutations we have tested change the structure of protein globally to disrupt both surfaces, and therefore, the wild type Bcl-2/Bax heterodimer may be stable in either front-front or rear-rear or both forms. However, this alternative explanation is less consistent with other experimental evidence. All of the mutant His6-tagged proteins were soluble after purification, similar to the wild type proteins (data not shown), and both Bcl-2-G145A and -V15E as well as Bax-I66E/D68R proteins could be stably expressed in cells (data not shown) (14, 24, 49, 50, 52), suggesting that the mutant proteins are properly folded. Bcl-2ΔTM-G145A forms homodimers as we reported previously (36), and Bax-I66E/D68R forms homo-oligomers as shown in Fig. 10B.
In addition to inhibiting Bax activation at the early stages as described above, Bcl-2 may inhibit Bax at a later stage. For example, Bcl-2 may bind Bax homodimers to inhibit further oligomerization. We recently demonstrated that the Bax homodimer is an intermediate from which tetramer and large oligomers can form (40). Furthermore, the Bax homodimer is formed through the front-front interface and would have two conformation-altered rear surfaces that could engage Bcl-2. Binding of Bcl-2 molecules to the two rear surfaces of the Bax homodimer may result in a hetero-tetramer with a Bax dimer sandwiched by two Bcl-2 molecules. Alternatively, binding of Bcl-2 to the Bax homodimer via one of the two rear surfaces may dissociate the Bax homodimer allowing another Bcl-2 to bind the front surface thereby forming a heterotrimer with one Bax sandwiched by two Bcl-2 molecules. It is also possible that the Bcl-2/Bax heterodimer may engage an active Bax protein via the front surface of Bcl-2 in the heterodimer to form a heterotrimer with one Bcl-2 sandwiched by two Bax. In any of these scenarios, conformational changes induced by binding of Bcl-2 to Bax homodimers would inhibit Bax oligomerization. Because large Bax and Bcl-2-containing oligomers were not detected when Bax was co-expressed with Bcl-2 in cells, it is likely that the Bax/Bcl-2 heterotrimer cannot stably engage other Bax and Bcl-2 proteins to form larger hetero-oligomers (14). Therefore, the interacting surfaces of Bcl-2 share common residues with the interacting surfaces of Bax (Fig. 7B), but it is presumably the differences in the binding interfaces in the hetero-complex formed by Bcl-2 and Bax that makes the complex not competent for further oligomerization.
The residues in α5-α6 regions of Bax and Bcl-2 that are involved in hetero- and homo-interactions are mostly different (Fig. 7B). The difference is intriguing because these α-helices of both multiple BH proteins are inserted into the MOM after activation by binding to BH3-only proteins (13, 14, 53, 55). Following the insertion into the membrane, Bax forms large oligomers that permeabilize the membrane releasing cytochrome c and other mitochondrial intermembrane space proteins. In contrast, the membrane-embedded Bcl-2 only forms small oligomers that release small molecules but do not release mitochondrial proteins (53, 56). Moreover, it is the membrane-embedded form of Bcl-2 that binds to the similarly embedded Bax thereby preventing it from forming the large oligomers that permeabilize membranes (14). Our homo- and heterodimer models suggest that the membrane-embedded regions in Bax and Bcl-2 may interface differently in homo- and hetero-complexes resulting in different structures in the membrane. In other words, Bcl-2 may function as a defective protomer to compete with the perfect Bax protomer thereby inhibiting Bax oligomerization. Interestingly, Bcl-xL was found to compete with Bax for activation of soluble monomeric Bax through interaction with membranes, tBid or tBid-activated Bax, thereby inhibiting Bax binding to membranes, oligomerization, and membrane permeabilization (11).
The effect of detergent on Bcl-2 conformation is unknown, but a large conformational change is expected. An NMR study of Bcl-xL lacking both C-terminal TM sequence and the loop between α1 and α2 in dodecylphosphocholine micelles suggests that Bcl-xL has a loosely packed, dynamic structure in micelles, with α1 and α6 and possibly α5 partially buried in the hydrophobic interior of the micelle. Other parts of the protein are located near the surface or on the outside of the micelle (57). Interestingly, in our hetero-oligomer model α1 and α6 of Bcl-2 are located in the rear surface that binds to the rear surface of Bax, which also includes α1 and α6, suggesting that the rear interface may form within the hydrophobic interior of the Triton X-100 micelle. Previously, we found that α5 of Bcl-2 moves into the interior of membrane bilayers in cultured cells after apoptotic stimuli, in isolated mitochondria, and liposomes after interaction with tBid or tBid-activated Bax, and this conformational alteration is required for Bcl-2 to inhibit active Bax (14, 53, 55).
In summary, our previous and current site-specific photocross-linking studies have provided the first set of comprehensive interface maps for Bax and Bcl-2 homo- and hetero-oligomers. All of the interfaces not only contain the canonical BH3 motif/BH1–3 groove interface but also a novel interface located on the opposite side of the molecules. In the past decade, effort has been focused on development of small chemical ligands (e.g. ABT-737) that bind to the BH1–3 groove of Bcl-2 to disrupt the binding with BH3-only proteins thereby inhibiting the anti-apoptotic activity of Bcl-2 in cancer cells (58). The novel interface we revealed in the Bcl-2/Bax hetero-oligomer may be the next target for developing a different kind of inhibitor that can block Bcl-2 binding with Bax through a different interface that is equally important for the anti-apoptotic activity that correlates with the cancer resistance to chemo- and radiation therapies (59–61).
Acknowledgment
We thank Gillian Air for help with editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM062964 (to J. L.). This work was also supported by Canadian Institutes of Health Research Grant FRN 12517 (to D. W. A.).
- BH
- Bcl-2 homology
- MOM
- mitochondrial outer membrane
- ϵANB-Lys
- Nϵ-(5-azido-2-nitrobenzoyl)-Lys
- ANB
- 5-azido-2-nitrobenzoyl
- PARP
- poly(ADP-ribose) polymerase
- tBid
- truncated Bid
- TM
- transmembrane.
REFERENCES
- 1.Leber B., Lin J., Andrews D. W. (2007) Apoptosis 12, 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 3.Kvansakul M., Yang H., Fairlie W. D., Czabotar P. E., Fischer S. F., Perugini M. A., Huang D. C., Colman P. M. (2008) Cell Death Differ. 15, 1564–1571 [DOI] [PubMed] [Google Scholar]
- 4.Petros A. M., Medek A., Nettesheim D. G., Kim D. H., Yoon H. S., Swift K., Matayoshi E. D., Oltersdorf T., Fesik S. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3012–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki M., Youle R. J., Tjandra N. (2000) Cell 103, 645–654 [DOI] [PubMed] [Google Scholar]
- 6.Janiak F., Leber B., Andrews D. W. (1994) J. Biol. Chem. 269, 9842–9849 [PubMed] [Google Scholar]
- 7.Tsujimoto Y., Ikegaki N., Croce C. M. (1987) Oncogene 2, 3–7 [PubMed] [Google Scholar]
- 8.Chen-Levy Z., Nourse J., Cleary M. L. (1989) Mol. Cell. Biol. 9, 701–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antonsson B., Montessuit S., Sanchez B., Martinou J. C. (2001) J. Biol. Chem. 276, 11615–11623 [DOI] [PubMed] [Google Scholar]
- 10.Hsu Y. T., Youle R. J. (1998) J. Biol. Chem. 273, 10777–10783 [DOI] [PubMed] [Google Scholar]
- 11.Billen L. P., Kokoski C. L., Lovell J. F., Leber B., Andrews D. W. (2008) PLoS Biol. 6, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. (2008) Cell 135, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 13.Annis M. G., Soucie E. L., Dlugosz P. J., Cruz-Aguado J. A., Penn L. Z., Leber B., Andrews D. W. (2005) EMBO J. 24, 2096–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dlugosz P. J., Billen L. P., Annis M. G., Zhu W., Zhang Z., Lin J., Leber B., Andrews D. W. (2006) EMBO J. 25, 2287–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan C., Dlugosz P. J., Peng J., Zhang Z., Lapolla S. M., Plafker S. M., Andrews D. W., Lin J. (2006) J. Biol. Chem. 281, 14764–14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruffolo S. C., Shore G. C. (2003) J. Biol. Chem. 278, 25039–25045 [DOI] [PubMed] [Google Scholar]
- 17.Eskes R., Desagher S., Antonsson B., Martinou J. C. (2000) Mol. Cell. Biol. 20, 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 19.Basañez G., Sharpe J. C., Galanis J., Brandt T. B., Hardwick J. M., Zimmerberg J. (2002) J. Biol. Chem. 277, 49360–49365 [DOI] [PubMed] [Google Scholar]
- 20.Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. (2002) Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 21.Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. (2005) Mol. Cell 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 22.Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. (2005) Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 23.Certo M., Del Gaizo Moore V., Nishino M., Wei G., Korsmeyer S., Armstrong S. A., Letai A. (2006) Cancer Cell 9, 351–365 [DOI] [PubMed] [Google Scholar]
- 24.Yin X. M., Oltvai Z. N., Korsmeyer S. J. (1994) Nature 369, 321–323 [DOI] [PubMed] [Google Scholar]
- 25.Lin B., Kolluri S. K., Lin F., Liu W., Han Y. H., Cao X., Dawson M. I., Reed J. C., Zhang X. K. (2004) Cell 116, 527–540 [DOI] [PubMed] [Google Scholar]
- 26.Wang K., Gross A., Waksman G., Korsmeyer S. J. (1998) Mol. Cell Biol. 18, 6083–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zha H., Aimé-Sempé C., Sato T., Reed J. C. (1996) J. Biol. Chem. 271, 7440–7444 [DOI] [PubMed] [Google Scholar]
- 28.Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., Fesik S. W. (1997) Science 275, 983–986 [DOI] [PubMed] [Google Scholar]
- 29.Smits C., Czabotar P. E., Hinds M. G., Day C. L. (2008) Structure 16, 818–829 [DOI] [PubMed] [Google Scholar]
- 30.Liu X., Dai S., Zhu Y., Marrack P., Kappler J. W. (2003) Immunity 19, 341–352 [DOI] [PubMed] [Google Scholar]
- 31.Petros A. M., Nettesheim D. G., Wang Y., Olejniczak E. T., Meadows R. P., Mack J., Swift K., Matayoshi E. D., Zhang H., Thompson C. B., Fesik S. W. (2000) Protein Sci. 9, 2528–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czabotar P. E., Lee E. F., van Delft M. F., Day C. L., Smith B. J., Huang D. C., Fairlie W. D., Hinds M. G., Colman P. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6217–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cartron P. F., Gallenne T., Bougras G., Gautier F., Manero F., Vusio P., Meflah K., Vallette F. M., Juin P. (2004) Mol. Cell 16, 807–818 [DOI] [PubMed] [Google Scholar]
- 34.Gavathiotis E., Suzuki M., Davis M. L., Pitter K., Bird G. H., Katz S. G., Tu H. C., Kim H., Cheng E. H., Tjandra N., Walensky L. D. (2008) Nature 455, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2009) Mol. Cell 36, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z., Lapolla S. M., Annis M. G., Truscott M., Roberts G. J., Miao Y., Shao Y., Tan C., Peng J., Johnson A. E., Zhang X. C., Andrews D. W., Lin J. (2004) J. Biol. Chem. 279, 43920–43928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neill J. W., Manion M. K., Maguire B., Hockenbery D. M. (2006) J. Mol. Biol. 356, 367–381 [DOI] [PubMed] [Google Scholar]
- 38.Denisov A. Y., Sprules T., Fraser J., Kozlov G., Gehring K. (2007) Biochemistry 46, 734–740 [DOI] [PubMed] [Google Scholar]
- 39.Feng Y., Lin Z., Shen X., Chen K., Jiang H., Liu D. (2008) J. Biochem. 143, 243–252 [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z., Zhu W., Lapolla S. M., Miao Y., Shao Y., Falcone M., Boreham D., McFarlane N., Ding J., Johnson A. E., Zhang X. C., Andrews D. W., Lin J. (2010) J. Biol. Chem. 285, 17614–17627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yethon J. A., Epand R. F., Leber B., Epand R. M., Andrews D. W. (2003) J. Biol. Chem. 278, 48935–48941 [DOI] [PubMed] [Google Scholar]
- 42.Fiebig A. A., Zhu W., Hollerbach C., Leber B., Andrews D. W. (2006) BMC Cancer 6, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin J., Liang Z., Zhang Z., Li G. (2001) J. Biol. Chem. 276, 41733–41741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krieg U. C., Walter P., Johnson A. E. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 8604–8608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu Y. T., Youle R. J. (1997) J. Biol. Chem. 272, 13829–13834 [DOI] [PubMed] [Google Scholar]
- 46.Zha H., Reed J. C. (1997) J. Biol. Chem. 272, 31482–31488 [DOI] [PubMed] [Google Scholar]
- 47.Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 48.Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 49.Sedlak T. W., Oltvai Z. N., Yang E., Wang K., Boise L. H., Thompson C. B., Korsmeyer S. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7834–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ottilie S., Diaz J. L., Chang J., Wilson G., Tuffo K. M., Weeks S., McConnell M., Wang Y., Oltersdorf T., Fritz L. C. (1997) J. Biol. Chem. 272, 16955–16961 [DOI] [PubMed] [Google Scholar]
- 51.Dewson G., Kratina T., Czabotar P., Day C. L., Adams J. M., Kluck R. M. (2009) Mol. Cell 36, 696–703 [DOI] [PubMed] [Google Scholar]
- 52.Hirotani M., Zhang Y., Fujita N., Naito M., Tsuruo T. (1999) J. Biol. Chem. 274, 20415–20420 [DOI] [PubMed] [Google Scholar]
- 53.Peng J., Tan C., Roberts G. J., Nikolaeva O., Zhang Z., Lapolla S. M., Primorac S., Andrews D. W., Lin J. (2006) J. Biol. Chem. 281, 35802–35811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walensky L. D., Pitter K., Morash J., Oh K. J., Barbuto S., Fisher J., Smith E., Verdine G. L., Korsmeyer S. J. (2006) Mol. Cell 24, 199–210 [DOI] [PubMed] [Google Scholar]
- 55.Kim P. K., Annis M. G., Dlugosz P. J., Leber B., Andrews D. W. (2004) Mol. Cell 14, 523–529 [DOI] [PubMed] [Google Scholar]
- 56.Peng J., Ding J., Tan C., Baggenstoss B., Zhang Z., Lapolla S. M., Lin J. (2009) Apoptosis 14, 1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Losonczi J. A., Olejniczak E. T., Betz S. F., Harlan J. E., Mack J., Fesik S. W. (2000) Biochemistry 39, 11024–11033 [DOI] [PubMed] [Google Scholar]
- 58.Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O'Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H. (2005) Nature 435, 677–681 [DOI] [PubMed] [Google Scholar]
- 59.Letai A. G. (2008) Nat. Rev. Cancer 8, 121–132 [DOI] [PubMed] [Google Scholar]
- 60.Yip K. W., Reed J. C. (2008) Oncogene 27, 6398–6406 [DOI] [PubMed] [Google Scholar]
- 61.Cory S., Huang D. C., Adams J. M. (2003) Oncogene 22, 8590–8607 [DOI] [PubMed] [Google Scholar]