Abstract
Although transcriptional effects of androgens have been extensively studied, mechanisms regulating transcription-independent (nongenomic) androgen actions are poorly understood. Previously, we have shown that paxillin, a multidomain adaptor protein, is a critical regulator of testosterone-induced MAPK-signaling during Xenopus oocyte maturation. Here we examine the nongenomic effects of dihydrotestosterone (DHT) in prostate cancer cells, focusing on how paxillin mediates Erk signaling and downstream physiologic actions. We show that in LnCAP cells DHT functions as a growth factor that indirectly activates the EGF-receptor (EGFR) via androgen receptor binding and matrix metalloproteinase-mediated release of EGFR ligands. Interestingly, siRNA-mediated knockdown of paxillin expression in androgen-dependent LnCAP cells as well as in androgen-independent PC3 cells abrogates DHT- and/or EGF-induced Erk signaling. Furthermore, EGFR-induced Erk activation requires Src-mediated phosphorylation of paxillin on tyrosines 31/118. In contrast, paxillin is not required for PKC-induced Erk signaling. However, Erk-mediated phosphorylation of paxillin on serines 83/126/130 is still needed for both EGFR and PKC-mediated cellular proliferation. Thus, paxillin serves as a specific upstream regulator of Erk in response to receptor-tyrosine kinase signaling but as a general regulator of downstream Erk actions regardless of agonist. Importantly, Erk-mediated serine phosphorylation of paxillin is also required for DHT-induced prostate-specific antigen mRNA expression in LnCAP cells as well as EGF-induced cyclin D1 mRNA expression in PC3 cells, suggesting that paxillin may regulate prostate cancer proliferation by serving as a liaison between extra-nuclear kinase signaling and intra-nuclear transcriptional signals. Thus, paxillin may prove to be a novel diagnostic or therapeutic target in prostate cancer.
Keywords: ERK, Growth Factors, MAP Kinases (MAPKs), Signal Transduction, Steroid Hormone, EGF Receptor, Paxillin, Non-genomic Androgen Signaling, Prostate Cancer
Introduction
Physiological functions of androgens are mediated via either nuclear (genomic) or extra-nuclear (nongenomic) actions of androgen receptors (ARs).2 Genomic actions involve binding of androgens to ARs, which then translocate to the nucleus, bind to androgen-response elements, and alter gene expression. In contrast, ARs also induce rapid nongenomic signals that are generally mediated by cross-talk between the AR and either G-proteins or growth factor receptors (1–4). Although transcriptional effects of androgens have been extensively studied, mechanisms regulating nongenomic actions of androgens are poorly understood.
One potential regulator of nongenomic androgen actions is paxillin. Paxillin is a multidomain adaptor protein that integrates many signals from integrins, cell surface receptors, and growth factors (10, 11). Through these interactions, paxillin regulates a variety of physiological functions, including matrix organization, cell motility, tissue remodeling, metastasis, gene expression, cell survival, and proliferation (10, 11). Paxillin is comprised of multiple structural domains that modulate protein-protein interactions (10) and numerous serine/threonine and tyrosine phosphorylation targets that act as docking sites for various signaling proteins. Phosphorylation of these sites by growth factor receptor-tyrosine kinases, Src kinases, and serine/threonine kinases regulate adaptor molecule binding that ultimately coordinates complex cell signaling pathways (10). The importance of paxillin in normal physiological functions is further evident from global paxillin knock-out studies, demonstrating that ablation of paxillin in mice is embryonic lethal (12, 13).
We previously demonstrated that in Xenopus oocytes, paxillin is essential for non-genomic androgen-induced Erk signaling and subsequent Erk-mediated oocyte maturation (5). Specifically, paxillin is required for synthesis and activation of MOS (the germ cell Raf homolog), which then promotes MEK and subsequently Erk signaling (5). Interestingly, Erk-mediated phosphorylation of paxillin is also required for androgen-induced oocyte maturation. Thus, in oocytes, paxillin is both an affector and effector of Erk signaling.
Here we significantly extend our findings in Xenopus oocytes to a mammalian somatic system. Given the well defined function of androgens and Erk signaling (3, 6–8) in prostate cancer (PCa) development (4, 9), we chose to examine the regulatory role of paxillin in androgen-induced Erk signaling and downstream physiologic actions. Androgen and epidermal growth factor (EGF) signaling are critical regulators of PCa development and progression (4, 9). In fact, paxillin association with focal adhesion molecules may be up-regulated in metastatic prostate carcinoma (14), and in PCa cell lines paxillin potentiates AR trans-activation by functioning as an AR co-activator (15). Here we report that paxillin is important for kinase signaling in response to multiple extra-nuclear signals in PCa cells, functioning as an upstream mediator of Erk activation and a downstream regulator of Erk signaling. Furthermore, we provide evidence that paxillin plays an important role in orchestrating cross-talk between extra-nuclear kinase signaling and intra-nuclear transcriptional events.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
LnCAP and PC3 cell lines were obtained from ATCC and cultured in RPMI 1640 medium (Invitrogen) containing 10% FBS and 1% penicillin-streptomycin. For experiments involving pharmacological inhibitors, cells were treated overnight with serum-free, phenol red-free RPMI 1640 media. Thereafter, cells were treated with vehicle (0.1% DMSO) or inhibitors Galardin, PP2, AG1478 (Calbiochem), flutamide, or erlotinib (Sigma) for 30 min before stimulation with 0.1% ethanol (vehicle) or 25 nm DHT for 30 min.
EGFR Transactivation Assay
A431 cells were used to detect DHT-mediated release of EGFR ligands from LnCAP cells. A431 cells (ATCC) were cultured in DMEM/F-12 (1:1) medium (Invitrogen) containing 10% FBS and 1% penicillin-streptomycin, serum-starved overnight, and then stimulated with medium from DHT-, DHT + galardin-, or vehicle (0.1% ethanol)-treated LnCAP cells for 60 min. As controls, A431 cells were stimulated with DHT or media alone. Thereafter, A431 cells were isolated for Western blot analysis to detect phosphorylated and total EGFR.
Transient Transfection
PC3 or LnCAP cells were treated with non-targeting siRNA pool (ThermoFisher Scientific) or paxillin-specific siRNA according to manufacturer's instructions. Two sets of human paxillin siRNAs were used: 1) human paxillin siRNA (Santa Cruz Biotechnology) containing three target-specific 20–25 nucleotide siRNAs or 2) human paxillin siRNA ON-TARGET plus SMARTpool (ThemoFisher Scientific) containing four siRNAs targeting the paxillin mRNA. The latter was used for all experiments here, although results were similar with both pools. For experiments involving constitutively active (ca) Raf (William Walker, University of Pittsburgh) or MEK (Melanie Cobb, University of Texas Southwestern Medical Center), cells were co-transfected with paxillin or nonspecific siRNAs and cDNAs encoding caRaf or caMEK. After 72 h, cells were treated overnight with serum-free, phenol red-free RPMI 1640 media and stimulated with 0.1% ethanol/DMSO (vehicle), 25 nm DHT (Steraloids), 20 ng/ml EGF (BD Biosciences), or 100 nm PMA (Sigma) for the times indicated for Western blots or 24 h for MTT assays.
Plasmids and Cloning
RNA was isolated from HEK293 cells using RNeasy mini kit (Qiagen) according to the manufacturer's instructions and reverse-transcribed to obtain cDNA. Paxillin was amplified from the cDNA with high fidelity Pfu Turbo (Stratagene) using primer pairs: 5′-ACCTTGAATTCATGGACGACCTCGACGCCCTGCTGGC-3′ and 5′-CTAAGCGGCCGCTTACTAGCAGAAGAGCTTGAGGAAGCA-3′. Wild-type paxillin was then cloned into pcDNA3.1(+) plasmid (Invitrogen) and confirmed by sequencing.
Site-directed Mutagenesis
Site-directed mutagenesis (Stratagene) was used to convert serines 83, 126, and 130 (S83A/S126A/S130A) or tyrosines 31 and 118 (Y31A/Y188A) to alanines. Clones were sequenced in entirety to confirm mutations. Residues were chosen based on previous studies (6, 7, 16, 17), demonstrating their importance for paxillin function.
Paxillin Rescue Experiments
PC3 cells were transfected with paxillin siRNA as described above. After 96 h, media was removed, and cells were transfected with Lipofectamine (Invitrogen) or Lipofectamine plus WT, S83A/S126A/S130A, or Y31A/Y118A paxillin cDNA. After 48 h, cells were treated overnight with serum-free, phenol red-free RPMI 1640 media and stimulated with the indicated ligands for 30 min for Western blots or 24 h for MTT assays.
Western Blot Analysis
Western blots were performed as described (18). Primary antibodies were: paxillin, paxillin-phospho-Tyr-118, paxillin-phospho-Tyr-31, Akt1/2/3, Akt1/2/3 phospho-Ser-473, EGFR at 1:1000 dilution, and EGFR-phospho-Tyr-845 (1:500) (Santa Cruz Biotechnology); paxillin-phospho-Ser-126 (Invitrogen); paxillin-phospho-Ser-83 (ECM Biosciences); p44/42-Erk1/2, phospho-p44/42-Erk1/2 (Thr-202/Tyr-04), phospho-Ser-217/221-MEK1/2, src, src-phospho-Tyr-416 (Cell Signaling Technology) at 1:1000 dilutions and β-actin (1:5000, Millipore).
MTT Assay
MTT assays were performed using a colorimetric assay cell proliferation kit (Roche Applied Science) according to the manufacturer's instructions.
Cell Migration/Invasion Assay
Cell migration/invasion assays were performed using a colorimetric QCM Cell invasion assay kit containing 24-well Boyden chambers with extracellular matrix-coated 8-μm pore size membranes (Millipore) according to the manufacturer's instructions.
RNA Extraction and Real Time PCR
RNA was isolated with the RNeasy mini kit (Qiagen) according to manufacturer's instructions. Levels of Kallikrein-related peptidase-3 (KLK3, or prostate-specific antigen (PSA)), paxillin, cyclin D1, and GAPDH expression were analyzed by the Δ/Δ-Ct method using Taqman gene expression assay primers (assay ID# Hs99999905_m1-GAPDH, Hs02576345_m1-KLK3, Hs99999004_m1-CCND1, and Hs01104424-m1-PXN; Applied Biosystems) on an ABI StepOne plus real-time PCR machine.
Statistical Analysis
Results for the MTT assay, cell migration-invasion assay, and real-time PCR were analyzed using Student's t test. A value of p ≤ 0.05 was considered significant.
RESULTS
DHT-induced Erk Signaling Occurs via Matrix Metalloproteinase (MMP)-mediated Transactivation of the EGFR
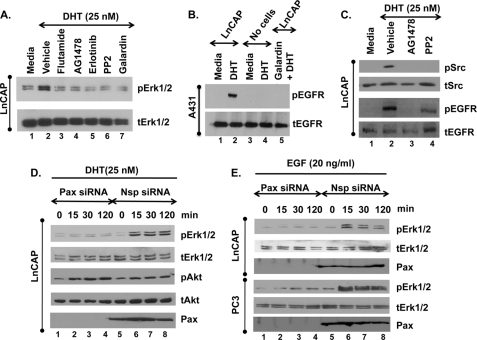
Because androgens induce Erk1/2 signaling in PCa cell lines (3, 10–12), we examined the underlying mechanism by which DHT activates Erk1/2 in LnCAP cells. DHT treatment of LnCAP cells for 30 min significantly induced Erk1/2 phosphorylation/activation (Fig. 1A, lane 2). Notably, saturating concentrations of DHT (25 nm) were used in these studies to maximize the significance of the inhibitor effects; however, lower concentrations (1–10 nm) promoted a similar magnitude response (not shown). DHT levels in the prostate have not been accurately determined but are reported to be at least 10–20 nm (19, 20). Pretreatment with the AR antagonist flutamide (Fig. 1A, lane 3) as well as EGFR inhibitors AG1478 and Erlotinib (Fig. 1A, lanes 4 and 5) blocked Erk1/2 phosphorylation, indicating that DHT induces EGFR trans-activation and subsequent Erk1/2 signaling via classical ARs. The concentrations of AG1478 (20 μm) and erlotinib (5 μm) used were based on concentration gradient experiments (supplemental Fig. S3, A and B) and previous studies in PCa cells (21–25) and other cell lines (18, 26–29). Of note, AG1478 at 20 μm specifically blocks EGF- but not FGF-induced Erk activation in LnCAP (supplemental Fig. S3C) and MEK activation in MLTC (18) cells, thus, demonstrating the specificity of AG1478 to EGFR inhibition and ruling out off target effects on the Ras/Raf/MEK/Erk pathway. Finally, the inhibitors alone (in absence of DHT) had no effect on Erk signaling (data not shown).
FIGURE 1.
DHT-induced Erk1/2 signaling occurs via MMP-mediated trans-activation of EGFR and is regulated by paxillin. A, serum-starved LnCAP cells were preincubated with vehicle (0.1% DMSO), 100 nm flutamide (AR inhibitor), 20 μm AG1478 (EGFR inhibitor), 5 μm Erlotinib (EGFR inhibitor), 20 μm PP2 (Src inhibitor), or 20 μm galardin (MMP inhibitor) for 30 min before stimulation with ethanol (Media) or 25 nm DHT for 30 min. Western blots of whole-cell extracts were performed for total and phosphorylated Erk1/2 (tERK1/2, pERK1/2). B, A431 cells cultured in DMEM/F-12 (1:1) medium (Invitrogen) containing 10% FBS and 1% penicillin-streptomycin were serum-starved overnight and then stimulated with medium from DHT (25 nm), DHT (25 nm) + galardin (20 μm), or vehicle (0.1% ethanol)-treated LnCAP cells for 60 min. As controls, A431 cells were stimulated with DHT (25 nm) or media alone (no cells). Thereafter, A431 cells were isolated for Western blot analysis to detect phosphorylated and total EGFR (pEGFR, tEGFR). C, serum-starved LnCAP cells were preincubated with vehicle (0.1% DMSO), 20 μm AG1478 (EGFR inhibitor), or 20 μm PP2 (Src inhibitor) for 30 min before stimulation with ethanol (Media) or 25 nm DHT for 30 min. Western blots of whole-cell extracts were performed for Src (tSrc, pSrc) or EGFR (tEGFR, pEGFR). D and E, LnCAP and PC3 cells were treated with non-targeting (Nsp) or paxillin-specific (Pax) siRNAs for 72 h followed by stimulation with 25 nm DHT (D) or 20 ng/ml EGF (E) for the indicated times. Western blots were performed for total and phosphorylated Erk1/2, Akt (tAkt and pAkt) or total paxillin (Pax). All experiments were performed at least three times with identical results.
The ability of the MMP inhibitor galardin to block DHT-induced Erk1/2 phosphorylation (Fig. 1, lane 7) and its rescue by EGF treatment (supplemental Fig. S1) suggests that DHT activates the EGFR through MMP activation, possibly by release of membrane-associated EGFR ligands (30, 31). In fact, medium from DHT (Fig. 1B, lane 2)- but not vehicle-treated (Fig. 1B, lane 1) LnCAP cells increased EGFR phosphorylation in A431 cells (known to express very high levels of EGFR), indicating that EGFR ligands were being released into the medium from DHT-treated LnCAP cells. Moreover, the addition of galardin to DHT-treated LnCAP cells (lane 5) blocked the ability of the medium to activate EGFR signaling in the A431 cells, demonstrating that the release of EGFR ligands is indeed MMP-dependent. Finally, DHT alone (lane 4) did not promote EGFR activation in the A431 cells.
Once activated, phosphorylated EGFR (Fig. 1C, lane 2) stimulates Src, as the EGFR inhibitor AG1478 blocked both Src (Fig. 1C, lane 3) and Erk (Fig. 1A, lane 4) phosphorylation, whereas the Src inhibitor PP2 blocked Erk (Fig. 1A, lane 6) but not EGFR (Fig. 1C, lane 4) phosphorylation. Thus, outside the nucleus, DHT functions just like EGF in LnCAP cells, only through indirect rather than direct activation of the EGFR.
Paxillin Regulates DHT- and EGF-induced ERK Activation in PCa Cells
Next we investigated whether paxillin regulated Erk1/2 activation in PCa cells. In androgen-dependent LnCAP cells, both DHT (Fig. 1D) and EGF (Fig. 1E) induced Erk1/2 activation from 15 to 120 min. Knockdown of paxillin abrogated DHT-induced Erk1/2, but not Akt, activation (Fig. 1D). Furthermore, paxillin knockdown in either LnCAP or androgen-independent PC3 cells inhibited EGF-induced Erk1/2 (Fig. 1E) but not Akt (not shown) activation. Similar effects were observed in DHT-treated LAPC-4 PCa cells (not shown) and FGF or EGF-treated HEK-293 cells (supplemental Fig. S2A). These observations suggest that paxillin is an important regulator of growth factor receptor-induced Erk1/2 signaling regardless of cell type or how the growth factor receptor is activated (EGFR either indirectly by DHT or directly by EGF, or FGFR directly by FGF). Furthermore, EGF-mediated Erk activation was attenuated in mouse embryonic fibroblasts from paxillin null mice (from Dr. Sheila Thomas, Harvard University), providing genetic confirmation of the siRNA experiments that paxillin is important for Erk1/2 signaling (supplemental Fig. S2B).
Paxillin Acts Downstream of the EGFR but Upstream of Raf/MEK
To investigate where paxillin functions in EGFR-induced signaling, we examined DHT- or EGF-induced activation of MEK and EGFR in paxillin-ablated LnCAP cells. Similar to Erk1/2 activation, knockdown of paxillin markedly lowered DHT (Fig. 2A) and EGF (not shown)-induced MEK1/2 phosphorylation. Furthermore, loss of DHT-induced Erk1/2 activation by paxillin knockdown could be rescued by overexpression of constitutively activated MEK (caMEK, Fig. 2B; lanes 7 and 8) or Raf (caRaf, Fig. 2C; lanes 7 and 8). The caMEK used has mutations substituting glutamic and aspartic acid for Ser-218 and Ser-222, significantly increasing the basal activity of MEK over the unphosphorylated wild-type enzyme (32, 33). The caRaf used is a fusion protein of the membrane localization signal of Ras to the carboxyl terminus of Raf that is constitutively activated independent of cellular Ras (34). Finally, knockdown of paxillin minimally affected DHT- or EGF-induced EGFR phosphorylation (Fig. 2D). Together these results demonstrate that paxillin functions downstream of the EGFR but upstream of Raf and MEK to regulate Erk1/2 activation.
FIGURE 2.
Paxillin functions upstream of Raf/MEK but downstream of the EGFR. A, non-targeting (Nsp) or paxillin (Pax)-specific siRNA-treated LnCAP cells were serum-starved and then stimulated with DHT (25 nm) for the indicated times. Western blots were performed for phosphorylated and total MEK1/2. Cell lysates were from the same experiments represented in Fig. 1C. B and C, paxillin- or non-targeting siRNA-treated LnCAP cells were co-transfected with cDNAs encoding caMEK or caRaf for 72 h. Serum-starved cells were then treated with 25 nm DHT for 30 min. Western blots detected total and phosphorylated Erk1/2 as well as paxillin. D, paxillin or non-targeting siRNA-treated LnCAP cells were stimulated with DHT (25 nm) or EGF (20 ng/ml) for 30 min and phosphorylated, and total EGFR was detected by Western blot. Cell lysates were from the same experiment in Figs. 1, C and D. Each experiment was performed at least three times with similar results.
Paxillin Knockdown Prevents Proliferation, Migration, and Invasion of PCa Cells
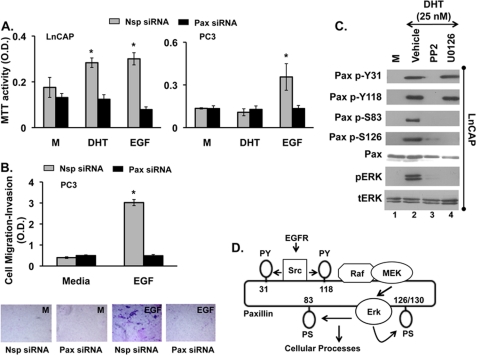
PCa progression correlates with proliferation, migration, and invasion, all of which are thought to require Erk1/2 signaling (2, 4, 11, 12). Thus, to establish the physiologic importance of paxillin in PCa cells, we treated LnCAP or PC3 cells with nonspecific or paxillin-specific siRNAs and measured DHT- or EGF-induced cell proliferation by MTT assay. DHT (25 nm) and EGF (20 ng/ml) significantly stimulated cell proliferation in LnCAP cells (Fig. 3A). Consistent with previous studies (35–39), we found that 0.1–100 nm DHT promoted LnCAP proliferation by both MTT and BrdU incorporation under conditions used here.3 Knockdown of paxillin markedly reduced DHT- and EGF-stimulated cell proliferation (Fig. 3A, left panel). Because PC3 cells are androgen-independent, EGF, but not DHT, induced cell proliferation (Fig. 3A, right panel) that was also abrogated by paxillin knockdown.
FIGURE 3.
Paxillin is required for DHT- or EGF-induced proliferation, migration, and invasion. LnCAP and PC3 cells were treated with non-targeting (Nsp) or paxillin-specific (Pax) siRNAs for 72 h, serum-starved overnight, and stimulated with ethanol (M), DHT (25 nm) or EGF (20 ng/ml) for 24 h. Proliferation was assayed by MTT assay (A), and cell migration-invasion was detected with a colorimetric QCM Cell invasion assay kit (B). Panel B demonstrates quantitative analysis of migration and invasion by absorbance after staining invading cells (upper panel) as well as images representing the underside of the extra-cellular matrix membrane containing migrated and invaded cells (lower panel). Data are represented as the means ± S.E. (n = 3). *, Student's t test, p ≤ 0.05 Nsp versus Pax siRNA. C, paxillin phosphorylation is shown. LnCAP cells were preincubated with vehicle (0.1% DMSO), 20 μm PP2 (Src inhibitor), or 20 μm U0126 (Erk1/2 inhibitor) for 30 min before stimulation with 0.1% ethanol or 25 nm DHT for 30 min. Western blots were performed for total and phosphorylated Erk1/2, total paxillin (Pax), phosphorylated paxillin at tyrosine-118 (Pax p-Y118), tyrosine-31 (Pax p-Y31), serine-83 (Pax p-S83), and serine-126 (Pax p-S126). All experiments were performed at least three times with similar results. D, a model of paxillin phosphorylation is shown. DHT or EGF via indirect or direct activation of the EGFR promotes Src-mediated phosphorylation of paxillin at tyrosines 31/118, leading to activation of Raf, MEK, and Erk1/2. Activated Erk1/2 in turn regulates phosphorylation of paxillin at serines 83/126.
We next used an in vitro cell migration/invasion assay consisting of a 24-well Boyden chamber with an extracellular matrix-coated membrane to investigate the importance of paxillin for EGF-induced cell migration and invasion of PC3 cells. PC3 cells migrated through the membrane and invaded the matrix in response to EGF, and paxillin knockdown abrogated this physiological process. Fig. 3B demonstrates quantitative analysis of migration/invasion by measuring absorbance after staining invading cells (upper panel) as well as qualitative images of the extracellular matrix-coated membrane underside containing migrated and invaded cells (lower panel). Collectively, these data demonstrate that paxillin is critical for proliferation, migration, and invasion of prostate cancer cell lines.
Phosphorylation of Paxillin at Tyrosines 31/118 and Serines 83/126/130 Is Essential for DHT- and EGF-mediated PCa Cell Proliferation
Receptor-tyrosine kinases are known to promote tyrosine and serine phosphorylation of paxillin (6, 16, 17, 40–43). Our results confirmed that, in LnCAP cells, DHT triggered phosphorylation of paxillin at tyrosines 31/118 and serines 83/126 (Fig. 3C, lane 2). Inhibition of the EGFR with AG1478 (not shown) or Src with PP2 (Fig. 3C, lane 3) abrogated phosphorylation at all four sites (Fig. 3C, lane 3). In contrast, the MEK inhibitor U0126 blocked DHT-induced phosphorylation of paxillin at serines 83/126 but not tyrosines 31/118 (Fig. 3C, lane 4). These results suggest that DHT, via trans-activation of the EGFR, promotes Src-mediated phosphorylation of paxillin at tyrosines 31/118, which is then required for subsequent Erk-mediated phosphorylation of serines 83/126 (Fig. 3D).
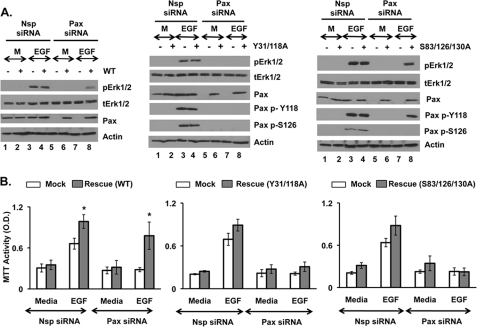
To confirm this proposed signaling sequence as well as the physiologic importance of these phosphorylation events, we mutated tyrosines 31 and 118 (Y31A/Y118A-paxillin) and serines 83, 126, and 130 (S83A/S126A/S130A-paxillin) to alanines. Serine 130 was included because it is phosphorylated in conjunction with serine 126 (11, 16, 17). Thereafter, we knocked down endogenous paxillin expression in PC3 cells by siRNA and re-expressed WT paxillin, S83A/S126A/S130A-paxillin, or Y31A/Y118A-paxillin. As expected, re-expression of WT paxillin rescued EGF-induced Erk1/2 phosphorylation (Fig. 4A, left panel, lane 8) and cell proliferation (Fig. 4B, left panel, right-most bar). In contrast, Y31A/Y118A-paxillin failed to rescue both EGF-induced Erk1/2 activation and cell proliferation (Fig. 4, A, middle panel, lane 8, and B, middle panel, last bar). These results confirm that Src-regulated paxillin phosphorylation of tyrosines is required for EGFR-mediated activation of Erk1/2 and downstream cellular functions. Notably, although endogenous paxillin was phosphorylated at Ser-126 in response to EGF (Fig. 4A, middle, lanes 3 and 4), re-expressed Y31A/Y118A-paxillin in the absence of endogenous wild-type paxillin was not (Fig. 4A, middle, lane 8), confirming that EGFR-mediated serine phosphorylation of paxillin requires prior tyrosine phosphorylation. Consistent with tyrosine phosphorylation being upstream of serine phosphorylation, S83A/S126A/S130A-paxillin was still phosphorylated at Tyr-118 in response to EGF (Fig. 4A, right, lane 8). Surprisingly, however, in the absence of endogenous paxillin, expression of S83A/S126A/S130A-paxillin rescued EGF-induced Erk1/2 activation (Fig. 4A, right panel, lane 8) but not cell proliferation (Fig. 4B, right panel, last bar). These results suggest that, although phosphorylation of paxillin at tyrosines 31/118 is essential for EGFR-mediated Erk1/2 activation and downstream cell proliferation, phosphorylation of paxillin at serines 83/126/130 is only necessary for cell proliferation.
FIGURE 4.
Phosphorylation of paxillin at tyrosines 31/118 and serines 83/126/130 is essential for proliferation. PC3 cells were initially transfected with Paxillin (Pax) or non-targeting (Nsp) siRNA. After 96 h, media was removed, and cells were then either mock-transfected or transfected with wild-type paxillin (left panel) or paxillin mutated at tyrosines 31/118 (Y31A/Y118A, middle panel) or serines 83/126/130 (S83A/S126A/S130A, right panel). After 48 h, cells were treated overnight with serum-free, phenol red-free RPMI 1640 media and stimulated with media (M) or 20 ng/ml EGF for 30 min (for Western blot) or 24 h (for MTT assay). Western blots (A) detected total and phosphorylated Erk1/2, total paxillin (Pax), phosphorylated paxillin at tyrosine-118 (Pax p-Y118), and serine-126 (Pax p-S126). Proliferation was detected by MTT assay (B). Data are represented as the mean ± S.E. (n = 3). *, Student's t test, p ≤ 0.05, rescue versus mock. Each experiment was performed at least three times with similar results.
Paxillin Specifically Regulates Receptor-tyrosine kinase- but Not PKC-induced Erk Activation
Because our data indicated that paxillin was a critical regulator EGFR/Src-induced Erk1/2 activation, we next determined whether paxillin was a universal modulator of Erk. As an alternative, receptor-tyrosine kinase/Src-independent means of activating Erk1/2, we used PMA to promote PKC-mediated Erk1/2 signaling. Paxillin-siRNA-treated LnCAP cells were stimulated with 0.1% DMSO (vehicle), EGF, or PMA for 30 min and Erk1/2 phosphorylation measured. Knockdown of paxillin abrogated EGF (Fig. 5A, lane 6)- but not PMA (Fig. 5A, lane 5)-induced Erk1/2 activation, demonstrating that the regulation of Erk activation by paxillin may be relatively specific to the receptor-tyrosine kinase/Src signaling pathway.
FIGURE 5.
PMA-mediated Erk1/2 activation is paxillin-independent, but PMA-mediated proliferation still requires serine phosphorylation of paxillin. A, Paxillin (Pax) or non-targeting (Nsp) siRNA-treated LnCAP cells were serum-starved and treated with PMA (100 nm) or EGF (20 ng/ml) for 30 min. Western blots were performed for paxillin and total and phosphorylated Erk1/2. B, serum-starved PC3 cells were pretreated with 0.1% DMSO or 20 μm U0126 (Erk inhibitor) for 30 min before stimulation with PMA (100 nm) or EGF (20 ng/ml) for another 30 min. Thereafter, levels of paxillin and phosphorylated paxillin at tyrosine-118 (Pax p-Y118) and serine-126 (Pax p-S126) were detected by Western blot. C, cell proliferation (MTT assay) of PC3 cells treated (24 h) with 0.1% DMSO (M) or 100 nm PMA in the presence or absence of 20 μm U0126. D, PC3 cells were initially transfected with Paxillin (Pax) or non-targeting (Nsp) siRNA. After 96 h, media were removed, and cells were then either mock-transfected or transfected with wild-type paxillin (left panel) or paxillin mutated at tyrosines 31/118 (Y31A/Y118A, middle panel) or serines 83/126/130 (S83A/S126A/S130A, right panel). After 48 h, cells were treated overnight with serum-free, phenol red-free RPMI 1640 media and stimulated with media containing 0.1% DMSO (M) or 100 nm PMA for 24 h. Cell proliferation was detected by MTT. Data are represented as the mean ± S.E. (n = 3). * Student's t test, p ≤ 0.05 PMA versus mock. Each experiment was performed at least three times with similar results.
PMA-mediated PCa Cell Proliferation Requires Erk-mediated Phosphorylation of Paxillin
In contrast to EGF treatment of PC3 cells, which promoted Src-mediated tyrosine as well as Erk-mediated serine phosphorylation of paxillin (Fig. 5B, lane 4), PMA only promoted serine phosphorylation (Fig. 5B, lane 2). This PMA-induced serine phosphorylation of paxillin was blocked by the MEK inhibitor U0126 (Fig. 5B, lane 3), demonstrating it to be MEK/Erk-dependent. Interestingly, PMA-induced proliferation of PC3 cells was also blocked by U0126 (Fig. 5C). Furthermore, siRNA-mediated ablation of paxillin abrogated PMA-induced cell proliferation of PC3 cells, which could be rescued by expression of either wild-type paxillin (Fig. 5D, left panel) or Y31A/Y118A-paxillin (Fig. 5D, middle panel) but not by S83A/S126A/S130A-paxillin (Fig. 5D, right panel). These results indicate that, although paxillin is not always required for Erk1/2 activation, Erk1/2-mediated phosphorylation of paxillin at serine residues seems critical for proliferation regardless of the agonist.
Erk-mediated Phosphorylation of Paxillin Is Required for Normal Transcription in LnCAP and PC3 Cells
Some studies suggest that extra-nuclear kinases activated by steroids or growth factors may regulate transcription (10, 11, 44–50). To examine the role of Erk and paxillin in regulating transcription in PCa cells, we first studied DHT-induced expression of PSA mRNA in LnCAP cells. Inhibition of Erk signaling by the MEK inhibitor U0126 or the EGFR inhibitor AG1478 as well as knockdown of paxillin expression abrogated DHT-induced expression of PSA mRNA (Fig. 6A). These data suggest that, in LnCAP cells, extra-nuclear DHT-mediated Erk1/2 activity (via EGFR and paxillin) is essential for normal intra-nuclear DHT-mediated transcription. Surprisingly, reduction of paxillin expression in PC3 cells similarly reduced EGF-mediated expression of cyclin D1 mRNA (Fig. 6B). Cyclin D1 mRNA expression could be rescued by re-expression of wild-type paxillin but not S83/126/20A-paxillin. These results, which mirror the proliferation data in Figs. 4 and 5, suggest that paxillin may regulate Erk-induced proliferation in part by enhancing Erk1/2-mediated transcription. Thus, paxillin may help mediate cross-talk between cytoplasmic kinase and nuclear transcriptional signaling.
FIGURE 6.
Paxillin regulates DHT or EGF-induced Erk-mediated gene expression in PCa cells. A, relative expression of PSA mRNA in LnCAP cells preincubated with vehicle (0.1% DMSO), 20 μm U0126 (Erk inhibitor), or 20 μm AG1478 (EGFR inhibitor) for 30 min or treated with paxillin (Pax)-specific siRNA for 72 h before stimulation with either ethanol (M) or 25 nm DHT for 24h. B, relative expression of Cyclin D1 mRNA in PC3 cells treated with nonspecific (Nsp)- or paxillin (Pax)-specific siRNA for 96 h followed by transfection with WT or mutated paxillin (S83A/S126A/S130A) before stimulation with either media or 20 ng/ml EGF for 24 h. All data are represented as the mean ± S.E. (n = 3) and normalized to GAPDH levels. *, Student's t test, p ≤ 0.05, stimulus versus media. All experiments were performed at least three times with similar results. C, shown is a proposed model describing the paxillin role in nongenomic AR or EGFR signaling in PCa cells.
DISCUSSION
This study reveals a remarkable conservation of paxillin function from frog germ cells to human PCa cells. However, although the general concepts of paxillin actions on Erk signaling are similar in lower versus higher vertebrates, this study in somatic cells highlights several novel regulatory roles of paxillin in Erk signaling that ultimately control important physiological functions in PCa cells such as transcription and proliferation.
To summarize our data, we propose the following model to describe extra-nuclear AR-mediated signaling in PCa cells (Fig. 6). Androgens bind to classical ARs, most likely located at or near the cell surface (3, 12, 51), to promote activation of MMPs and release of membrane-associated EGFR ligands. Although we have not identified specific EGFR ligands being released in LnCAP cells, previous studies implicate heparin bound-EGFs as meditators of G-protein-coupled or steroid receptor cross-talk with EGF receptors (30, 31). These ligands bind to and activate the EGFR, which then activates Src, Akt, and MEK/Erk1/2 (Fig. 1). Notably, prior co-immunoprecipitation studies in LnCAP cells suggested that DHT-induced Erk1/2 activation (10, 11) might be mediated by extra-nuclear steroid receptors directly binding to and activating Src (52) followed by EGFR phosphorylation (3, 12). However, here EGFR inhibition blocked DHT-induced Src phosphorylation, whereas Src inhibition had minimal effect on DHT-induced EGFR phosphorylation (Fig. 1B). Thus, similar to EGFR signaling in other cell types (53–58), Src actions appear downstream of EGFR activation in DHT-stimulated LnCAP cells.
Irrespective of the underlying mechanism, the rapid and robust trans-activation of the EGFR by DHT highlights the novel concept that, outside of the nucleus, androgen actions are just like EGF with respect to activation of cytoplasmic kinase cascades (3, 4, 44). EGFR-induced kinase pathways are known to modulate steroid receptor-mediated transcriptional signaling by altering both receptor and co-regulator activities (10, 11, 44, 45, 49, 50, 59, 60). Thus, indirect activation of the EGFR by DHT similarly leads to “outside-inside” cross-talk whereby rapid activation of extra-nuclear kinases enhances intra-nuclear transcriptional signaling. Data that MEK and EGFR inhibition block DHT-induced PSA mRNA expression (Fig. 6A) support this model.
What mediates this outside-inside cross-talk between extra-nuclear kinases and intra-nuclear transcription? Because paxillin knockdown abrogated both EGF- and DHT-induced Erk1/2 activation (Fig. 1, C and D) as well as DHT-induced PSA mRNA (Fig. 6A) and EGF-induced cylin D1 mRNA expression (Fig. 6B), paxillin appears to be at least one key regulator of outside-inside signaling in response to both direct (EGF) and indirect (DHT) activation of the EGFR. Our data further suggest that paxillin-mediated regulation of extra-nuclear kinases and intra-nuclear transcription in turn controls PCa cell proliferation, invasion, and migration. Thus, paxillin is a critical regulator of multiple EGFR/Erk-regulated processes in PCa cells.
How does paxillin mediate these EGFR/Erk-regulated functions? One mechanism is at the level of EGFR/Src-induced activation of the Raf/MEK/Erk signaling pathway (Fig. 6C). Prior studies suggested that Src-mediated phosphorylation of tyrosines 31/118 on paxillin was important for Erk activation in response to FAK or integrin-mediated signaling (6, 40, 41). Our paxillin knockdown and rescue experiments with Y31A/Y118A-paxillin in PC3 cells unequivocally confirm that tyrosine phosphorylation of paxillin at these residues is required for EGFR-mediated Erk1/2 activation and downstream proliferation (Fig. 4). Previous studies (40, 41, 61) also suggested that paxillin might function as a scaffold to hold Raf, MEK, and Erk1/2 in a signaling complex. Most of these studies (17, 40–42, 61, 62) focused on the association of paxillin with focal adhesion molecules, using overexpression and co-precipitation studies to show interactions. In our study, the morphology of PCa cells with reduced paxillin expression appeared grossly normal, although we did not specifically examine cell-cell adhesions. However, in paxillin knockdown cells, expression of caRaf or caMEK was sufficient to promote Erk1/2 signaling (Fig. 2, B and C). Thus, although paxillin may form a complex with Raf, MEK and Erk, these interactions are not necessary for signaling by or downstream of Raf. In fact, paxillin appears to function between EGFR and Raf (Fig. 6C), as EGFR phosphorylation is minimally affected by paxillin knockdown (Fig. 2D).
Interestingly, in androgen-induced maturation of Xenopus oocytes, paxillin also functions just upstream of MOS, the germ cell homologue of Raf, again demonstrating the remarkable conservation of paxillin function from lower to higher vertebrates. However, the requirement for initial Src-mediated tyrosine phosphorylation of paxillin, the ability of paxillin to regulate downstream Erk functions regardless of the agonist, and the ability of paxillin to mediate cross-talk between cytoplasmic kinase and nuclear transcriptional signaling are all specific to somatic cells, as they are not seen in frog oocytes.
Our study suggests that paxillin is a relatively specific regulator of receptor-tyrosine kinase/Src-mediated Erk1/2 activation in PCa cells, as paxillin knockdown had no effect on PKC-mediated Erk activation in PC3 cells (Fig. 5A). In contrast, paxillin appears to be a general regulator of Erk-mediated cellular processes such as transcription or proliferation, irrespective of the stimulus, as it was required for both PKC and EGFR-mediated proliferation in PC3 cells (Fig. 5). Importantly, serine phosphorylation of paxillin appears critical for these Erk-mediated processes, as in paxillin-knockdown PC3 cells S83A/S126A/S130A-paxillin was unable to rescue proliferation or induction of cyclin D1 mRNA in response to either EGF- or PKC-mediated Erk1/2 activation (Figs. 5 and 6). In fact, our results (Figs. 3C and 5B) and previous evidence (5, 16, 17, 40, 41) indicate that Erk itself may directly phosphorylate paxillin at these serine residues.
To summarize, paxillin regulates Erk-mediated processes by two means (Fig. 6C); 1) by specifically regulating receptor-tyrosine kinase-mediated Erk1/2 activation via Src-mediated tyrosine phosphorylation and 2) more broadly by regulating of Erk-mediated downstream processes like intra-nuclear transcription and proliferation via Erk-mediated serine phosphorylation. Thus, paxillin can be both an affector and an effector of Erk signaling in PCa cells, depending upon the stimulus. How paxillin act as an effector to regulate transcription in PCa cells is unknown; however, one possibility is that paxillin constitutively binds to inactive Erk1/2 to keep it sequestered. Consistent with this hypothesis, the binding affinity of inactive Erk1/2 to paxillin appears higher than that of activated Erk1/2 (41, 50). Irrespective of how it is triggered, activated Erk1/2 might then promote serine phosphorylation of paxillin, releasing Erk from paxillin and permitting Erk-mediated transcription, cell proliferation, migration, and invasion. Further work is needed to investigate the details of this pathway. However, these studies underscore the importance of paxillin in regulating proliferation in both androgen-dependent and -independent PCa regardless of the stimulus (steroids or other growth factors). In fact, understanding how paxillin regulates Erk-mediated proliferation may lead to novel therapeutic targets for cancer treatment as well possible diagnostic markers for tumor aggressiveness.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of constitutively active Raf from Dr. William Walker (University of Pittsburgh) and constitutively active MEK from Dr. Melanie Cobb (University of Texas Southwestern Medical Center). We also thank Dr. Sheila Thomas (Harvard University) for the mouse embryonic fibroblasts from paxillin null mice.
This work was supported, in whole or in part, by National Institutes of Health Grant DK59913 (to S. R. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
A. Sen, K. O'Malley, Z. Wang, G. V. Raj, D. B. DeFranco, and S. R. Hammes, unpublished data.
- AR
- androgen receptor
- DHT
- dihydrotestosterone
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ca
- constitutively active
- EGFR
- EGF receptor
- PSA
- prostate-specific antigen
- MMP
- matrix metalloproteinase
- PMA
- phorbol 12-myristate 13-acetate.
REFERENCES
- 1.Lange C. A., Gioeli D., Hammes S. R., Marker P. C. (2007) Annu. Rev. Physiol. 69, 171–199 [DOI] [PubMed] [Google Scholar]
- 2.Hammes S. R., Levin E. R. (2007) Endocr. Rev. 28, 726–741 [DOI] [PubMed] [Google Scholar]
- 3.Migliaccio A., Castoria G., Di Domenico M., Ciociola A., Lombardi M., De Falco A., Nanayakkara M., Bottero D., De Stasio R., Varricchio L., Auricchio F. (2006) Ann. N.Y. Acad. Sci. 1089, 194–200 [DOI] [PubMed] [Google Scholar]
- 4.Zhu M. L., Kyprianou N. (2008) Endocr. Relat. Cancer 15, 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasar M., DeFranco D. B., Hammes S. R. (2006) J. Biol. Chem. 281, 39455–39464 [DOI] [PubMed] [Google Scholar]
- 6.Brown M. C., Turner C. E. (2004) Physiol. Rev. 84, 1315–1339 [DOI] [PubMed] [Google Scholar]
- 7.Deakin N. O., Turner C. E. (2008) J. Cell Sci. 121, 2435–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagel M., George E. L., Kim A., Tamimi R., Opitz S. L., Turner C. E., Imamoto A., Thomas S. M. (2002) Mol. Cell. Biol. 22, 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wade R., Bohl J., Vande Pol S. (2002) Oncogene 21, 96–107 [DOI] [PubMed] [Google Scholar]
- 10.Peterziel H., Mink S., Schonert A., Becker M., Klocker H., Cato A. C. (1999) Oncogene 18, 6322–6329 [DOI] [PubMed] [Google Scholar]
- 11.Unni E., Sun S., Nan B., McPhaul M. J., Cheskis B., Mancini M. A., Marcelli M. (2004) Cancer Res. 64, 7156–7168 [DOI] [PubMed] [Google Scholar]
- 12.Migliaccio A., Castoria G., Di Domenico M., de Falco A., Bilancio A., Lombardi M., Barone M. V., Ametrano D., Zannini M. S., Abbondanza C., Auricchio F. (2000) EMBO J. 19, 5406–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki H., Ueda T., Ichikawa T., Ito H. (2003) Endocr. Relat. Cancer 10, 209–216 [DOI] [PubMed] [Google Scholar]
- 14.Tremblay L., Hauck W., Aprikian A. G., Begin L. R., Chapdelaine A., Chevalier S. (1996) Int. J. Cancer 68, 164–171 [DOI] [PubMed] [Google Scholar]
- 15.Kasai M., Guerrero-Santoro J., Friedman R., Leman E. S., Getzenberg R. H., DeFranco D. B. (2003) Cancer Res. 63, 4927–4935 [PubMed] [Google Scholar]
- 16.Woodrow M. A., Woods D., Cherwinski H. M., Stokoe D., McMahon M. (2003) Exp. Cell Res. 287, 325–338 [DOI] [PubMed] [Google Scholar]
- 17.Cai X., Li M., Vrana J., Schaller M. D. (2006) Mol. Cell. Biol. 26, 2857–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evaul K., Hammes S. R. (2008) J. Biol. Chem. 283, 27525–27533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Titus M. A., Schell M. J., Lih F. B., Tomer K. B., Mohler J. L. (2005) Clin. Cancer Res. 11, 4653–4657 [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama T., Ikarashi T., Hashimoto Y., Wako K., Takahashi K. (2007) J. Urol. 178, 1282–1289 [DOI] [PubMed] [Google Scholar]
- 21.Qi W., Cooke L. S., Stejskal A., Riley C., Croce K. D., Saldanha J. W., Bearss D., Mahadevan D. (2009) BMC Cancer 9, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Festuccia C., Gravina G. L., Biordi L., D'Ascenzo S., Dolo V., Ficorella C., Ricevuto E., Tombolini V. (2009) Prostate 69, 1529–1537 [DOI] [PubMed] [Google Scholar]
- 23.Iwata K. K., Mantis C. (2004) Proc. Am. Assoc. Cancer Res. 9, 142 [Google Scholar]
- 24.Zhang L., Davis J. S., Zelivianski S., Lin F. F., Schutte R., Davis T. L., Hauke R., Batra S. K., Lin M. F. (2009) Cancer Lett. 285, 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sastry K. S., Karpova Y., Kulik G. (2006) J. Biol. Chem. 281, 27367–27377 [DOI] [PubMed] [Google Scholar]
- 26.Li Z., Xu M., Xing S., Ho W. T., Ishii T., Li Q., Fu X., Zhao Z. J. (2007) J. Biol. Chem. 282, 3428–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamnongjit M., Gill A., Hammes S. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16257–16262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campos L. S., Leone D. P., Relvas J. B., Brakebusch C., Fässler R., Suter U., ffrench-Constant C. (2004) Development 131, 3433–3444 [DOI] [PubMed] [Google Scholar]
- 29.Askari M. D., Tsao M. S., Schuller H. M. (2005) J. Cancer Res. Clin. Oncol. 131, 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Razandi M., Pedram A., Park S. T., Levin E. R. (2003) J. Biol. Chem. 278, 2701–2712 [DOI] [PubMed] [Google Scholar]
- 31.Filardo E. J., Quinn J. A., Frackelton A. R., Jr., Bland K. I. (2002) Mol. Endocrinol. 16, 70–84 [DOI] [PubMed] [Google Scholar]
- 32.Robinson M. J., Cheng M., Khokhlatchev A., Ebert D., Ahn N., Guan K. L., Stein B., Goldsmith E., Cobb M. H. (1996) J. Biol. Chem. 271, 29734–29739 [DOI] [PubMed] [Google Scholar]
- 33.Mansour S. J., Matten W. T., Hermann A. S., Candia J. M., Rong S., Fukasawa K., Vande Woude G. F., Ahn N. G. (1994) Science 265, 966–970 [DOI] [PubMed] [Google Scholar]
- 34.Leevers S. J., Paterson H. F., Marshall C. J. (1994) Nature 369, 411–414 [DOI] [PubMed] [Google Scholar]
- 35.Arnold J. T., Le H., McFann K. K., Blackman M. R. (2005) Am. J. Physiol. Endocrinol. Metab. 288, E573–E584 [DOI] [PubMed] [Google Scholar]
- 36.Zheng Z., Cai C., Omwancha J., Chen S. Y., Baslan T., Shemshedini L. (2006) J. Biol. Chem. 281, 4002–4012 [DOI] [PubMed] [Google Scholar]
- 37.Fu M., Liu M., Sauve A. A., Jiao X., Zhang X., Wu X., Powell M. J., Yang T., Gu W., Avantaggiati M. L., Pattabiraman N., Pestell T. G., Wang F., Quong A. A., Wang C., Pestell R. G. (2006) Mol. Cell. Biol. 26, 8122–8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gamble S. C., Odontiadis M., Waxman J., Westbrook J. A., Dunn M. J., Wait R., Lam E. W., Bevan C. L. (2004) Oncogene 23, 2996–3004 [DOI] [PubMed] [Google Scholar]
- 39.Ripple M. O., Henry W. F., Rago R. P., Wilding G. (1997) J. Natl. Cancer Inst. 89, 40–48 [DOI] [PubMed] [Google Scholar]
- 40.Ishibe S., Joly D., Liu Z. X., Cantley L. G. (2004) Mol. Cell 16, 257–267 [DOI] [PubMed] [Google Scholar]
- 41.Ishibe S., Joly D., Zhu X., Cantley L. G. (2003) Mol. Cell 12, 1275–1285 [DOI] [PubMed] [Google Scholar]
- 42.Chen H. Y., Shen C. H., Tsai Y. T., Lin F. C., Huang Y. P., Chen R. H. (2004) Mol. Cell. Biol. 24, 10558–10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb D. J., Schroeder M. J., Brame C. J., Whitmore L., Shabanowitz J., Hunt D. F., Horwitz A. R. (2005) J. Cell Sci. 118, 4925–4929 [DOI] [PubMed] [Google Scholar]
- 44.Cheng J., Watkins S. C., Walker W. H. (2007) Endocrinology 148, 2066–2074 [DOI] [PubMed] [Google Scholar]
- 45.Fix C., Jordan C., Cano P., Walker W. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10919–10924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carey A. M., Pramanik R., Nicholson L. J., Dew T. K., Martin F. L., Muir G. H., Morris J. D. (2007) Int. J. Cancer 121, 520–527 [DOI] [PubMed] [Google Scholar]
- 47.Franco O. E., Onishi T., Yamakawa K., Arima K., Yanagawa M., Sugimura Y., Kawamura J. (2003) Prostate 56, 319–325 [DOI] [PubMed] [Google Scholar]
- 48.Xu Y., Chen S. Y., Ross K. N., Balk S. P. (2006) Cancer Res. 66, 7783–7792 [DOI] [PubMed] [Google Scholar]
- 49.O'Malley B. W., Kumar R. (2009) Cancer Res. 69, 8217–8222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lidke D. S., Huang F., Post J. N., Rieger B., Wilsbacher J., Thomas J. L., Pouysségur J., Jovin T. M., Lenormand P. (2010) J. Biol. Chem. 285, 3092–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cinar B., Mukhopadhyay N. K., Meng G., Freeman M. R. (2007) J. Biol. Chem. 282, 29584–29593 [DOI] [PubMed] [Google Scholar]
- 52.Varricchio L., Migliaccio A., Castoria G., Yamaguchi H., de Falco A., Di Domenico M., Giovannelli P., Farrar W., Appella E., Auricchio F. (2007) Mol. Cancer Res. 5, 1213–1221 [DOI] [PubMed] [Google Scholar]
- 53.Mao W., Irby R., Coppola D., Fu L., Wloch M., Turner J., Yu H., Garcia R., Jove R., Yeatman T. J. (1997) Oncogene 15, 3083–3090 [DOI] [PubMed] [Google Scholar]
- 54.Goi T., Shipitsin M., Lu Z., Foster D. A., Klinz S. G., Feig L. A. (2000) EMBO J. 19, 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dimri M., Naramura M., Duan L., Chen J., Ortega-Cava C., Chen G., Goswami R., Fernandes N., Gao Q., Dimri G. P., Band V., Band H. (2007) Cancer Res. 67, 4164–4172 [DOI] [PubMed] [Google Scholar]
- 56.Frame M. C. (2004) J. Cell Sci. 117, 989–998 [DOI] [PubMed] [Google Scholar]
- 57.Lu S., Ouyang M., Seong J., Zhang J., Chien S., Wang Y. (2008) PLoS Comput. Biol. 4, e1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seong J., Lu S., Ouyang M., Huang H., Zhang J., Frame M. C., Wang Y. (2009) Chem. Biol. 16, 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker W. H., Cheng J. (2005) Reproduction 130, 15–28 [DOI] [PubMed] [Google Scholar]
- 60.Xu J., Wu R. C., O'Malley B. W. (2009) Nat. Rev. Cancer 9, 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobkin-Bekman M., Naidich M., Rahamim L., Przedecki F., Almog T., Lim S., Melamed P., Liu P., Wohland T., Yao Z., Seger R., Naor Z. (2009) Mol. Endocrinol. 23, 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ku H., Meier K. E. (2000) J. Biol. Chem. 275, 11333–11340 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.