Abstract
C1qs are key components of the classical complement pathway. They have been well documented in human and mammals, but little is known about their molecular and functional characteristics in fish. In the present study, full-length cDNAs of c1qA, c1qB, and c1qC from zebrafish (Danio rerio) were cloned, revealing the conservation of their chromosomal synteny and organization between zebrafish and other species. For functional analysis, the globular heads of C1qA (ghA), C1qB (ghB), and C1qC (ghC) were expressed in Escherichia coli as soluble proteins. Hemolytic inhibitory assays showed that hemolytic activity in carp serum can be inhibited significantly by anti-C1qA, -C1qB, and -C1qC of zebrafish, respectively, indicating that C1qA, C1qB, and C1qC are involved in the classical pathway and are conserved functionally from fish to human. Zebrafish C1qs also could specifically bind to heat-aggregated zebrafish IgM, human IgG, and IgM. The involvement of globular head modules in the C1q-dependent classical pathway demonstrates the structural and functional conservation of these molecules in the classical pathway and their IgM or IgG binding sites during evolution. Phylogenetic analysis revealed that c1qA, c1qB, and c1qC may be formed by duplications of a single copy of c1qB and that the C1q family is, evolutionarily, closely related to the Emu family. This study improves current understanding of the evolutionary history of the C1q family and C1q-mediated immunity.
Keywords: Complement, Evolution, Gene Structure, Protein Structure, Zebra Fish
Introduction
Complement plays a central role in innate and adaptive immunity (1). Its activation can be initiated via three distinct pathways: the classical pathway involving binding of C1 to Ag-Ab complex, the alternative pathway initiated by spontaneous deposition of complement factor C3b, and the mannose-binding lectin pathway dependent on the binding of mannose-binding lectin to carbohydrates (2). The original opsonic complement system generally was believed to be composed of the lectin or alternative pathway, a primitive state that was conserved for a long time, at least from the echinoderms up to the cyclostomes (3). Considerable research has been conducted on alternative and lectin pathway components (4–8). However, very few studies have focused on classical pathway molecules in lower vertebrate species. Classical complement components also have been studied best in mammalian vertebrate species, whereas only a few studies in nonmammalian vertebrate species have been attempted. Most of the known classical pathway components in lower vertebrate species were characterized only at the cDNA level; very few molecules have been purified and functionally characterized.
In human and mammals, C1q is the first subcomponent of the classical pathway of complement activation and a major connecting link between classical pathway-driven innate immunity and IgG- or IgM-mediated acquired immunity. The human C1q molecule (460 kDa) is composed of 18 polypeptide chains (six A, six B, and six C chains). The A chain (223 residues), B chain (226 residues), and C chain (217 residues) each have a short (3–9 residues) N-terminal region, followed by a collagen domain (∼81 residues) and a C-terminal globular domain (∼135 residues) (9). The globular domain of each C1q stalk is composed of the C-terminal halves of one A (ghA), one B (ghB), and one C (ghC) chain, which are organized as a heterotrimer. A previous study has demonstrated that human ghA, ghB, and ghC have distinct ligand-binding properties, with each able to bind to heat-aggregated IgG or IgM (10).
In teleost fish, many components involved in the activation of complement pathways have been cloned, including factor Bf/C2 (11–13), factor D (Df), C3 (6, 14), C1r/C1s (15), and C4 (14). The classical pathway has been identified at the functional and biochemical levels, but the structural and functional characterization of teleost c1q genes is still lacking. Therefore, the role of these molecules in the classical pathways of these animals is still under speculation (16). Cartilaginous fish (sharks) possess Igs, C1q, C1r/s, C4, and C2, which suggests that the classical pathway may be established by the emergence of jawed vertebrates (17). Recently, C1q-like molecules have been identified in both lamprey (18) and amphioxus (19). These molecules have been demonstrated to act as lectins and function as initial recognition molecules that connect the C1q to the lectin pathway and the innate immunity. These findings indicate that C1q may have a much longer evolutionary history than thought previously, and C1q molecules may have undergone a functional transition (from involvement in innate immunity to adaptive immunity) from lower invertebrates to higher vertebrates throughout evolution. Therefore, the evolutionary history of C1q is yet to be discovered. As an evolutionary linker between invertebrates and higher vertebrates, fish is believed to be an important model in the evolutionary study and may help reveal many evolution mechanisms. The present study is the first to report the identification and functional characterization of c1qA, c1qB, and c1qC genes, as well as the evolution of the C1q family using the teleost model. Results of this study can provide insight into the molecular and functional evolutionary history of the C1q family and the classical pathway in early vertebrates.
EXPERIMENTAL PROCEDURES
Experimental Fish
Zebrafish (Danio rerio, weighing ∼0.5–1.0 g) and Crucian carp (Carassius auratuses, weighing ∼250–500 g) were kept in recirculating water at 26 °C and fed with commercial pellets at a daily ration of 0.7% of their body weight. All fish were held in the laboratory for at least 2 weeks prior to the experiments for acclimatization and evaluation of overall fish health. Only healthy fish, as determined by general appearance and activity level, were used for the study.
Sequence Retrieval
Genomic and expressed sequence tag databases maintained by the NCBI (http://www.ncbi.nlm.nih.gov/), Ensembl, the University of California, Santa Cruz Genome Browser, the D. rerio Sequencing Project, and the TIGR Gene Indices were employed to retrieve C1q family members, including C1qA, C1qB, and C1qC in zebrafish. Known human and mouse C1q proteins or their truncated sequences containing the C1q domain were used as queries. After a number of C1q-like genomic sequence segments were obtained from the databases by BLAST (20), complete sequences of C1q domain-containing genes were obtained by retrieving neighboring regions, which was performed by zooming and scrolling over chromosomes in the Genome Browser. These genomic sequences were used to search coding exons or open reading frames (ORFs) using the GENSCAN program (Massachusetts Institute of Technology) or the ORF finder programs at the NCBI Web site (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Finally, the translated proteins from predicted transcripts were verified by BLAST (21) in the Swiss-Prot database. Using this method, a sequence similar to the c1qC gene was obtained and was further analyzed using the GENSCAN (22), BLAST (23), and FASTA (24) programs. A possible coding sequence was found and exploited to design primers for obtaining the full-length zebrafish c1qC. According to the synteny comparative analysis approach (25), the genes surrounding c1qC are conserved between human, mouse, and zebrafish. Thus, sequences surrounding c1qC in the zebrafish genome were analyzed using the GENSCAN program (Massachusetts Institute of Technology). A c1qA and a c1qB gene of zebrafish homologous to human c1qA and c1qB were identified by performing a BLAST search on all potential proteins in the Swiss-Prot database.
Cloning and Sequencing of Zebrafish c1qA, c1qB, and c1qC cDNAs
Total RNA was isolated from spleen and head kidney using TRIzol reagent (Invitrogen) and was reverse-transcribed using 3′-full RACE core set (Takara Bio, Inc.) following the manufacturer's instructions. The cDNAs of zebrafish c1qA, c1qB, and c1qC were generated by RT-PCR. (The primers are shown in supplemental Table S1.) The 5′- and 3′-full RACE core set (Takara Bio, Inc.) were utilized to obtain 5′ and 3′ unknown regions, respectively. Finally, full-length cDNA sequences containing the 5′ untranslated region (UTR) and 3′ UTR were assembled. PCR products were loaded on a 1.2% (w/v) agarose gel and visualized by staining with 0.1 mg/ml ethidium bromide. All PCR products were purified using a gel extraction kit (Qiagen), cloned into the pUCm-T vector (Takara Bio, Inc.), and then sequenced on MegaBACE 1000 Sequencer (GE Healthcare) using a DYEnamic ET dye terminator cycle sequencing kit (Pharmacia).
Bioinformatics Analysis
A search for potential functional motifs was done in the PROSITE database (26). Signal peptide prediction was performed using the SignalP program, and multiple alignments of sequences were conducted using the ClustalW program (version 1.83) (27). Phylogenetic trees were constructed with the MEGA4.0 program (28) using a neighbor-joining method, and the statistical significance of each branch was examined by the bootstrapping method. The veracity of these trees was studied using the bootstrapping method by executing 1000 replicates. Gene organizations (intron/exon boundaries) were elucidated by comparing zebrafish c1qA, c1qB, and c1qA cDNAs with their genome sequences, and figures were drawn using GeneMaper 2.5.
Tissue Expressions of c1qA, c1qB, and c1qC Genes
Total RNAs from the head kidney, liver, heart, spleen, intestine, brain, gill, skin, and muscle were isolated as described above and then treated with DNaseI (Qiagen). RNA concentrations were measured by spectrophotometry. Once integrity was certified by analysis on a 1.5% (w/v) agarose gel, RNA was reverse-transcribed into cDNA using an ExScript RT Reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol. The PCR program was as follows: 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s to amplify transcripts of c1qA, c1qB, c1qC, and β-actin (as a standard) using the primers shown in supplemental Table S1. PCR products were analyzed by 1% agarose gel electrophoresis and stained with ethidium bromide.
Production of Recombinant ghA, ghB, and ghC Proteins
Recombinant globular head regions of the A chain (ghA, residues 110–245), the B chain (ghB, residues 118–238), and the C chain (ghC, residues 112–240) were expressed as fusion proteins linked to thioredoxin (Trx)2 in Escherichia coli BL21 cells and purified on a Ni2+-chelating Sepharose column. Protein concentrations were determined using the Bradford method. Protein sizes and purities were confirmed by SDS-PAGE.
Preparation of anti-ghA, anti-ghB, and anti-ghC Antibodies
Male New Zealand white rabbits (6 weeks old, weighing ∼3 kg) were immunized with 100 μg purified recombinant ghA, ghB, and ghC proteins in complete Freund's adjuvant eight times at biweekly intervals. One week after final immunization, the rabbits were bled when antibody titers were above 1:10,000 as determined by microplate-based enzyme linked immunosorbent assay (ELISA) using recombinant proteins adsorbed to the solid phase. Antibodies were affinity-purified into the IgG isotype by a protein A agarose column and immunosorbent-based protocol using recombinant protein absorbed to the nitrocellulose membrane phase (Qiagen).
Western Blot
Carp serum was collected and purified according to the method described by Sasaki et al. (29) with some modifications. Briefly, 10 ml of carp serum was dialyzed successively against 80 and 120 ml of 30 mm EGTA (pH 7.5) for 7 and 13 h at 4 °C, respectively. It was collected by 10,000 × g centrifugation at 4 °C and then washed twice with the same EGTA solution. Precipitate was dissolved in 1 ml 500 mm NaC1, 10 mm EDTA (pH 5.8), and the resulting solution was electrophoresed on 12% SDS-PAGE gel using Laemmli buffer (30). The proteins separated were blotted on nitrocellulose membrane (Hybond, Amersham Biosciences Pharmacia), which was then blocked in TBS containing 2% BSA and 0.05% Tween 20 overnight at 4 °C and then incubated with rabbit anti-zebrafish ghA, ghB, or ghC antibody (1:2000) for 2 h at 37 °C. This was followed by staining with HRP-labeled anti-rabbit IgG. After three washings with TBS, the membrane was incubated with a horseradish peroxidase-conjugated secondary antibody and visualized with an enhanced chemiluminescence detection system and x-ray film (31).
Involvement of Zebrafish C1q in Classical Pathway
The involvement of zebrafish C1q in the fish classical pathway was examined by a hemolytic inhibitory assay using anti-zebrafish C1qA, C1qB, and C1qC antibodies as described previously (10) using the carp serum complement system. Briefly, carps were immunized with sheep red blood cells (SRBCs) by weekly intraperitoneal injection of 0.5 ml cell suspension (109 cells/ml) for 4 weeks. Three days after the last injection, carps were bled by caudal vein puncture. The anti-SRBC antiserum was prepared as described previously (32) and incubated at 50 °C for 30 min to inactivate complement activity. In parallel, the nonimmunized carp serum was used as complement source. For hemolytic assay, SRBCs were prepared in dextrose gelatin Veronal buffer (DGVB; 2.5 mm sodium barbital, 71 mm NaCl, 0.15 mm CaCl2, 0.5 mm MgCl2, 2.5% w/v glucose, 0.1% w/v gelatin, pH 7.4) and sensitized with carp anti-SRBC antiserum. Then, complement-source sera were incubated with an equal amount of sensitized SRBC at 37 °C for 30 min. After centrifugation, the sera were tested for hemolytic activities by measuring the A405 values. Total hemolysis (100%) was assessed as the amount of hemoglobin released upon cell lysis with water. The percentage of hemolysis was expressed by comparing the A405 values with the A405 values of 100% hemolysis. All experiments were conducted in duplicate.
Inhibition of C1q-dependent Hemolysis by C1q Modules
Competitive inhibition assay of C1q-dependent hemolysis by Trx-ghA, -ghB, and -ghC was performed as described previously (10) with minor modifications. Briefly, SRBC sensitized with rabbit anti-SRBC antibody (EAIgG), anti-SRBC antibody (EAIgM), or fish anti-SRBC antibody (EAIgM(fish)) were prepared in dextrose gelatin Veronal buffer (DGVB2+). Diluted guinea pig serum (1:20) was used as a source of complement. The amount was used to cause 50% hemolysis of sensitized SRBC determined by co-incubation of different amounts of 1:20 diluted guinea pig serum and DGVB2+, with 100 μl EAIgG, EAIgM, or EAIgM(fish) at 37 °C for 30 min. The EAIgG, EAIgM, or EAIgM(fish) samples (107 cells per 100 μl) were pretreated with a range of concentrations of Trx-ghA, -ghB, or -ghC (1.25, 2.5, 5, 10, 20, and 40 μg each) for 1 h at 37 °C. Cells were centrifuged, and the pellet was washed and resuspended in 100 μl DGVB2+. Each aliquot of pretreated EA cells was added to the determined amount of 1:20 diluted guinea pig serum that can cause 50% hemolysis. Following a 1-h incubation at 37 °C, unlysed cells were pelleted by centrifugation, and the A405 of the supernatant was read. The EAIgG, EAIgM, or EAIgM(fish) treated with PBS was used as control, and Trx was used as a negative control protein. Percentage inhibition was calculated as % hemolysis (%) = (A405 (control) − A405 (Treated))/A405 (control).
Interaction of C1q Modules with Human IgG or IgM
Quantitative ELISA was performed to detect the interactions of recombinant ghA, ghB, and ghC modules with heat-aggregated human IgG or IgM. Microtiter wells were coated with different concentrations of Trx-ghA, -ghB, -ghC, and Trx in carbonate buffer (pH 9.6) overnight at 4 °C. Any nonspecific binding sites were blocked with 200 μl/well 1% w/v BSA for 1 h at 37 °C. Wells were washed with PBS containing 0.05% Tween 20 (PBST) three times and then incubated with heat-aggregated IgG (10 μg/ml) or IgM (20 μg/ml) in TBS-NTC (50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.05% w/v NaN3, 0.05% v/v Tween 20, and 5 mm CaCl2) at 37 °C for 1 h. After three rounds of washing with PBST, wells were incubated with either goat anti-human IgG or IgM (1:1000) conjugated to HRP. Color was developed using p-nitrophenyl phosphate, and A450 was measured. Data are given as an average of three repetitions ± S.D.
Statistical Analysis
Statistical evaluation of differences between experimental group means was done by ANOVA and multiple Student's t tests. A value of p < 0.05 was considered significant. Sample numbers of each group were more than six fish of equal mean body weight. Data points were from at least three independent experiments.
RESULTS
Identification of Zebrafish c1qA, c1qB, and c1qC Genes
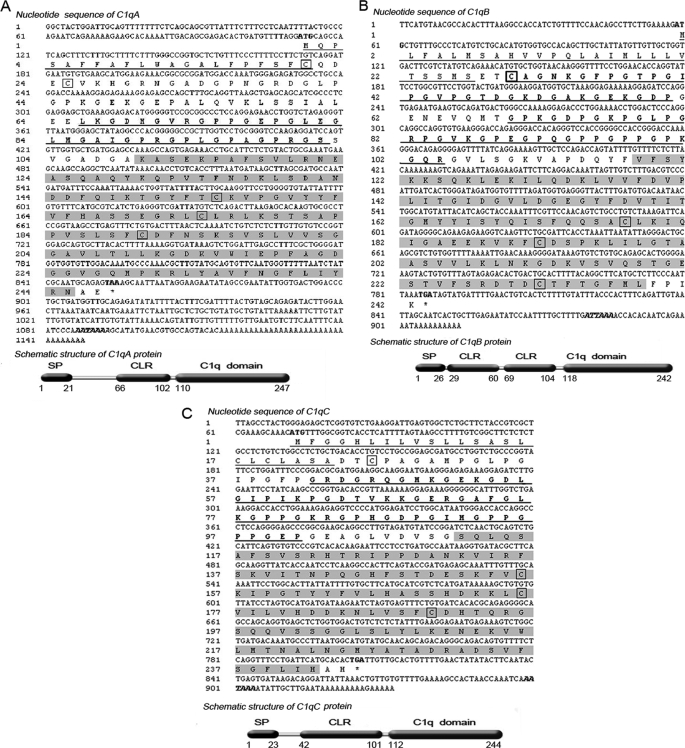
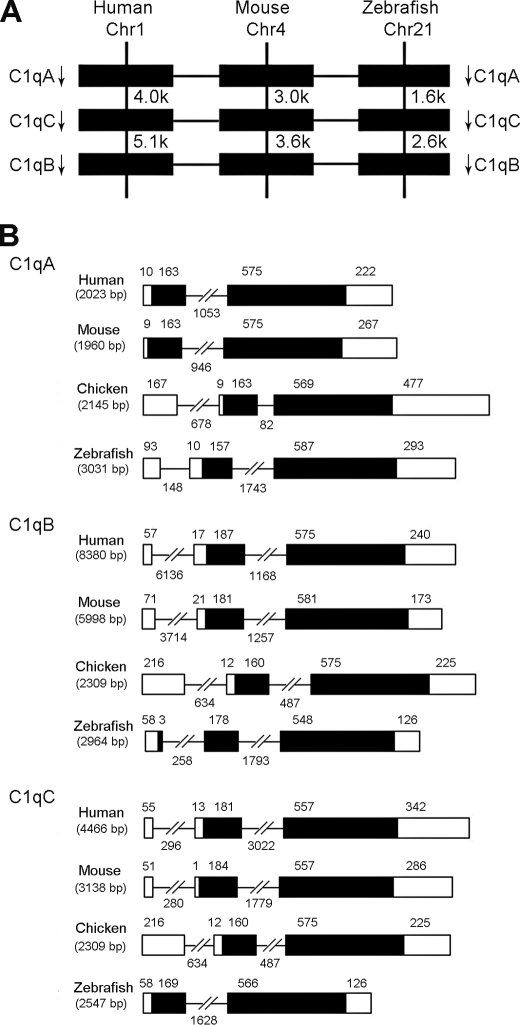
Using cDNA from spleen and head kidney as template, the full-length zebrafish c1qA, c1qB, and c1qC cDNAs were cloned. Nucleotide and predicted amino acid sequences of these three C1q molecules are shown in Fig. 1. The c1qA cDNA was 1148 bp in size, containing an ORF of 741 bp that translates to a 247-amino acid peptide, a 5′ UTR of 111 bp, and a 3′ UTR of 293 bp. The predicted peptide shared 32–39% identity with other known C1qA sequences. The isolated 913-bp full-length cDNA of c1qB contained a 729-bp ORF encoding a putative C1qB protein of 242 amino acids, a 5′ UTR of 58 bp, and a 3′ UTR of 126 bp. The deduced peptide shared 25–35% identity with other known C1qB sequences. Finally, the cloned c1qC cDNA was 932 bp in size, containing an ORF of 735 bp that translates to a 244-amino acid peptide, a 5′ UTR of 71 bp, and a 3′ UTR of 126 bp. This C1qC peptide shared 43–47% identity with other known C1qC homologies. In addition, these three zebrafish C1qs shared 30–33% identity with one another. In zebrafish, c1qA, c1qB, and c1qC genes are clustered in the same strand orientation within a 12.3-kb region of chromosome 21 in the order of c1qA–c1qC–c1qB. This is slightly different from that of human c1qA, c1qC, and c1qB genes at chromosome 1 spanning 23.8 kb and mouse at chromosome 4 spanning 17.5 kb. However, the synteny and organization of these genes were well conserved among zebrafish, mouse, and human (Fig. 2A). The genomic structures of these genes consisted of two to three exons and one to two introns, consistent with those of many other species, including human, mouse, and chicken (Fig. 2B). The sizes of exons and introns of c1qA and c1qC are almost similar among different species. The c1qB of zebrafish has three exons in its coding region, whereas the c1qB of human, mouse, and chicken has only two exons.
FIGURE 1.
Full-length nucleotide sequence with predicted amino acid sequence schematic structure of zebrafish c1qA (accession no. FJ713133.1) (A), c1qB (accession no. FJ713134.1) (B), and c1qC (accession no. FJ713135.1) (C). The start and stop codons are in boldface type. The polyadenylation signal (AATAAA) is in boldface and italic type. The putative signal peptide is underlined. The collagen-like region (CLR) is in boldface type and underlined, and the C1q domain is highlighted. The cysteines are boxed. C1q protein consists of a signal peptide (SP), a collagen-like region, and a C-terminal C1q domain.
FIGURE 2.
A, comparison of gene locations of c1qA, c1qB, and c1qC among human, mouse, and zebrafish. Arrows indicate gene orientation. Numbers below gene names indicate gene positions. B, comparison of genomic organizations of c1qA, c1qB, and c1qC genes among human, mouse, chicken, and zebrafish. Rectangles represent the exons, whereas the lines between them indicate the introns. Exons are indicated by black boxes, and untranslated regions are indicated by white boxes. The sizes of exons and introns are indicated above them.
Structural Characterization of Zebrafish: C1qA, C1qB, and C1qC
Predicted zebrafish C1qA, C1qB, and C1qC proteins consisted of a leading signal peptide, a collagen-like region, and a C-terminal C1q domain (Fig. 1). They have predicted molecular weights of 26.1, 25.4, and 25.8 kDa, respectively. Sequence alignment showed that zebrafish C1qA, C1qB, and C1qC have 25–50% amino acid identities with human and other vertebrates overall. Four cysteine residues conserved in mammalian and avian C1q molecules were also seen in zebrafish C1qA, C1qB, and C1qC, and they were conserved completely among fish and other vertebrates (supplemental Fig. S1). Potential functional motifs of zebrafish C1qA, C1qB, and C1qC searched in the PROSITE database revealed the presence of a C1q domain in the C-terminal region (residues approximately from 110 to 240) of the molecules. To evaluate the implications of sequence conservation between human gC1q and zebrafish gC1q, molecular modeling studies were performed using human ghA, ghB, and ghqC as templates. The modeled tertiary structures of zebrafish ghA, ghB, and ghC are similar to human ghA, ghB, and ghC, respectively (supplemental Fig. S2). Each individual globular head module, with its N and C termini emerging at the base of the trimer, has a jelly-roll topology consisting of a 10-stranded β-sandwich made up of two five-stranded anti-parallel β-sheets. Such folding also is seen in TNF family proteins, which control many aspects of inflammation, adaptive immunity, apoptosis, energy homeostasis, and organogenesis (30). The predicted zebrafish ghA, ghB, and ghC tertiary structure showed a jelly-roll topology consisting of a 10-stranded β-sandwich, indicating that it belongs to the TNF family.
Of four conserved cysteines in each chain (at positions 4, 135, 154, and 171, as per B chain numbering), the cysteine at position 4 is involved in inter-chain disulfide bridges, yielding the A–B and C–C subunits. The other three cysteines are considered to yield one intra-chain disulfide bond and one free thiol group per C-terminal globular region in human C1q (10). The conservation of these four cysteines in zebrafish implied that the crystal structure of zebrafish C1q is similar to that of human. Mutational study revealed the potentially important residues of human C1q for IgG interaction are Arg114, Arg129, and Glu162 of B chain (10). However, these residues were not found in zebrafish, which indicates that C1q in zebrafish may have different binding specificities.
Multiple Alignment and Phylogenetic Analysis
Multiple alignment of C1qA, C1qB, and C1qC family members was performed to evaluate the phylogenetic relationships between zebrafish and other species. Results showed that zebrafish C1qA, C1qB, and C1qC have stretch similar to those of different species. The important functional amino acid residues and domains in the molecules, such as the collagen-like region, C1q domain, and four cysteine residues are well conserved from human to fish, and the sequence identities among the family members were 25–99% (supplemental Table S2). The constructed phylogenetic tree of all C1q molecules identified, including C1qs in human (Homo sapiens), chimpanzee (Pan troglodytes), mouse (Mus musculus), dog (Canis lupus), chicken (Gallus domestiaus), zebrafish (D. rerio), lamprey (Lethenteron japonicum), shark (Nurse shark), and amphioxus (Branchiostoma belcheri tsingtauense), showed that zebrafish C1qA and C1qC are clustered with the C1qA and C1qC of other species, respectively. However, zebrafish C1qB is clustered with shark C1q and lamprey C1q to form an exclusive group and is in a more original position. A phylogenetic tree based on the gC1q domains also was constructed. This also showed that zebrafish C1qB is clustered with amphioxus C1q (supplemental Figs. S3 and S4). Thus, zebrafish C1qB may have maintained the primitive form during early evolution.
Tissue Distribution Analysis
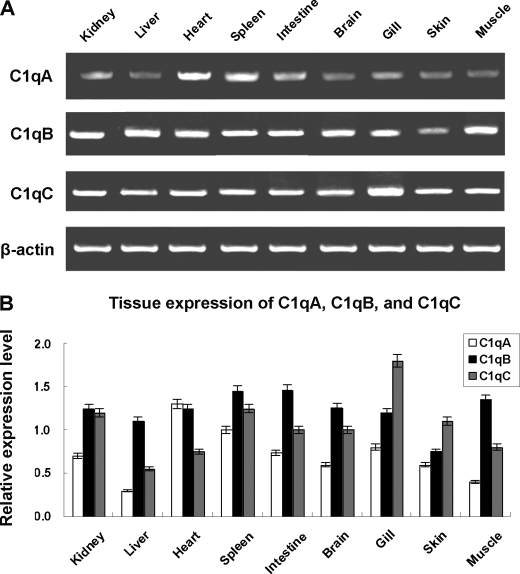
Tissue distribution analysis showed that zebrafish c1qA, c1qB, and c1qC transcripts are expressed broadly in all the tissues examined, including head kidney, liver, heart, spleen, intestine, brain, gill, skin, and muscle (Fig. 3). In addition, the expression of zebrafish c1qA was selectively examined during the embryonic development and fry. Results show that the c1qA transcript was also strongly detectable from 0 h (fertilized egg) to 28 days (fry), indicating maternal transfer of C1q to the egg (data not shown). Similar results also have been seen in amphioxus C3 (33).
FIGURE 3.
Tissue distributions of zebrafish c1qA, c1qB, and c1qC. A, detection of c1qA, c1qB, and c1qC transcripts in different tissues by RT-PCR. B, semiquantitative analysis of the expression of zebrafish c1qA, c1qB, and c1qC. Expression levels are expressed as a ratio to β-actin mRNA levels after densitometric scanning of gels stained with ethidium bromide. Values are mean ± S.D. The relative expression values were averaged from three fish.
Preparation of Recombinant C1qA, C1qB, and C1qC Antibodies
Globular head regions of zebrafish C1qA, C1qB, and C1qC were expressed in E. coli BL21 cells as soluble proteins for antibody preparation and functional analysis. Recombinant proteins were purified using nickel-nitrilotriacetic acid-agarose affinity chromatography and examined by SDS-PAGE under reducing conditions (supplemental Fig. S5). Antibodies for C1qA, C1qB, and C1qC were affinity-purified from the immunized rabbit serum into IgG isotypes by protein A-agarose column. ELISA analyses showed that the purified antibodies had an average titer above 1:10,000 and were specific to C1qA, C1qB, and C1qC, respectively, because no cross reactions among C1qA, C1qB, C1qC, and other proteins from tissue extracts were observed (data not shown).
Involvement of C1q in Classical Pathway
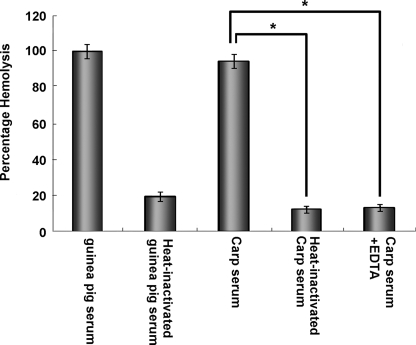
Hemolysis inhibitory assay using anti-zebrafish C1qA, C1qB, and C1qC antibodies was used to demonstrate the involvement of zebrafish C1q molecules in the classical complement pathway. Carp serum was used because carp is genetically close to zebrafish, and its serum is easy to collect for obtaining enough quantities for experiments. To determine whether C1q in zebrafish and carp could be substituted with each other, Western blot was performed. Results showed that strong cross reactions were detected between zebrafish anti-ghA, anti-ghB, and anti-ghC and carp C1qA, C1qB, and C1qC molecules, respectively. Remarkably, the molecular weights of C1qA, C1qB, and C1qC (26, 25, and 25 kDa, respectively) detected in carp serum were similar to those estimated in zebrafish. This indicates the structural conservation of complements between the two species (Fig. 4). Results of the hemolysis assay showed almost complete hemolysis of SRBC sensitized with anti-SRBC antiserum after the addition of normal carp serum. In addition, SRBC hemolysis decreased with heat inactivation of carp serum or EDTA chelating of the Ca2+ in the serum. These are consistent with the results from the control group (guinea pig) and conform to the characterization of classical complement pathway (Fig. 5). Furthermore, the C1q-dependent hemolysis inhibitory assay showed that the hemolytic activities in carp serum could be inhibited by anti-ghA, anti-ghB, and anti-ghC antibodies in a dose-dependent manner. Results suggest the functional conservation of C1qA, C1qB, and C1qC and their involvement in the classical pathway (Fig. 6C).
FIGURE 4.
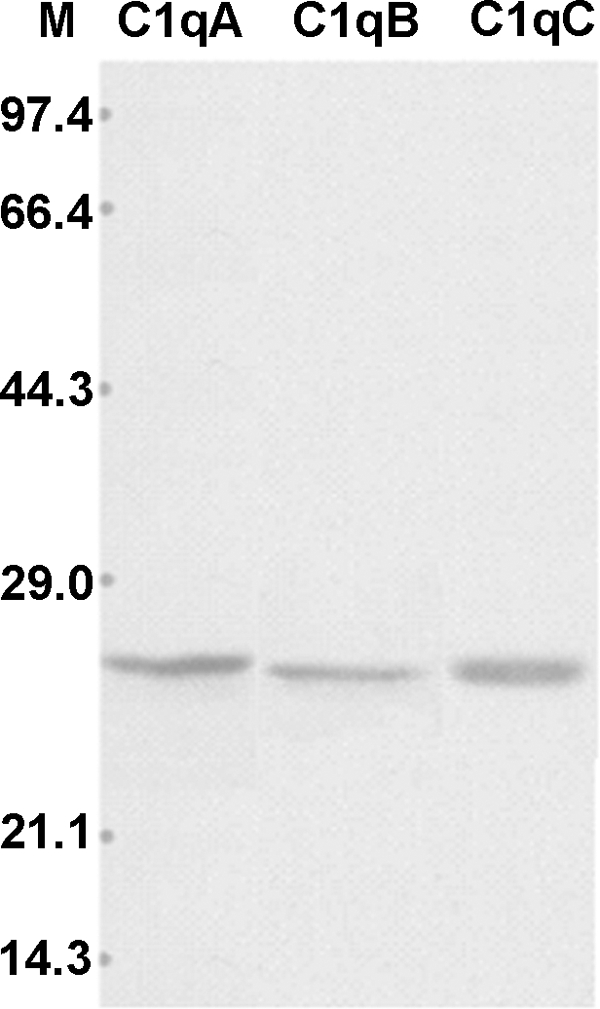
Western blot analysis of C1q in carp serum (lane M: molecular marker). Carp serum was electrophoresed on 12% SDS-PAGE gel using Laemmli buffer. The proteins separated were blotted on nitrocellulose membrane and immunostained with rabbit anti-fish ghA, ghB, or ghC antibodies, followed by staining with HRP-labeled anti-rabbit IgG.
FIGURE 5.
Hemolytic study of carp serum using SRBC sensitized with carp anti-SRBC antibody. 100 μl of serum was incubated with an equal amount of sensitized SRBC at 37 °C for 30 min. After centrifugation, the sera were tested for hemolytic activity by measuring the A405 values. The guinea pig serum (1:20 diluted) was used as a positive control. Vertical bars represent the mean ± S.D. (n = 4). Significant differences are indicated with asterisks at p < 0.05.
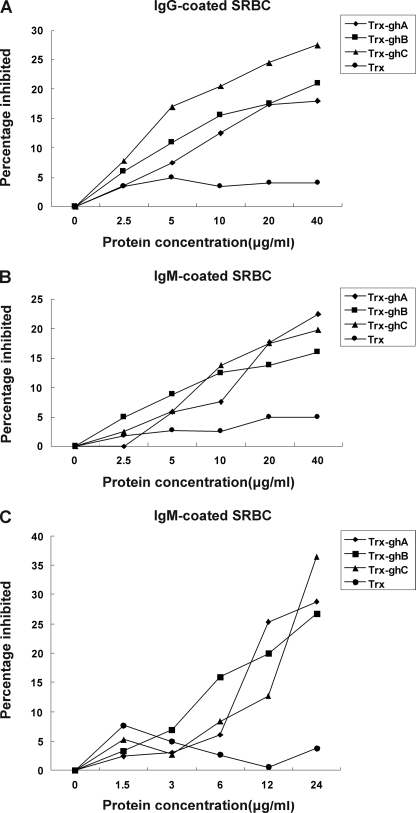
FIGURE 6.
Inhibition by Trx-ghA, Trx-ghB, or Trx-ghC of C1q-dependent hemolysis of SRBC sensitized with rabbit anti-SRBC antibody (IgG) (A), mouse anti-SRBC antibody (IgM) (B), and carp anti-SRBC antibody (C). In A and B, SRBC sensitized with IgG (SRBCIgG) or IgM (SRBCIgM) were pretreated with various concentrations of Trx-ghA, Trx-ghB, or Trx-ghC for 1 h at 37 °C. 1:20 diluted guinea pig serum was then added to pretreated SRBC to initiate hemolysis. In C, nonimmunized zebrafish serum diluted 1:3 was alternatively used; nonimmunized zebrafish serum has been proven previously to not lead to hemolysis). The unlysed cells were centrifuged, and A405 values of the supernatants were measured. The percentage inhibited was determined relative to the untreated sample. The means of three experiments are presented for each set of experiments.
The ghA, ghB, and ghC Modules Involved in C1q-dependent Classical Pathway
To give direct evidence of the involvement of zebrafish C1q molecules in C1q-dependent classical pathway and to examine the participation of ghA, ghB, and ghC modules in this process, the inhibitory effects of recombinant Trx-tagged ghA, ghB, and ghC on C1q-dependent hemolysis were analyzed. SRBC were sensitized with anti-sheep erythrocyte IgG, anti-sheep erythrocyte IgM, or anti-fish erythrocyte IgM to yield EAIgG, EAIgM, or EAIgM(fish) EA cells. Then, EAIgG, EAIgM, or EAIgM(fish) cells were pretreated with various concentrations of Trx-ghA, Trx-ghB, ghC, or Trx (negative control) alone (1.25–40 μg) before they were added to the determined amount of 1:20 diluted guinea pig serum. As shown in Fig. 6 (A–C), all three modules could inhibit hemolysis in a dose-dependent manner. Based on the C1q-dependent hemolysis of sensitized SRBCs, the potencies of the three recombinant proteins as inhibitors of C1q follow the order ghC > ghB > ghA for EAIgG, and ghC ∼ ghB ∼ ghA for EAIgM and EAIgM(fish). In contrast, Trx did not interfere with C1q-dependent hemolysis.
Conserved Functional Binding Sites in Globular Head Modules
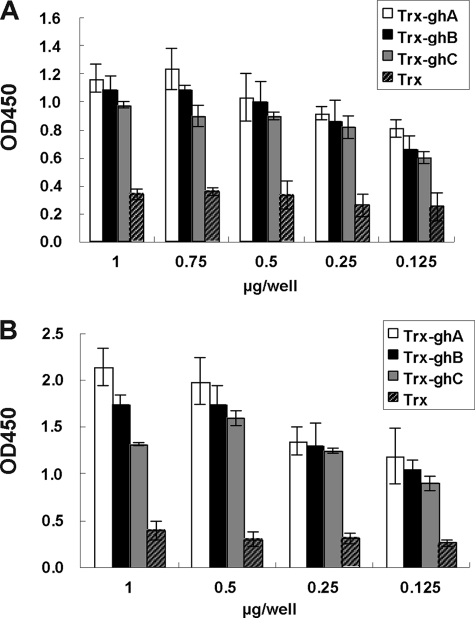
To further evaluate the functional conservation of globular head modules in C1q molecules in the classical pathway and of their binding sites to IgM or IgG isotypes during evolution, a direct binding assay by quantitative ELISA and a competitive C1q-dependent hemolysis inhibitory assay were performed. In binding assay, different concentrations of recombinant zebrafish Trx-ghA, Trx-ghB, and Trx-ghC tagged proteins were used to coat the microtiter wells and incubated with heat-aggregated human IgM and IgG. This was followed by detection with HRP-conjugated anti-human IgM or IgG. Results showed that zebrafish Trx-ghA, Trx-ghB, and Trx-ghC could all bind with human IgM and IgG in a dose-dependent manner, whereas Trx (negative control) showed weaker binding background (Fig. 7). In a competitive inhibition assay, the inhibitory effects of Trx-ghA, Trx-ghB, and Trx-ghC on C1q-dependent hemolysis were examined. The SRBCs were sensitized with hemolysin (anti-sheep erythrocyte IgG or IgM or anti-fish erythrocyte IgM) to yield EAIgG, EAIgM, or EAIgM(fish) cells. These cells were then pretreated with various concentrations of Trx-ghA, Trx-ghB, Trx-ghC, or Trx, and incubated with 1:20 diluted guinea pig serum and DGVB2+ solution. The incubation of 17 μl 1:20 diluted guinea pig serum with 133 μl DGVB2+ solutions and 100 μl EAIgG or EAIgM was found to cause 50% hemolysis and was used throughout the experiments. Trx-ghA, Trx-ghB, or Trx-ghC could inhibit the hemolysis of EAIgG or EAIgM (Fig. 6). Based on C1q-dependent hemolysis of SRBCs, the potencies of the three recombinant proteins as inhibitors of C1q are in the order of ghC > ghB > ghA for EAIgG. There were no significant differences between these three recombinant proteins in the inhibition of hemolysis for EAIgM. As a control, Trx did not interfere with the C1q-dependent hemolysis.
FIGURE 7.
Binding of heat-aggregated IgG and IgM to Trx-ghA, Trx-ghB, and Trx-ghC. Various concentrations of Trx-ghA, Trx-ghB, or Trx-ghC were coated to the microtiter wells and a fixed concentration of either heat-aggregated human IgG (10 μg/well) (A) or heat-aggregated human IgM (20 μg/well) (B) was added and probed with HRP-conjugated anti-human IgG (A) or anti-human IgM (B), respectively. Color was developed using p-nitrophenyl phosphate, and A450 values were measured. All experiments were performed in duplicate, and Trx was used as a negative control.
Global Evolutionary Analysis of C1q Family
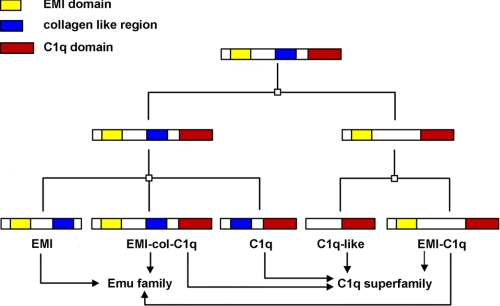
The globular (gC1q) domain similar to C1q was also found in many non-complement C1q domain-containing (C1qDC) proteins that have a similar crystal structure to the multifunctional TNF ligand family (34) as well as diverse functions. The predominant domain organization of C1qDC proteins includes a leading signal peptide, a collagen-like region of variable length, and a C-terminal C1q domain. This organization is similar to the domain structures of the A, B, and C chains of C1q. A total of 31 independent C1qDC gene sequences, including adiponectin, AQL1, AQL2, C1qTNF, and CBLN, have been screened from the human genome. To illustrate the evolutionary relationships in C1q superfamily, whole C1q family members containing gC1q domain were identified from different teleost fish, including zebrafish, Fugu (Fugu rubripes), and Tetraodon (Tetraodon nigroviridis), as well as other vertebrate species. Sixty-eight C1qDC gene sequences, including collagen, C1qTNF, adiponectin, cereberlin, C1q, C1q-like, multimerin, and EMILIN, were identified from teleost fish. Based on sequence homology, functional relatedness, and similarity in domain structure, these C1qDC proteins can be classified into three major subfamilies: the C1q-colDC subfamily (the adiponectin/short collagen group), the C1q-like subfamily (the precerebellin/C1qL group), and the EMI-C1qDC subfamily (the EMILIN/multimerin group) (supplemental Fig. S6). The EMI-C1qDC subfamily contains an EMI domain, which is a novel cysteine-rich domain of EMILINs, and also is classified as a member of the Emu family. The predominant protein structure of the Emu family includes an N-terminal signal peptide followed by the EMI domain, an interrupted collagen stretch, and a conserved C-terminal domain of unknown function. A phylogenetic tree using all the EMI domain-containing sequences from human, mouse, and zebrafish was constructed to reveal the phylogenetic relationship and domain structure of these Emu members (supplemental Fig. S7). Among these sequences, some molecules, such as mouse EMILIN 1 and human multimerin 1, were found to contain an EMI domain, a collagen region, and a C1q domain and were designated as EMI-collagen-C1q domain-containing molecule. Phylogenetic and domain organization analysis revealed an evolutionary relationship between C1q and Emu superfamily that has never been reported (Fig. 8). These two families may have both originated from an EMI-collagen-C1q domain-containing molecule. This can be elucidated via three points. First, EMI-collagen-C1q domain-containing molecules possess all the three motifs characteristic of Emu family members (an EMI domain exclusive to the family) and C1q family members (a collagen-like region and a C1q domain). In contrast, C1q-like and C1qDC molecules only have one or two motifs. Second, two phylogenetic trees (supplemental Figs. 6 and 7) demonstrate that the EMI-collagen-C1q domain-containing molecules are in an original position. Finally, the gene structures of these C1q domain-containing molecules revealed that EMI-collagen-C1q domain-containing molecules have multiple exons (35), whereas C1q-like and C1qDC molecules were encoded by only three or four exons (data not shown). These indicate that EMI-collagen-C1q domain-containing molecules may be the primitive form of C1q-like and C1qDC molecules. Thus, this study hypothesizes that the C1q and Emu superfamilies both may have originated from a common ancestor containing three domains: an EMI domain, a C1q domain, and a collagen domain. After some evolutionary events, such as gene duplication and segmentation, five subsets with different motifs were formed (Fig. 8). These subsets became divided into two families, Emu and C1q, according to the different motifs they contained.
FIGURE 8.
The evolution of the C1q and Emu families. □, a gene duplication event accompanied by loss of some gene segment. Finally, five subsets were formed in the two families. The topmost subset is the ancestor containing the three domains.
DISCUSSION
C1q is the first component of the classical complement pathway in mammals. It plays a key role in the recognition of immune complexes to initiate the classical complement and is a major connecting link between classical pathway-driven innate immunity and acquired immunity (36). Although C1q has been found in lower vertebrates, such as jawless vertebrates (lampreys and hagfishes) and invertebrates, it was found to be uninvolved in the classical pathway. Instead, it acts as a lectin with a role in innate immunity, mainly due to the lack of Igs and the related adaptive immune systems in these species. For example, C1q is an N-acetylglucosamine (GlcNAc)-binding lectin homologue in lamprey serum (18). Therefore, C1q experienced a functional evolution from an innate immunity-related molecule to a molecule participating in adaptive immunity. Investigations of the evolutionary pattern of C1q would be of interest.
Fish is the oldest species possessing typical IgM, so this study speculates that the functional evolution, from involvement in innate immunity to adaptive immunity, of C1q may have occurred in fish. Some previous studies have shown that the classical pathway first appeared in fish (8, 16), although the exact molecular composition and characterization of the molecules in this pathway are still lacking. Specifically, there is a lack of sufficient evidence for the molecular properties and functional characterization of C1q as a key component that initiates the classical pathway.
The present study is the first to attempt the cloning of c1qA, c1qB, and c1qC in zebrafish and prove that the C1q molecule could participate in the classical pathway. This could provide direct evidence that the classical pathway first appeared in teleost fish and is triggered by the binding of C1q to Ag-Ab complex. The C1q molecules identified were completely conserved; not only in the molecular structure, such as four Cys, spatial structure of globular head, and domain organization but also in functional characterization in different species. Functional analysis revealed that C1q proteins could replace one another between different fish species or between fish and mammals, indicating that the functional characterization and mechanism of the classical pathway mediated by C1q are conserved in the evolutionary history. This conservation included the structure of the binding sites. In mammals, C1qA, C1qB, and C1qC could bind to an Ig molecule through its globular head (10), and the CH3 domain of IgM or CH2 domain of IgG is recognized as the C1q binding site (37, 38). This study demonstrated that fish C1qA, C1qB, and C1qC could inhibit C1q-dependent hemolysis in a mammalian experimental model through competitive inhibition study. Results suggest that fish C1qA, C1qB, and C1qC could bind to the same sites of IgM and IgG as mammal C1q. Although fish do not possess IgG molecules, the potential IgG binding sites are already present in fish C1q. Therefore, the interaction between C1qA, C1qB, C1qC, and IgG can be reasonably assumed to be due to the structural adaption of IgG to C1q molecule during later evolution. Findings also indicate that IgM and IgG do not share the same binding sites when interacting with C1q. Extensive studies are required to explore the biological significance of the results. For example, this may be responsible for avoiding competition between IgM and IgG molecular interaction with C1qA, C1qB, and C1qC to maximize the efficiency of the classical pathway in adaptive immunity.
Evolutionary analysis led this study to propose that c1qA, c1qB, and c1qC may have come from one ancestor molecule through gene duplication, because these three genes are linearly distributed on the same chromosome, and only one C1q-like gene was found in early lower vertebrates. To ascertain the origin and formation of C1qA, C1qB, and C1qC, the C1q molecules of different vertebrates were analyzed and used to construct a phylogenetic tree. Analysis showed that C1qB is the potential ancestor molecule because it is located in the same branch as amphioxus C1q, an earlier lower vertebrate than zebrafish. Furthermore, these two molecules are in a more original position in the phylogenetic tree compared with C1qA and C1qC. Three C1q molecules were identified in zebrafish, whereas only two were found in sharks (39) and only one in lower invertebrates such as the lamprey. The C1q molecule consists of identical 24-kDa subunits that form a dimeric structural unit linked by disulfide bond(s) and nine dimeric structural units (18 chains), which may be noncovalently bonded to give a hexameric structure (18). Thus, this study hypothesizes that the lamprey C1q is a homologue of zebrafish C1qA, C1qB, or C1qC and that the other two molecules were formed by duplication of the lamprey C1q homologue.
The recent identification of more C1q family members have led to the formation of a C1q superfamily, which makes studies of C1qA, C1qB, and C1qC molecules and the evolutionary relationships between C1q superfamily members important areas of interest. In this study, ∼68 C1q family members were identified from teleost fish. Evolutionary analysis revealed a relationship between the C1q and Emu superfamilies, which may have both come from an EMI-collagen-C1q domain-containing molecule.
Supplementary Material
This work was supported by grants from the National Basic Research Program of China (973) (2006CB101805), the Hi-Tech Research and Development Program of China (863) (2008AA09Z409), the National Natural Science Foundation of China (30871936 and 30571423), and the Science and Technology Foundation of Zhejiang Province (2006C12038, 2006C23045, 2006C12005, and 2007C12011).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S7.
- Trx
- thioredoxin
- SRBC
- sheep red blood cell
- C1qDC
- C1q domain-containing.
REFERENCES
- 1.Hoffmann J. A., Kafatos F. C., Janeway C. A., Ezekowitz R. A. (1999) Science 284, 1313–1318 [DOI] [PubMed] [Google Scholar]
- 2.Thiel S., Vorup-Jensen T., Stover C. M., Schwaeble W., Laursen S. B., Poulsen K., Willis A. C., Eggleton P., Hansen S., Holmskov U., Reid K. B., Jensenius J. C. (1997) Nature 386, 506–510 [DOI] [PubMed] [Google Scholar]
- 3.Nonaka M., Smith S. L. (2000) Fish Shellfish Immunol. 10, 215–228 [DOI] [PubMed] [Google Scholar]
- 4.Fujita T. (2002) Nat. Rev. Immunol. 2, 346–353 [DOI] [PubMed] [Google Scholar]
- 5.Smith L. C., Azumi K., Nonaka M. (1999) Immunopharmacology 42, 107–120 [DOI] [PubMed] [Google Scholar]
- 6.Sunyer J. O., Lambris J. D. (1998) Immunol. Rev. 166, 39–57 [DOI] [PubMed] [Google Scholar]
- 7.Gross P. S., Al-Sharif W. Z., Clow L. A., Smith L. C. (1999) Dev. Comp. Immunol. 23, 429–442 [DOI] [PubMed] [Google Scholar]
- 8.Nonaka M. (2000) Curr. Top. Microbiol. Immunol. 248, 37–50 [DOI] [PubMed] [Google Scholar]
- 9.Sellar G. C., Blake D. J., Reid K. B. (1991) Biochem. J. 274, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kishore U., Gupta S. K., Perdikoulis M. V., Kojouharova M. S., Urban B. C., Reid K. B. (2003) J. Immunol. 171, 812–820 [DOI] [PubMed] [Google Scholar]
- 11.Sunyer J. O., Zarkadis I., Sarrias M. R., Hansen J. D., Lambris J. D. (1998) J. Immunol. 161, 4106–4114 [PubMed] [Google Scholar]
- 12.Kuroda N., Wada H., Naruse K., Simada A., Shima A., Sasaki M., Nonaka M. (1996) Immunogenetics 44, 459–467 [DOI] [PubMed] [Google Scholar]
- 13.Nakao M., Fushitani Y., Fujiki K., Nonaka M., Yano T. (1998) J. Immunol. 161, 4811–4818 [PubMed] [Google Scholar]
- 14.Kuroda N., Naruse K., Shima A., Nonaka M., Sasaki M. (2000) Immunogenetics 51, 117–128 [DOI] [PubMed] [Google Scholar]
- 15.Nakao M., Osaka K., Kato Y., Fujiki K., Yano T. (2001) Immunogenetics 52, 255–263 [DOI] [PubMed] [Google Scholar]
- 16.Boshra H., Li J., Sunyer J. O. (2006) Fish Shellfish Immunol. 20, 239–262 [DOI] [PubMed] [Google Scholar]
- 17.Dodds A. W., Matsushita M. (2007) Immunobiology 212, 233–243 [DOI] [PubMed] [Google Scholar]
- 18.Matsushita M., Matsushita A., Endo Y., Nakata M., Kojima N., Mizuochi T., Fujita T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10127–10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y., Huang H., Wang Y., Yu Y., Yuan S., Huang S., Pan M., Feng K., Xu A. (2008) J. Immunol. 181, 7024–7032 [DOI] [PubMed] [Google Scholar]
- 20.Kent W. J. (2002) Genome Res. 12, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burge C. B., Karlin S. (1998) Curr. Opin. Struct. Biol. 8, 346–354 [DOI] [PubMed] [Google Scholar]
- 23.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 24.Pearson W. R., Lipman D. J. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou J., Yoshiura Y., Dijkstra J. M., Sakai M., Ototake M., Secombes C. (2004) Fish Shellfish Immunol. 17, 403–409 [DOI] [PubMed] [Google Scholar]
- 26.Falquet L., Pagni M., Bucher P., Hulo N., Sigrist C. J., Hofmann K., Bairoch A. (2002) Nucleic Acids Res. 30, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S., Tamura K., Nei M. (2004) Brief Bioinform. 5, 150–163 [DOI] [PubMed] [Google Scholar]
- 29.Sasaki T., Ueda M., Yonemasu K. (1982) J. Immunol. Methods 48, 121–131 [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 31.Towbin H., Staehelin T., Gordon J. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bag M. R., Makesh M., Rajendran K. V., Mukherjee S. C. (2009) Fish Shellfish Immunol. 26, 275–278 [DOI] [PubMed] [Google Scholar]
- 33.Liang Y., Zhang S., Wang Z. (2009) PLoS One 4, e4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro L., Scherer P. E. (1998) Curr. Biol. 8, 335–338 [DOI] [PubMed] [Google Scholar]
- 35.Leimeister C., Steidl C., Schumacher N., Erhard S., Gessler M. (2002) Dev. Biol. 249, 204–218 [DOI] [PubMed] [Google Scholar]
- 36.Kishore U., Reid K. B. M. (1999) Immunopharmacol. 41, 15–21 [DOI] [PubMed] [Google Scholar]
- 37.Duncan A. R., Winter G. (1988) Nature 332, 738–740 [DOI] [PubMed] [Google Scholar]
- 38.Shulman M. J., Collins C., Pennell N., Hozumi N. (1987) Eur. J. Immunol. 17, 549–554 [DOI] [PubMed] [Google Scholar]
- 39.Smith S. L. (1998) Immunol. Rev. 166, 67–78 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.