Abstract
Previous work has shown that several nucleoporins, including Nup62 are degraded in cells infected with human rhinovirus (HRV) and poliovirus (PV) and that this contributes to the disruption of certain nuclear transport pathways. In this study, the mechanisms underlying proteolysis of Nup62 have been investigated. Analysis of Nup62 in lysates from HRV-infected cells revealed that Nup62 was cleaved at multiple sites during viral infection. The addition of purified HRV2 2A protease (2Apro) to uninfected HeLa whole cell lysates resulted in the cleavage of Nup62, suggesting that 2Apro is a major contributor to Nup62 processing. The ability of purified 2Apro to cleave bacterially expressed and purified Nup62 demonstrated that 2Apro directly cleaves Nup62 in vitro. Site-directed mutagenesis of putative cleavage sites in Nup62 identified six different positions that are cleaved by 2Apro in vitro. This analysis revealed that 2Apro cleavage sites were located between amino acids 103 and 298 in Nup62 and suggested that the N-terminal FG-rich region of Nup62 was released from the nuclear pore complex in infected cells. Analysis of HRV- and PV-infected cells using domain-specific antibodies confirmed that this was indeed the case. These results are consistent with a model whereby PV and HRV disrupt nucleo-cytoplasmic trafficking by selectively removing FG repeat domains from a subset of nuclear pore complex proteins.
Keywords: Nuclear Pore, Nuclear Transport, Positive Strand RNA Viruses, Proteolytic Enzymes, Viral Protease, Host-Pathogen Interactions, Picornavirus, Poliovirus, Rhinovirus
Introduction
The nuclear pore complex (NPC)2 is a dynamic gateway between the nucleus and the cytoplasm that mediates nucleo-cytoplasmic transport via interaction with numerous soluble transport factors (1). In vertebrates, the NPC is an ∼60-MDa structure composed of multiple copies of nearly 30 different nucleoporins (Nups) (2, 3). Analysis of the yeast NPC showed that ∼40% of Nups contain unstructured domains rich in phenylalanine and glycine (FG) residues (FG-Nups) (2, 3). These FG-rich regions are thought to play an import role in regulating nucleo-cytoplasmic trafficking by serving as docking sites for soluble nuclear transport factors and providing a physical barrier that prevents diffusion across the NPC (4–6).
Nup62 is a glycosylated FG-Nup located in the central channel of the NPC. The N-terminal FG-rich region of Nup62 serves as a docking site for NTF2 (nuclear transport factor 2) (7, 8), which is the transport receptor for Ran, a small GTPase that provides directionality to the transport process (Fig. 1A) (9). The FG-rich region of Nup62 is connected to the C-terminal region via a serine/threonine-rich linker that is thought to be the site of glycosylation (10). The C terminus of Nup62 is predicted to adopt a coiled-coil structure and to facilitate the anchoring of Nup62 to the NPC (11, 12). The C terminus of Nup62 has been shown to interact with the transport receptor importin-β in vitro (13) and to mediate interaction with the NPC proteins Nup58, Nup54, and Nup45 that together constitute the Nup62 complex (14, 15).
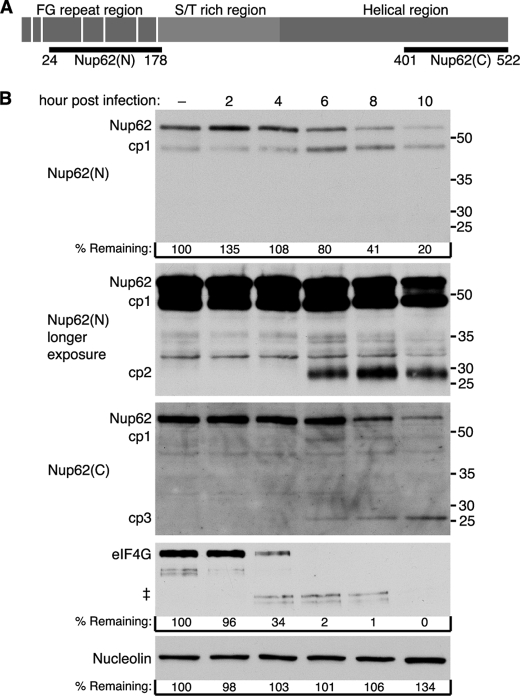
FIGURE 1.
Degradation of Nup62 during HRV2 infection. A, schematic representation of Nup62 and regions recognized by anti-Nup62 antibodies. Vertical white bars indicate the location of FG repeats. Nup62(N) and Nup62(C) indicate the regions of Nup62 used to raise antibodies to the N and C terminus, respectively. B, HRV2 infection induces Nup62 degradation. Twenty-five μg of whole cell lysates prepared from mock-infected cells or cells that had been infected with HRV2 for the indicated length of time were analyzed by immunoblotting with Nup62(N) or Nup62(C). Immunoblots were stripped and reprobed to detect nucleolin and eIF4GI. Molecular mass markers are indicated in kilodaltons. % remaining, band intensities were quantitated by densitometry, and the amounts relative to mock-infected cells are indicated. ‡, eIF4GI cleavage products. longer exposure, a longer exposure of the Nup62(N) blot.
Human rhinovirus (HRV), along with poliovirus, is a member of the Picornaviridae family. Picornaviruses are characterized by single-stranded RNA genomes of positive polarity. After entry, the viral RNA genome is translated and then replicated in the host cytoplasm. Interestingly, during viral replication, a number of host nuclear proteins relocalize to the cytoplasm and interact with viral RNA or gene products (16–18). This abnormal localization of nuclear proteins has been explained by inhibition of nuclear import during HRV and poliovirus infection along with alteration of the NPC through degradation of Nup62, Nup98, and Nup153 (19–21). Consistent with the loss of material from the NPC in infected cells, Belov et al. (22) observed reduced staining of the NPC in electron micrographs of poliovirus-infected cells. Despite these apparent alterations to the composition of the NPC, certain import and export pathways were still functional in poliovirus-infected cells, indicating that the NPC is not completely destroyed and that it retains at least some functionality (20). Prior work has implicated the viral protease, 2Apro, in the alterations to the NPC that occur in infected cells. For example, expression of 2Apro in HeLa cells results in increased permeability of the nuclear envelope, relocalization of nuclear proteins to the cytoplasm, and inhibition of mRNA export (22, 23). In addition, 2Apro is capable of cleaving Nup98 in vitro (21). However, the contribution of 2Apro in the degradation of other NPC proteins, including Nup62, is not known.
In this study, the mechanism of Nup62 degradation during HRV infection was analyzed. The results indicate that 2Apro is the major viral protease responsible for degradation of Nup62 in infected cells. We find that 2Apro cleaves Nup62 directly and identify multiple 2Apro cleavage sites in Nup62 that are clustered within or adjacent to the central serine/threonine-rich region of the protein. Examination of Nup62 in poliovirus and HRV-infected cells indicates that although these viruses differentially target Nup62 for proteolysis, infection with either virus results in the removal of the N-terminal domain of Nup62 containing the FG repeats (24).
EXPERIMENTAL PROCEDURES
Cell Culture and Virus
HeLa cells were maintained in a monolayer in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mm l-glutamine, and penicillin/streptomycin at 37 °C in 5% CO2. The HGP strain of human rhinovirus type 2 (HRV2) was purchased from the ATCC, and viral stocks were amplified by infection of HeLa monolayers. Mahoney type 1 poliovirus (PV) stocks were prepared as described previously (18). HeLa cells at 80% confluence were either mock-infected or infected at a multiplicity of infection of 50 for the indicated time. Virus was adsorbed for 30 min at 32 °C (HRV2) or 37 °C (PV) in phosphate-buffered saline (PBS) supplemented with 1 mm MgCl2 and 1 mm CaCl2. After adsorption, unbound virus was removed, and DMEM with 10% fetal bovine serum (FBS), 2 mm l-glutamine, and penicillin/streptomycin was added.
Protein Purification
The full-length human Nup62 open reading frame in pcDNA3.1/HisB (a kind gift from Dr. N. R. Yaseen) was isolated by digestion with BamHI and XhoI and subcloned into the corresponding sites of pET28b(+) vector (Novagen) to create pET28b(+)-Nup62, which encodes a full-length Nup62 with an N-terminal His6 tag. The pET28b(+)-Nup62 construct was transformed into BL21(DE3)RIPL, and Nup62 protein expression was induced by the addition of 1 mm isopropyl-β-d-thiogalactoside when cultures reached an A600 = 0.6. Cultures were incubated for an additional 4 h at 37 °C, sonicated, and then clarified by centrifugation at 38,500 × g. Nup62 localized to inclusion bodies was solubilized with buffer B (8 m urea, 0.1 m NaH2PO4, and 0.01 m Tris-Cl (pH 8.0)) and then purified on Ni2+-NTA spin columns as described by the manufacturer (Qiagen). Purified Nup62 was dialyzed twice for 4 h and then overnight at 4 °C against 1 liter of refolding buffer (400 mm l-arginine, 0.4 m Tris-Cl (pH 8.0), 0.2 mm EDTA, 5 mm reduced glutathione, 0.5 mm oxidized glutathione). Nup62 was concentrated by placing the dialysis bag on a bed of polyethylene glycol 20,000 for 1 h, quantified using the Bio-Rad protein assay kit, and stored at −20 °C.
Immunoblotting
HeLa whole cell lysates were prepared by washing cells twice with PBS, followed by a 20-min incubation on ice in Tx lysis buffer (50 mm triethanolamine, pH 7.4, 500 mm NaCl, 0.5% Triton X-100, and 1 mm dithiothreitol (DTT)) (25). Lysates were cleared by centrifugation at 20,000 × g for 5 min and quantified using the Bio-Rad protein assay kit. Equal quantities of protein were separated by SDS-PAGE, followed by transfer to a PVDF membrane (Millipore). Nup62 was detected by mAb414 (Covance Inc., catalog no. MMS-120P), Nup62(N) raised against N-terminal amino acids 24–178 of Nup62 (BD Transduction Laboratories, catalog no. 610498), and Nup62(C) raised against C-terminal amino acids 401–522 of Nup62 (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), catalog no. sc-1915). Nup155 was detected using a rabbit polyclonal antibody kindly provided by Susan Wente (Vanderbilt). Mouse monoclonal antibodies were used to detect nucleolin (MS3 (26) and GFP (Clontech, catalog no. 632381), whereas rabbit polyclonal antiserum was used to detect eIF4G (27) and NF-κB (Santa Cruz Biotechnology, Inc., catalog no. sc-109). Antibody-antigen complexes were detected using an HRP-conjugated secondary antibody and chemiluminescence.
In Vitro Cleavage Assay
Uninfected HeLa whole cell lysates were prepared as described above. HRV2 2Apro was purified from bacteria as described (28). For cleavage assays, the indicated amount of HRV2 2Apro was incubated with 25 μg of uninfected HeLa whole cell lysates, in vitro translated Nup62, or the indicated amount of purified Nup62 at 30 °C for the indicated amount of time in 2A reaction buffer (50 mm Tris-Cl (pH 8.0), 50 mm NaCl, 5 mm DTT, and 1 mm EDTA) (29). For cleavage reactions with rhinovirus 3C protease (3Cpro), commercially available HRV14 3Cpro (GE Healthcare) was incubated with 25 μg of uninfected HeLa whole cell lysates at 30 °C in 3C reaction buffer (50 mm Tris-Cl, 150 mm NaCl, 1 mm DTT, and 1 mm EDTA, pH 7.0, at 25 °C). After the incubation, samples were electrophoresed and analyzed by immunoblotting as described above.
N-terminal Sequencing
Fifteen μg of purified Nup62 was incubated with purified HRV2 2Apro at 30 °C for 4 h. Following incubation, the sample was loaded on a 10% NEXT GEL (Amresco) and transferred to a PVDF membrane. Cleavage products on the membrane were visualized by Coomassie Brilliant Blue staining and cut out for sequencing. Automated Edman degradation was performed using an ABI Procise 494 at the Protein Structure Core Facility, University of Nebraska Medical Center.
Mutagenesis and in Vitro Translation
Point mutants were constructed at putative cleavage sites in Nup62 using the QuikChange II site-directed mutagenesis kit (Stratagene) and confirmed by sequencing. For in vitro translation, 0.5 μg of plasmid containing wild-type or mutant Nup62 open reading frame was incubated with 1 μl of TNT reaction buffer (Promega), 12.5 μl of rabbit reticulocyte lysate, 0.5 μl of T7 RNA polymerase, 0.5 μl of 1 mm amino acid mixture without methionine, and 10 μCi of [35S]methionine (>1,000 Ci/mmol; GE Healthcare) for 90 min at 30 °C. After incubation, samples were immediately stored at −80 °C. Relative amounts of wild-type and mutant Nup62 in reactions were determined by SDS-PAGE and autoradiography, followed by densitometry.
Nup62 Overexpression in Vivo
pAcGFP-Nup62WT was constructed by subcloning the PCR-amplified Nup62 open reading frame with introduced SalI and KpnI restriction sites into the corresponding sites of pAcGFP1-C1 (Clontech). pAcGFP-Nup62(A103D) was constructed by site-directed mutagenesis of pAcGFP-Nup62WT as described above. Transfection was performed using TransIT-LT1 reagent (Mirus Bio) as recommended by the manufacturer. One day after transfection, cells were infected with HRV2. Eighteen h after infection, cells were harvested for immunoblotting as described above.
Indirect Immunofluorescence
The anti-Nup62 antibodies described above were used for indirect immunofluorescence analysis. Mock- or virus-infected cells growing on 12-mm glass coverslips were washed three times with PBS and immunostained for Nup62 by fixing in 3% formaldehyde for 20 min at 25 °C, washing three times with PBS, and permeabilizing in methanol at −20 °C for 5 min. Following fixation/permeabilization, cells were washed three times with PBS and then blocked in base solution (PBS containing 2% bovine serum albumin and 0.05% Triton X-100) for 15 min at 25 °C. Coverslips were inverted into 40 μl of base solution containing diluted primary antibody and incubated overnight at 4 °C. Coverslips were washed three times in base solution at 25 °C and then inverted into 40 μl of base solution containing either a 1:2000 dilution of AlexaFluor 555-conjugated donkey anti-goat or a 1:1000 dilution of AlexaFluor 488-conjugated goat anti-mouse immunoglobulin (Invitrogen) and incubated for 1 h at 25 °C. Coverslips where washed twice in PBS, once in PBS containing 0.2 μg/ml Hoechst 33258, drained, and mounted in Vectashield mounting medium (Vector Laboratories) onto glass slides. Cells were observed using a Nikon Eclipse E1000 fluorescent microscope with a ×60 objective, and images were acquired using a Hamamatsu Orca digital camera and Metamorph software.
RESULTS
Nup62 Is Cleaved at Multiple Sites in Cells Infected with HRV Type 2
Human rhinoviruses are subdivided into three species, designated group A, B, and C. Previously, we demonstrated that infection of cells with HRV14, a group B rhinovirus, results in the specific cleavage of Nup62 (19). To determine whether this also occurred with group A rhinoviruses, the status of Nup62 in cells infected with HRV2 was examined. Fig. 1B shows that levels of Nup62 began to decline by 6 h postinfection and that by 10 h postinfection, levels of Nup62 were 20% of the levels seen in uninfected cells. Analysis of eIF4GI confirmed that the cells were efficiently infected with HRV2 and revealed that eIF4GI was degraded much more rapidly following infection than was Nup62 (Fig. 1B). As expected, levels of nucleolin were unchanged during the course of infection (Fig. 1B). These results indicate that Nup62 is targeted for proteolysis by both group A and B rhinoviruses.
This analysis also revealed novel bands that may represent Nup62 cleavage products (cp). Using an antibody specific for the N terminus of Nup62 (Nup62(N); Fig. 1A) the intensity of an ∼48 kDa band (cp1) increased at 6 h postinfection, the time when full-length Nup62 began to decline (Fig. 1B). Although a cellular protein of the same size appeared to cross-react with this antibody, analysis with an antibody that reacts with the C-terminal region of Nup62 (Nup62(C); Fig. 1A) confirmed the loss of full-length Nup62 beginning at 6 h postinfection and also revealed an ∼48-kDa cleavage product that appeared similar in size to cp1, indicating that this probably represents a bona fide Nup62 cleavage product (Fig. 1B). A longer exposure of the Nup62(N) immunoblot revealed an ∼29 kDa band (cp2) that became detectable at 6 h postinfection. Similarly, a second reactive band observed with Nup62(C) migrated at ∼26 kDa (cp3) and may represent a third Nup62 cleavage product. Interestingly, levels of cp1 declined from 6 to 10 h postinfection, whereas levels of cp3 appeared to increase over this same time frame, raising the possibility that cp3 derives from further proteolysis of cp1. If these bands do represent bona fide cleavage products of Nup62, it is notable that their combined size (103 kDa) would be greater than that of intact Nup62 (62 kDa). This suggests that multiple sites in Nup62 are targeted for proteolysis in infected cells.
2A Protease Cleaves Nup62 in Vitro
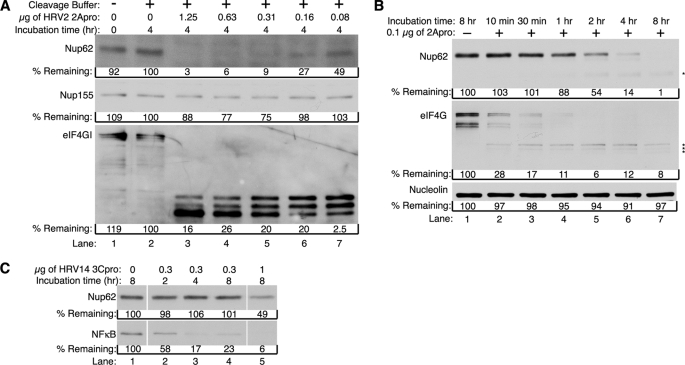
Previously, we found that the viral 2A protease (2Apro) was responsible for cleavage of Nup98 in infected cells (21). To determine whether 2Apro was also responsible for the cleavage of Nup62, we incubated uninfected HeLa whole cell lysates with increasing amounts of purified HRV2 2Apro and analyzed the resulting products by immunoblotting. The results indicate that the addition of as little as 0.08 μg of 2Apro resulted in a 50% decline in Nup62 levels within 4 h, and 0.31 μg of 2Apro caused a greater than 90% reduction in Nup62 levels over this same time period (Fig. 2A, lanes 7 and 5, respectively). This was not due to Nup62 being unstable in the lysates under these conditions because incubation in buffer alone had no effect on Nup62 levels (Fig. 2A, lane 2). The translation factor eIF4GI, a known target of 2Apro (28, 30), appeared to be much more sensitive to the addition of protease because incubation with as little as 0.08 μg of 2Apro was sufficient to induce complete cleavage of eIF4GI. The addition of 2Apro did not cause nonspecific cleavage of NPC proteins because levels of Nup155, which is not degraded in poliovirus-infected cells,3 remained relatively unchanged even at the highest concentration of 2Apro tested (Fig. 2A). These results indicate that the addition of 2Apro to whole cell lysates is sufficient to induce the cleavage of Nup62.
FIGURE 2.
HRV2 2Apro induces cleavage of Nup62. A, 2Apro causes cleavage of Nup62 in cell lysates. Uninfected HeLa whole cell lysates were incubated in the presence or absence of the indicated amount of purified HRV2 2Apro for 4 h at 30 °C. After incubation, extracts were analyzed by immunoblotting to detect Nup62 using mAb414. Immunoblots were sequentially stripped and reprobed to detect Nup155 and eIF4GI. B, cleavage kinetics of in vitro Nup62 degradation. Uninfected HeLa whole cell lysates were incubated with or without 0.1 μg of purified HRV2 2Apro for the indicated times and analyzed for cleavage of Nup62 by immunoblotting with Nup62(N). Immunoblots were sequentially stripped and reprobed with anti-eIF4G antibody and anti-nucleolin antibody. Cleavage products are indicated with an asterisk. C, Nup62 is resistant to cleavage by 3Cpro. Uninfected HeLa whole cell lysates were incubated in the presence or absence of the indicated amount of purified HRV14 3Cpro at 30 °C for the indicated amount of time. After incubation, extracts were analyzed by immunoblotting to detect Nup62 using Nup62(N). The immunoblot was then stripped and reprobed to detect NF-κB. % remaining, band intensities were quantitated by densitometry, and the amounts relative to mock-infected cells are indicated.
To further examine the kinetics of Nup62 cleavage using this in vitro cleavage assay, proteolysis in the presence of a low concentration of protease was examined over time. When 0.1 μg of 2Apro was added to whole cell lysates, levels of Nup62 were reduced to ∼50% of their starting values within 2 h (Fig. 2B, lane 5). Levels of Nup62 dropped to 14% by 4 h and were undetectable following an 8-h incubation (Fig. 2B, lanes 6 and 7). Proteolysis required the addition of viral protease because no degradation of Nup62 was seen when lysates were incubated for 8 h in cleavage buffer lacking 2Apro (Fig. 2B, lane 1). Under these conditions levels of eIF4GI began to decline very rapidly and were reduced to 30% within 10 min of 2Apro addition (Fig. 2B, lane 2). These findings confirm that eIF4GI is more sensitive to cleavage by 2Apro than is Nup62 and are in good agreement with the results seen in infected cells (Fig. 1B).
To determine if other rhinovirus proteases might also induce proteolysis of Nup62, we examined the effect of 3Cpro on levels of Nup62 in whole cell lysates. For this assay, we used purified HRV14 3Cpro because purified HRV2 3Cpro was not available. When whole cell lysates were incubated with 0.3 μg of 3Cpro for up to 8 h, no change in the levels of Nup62 was observed (Fig. 2C, lanes 2–4). To confirm that the 3Cpro used in these assays was active, we assayed for cleavage of the p65 subunit of NF-κB, a known target of 3Cpro (31). Levels of NF-κB/p65 were reduced by more than 40% within 2 h of 3Cpro addition and by nearly 80% following a 4-h incubation, indicating that the 3Cpro was active (Fig. 2C, lanes 2 and 3). Interestingly, when Nup62 was exposed to high concentrations of 3Cpro for extended periods, partial proteolysis of Nup62 was evident (Fig. 2C, lane 5). Although we cannot rule out the possibility that 3Cpro contributes to cleavage of Nup62 in vivo, Ghildyal et al. (32) did not observe proteolysis of Nup62 in cells overexpressing HRV 16 3Cpro or 3CDpro, making is seem likely that the partial cleavage shown here is due to the high concentration of 3Cpro used in these assays. Regardless, because much lower amounts of 2Apro and shorter incubation times induced essentially complete degradation of Nup62 (Fig. 2A), it seems likely that 2Apro is the major protease responsible for cleavage of Nup62 in infected cells.
In Vitro Translated Nup62 Is Cleaved by 2Apro
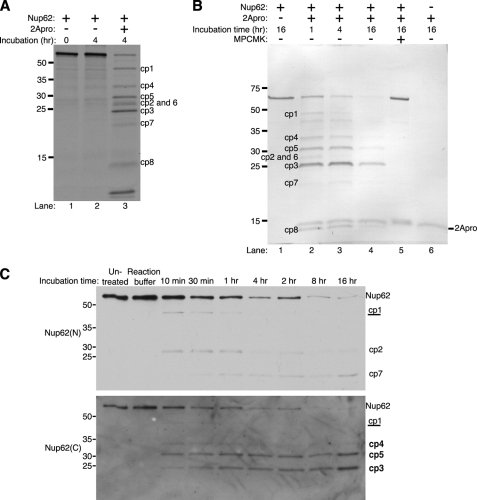
The cleavage assay with HeLa whole cell lysates implied that HRV2 2Apro induced the proteolysis of Nup62. However, it was possible that other host cellular factors contributed to the degradation of Nup62 in these assays. To determine whether 2Apro could cause the degradation of Nup62 in the absence of HeLa cell factors, we examined the ability of HRV2 2Apro to cleave Nup62 that had been translated and radiolabeled in rabbit reticulocyte lysates. Additionally, the use of a radiolabeled Nup62 substrate should allow the detection of additional cleavage products that would have been missed if they lacked epitopes reactive with the antibodies used in Fig. 1B. Indeed, autoradiography revealed that 2Apro could cleave Nup62 under these conditions and that this resulted in the appearance of several additional cleavage products (Fig. 3A, lane 3). Comparison of the molecular weights of these cleavage products revealed that several could correspond to those detected by immunoblot in lysates from virus-infected cells (compare Figs. 1B and 3A, cp1, cp2, and cp3). Overnight incubation under these conditions showed almost complete cleavage of full-length Nup62 and the accumulation of a major cleavage product that corresponded to cp3. It is interesting to note that cp3 was also observed to increase in abundance during the course of infection (Fig. 1B). These findings indicate that 2Apro is able to cleave Nup62 in the absence of HeLa cell factors and are consistent with 2Apro recognizing multiple cleavage sites within Nup62.
FIGURE 3.
2Apro cleaves Nup62 directly. A, cleavage of in vitro translated Nup62 by 2Apro. Radiolabeled Nup62 was generated by in vitro translation in the presence of [35S]methionine. 1 μl of the translation reaction was either prepared for loading immediately (0 h incubation) or incubated for 4 h at 30 °C in the presence or absence of 0.3 μg of purified 2Apro. After incubation, samples were electrophoresed, transferred to PVDF membrane, and analyzed by autoradiography. B, 2Apro can cleave purified Nup62. Nup62 was expressed and purified from bacteria and then incubated with or without 0.3 μg of purified 2Apro for the indicated times at 30 °C. In some cases, 100 μm N-(methoxysuccinyl)-Ala-Ala-Pro-Val-chloromethyl ketone (MPCMK) was included to inhibit 2Apro activity. After incubation, samples were electrophoresed, transferred to PVDF membrane, and stained with Coomassie Brilliant Blue. C, characterization of in vitro Nup62 cleavage products. Nup62 was purified from bacteria and incubated in the presence or absence of 0.3 μg of purified HRV2 2Apro for the indicated times at 30 °C. After incubation, immunoblotting was performed with Nup62(N), followed by striping and reprobing with Nup62(C) to detect N- and C-terminal cleavage products, respectively. Untreated, 16-h incubation of purified Nup62 in PBS. Reaction buffer, 16-h incubation of purified Nup62 in 2Apro reaction buffer without protease. Cleavage products reactive with Nup62(N) are indicated by normal type, those reactive with Nup62(C) are indicated in boldface type, and those reactive with both antibodies are underlined. Molecular mass markers are indicated in kilodaltons.
2Apro Cleaves Nup62 Directly
Although the previous results indicated that 2Apro could induce the cleavage of Nup62 in whole cell lysates or in rabbit reticulocyte lysates, it was not possible to determine if this was direct or due to activation of latent cellular proteases. To determine if 2Apro could directly cleave Nup62, we expressed and purified from bacteria full-length human Nup62 with a histidine tag at its N terminus. Incubation of purified HRV2 2Apro with purified Nup62 for 1 or 4 h revealed a cleavage pattern that was very similar to that seen with in vitro translated Nup62 (Fig. 3, compare B (lanes 2 and 3) with A (lane 3)). Incubation for 16 h resulted in almost complete cleavage of full-length Nup62 and a predominant band that corresponded to cp3 (Fig. 3B, lane 4), very similar if not identical to what was seen with the in vitro translated Nup62 and again perhaps mirroring the accumulation of cp3 seen in infected cells. Cleavage in this assay was mediated by 2Apro because no degradation was seen in the absence of protease, and cleavage could be completely prevented by the addition of N-(methoxysuccinyl)-Ala-Ala-Pro-Val-chloromethyl ketone, a known inhibitor of 2Apro (Fig. 3B, lanes 1 and 5, respectively) (33). These results indicate that 2Apro is capable of directly cleaving Nup62 at multiple sites.
To determine if the cleavage products seen with bacterially expressed Nup62 corresponded to those seen with endogenous Nup62 in lysates from virus-infected cells (Fig. 1B), reaction products were probed with the anti-Nup62 antibodies used in Fig. 1 that react with the N and C terminus (Nup62(N) and Nup62(C), respectively). As was the case in infected cell lysates, Nup62(N) detected both cp1 and cp2, whereas Nup62(C) detected cp1 and cp3. Also, as seen in infected cell lysates, levels of cp1 and cp2 gradually declined over the incubation period, whereas levels of cp3 increased, suggesting that there may be a precursor/product relationship between these cleavage products. In addition, Nup62(N) detected one additional cleavage product (cp7), whereas Nup62(C) detected two additional cleavage products (cp4 and cp5) that were not visible in infected cell lysates (compare Figs. 1B and 3C). Currently, it is not clear if these represent bona fide cleavage products that are rapidly degraded by 2Apro or other viral or cellular factors in infected cells. Because a very similar cleavage pattern was seen with in vitro translated Nup62, it seems unlikely that these cleavage products arise due simply to inappropriate folding of Nup62 when expressed and purified from bacteria. Cumulatively, these results indicate that bacterially expressed Nup62 is probably cleaved at sites that correspond to those cleaved by 2Apro in infected cells.
Identification of 2Apro Cleavage Sites in Nup62
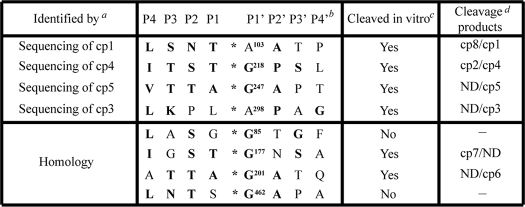
Protease cleavage sites are typically designated using the format, Pn … P2-P1-P1′-P2′ … Pn′, where the scissile bond is located between P1 and P1′ (34). Analysis of 2Apro cleavage sites reveals a strong preference for Gly at P1′; Thr, Ser, or Asn at P2; and a hydrophobic amino acid, such as Ile or Leu, at P4 (35–37). To identify potential 2Apro cleavage sites in Nup62, a combination of N-terminal sequencing of cleavage products and sequence analysis to identify sequence motifs in Nup62 with reasonable homology to known 2Apro cleavage sites was undertaken. N-terminal sequencing of cleavage products corresponding to cp1, -3, -4, and -5 identified four potential cleavage sites (Table 1). The amino acid sequence surrounding the cleavage sites at Gly218 and Gly247 conforms well to the consensus 2Apro cleavage site. Surprisingly, the other two cleavage sites identified by N-terminal sequencing contained Ala instead of Gly at the P1′-position (Table 1, A103 and A298). Based on the molecular weight of observed cleavage products, we identified four other potential cleavage sites in Nup62 with good homology to the consensus 2Apro recognition sequence (Table 1).
TABLE 1.
Analysis of 2Apro cleavage sites in Nup62
a Cleavage sites in Nup62 were identified by N-terminal sequencing or by homology with known 2Apro cleavage sites.
b Nomenclature is as previously described (32). Boldface amino acids indicate residues found at that position in known 2Apro cleavage sites. *, cleavage site.
c This column indicates whether cleavage was inhibited by mutation of P1′ to Lys. See Fig. 4A.
d Cleavage products that arise due to cleavage at the indicated sites. ND, none detected.
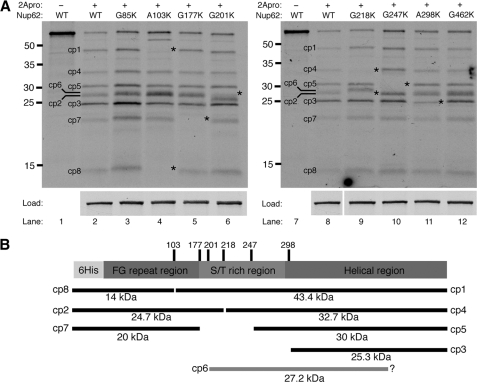
To determine whether these represent authentic cleavage sites, mutations were introduced into the Nup62 open reading frame that were predicted to disrupt cleavage by 2Apro at these sites. Previous work has shown that substitution of Gly with Lys at P1′ prevents cleavage by 2Apro (35, 38). Consequently, we replaced Gly or Ala at this position with Lys and compared the cleavage of these mutants with wild-type Nup62. Mutation of all four 2Apro cleavage sites identified by N-terminal sequencing resulted in an altered cleavage pattern compared with wild-type Nup62, confirming that these are recognized and cleaved by 2Apro in vitro (Fig. 4A and Table 1). As expected, mutation of Ala103 to Lys resulted in the disappearance of cp1 (Fig. 4A, compare lanes 2 and 4). In addition, this mutation also resulted in the loss of cp8, indicating that cleavage at Ala103 gives rise to both cp1 and cp8. Mutation of the cleavage site sequence obtained by sequencing cp4 (G218K) resulted in the loss of cp4 as expected and identified cp2 as the N-terminal product released by cleavage at this position (Fig. 4A, compare lanes 8 and 9). Mutation of the cleavage sites identified in cp5 and cp3 (G247K or A298K, respectively) resulted in a dramatic decrease in the intensity of these cleavage products but did not identify any additional N-terminal products that arise due to cleavage at these positions (Fig. 4A, compare lanes 10 and 11 with lane 8). Cumulatively, these results demonstrate that 2Apro cleaves Nup62 at amino acids 103, 218, 247, and 298 in vitro.
FIGURE 4.
Identification of 2Apro cleavage sites in Nup62. A, analysis of 2Apro cleavage sites identified by N-terminal sequencing or sequence homology. In vitro translated radiolabeled WT or mutant Nup62s were incubated with 0.3 μg of purified 2Apro for 4 h at 30 °C. Constructs are designated by the P1′ amino acid and its location in the Nup62 primary sequence followed by the mutant amino acid inserted at that position. Cleavage products that change in abundance due to a mutation are indicated with an asterisk. Load, the amount of full-length protein used in each cleavage assay. Molecular mass markers are indicated in kilodaltons. B, schematic representation of Nup62 showing the location of cleavage sites and corresponding cleavage products. The amino acid position of identified cleavage sites are indicated along with corresponding cleavage products and their predicted molecular mass and cp designation (as shown in Fig. 3A). Cleavage products that arise due to processing at unidentified cleavage sites are indicated by a gray bar.
A similar analysis of the four cleavage sites identified based on their homology to known 2Apro cleavage sites was undertaken. One potential cleavage site was identified at Gly177 that was predicted to give rise to cp7. Indeed, mutation of Gly177 to Lys caused a decrease in the amount of cp7 (Fig. 4A, compare lanes 2 and 5). Similarly, mutation of Gly201 to Lys abrogated the production of cp6 (Fig. 4A, compare lanes 2 and 6). Surprisingly, although the amino acid sequence surrounding both Gly85 and Gly462 conformed well to the consensus 2Apro cleavage sequence, mutation of either of these glycine residues did not result in any alteration in cleavage pattern compared with wild-type Nup62 (Fig. 4A, compare lanes 3 and 12 with lanes 2 and 8). Nevertheless, these results indicate that cleavage at amino acids Gly177 and Gly201 is responsible for production of cp7 and cp6, respectively, and demonstrate that the in vitro cleavage of Nup62 by 2Apro is specific for a subset of the potential 2Apro cleavage sites. Table 1 summarizes the results of the analysis of 2Apro cleavage sites in Nup62 and indicates the cleavage products that are produced following proteolysis at each site. In addition, Fig. 4B provides a schematic representation of this information, showing the location of identified 2Apro cleavage sites within Nup62 and the corresponding products that arise when proteolysis occurs at these positions.
Nup62 Is Cleaved at Ala103 in Infected Cells
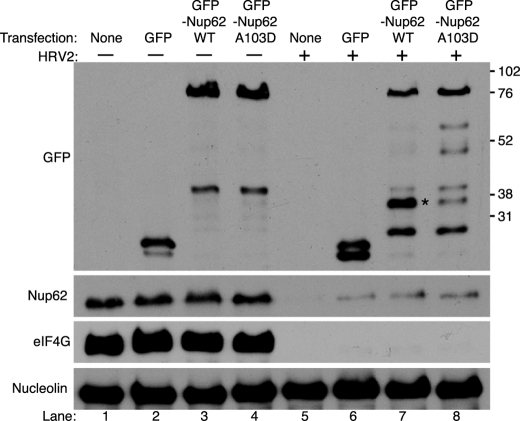
Cp1 is one of the earliest detectable cleavage products in vivo (Fig. 1B), and in vitro analysis and mutagenesis indicated that both cp1 and cp8 derived from cleavage of Nup62 at Ala103 (Fig. 4B). To confirm that Ala103 represents a bona fide cleavage site in vivo, proteolysis of GFP-tagged wild type or mutant Nup62 containing Asp at position 103 was examined. Substrates containing Asp at the P1′-position are not cleaved efficiently by 2Apro in vitro (35). Following transfection, immunoblot analysis using antibodies to GFP revealed no differences in uninfected cells between the wild type and A103D forms of Nup62 (Fig. 5, lanes 3 and 4). HRV2 infection resulted in the partial proteolysis of wild type GFP-Nup62 and the appearance of a prominent cleavage product of ∼37 kDa (compare lanes 3 and 7) that reacted with anti-GFP antibodies. The predicted molecular weight of this proteolytic product corresponds to that of GFP (∼27 kDa) and cp8 (∼10 kDa) and suggests that it was produced by cleavage at Ala103. Consistent with this interpretation, mutation of Ala103 to Asp significantly reduced the amount of the 37-kDa cleavage product and instead resulted in the appearance of slower migrating products that probably result from cleavage of Nup62 at sites C-terminal to Ala103, probably at positions Gly177, Gly201, Gly218, Gly247, or Ala298 (compare lanes 7 and 8). Interestingly, these same cleavage products were present, although much less abundant, in infected cells expressing wild type GFP-Nup62. These results confirm that Ala103 is recognized by HRV2 2Apro in infected cells.
FIGURE 5.
Nup62 is cleaved at Ala103 in HRV2-infected cells. Plasmids encoding GFP or GFP fused to wild type or mutant Nup62 (GFP-Nup62Wt or GFP-NUP62A103D, respectively) were transfected into HeLa cells, and 24 h later were infected with HRV2. Whole cell lysates prepared 18 h later were electrophoresed, transferred to PVDF membrane, and analyzed by immunoblotting with antibodies recognizing GFP, Nup62(N), eIF4GI, or nucleolin. Molecular mass markers are indicated in kilodaltons. None, cells were treated with transfection reagent without plasmid. Cleavage product arising due to proteolysis at Ala103 is indicated with an asterisk.
The FG-rich Region of Nup62 Is Released from the NPC of Infected Cells
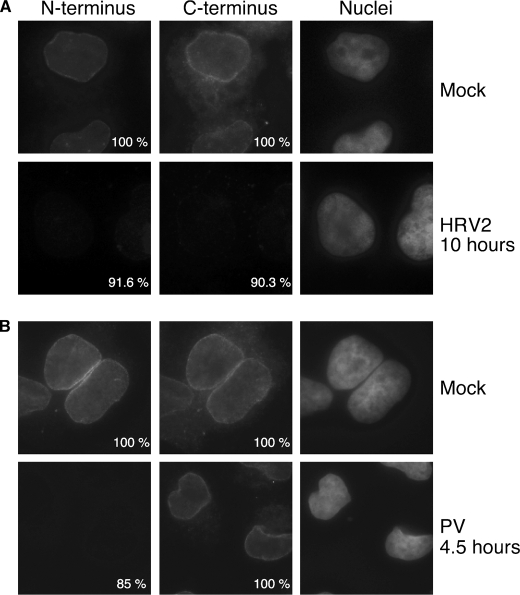
Previous work has shown that poliovirus and rhinovirus infection alters NPC composition and function (19, 20, 22). The identification of 2Apro cleavage sites between amino acids 103 and 298 in Nup62 raised the possibility that cleavage might result in the release of the FG-rich region of Nup62 from the NPC (Fig. 4B). To investigate this possibility and to examine the biological consequences of Nup62 cleavage, we stained infected cells with antibodies that react with the N terminus (mAb414) or C terminus (Nup62(C)) of Nup62. Although the epitope in Nup62 recognized by mAb414 has not been well defined, it is thought to react with the FG-rich region located in the N terminus of Nup62 as well as several other NPC proteins (39). The region containing the epitope recognized by Nup62(C) is indicated in Fig. 1A. In mock-infected cells, mAb414 and Nup62(C) stained the nuclear rim in a punctate pattern typical of nuclear pore proteins (Fig. 6A). Following HRV2 infection, however, the staining with antibodies against either the N terminus or C terminus of Nup62 was drastically reduced in more than 90% of infected cells (Fig. 6A). These data suggest that proteolysis of Nup62 in HRV2-infected cells results in the release of both N- and C-terminal domains of Nup62 from the NPC. In addition, the reduced staining with mAb414 confirms the loss of the FG-rich regions of Nup153, Nup214, and Nup358 from the NPC of rhinovirus-infected cells.
FIGURE 6.
Nup62 association with the NPC in infected cells. A, HRV2-infected cells. Indirect immunofluorescence was performed with mAb414 (N terminus) and Nup62(C) (C terminus) on cells that had been infected with HRV2 for the indicated amount of time. Seven different fields were analyzed, and the percentage of cells showing the indicated pattern of N- and C-terminal staining is indicated. B, poliovirus-infected cells. Cells infected with poliovirus were analyzed as described above. mAb414 and Nup62(C) were detected with FITC and TRITC filters, respectively. Hoechst-stained DNA (Nuclei) was visualized with a UV filter. All images were acquired with identical exposure times and adjustments.
To determine if infection with other picornavirus also induces the release of Nup62 from the NPC, we examined poliovirus-infected cells in a similar fashion. As was the case following HRV2 infection, 85% of poliovirus-infected cells showed very little staining with mAb414 (Fig. 6B), suggesting that the FG-rich regions of Nup62 and other FG-containing Nups are removed from the NPC. In contrast to what was seen in cells infected with HRV, however, the number of poliovirus-infected cells staining with the C-terminal antibody was indistinguishable from mock-infected controls (Fig. 6B), indicating that this portion of Nup62 is retained at the NPC in poliovirus-infected cells. Thus, it appears that although PV and HRV may cleave different sites in Nup62, all of these viruses bring about the removal from the NPC of the FG repeats contained in the N terminus of Nup62 and that of other NPC proteins.
DISCUSSION
This study was designed to examine the mechanisms responsible for cleavage of Nup62 in rhinovirus-infected cells. Our finding that Nup62 is cleaved in cells infected with HRV2, along with prior work showing cleavage in HRV14-infected cells (19), suggests that Nup62 is a proteolytic target in all rhinovirus-infected cells. Our results clearly indicate that HRV2 2Apro directly cleaves Nup62 at multiple sites. This, along with previous work showing that HRV2 2Apro cleaves Nup98 (21) and that poliovirus 2Apro can cause alterations in nuclear envelope permeability (22) make it seem likely that 2Apro is a major contributor to the alterations in the NPC that occur in poliovirus and rhinovirus-infected cells.
The addition of 2Apro to uninfected cell lysates was sufficient to induce the degradation of Nup62. Subsequent analysis of Nup62 produced in rabbit reticulocyte lysate or bacteria indicated that 2Apro could proteolyze Nup62 directly and that this did not require other HeLa cell factors. These analyses also revealed the presence of multiple cleavage products, indicating that 2Apro could cleave Nup62 at multiple sites. Interestingly, Coomassie staining and immunoblot analysis of cleavage assays using purified bacterial Nup62 revealed more cleavage products than were detected in infected cell lysates. As the Nup62 used in these assays was purified under denaturing conditions and subsequently refolded, it is certainly possible that some of these cleavage products arise due to improper folding and inappropriate exposure of 2Apro cleavage sites. However, several observations suggest that this is not the case. First, the observation that several of the cleavage products observed in the in vitro assay appeared to comigrate with cp1, -2, and -3 that were identified in infected cell lysates suggests that these assays faithfully reproduced at least some of the cleavage events that occur in vivo. Second, the finding that good consensus 2Apro cleavage sites at Gly85 or Gly462 were not proteolyzed by 2Apro indicated that not all possible sites are recognized and that there was some selectivity in the choice of proteolytic sites by 2Apro. Finally, a very similar cleavage pattern was detected when Nup62 derived from rabbit reticulocyte lysates was assayed, suggesting that the “folding” of Nup62 and recognition by 2Apro in these diverse systems is very similar. The inability to detect these additional cleavage products in infected cells may be due to their being less stable in infected cells, perhaps due to subsequent targeting by viral or cellular encoded proteases. In addition, the in vitro analysis was not limited to detection of only those bands that were reactive with available antibodies. Thus it seems likely that the cleavage products identified in vitro may also be produced in infected cells.
Our in vitro analysis of Nup62 cleavage has identified eight distinct cleavage products (cp1 to -8) and six 2Apro cleavage sites clustered between amino acids 103 and 298 (Fig. 4B). Significantly, cleavage at these positions can explain the appearance of cp1, cp2, and cp3 seen in infected cell lysates (Fig. 1B). For example, when Ala103 is mutated to Lys, levels of cp1 along with cp8 were diminished. Similarly, mutation of Gly218 resulted in a decrease in both cp2 and cp4. Interestingly, although cp1 and cp2 were the first cleavage products detected in both infected cells and in time courses with bacterially expressed Nup62, levels of these cleavage products declined at later time points, suggesting that they were further proteolyzed. This was in contrast to cp3, whose levels appeared to increase as those of cp1 and cp2 declined. The identification of a 2Apro cleavage site at Ala298 that is responsible for production of cp3 in vitro indicates that cp3 could arise due to subsequent proteolysis of cp1 or other Nup62 fragments encompassing amino acids 298–522. Similarly, the identification of 2Apro cleavage sites at Gly177 and Gly201 could result in further proteolysis of cp1 and cp2 and contribute to the decrease in levels of these cleavage products. Cleavage at Gly247 appears to give rise to cp5, a cleavage product that, like cp3, increased at later times in the in vitro cleavage assays. Although cp5 was not detected in infected cell lysates, it is possible that cleavage at Gly247 may also contribute to the decrease in cp1 levels seen in infected cells. The finding that cp7 was produced by cleavage at position Gly177 and was reactive with an antibody raised against amino acids 24–178 of Nup62 (Nup62(N); see Fig. 1A) indicated that this fragment extends from Gly177 toward the N terminus of Nup62. Mutation of Gly201 caused a decrease in cp6, a cleavage product that was not detected by either Nup62(N) or Nup62(C) (Fig. 3C). This suggests that cp6 is produced by cleavage at Gly201 and another, as yet unidentified 2Apro cleavage site located in the C terminus of Nup62 (Fig. 4B, cp6).
Comparison of 2Apro cleavage sites in viral and cellular substrates has not resulted in the emergence of a strong consensus sequence for this protease. In general, however, 2Apro cleavage sites exhibit preferences for Gly at P1′; hydrophobic amino acids at P4 and P2′; and Thr, Ser, or Asn at P2 (37, 40). Of the six 2Apro cleavage sites identified in Nup62, most conform rather well to this pattern (Table 1). All contain a hydrophobic amino acid at the P4-position, and all, except for the Gly177 site, have a hydrophobic amino acid at P2′. Skern et al. (40) have proposed that the P2 residue forms a hydrogen bond with Ser83 of the HRV2 2Apro or its equivalent in other HRVs. The only residues that can do this are Ser, Thr, and Asn. Pertinently, only these residues are found at the 2Apro cleavage sites on the HRV polyproteins or in the eIF4G cleavage sites (41, 42). All of the sites identified in Nup62 except one (Ala298) contain Thr (Gly247 and Gly201), Ser (Gly218 and Gly177), or Asn (Ala103) at this position. Pro is found at P2 in the Ala289 site, and previous work has shown that 2Apro can indeed tolerate Pro at this position, albeit resulting in reduced cleavage efficiency (35). Less efficient recognition of this site may explain the delayed appearance of cp3 in infected cells and in the in vitro cleavage assays.
Although four of the cleavage sites contained the expected Gly at P1′, two sites were found to contain Ala at this position (Ala103 and Ala298; Table 1). This was unexpected, given that no 2Apro cleavage sites have been identified to date with anything other than Gly at P1′. The amino acid sequences around Ala103 and Ala298 conform to reasonable 2Apro consensus sequences with hydrophobic residues at P4 and P2′. The structure of the HRV2 2Apro active site indicates that bulky amino acids are unlikely to be accepted at P1′ due to the proximity of the imidazole ring on His18 in 2Apro (43). However, the small size of Ala might allow it to be tolerated in place of Gly at the P1′-position of an HRV2 2Apro substrate. In support of this possibility, Hellen et al. (36) showed that trans substrates with Ala at P1′ were still cleaved by poliovirus 2Apro with reasonable efficiency. Our finding that mutation of either of these residues to Lys resulted in distinct changes to the processing pattern indicates that these sites are recognized by HRV2 2Apro in vitro. Furthermore, mutation of Ala103 to Asp prevented cleavage at this site in vivo, confirming that 2Apro can utilize Ala in the P1′-position.
Both poliovirus and rhinovirus infection resulted in a loss of staining at the NPC with antibodies reactive with the N-terminal FG-rich region of Nup62. In contrast, staining with a C-terminal antibody was reduced in RV-infected cells but not in cells infected with poliovirus. These results indicate that cleavage by HRV 2Apro induces the release of the entire Nup62 protein from the NPC, whereas cleavage with PV 2Apro appears to selectively remove the N terminus of Nup62. In addition, these results indicate that FG repeat domains found in Nup153, Nup214, and Nup358 are also released from the NPC of infected cells. The FG repeats found in Nup62, Nup98, Nup153, and several other NPC proteins are thought to be natively unfolded and to provide docking sites for cargo-transport receptor complexes in transit through the NPC while at the same time serving as barriers against passive diffusion (3, 4, 44, 45). Patel et al. (4) found that removal of the FG repeats from the yeast homologues of Nup98 and Nup153 resulted in an increase in the permeability barrier of the NPC. Thus, the loss of Nup62, Nup153, and Nup98 and perhaps other FG-containing Nups could explain the increased permeability of the nuclear envelope that has been reported in infected cells (22).
Despite the specific cleavage of Nups in infected cells, NPCs remain functional and can support certain trafficking pathways (20, 21). This is perhaps not surprising, given that experiments in Saccharomyces cerevisiae and Xenopous laevis have shown that NPCs can still function even when a significant mass of the FG repeats has been removed (46–48). Further experiments will be necessary to determine how extensively the NPC is modified in infected cells and what the consequences of these modifications are on NPC architecture and function.
Acknowledgments
We thank Carla Sousa for purified HRV2 2Apro, Susan Wente for Nup155 antibody, Nabeel Yaseen for the Nup62 cDNA, and members of the Gustin laboratory for help throughout the course of this study.
This work was supported, in whole or in part, by National Institutes of Health Grants AI059467 and AI064432 and by National Institutes of Health/NCRR COBRE Grant P20 RR15587 and INBRE Grant P20 RR016454 (to K. E. G.). This work was also supported by Austrian Science Fund Grant P20889 (to T. S.) and by American Cancer Society Grant RSG 109705 (to K. E. G.).
K. E. Gustin, unpublished results.
- NPC
- nuclear pore complex
- HRV
- human rhinovirus
- PV
- poliovirus
- eIF4GI
- eukaryotic translation initiation factor 4G1
- cp
- cleavage product(s)
- Nup
- nucleoporin
- TRITC
- tetramethylrhodamine isothiocyanate
- 3Cpro
- rhinovirus 3C protease.
REFERENCES
- 1.Tran E. J., Wente S. R. (2006) Cell 125, 1041–1053 [DOI] [PubMed] [Google Scholar]
- 2.Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. (2002) J. Cell Biol. 158, 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. (2000) J. Cell Biol. 148, 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel S. S., Belmont B. J., Sante J. M., Rexach M. F. (2007) Cell 129, 83–96 [DOI] [PubMed] [Google Scholar]
- 5.Bayliss R., Littlewood T., Stewart M. (2000) Cell 102, 99–108 [DOI] [PubMed] [Google Scholar]
- 6.Bayliss R., Ribbeck K., Akin D., Kent H. M., Feldherr C. M., Görlich D., Stewart M. (1999) J. Mol. Biol. 293, 579–593 [DOI] [PubMed] [Google Scholar]
- 7.Paschal B. M., Gerace L. (1995) J. Cell Biol. 129, 925–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarkson W. D., Kent H. M., Stewart M. (1996) J. Mol. Biol. 263, 517–524 [DOI] [PubMed] [Google Scholar]
- 9.Ribbeck K., Lipowsky G., Kent H. M., Stewart M., Görlich D. (1998) EMBO J. 17, 6587–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lubas W. A., Smith M., Starr C. M., Hanover J. A. (1995) Biochemistry 34, 1686–1694 [DOI] [PubMed] [Google Scholar]
- 11.Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B. T., Rout M. P., Sali A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 13.Percipalle P., Clarkson W. D., Kent H. M., Rhodes D., Stewart M. (1997) J. Mol. Biol. 266, 722–732 [DOI] [PubMed] [Google Scholar]
- 14.Buss F., Stewart M. (1995) J. Cell Biol. 128, 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finlay D. R., Meier E., Bradley P., Horecka J., Forbes D. J. (1991) J. Cell Biol. 114, 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meerovitch K., Svitkin Y. V., Lee H. S., Lejbkowicz F., Kenan D. J., Chan E. K., Agol V. I., Keene J. D., Sonenberg N. (1993) J. Virol. 67, 3798–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride A. E., Schlegel A., Kirkegaard K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2296–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waggoner S., Sarnow P. (1998) J. Virol. 72, 6699–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustin K. E., Sarnow P. (2002) J. Virol. 76, 8787–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustin K. E., Sarnow P. (2001) EMBO J. 20, 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park N., Katikaneni P., Skern T., Gustin K. E. (2008) J. Virol. 82, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belov G. A., Lidsky P. V., Mikitas O. V., Egger D., Lukyanov K. A., Bienz K., Agol V. I. (2004) J. Virol. 78, 10166–10177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castelló A., Izquierdo J. M., Welnowska E., Carrasco L. (2009) J. Cell Sci. 122, 3799–3809 [DOI] [PubMed] [Google Scholar]
- 24.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Rout M. P., Sali A. (2007) Nature 450, 683–694 [DOI] [PubMed] [Google Scholar]
- 25.Panté N., Bastos R., McMorrow I., Burke B., Aebi U. (1994) J. Cell Biol. 126, 603–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson M. O., Guetzow K., Busch H. (1981) Exp. Cell Res. 135, 259–265 [DOI] [PubMed] [Google Scholar]
- 27.Aldabe R., Feduchi E., Novoa I., Carrasco L. (1995) Biochem. Biophys. Res. Commun. 215, 928–936 [DOI] [PubMed] [Google Scholar]
- 28.Liebig H. D., Ziegler E., Yan R., Hartmuth K., Klump H., Kowalski H., Blaas D., Sommergruber W., Frasel L., Lamphear B. (1993) Biochemistry 32, 7581–7588 [DOI] [PubMed] [Google Scholar]
- 29.Skern T., Liebig H. D. (1994) Methods Enzymol. 244, 583–595 [DOI] [PubMed] [Google Scholar]
- 30.Lamphear B. J., Yan R., Yang F., Waters D., Liebig H. D., Klump H., Kuechler E., Skern T., Rhoads R. E. (1993) J. Biol. Chem. 268, 19200–19203 [PubMed] [Google Scholar]
- 31.Neznanov N., Chumakov K. M., Neznanova L., Almasan A., Banerjee A. K., Gudkov A. V. (2005) J. Biol. Chem. 280, 24153–24158 [DOI] [PubMed] [Google Scholar]
- 32.Ghildyal R., Jordan B., Li D., Dagher H., Bardin P. G., Gern J. E., Jans D. A. (2009) J. Virol. 83, 7349–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molla A., Hellen C. U., Wimmer E. (1993) J. Virol. 67, 4688–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schechter I., Berger A. (1967) Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 35.Sommergruber W., Ahorn H., Zöphel A., Maurer-Fogy I., Fessl F., Schnorrenberg G., Liebig H. D., Blaas D., Kuechler E., Skern T. (1992) J. Biol. Chem. 267, 22639–22644 [PubMed] [Google Scholar]
- 36.Hellen C. U., Lee C. K., Wimmer E. (1992) J. Virol. 66, 3330–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blom N., Hansen J., Blaas D., Brunak S. (1996) Protein Sci. 5, 2203–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gradi A., Foeger N., Strong R., Svitkin Y. V., Sonenberg N., Skern T., Belsham G. J. (2004) J. Virol. 78, 3271–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis L. I., Blobel G. (1986) Cell 45, 699–709 [DOI] [PubMed] [Google Scholar]
- 40.Skern T., Hampolz B., Guarne A., Fita I., Bergman E., Petersen J., James M. N. (2002) in Molecular Biology of Picornaviruses (Semler B. L., Wimmer E. eds) pp. 199–212, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
- 41.Gradi A., Svitkin Y. V., Sommergruber W., Imataka H., Morino S., Skern T., Sonenberg N. (2003) J. Virol. 77, 5026–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sousa C., Schmid E. M., Skern T. (2006) FEBS Lett. 580, 5713–5717 [DOI] [PubMed] [Google Scholar]
- 43.Petersen J. F., Cherney M. M., Liebig H. D., Skern T., Kuechler E., James M. N. (1999) EMBO J. 18, 5463–5475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rexach M., Blobel G. (1995) Cell 83, 683–692 [DOI] [PubMed] [Google Scholar]
- 46.Walther T. C., Pickersgill H. S., Cordes V. C., Goldberg M. W., Allen T. D., Mattaj I. W., Fornerod M. (2002) J. Cell Biol. 158, 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R. (2004) Nat. Cell Biol. 6, 197–206 [DOI] [PubMed] [Google Scholar]
- 48.Zeitler B., Weis K. (2004) J. Cell Biol. 167, 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]