Abstract
In the vertebrate central nervous system, maturation of oligodendrocytes is accompanied by a dramatic transformation of cell morphology. Juxtanodin (JN) is an actin cytoskeleton-related oligodendroglial protein that promotes arborization of cultured oligodendrocytes. We performed in vitro and in culture experiments to further elucidate the biochemical effects, molecular interactions, and activity regulation of JN. Pulldown and co-sedimentation assays confirmed JN binding to filamentous but not globular β-actin largely through a C-terminal domain of 14 amino acid residues. JN had much lower affinity to F-α-actin than to F-β-actin. Bundling and actin polymerization assays revealed no JN influence on F-β-actin cross-linking or G-β-actin polymerization. Sedimentation assay, however, demonstrated that JN slowed the rate of F-β-actin disassembly induced by dilution with F-actin depolymerization buffer. JN-S278E mutant, a mimic of phosphorylated JN at serine 278, exhibited a much diminished affinity/stabilizing effect on F-β-actin. Immunoblotting revealed both phosphorylated and dephosphorylated native JN of the brain, with the former migrating slightly slower than the latter and becoming undetectable when brain lysate was subjected to in vitro dephosphorylation prior to being loaded for electrophoresis. In cultured OLN-93 cells, overexpression of JN promoted the formation of actin fibers and inhibited F-actin disassembly induced by latrunculin A. S278E phosphomimetic mutation resulted in loss of JN activity in cultured cells, whereas S278A, T258A, and T258E dephospho-/phosphomimetic mutations did not. These findings establish JN as an actin cytoskeleton-stabilizing protein that may play active roles in oligodendroglial differentiation and myelin formation. Specific phosphorylation of JN might serve as an important mechanism regulating JN functions.
Keywords: Actin, Cytoskeleton, Molecular Dynamics, Protein Phosphorylation, Protein-Protein Interactions, Central Nervous System, ERM Proteins, Juxtanodin, Myelin, Oligodendrocyte
Introduction
During the development of the central nervous system, oligodendrocytes (OLs)3 elaborate highly branched processes that target and wrap around neuronal axons to form myelin sheaths that electrically insulate axons and enable rapid saltatory conduction of action potentials (1–3). The functional roles of OLs are critically dependent on the establishment of their arborized morphology, which in turn is supported by cytoskeletal microtubules and microfilaments but not intermediate filaments (4). Recent studies have provided important insights into actin dynamics during oligodendrocyte maturation. Microfilaments are found to guide the local reorganization of microtubules for the elongation of oligodendrocyte processes and formation of new branches in culture (5, 6). Microfilaments are also suggested to play an active role in the expression of myelin-specific proteins such as 2′,3′-cyclic nucleotide phosphodiesterase, myelin-associated glycoprotein, and P0 during myelination (7). A large number of actin-binding proteins such as Arp2/3 (actin-related proteins), WASP (Wiskott-Aldrich syndrome protein), WAVE (Wiskott-Aldrich syndrome protein family verprolin homologous protein), and vinculin are present in OLs, and participate in OL maturation and myelination by spatiotemporal regulation of actin reorganization (8). The importance of these molecules can be glimpsed from, for example, knock-out mice of WAVE1, a protein promoting actin polymerization through its ability to activate the Arp2/3 complex, that exhibit regional hypomyelination in the central nervous system. Oligodendrocyte precursor cells from WAVE1-null mice have fewer processes and are defective in lamella formation in culture (9).
Juxtanodin (JN) was first identified as an actin cytoskeleton-associated oligodendroglial protein by screening cell type-specific genes of the central nervous system (10). It has similarity in amino acid sequence with ERM (ezrin, radixin, and moesin) family proteins. Although it lacks the N-terminal FERM (4.1, ezrin, radixin, moesin) domain, which is conserved in ERM proteins and has a weak similarity along the central helical coiled-coil region, JN shares 34 almost identical amino acid residues at the C terminus with the putative F-actin-binding site of the ERM proteins (11). Two known phosphorylation sites of ERM proteins are also conserved in JN, including the well characterized threonine residue (threonine 557 in mouse moesin) and a serine site (serine 575 in mouse moesin) (12, 13). Previous studies in our laboratory have shown that JN expression parallels temporally and spatially the onset of myelination in vivo. Furthermore, overexpression of JN promotes arborization of cultured OLN-93 cells and primary oligodendrocytes (10). To further clarify the molecular interaction, possible biochemical effects, and mechanisms of activity regulation of JN, we carried out a series of in vitro and in culture experiments. It is found that JN binds F-actin, stabilizes F-actin in vitro and in vivo, and the activity of JN on F-actin is dependent on JN dephosphorylation at serine 278.
EXPERIMENTAL PROCEDURES
Fusion Proteins
JN268 and JN170, which lacked the C-terminal 14-amino acid (aa) or 112-aa residues of JN, and the full-length Juxtanodin were subcloned in PET41a(+) vector (Novagen, Germany). The glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli strain BL21(DE3) cells and purified according to standard protocols using glutathione-Sepharose 4B (Amersham Biosciences). Polyhistidine fusion proteins of JN and its mutants were also created. JN cDNA tagged with a coding sequence of 6 histidines at the N terminus was constructed by polymerase chain reaction (PCR) using the following primers: 5′-gatcatatgcatcaccatcaccatcacacagatactcct-3′ and 5′-gctctcgagttataagtgcatcattgac-3′, JN with the N-terminal polyhistidine tag was cloned at PET41a(+) between the NdeI and XhoI sites. The proteins were expressed in BL21(DE3) cells and purified according to standard protocols using nickel-nitrilotriacetic acid Superflow cartridges (Qiagen, Germany). All the resulting constructs were fully sequenced to ensure the correct reading frame before induction of expression.
The affinity purified proteins were further purified by gel filtration chromatography using Sephadex G-75 (GE Healthcare) and buffer-exchanged to G or F buffer (see below) for further studies using 10-kDa cut-off Amicon ultracentrifugal filters (Millipore). Prior to experiments, protein solutions were pre-cleared at 160,000 × g, sized by SDS-PAGE, and their concentrations were determined by Bradford assay (Pierce) using bovine serum albumin as a standard.
For transfection and overexpression in cultured mammalian cells, constructs of JN, JNc (encoding aa residues 101–282 of JN), and JN141 (encoding aa residues 1–141 of JN) were inserted into the mammalian cell expression vector pXJ40 in-frame with an FLAG tag-coding sequence at the 5′-end as described previously (10).
Site-directed Mutagenesis
Plasmids pXJ40-JN or PET41a-His6JN containing the insert of full-length cDNA of JN were used as templates to create a series of single point mutations (T258A, T258E, S278A, and S278E) using the GeneTailorTM Site-directed Mutagenesis System according to instructions provided by the manufacturer (Invitrogen).
GST Pull-down Assay and Mass Spectrometry
Approximately 1.5 g of brain tissue from 21- or 30-day old Wistar rats was homogenized in 15 ml of T-PER Tissue Protein Extraction buffer (Pierce) using a tissue homogenizer. The homogenate was centrifuged for 20 min at 15,000 × g at 4 °C. The supernatant was collected and further centrifuged for another 30 min at 15,000 × g at 4 °C. The brain lysate was pre-cleared by incubation with glutathione-Sepharose 4B (GSH) beads for 4 h at 4 °C with gentle rocking. Three aliquots of pre-cleared brain lysate were incubated with 50 μl of GST-JN-conjugated GSH beads, GST-conjugated GSH beads, and GSH beads alone, respectively, for 4 h at 4 °C with gentle rocking. The beads were collected by centrifugation at 500 × g at 4 °C, followed by six washes with ice-cold phosphate-buffered saline (PBS) buffer or T-PER Tissue Protein Extraction buffer (Pierce). The beads were resuspended in 100 μl of 1× SDS-PAGE sample loading buffer and heated for 5 min at 95 °C. Samples were resolved by SDS-PAGE and visualized by Coomassie Blue staining. The desired protein bands were excised from the polyacrylamide gel, reduced, alkylated, and in-gel digested with trypsin. The extracted peptides were analyzed on a matrix-assisted laser desorption/ionization mass spectrometer equipped with a time-of-flight analyzer (MALDI-TOF, Voyager STR Biospectrometry work station, Applied System). The obtained mass fingerprints were used to perform a MS-Fit search against NCBI databases to identify the proteins.
G-actin Pull-down Assay
G-actin pull-down assays were performed as described by James and co-workers (14), with minor modifications. Briefly, equal amounts of GST control or fusion proteins (GST, GST-JN, and GST-JN170) were immobilized on glutathione-Sepharose beads, respectively, at a concentration of ∼1 mg/ml of packed beads. Glutathione beads conjugated with or without GST fusion proteins were washed three times in 400 μl of G buffer (5 mm Tris-HCl, pH 8.0, 0.2 mm CaCl2, and 0.2 mm ATP). G-β-actin (0.1 mg/ml) (Cytoskeleton Inc.) in G buffer was precleared at 160,000 × g to remove any polymerized actin. Approximately 40 μl of the beads conjugated with or without GST fusion proteins were incubated with 100 μl of 0.1 mg/ml of G-β-actin in G buffer. After gentle tumbling for 1 h at 4 °C, the beads were washed three times in 400 μl of G buffer. Proteins bound to the beads were eluted with SDS-PAGE sample buffer, resolved on a SDS-PAGE gel, and analyzed by Coomassie Blue staining.
Actin Filament Co-sedimentation Assays
The F-actin binding ability of JN and truncated JN molecules was measured using centrifugation conditions that pellet F-actin. Actin filament co-sedimentation assay was also used to detect the F-actin stabilizing ability of polyhistidine-tagged JN wild type (WT) and JN-S278E mutant. GST or polyhistidine fusion proteins in F buffer (5 mm Tris-HCl, pH 8.0, 0.2 mm CaCl2, 50 mm KCl, 2 mm MgCl2, and 1 mm ATP) were added to ∼18 μm polymerized actin (Cytoskeleton Inc.) in a total volume of 50 μl of F buffer in ultracentrifuge tubes. The samples were incubated at room temperature for 30 min to allow for interaction between the fusion proteins and F-actin. Negative control reactions containing all the components of the assay, except F-actin or GST fusion proteins, were performed in parallel. After incubation, the samples were centrifuged at 160,000 × g for 1.5 h at room temperature in a TLA-100.3 rotor in a Beckman Optima L-90K ultracentrifuge.
We also tested actin binding of FLAG-tagged JN, JNc, JN141, and JN-S278E that were generated by transfection and overexpression in cultured OLN-93 cells. Lysates of the transfected OLN-93 cells were first mixed with actin polymerizing buffer (50 mm KCl, 2 mm MgCl2, and 1 mm ATP, Cytoskeleton Inc.), and then incubated at room temperature for 1 h to facilitate polymerization of endogenous actin of the cells. The F-actin and any associated JN molecules in the cell lysates were then removed by centrifugation at 160,000 × g for 1.5 h. The supernatants were then mixed with ∼18 μm polymerized β-actin (Cytoskeleton Inc.) in a total volume of 50 μl of F buffer, and incubated at room temperature for 1 h. Parallel reactions without addition of polymerized actin served as controls. The reactions were finally centrifuged at 160,000 × g for 1.5 h. The supernatant and pellet were resolved by SDS-PAGE and electrotransferred onto polyvinylidene fluoride (PVDF) membrane. Actin was visualized by Ponceau S staining of the PVDF membrane. JN, JNc, JN141, or JN-S278E were detected by immunoblotting (see below).
F-actin Depolymerization Assay
For actin depolymerization studies, F-actin pellet alone or in the presence of polyhistidine-tagged JN WT or JN-S278E mutant was resuspended at a concentration of 2 μm in G buffer at room temperature. After 1.5 or 3 h, the samples were ultracentrifuged at 160,000 × g for 1.5 h. The pellets were then solubilized in SDS-PAGE sample buffer and analyzed by Coomassie Blue staining.
F-actin Bundling Assay
For F-actin bundling assay, excessive JN was incubated with ∼18 μm polymerized β-actin in a total volume of 50 μl of F buffer for 30 min at room temperature and centrifuged in an Eppendorf centrifuge at 15,000 × g for 15 min to sediment bundled actin filaments. Equal volumes of pellet and supernatant were solubilized in SDS-PAGE sample buffer and analyzed by Coomassie Blue staining.
Measurement of Actin Assembly
The effect of GST-JN and GST on the rate and extent of actin polymerization was determined using the Actin Polymerization Biochem KitTM (Cytoskeleton) according to instructions provided by the manufacturer. Nine μm monomeric β-actin mixed with pyrene-labeled rabbit skeletal muscle actin (Cytoskeleton) at a ratio of 10:1 in general actin buffer (5 mm Tris-HCl, pH 8.0, and 0.2 mm CaCl2) was polymerized alone or in the presence of either GST-JN or GST by adding 1/10 volume of actin polymerization buffer (500 mm KCl, 20 mm MgCl2, and 10 mm ATP). To increase the sensitivity of polymerization assay by reducing the starting background fluorescence, the samples were centrifuged at 160,000 × g for 2 h at 4 °C before induction of polymerization. The increase in the rate of fluorescence was monitored using a SpectraMAX Gemini EM fluorometer (Molecular Devices) with excitation and emission at 365 and 407 nm, respectively, over a period of ∼30 min with sampling intervals of 30 s.
Transfection, Latrunculin A Treatment, and Immunofluorescence
OLN-93 cells were transfected using a microporator as instructed by the manufacturer (Digital Bio, South Korea). To treat OLN-93 with latrunculin A (LatA), we applied a concentrated stock of LatA (Sigma) in prewarmed DMEM to the wells at final concentrations of 0.5 or 0.25 μm. Forty-eight hours after transfection (4 h after LatA treatment), cells were fixed with 3% paraformaldehyde. The fixed cells were preincubated in PBS-T-NGS (1 × PBS, 0.3% Triton X-100, and 6% normal goat serum) before overnight incubation in primary antibody (monoclonal anti-FLAG, Sigma, 1:1000, or polyclonal anti-JN, made in house, 1:100). A secondary antibody conjugated to Alexa Fluor 488 (Invitrogen) was used for detecting the primary antibody. Rhodamine-conjugated phalloidin was used for detecting F-actin in concentrations as instructed by the manufacturer (Cytoskeleton Inc.).
In Vitro Dephosphorylation
The phosphorylation status of native JN of rat brain and overexpressed JN from OLN-93 cells was examined by in vitro dephosphorylation. Brain or transfected cell lysates were mixed with 3 units of calf intestinal alkaline phosphatase (CIAP; Promega) in a 20-μl alkaline phosphatase reaction buffer (50 mm Tris-HCl, pH 9.3, 1 mm MgCl2, 0.1 mm ZnCl2, and 1 mm spermidine). The reaction lasted for 60 min at 37 °C. Controls consisted of reactions with CIAP replaced by H2O. The reaction samples were then subjected to SDS-PAGE and immunoblotting (see below) for detection and differentiation of phosphorylated and dephosphorylated JN molecules.
Native PAGE
Discontinuous native PAGE was performed as previously described (15). Briefly, G-β-actin or lysate of OLN-93 cells overexpressing FLAG-tagged JN was mixed with an equal volume of 2× Tris glycerol sample buffer (2.5 ml of 0.5 m Tris-HCl, pH 6.8, 2 ml of glycerol, 10 μl of 1% bromphenol blue). The samples were loaded onto native PAGE minigel either directly or after being heated to 80 °C, resolved by electrophoresis, and then subjected to immunoblotting.
Immunoblotting
The electrophoresis resolved protein samples were electrotransferred onto PVDF membrane. After washes and an incubation in nonfat milk (Bio-Rad) to block nonspecific binding, the membrane was probed with rabbit anti-JN antibody (1:200), mouse anti-FLAG antibody (Sigma, 1:2000), or mouse anti-β-actin antibody (Sigma, 1:2000). The membrane was then washed and incubated with alkaline phosphatase-conjugated secondary antibody. Immunodetection was performed using CDP-Star chemiluminescence reagent (Roche Applied Science).
Data Analyses
All data were verified by at least two repeats of the experiments. SDS-PAGE gels were scanned with a GS-710 calibrated imaging densitometer (Bio-Rad), and densitometric analyses of immunofluorescence signals of cells were performed using ImageJ software. Cellular F-actin content was calculated by multiplying the mean OD (after subtracting background) of rhodamine-conjugated phalloidin staining with the cellular area. Cellular immunofluorescence results were also analyzed using a laser scanning confocal microscope (Olympus Fluoview FV1000, Olympus, Tokyo, Japan).
RESULTS
F-actin as an Interacting Partner of Juxtanodin
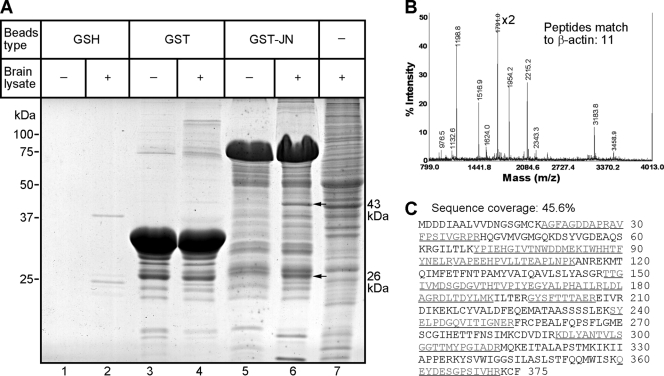
To identify possible proteins binding with JN, a GST pulldown assay was performed. Because the expression of JN in the rat brain as detected by Western blot rapidly increases from postnatal day 14 to 21 and thereafter remains at the relatively high level of expression up to at least postnatal day 60 (10), rat brain lysate of postnatal days 21 or 30 was used for pulling down GST-JN interacting proteins. The brain lysate was pre-cleared by incubation with GSH beads that retained proteins nonspecifically interacting with the matrix, and then incubated with GSH beads conjugated with the GST-JN fusion protein expressed in vitro. Two protein bands specific for the GST-JN pull-down (Lane 6 in Fig. 1A) were excised from the gels and analyzed by using MALDI-TOF MS. The 43-kDa band was identified as actin, probably a candidate interacting partner of JN; the MALDI-TOF MS peptide mass map of the band is shown in Fig. 1B. It revealed 11 matching peptides with 45.6% sequence coverage of actin (Fig. 1C). Another protein band localized around 26 kDa (Fig. 1A) was identified as glutathione S-transferase (data not shown). Because GST has the ability to form dimer, the endogenous GST was most possibly pulled down by the GST tag of the GST-JN. As controls, GST-conjugated beads or GSH beads alone were unable to pulldown brain actin (Fig. 1A, lanes 1–4).
FIGURE 1.
Identification of actin as an interacting partner of JN by GST pulldown assay. A, GST-JN fusion protein coupled to GSH beads (lanes 5 and 6) was used to pull down JN-interacting proteins from postnatal day 30 brain lysate. Bound proteins were separated by SDS-PAGE and stained with Coomassie Blue. The two labeled bands (arrows) specific for the GST-JN pull-down were excised from the gel and analyzed by MALDI-TOF mass spectrometry. As controls, GSH beads alone (lanes 1 and 2) and GST tag protein (lanes 3 and 4) were both unable to pull down rat brain actin to a level above detection. Lane 7 displays the gel electrophoresis pattern of input brain lysate. B, MALDI-TOF peptide mass spectrum identified the major band at 43 kDa as actin that was pulled down by JN. Note that only the lower half of the highest peak at 1791.0 is shown. The upper half is cut off from the graph and indicated with ×2. Peaks indicated with values were matched to actin by peptide mass fingerprinting. C, MALDI-TOF/MS revealed 11 matching peptides with 45.6% sequence coverage (underlined) of actin.
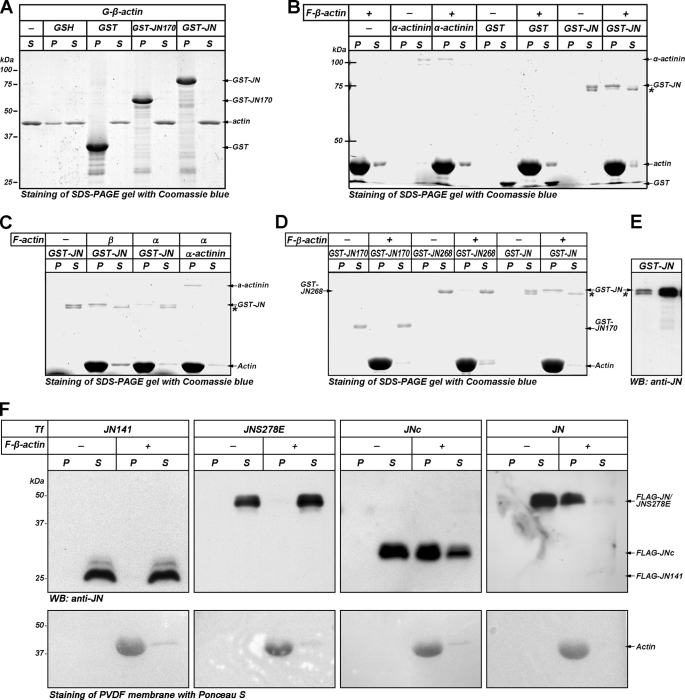
Our previous work indicates that JN might affect actin dynamics (10). G-actin pulldown assays were performed to examine the ability of JN to bind to monomeric G-β-actin in vitro. To ensure its monomeric form, G-β-actin was precleared by ultracentrifugation to remove any polymerized actin before being incubated together with GSH beads or GST-JN fusion protein beads in a buffer lacking KCl and MgCl2. As shown in Fig. 2A, no significant actin was detected on SDS-PAGE in eluted samples from both the experimental (GST-JN) and control (beads alone or GST) groups, suggesting that JN did not bind to G-β-actin. As expected, both GST-tagged JN268 and JN170, lacking of the last 14 and 112 amino acids of JN C terminus, respectively, also failed to capture monomeric β-actin (Fig. 2A and data not shown).
FIGURE 2.
Biochemical characterization of interaction of JN with actin. A, G-actin pulldown assays were performed to examine the ability of JN to bind G-β-actin in vitro. G-β-actin was incubated together with GSH beads alone, GST, GST-JN170, or GST-JN fusion protein bound beads in G-buffer, respectively. Samples eluted from the beads (P) were resolved by SDS-PAGE and stained with Coomassie Blue. Input G-actin (lane 1) and the sample fractions remaining in the supernatants (S) served as controls. B, JN binding to F-β-actin was investigated by F-actin co-sedimentation assay. GST-JN, α-actinin (positive control), or GST (negative control) were incubated in F buffer with (+) or without (−) pre-polymerized F-β-actin. The mixtures were ultracentrifuged, and the supernatants (S) and pellets (P) were resolved by SDS-PAGE and stained with Coomassie Blue. Each S and P represent the soluble and pellet fractions from the same sample. C, F-actin co-sedimentation assays to test binding specificity of JN to skeletal muscle α-actin and platelet β-actin. D, F-actin co-sedimentation assays to determine F-β-actin-binding site of JN. Truncated GST-JN proteins were compared with wild-type JN for their F-β-actin binding abilities. GST-JN268 lacks the C-terminal 14 aa residues of JN, whereas GST-JN170 has a deletion of the last 112 aa residues of JN. E, Western blotting (WB) confirmed that both bands of the GST-JN doublet (marked by an arrow and an asterisk, respectively) were JN immunoreactive. The lanes were loaded with ∼5.5 and 11 ng of GST-JN, respectively. F, co-sedimentation assays to compare F-β-actin binding capability of FLAG-tagged JN, JNc, JN-S278E, and JN141. Lysates of cultured OLN-93 cells overexpressing the FLAG-tagged JN, JNc, JN-S278E, or JN141 were first subjected to actin polymerization reaction and ultracentrifugation to remove endogenous F-actin and associated JN/mutants. The supernatants containing the overexpressed JN/mutants were then incubated together with purified F-β-actin. Controls consist of reactions without the addition of F-β-actin (F-actin (−)). The reaction samples were then ultracentrifuged. The supernatants and pellets were loaded for SDS-PAGE and analyzed for JN binding by Western blotting. F-actin was revealed by staining the PVDF membrane with Ponceau S. The positions of various proteins are indicated by arrows on the side. Molecular mass markers in kDa are indicated on the left. Asterisks in B–E mark a GST-JN band with possible modification or C-terminal degradation.
F-actin binding capabilities of WT and truncated GST-JN proteins were examined by F-actin co-sedimentation assays. The GST fusion proteins were incubated with a fixed amount of polymerized F-β-actin and the filaments were pelleted by ultracentrifugation. Actin filaments longer than 10 subunits in length will sediment under these conditions (16). As shown in Fig. 2B, almost all actin sedimented in the pellet, indicating the effectiveness of filamentous actin assembling under the present polymerization condition. As a negative control, GST protein was not found in the pellet together with F-actin. In contrast, a considerable amount of GST-JN co-sedimented with pre-polymerized F-actin. Under the same condition, however, GST-JN268 mostly remained in the supernatant, suggesting a critical role of the C-terminal 14 amino acid residues of JN in its F-actin binding (Fig. 2D). Deletion of the last 112 amino acids of JN (GST-JN170) eliminated the F-actin binding property of JN completely (Fig. 2D). GST-JN protein alone (without F-β-actin) also did not sediment by itself and remained soluble (Fig. 2, B and D). It was apparent that GST-JN did not affect the amount of β-actin that sedimented (Fig. 2, B and D). α-Actinin, a known actin-binding protein that decorates actin filaments sideways, was used as a positive control for actin binding here (Fig. 2B).
To further elucidate the relationship between the JN structure and its actin-binding activity, F-actin co-sedimentation was also performed using FLAG-tagged JN, JNc (a JN deletion mutant lacking the N-terminal 100 aa residues), JN141 (another JN deletion mutant lacking the C-terminal 141 aa residues), and JN-S278E, which were produced by transfection and overexpression in cultured OLN-93 cells (an oligodendroglia cell line derived from primary rat brain glia culture (23) and showing no endogenous JN immunoreactivity under normal culture conditions (10)). The results indicated that FLAG-tagged JN bound F-β-actin, so did JNc, although the binding affinity by the latter seemed weaker than the former. JN141 C-terminal deletion mutant and the JN-S278E point mutant did not show significant binding to F-actin under the current in vitro experimental condition (Fig. 2F). Thus, it appears that the N-terminal half of JN was nonessential for F-actin binding of JN. The S278E mutation represented a Ser278-phosphorylated JN mimic that apparently also disrupted F-actin binding and other biological functions of JN (see below).
Some actin-related proteins exhibit isoform preference in their actin binding (17, 18). We compared binding specificity of JN to skeletal muscle α-actin and to platelet β-actin by the actin filament co-sedimentation assay described above. Following ultracentrifugation, the amount of JN associated with the actin filament pellet was measured on SDS-PAGE (Fig. 2C). Coomassie Blue staining showed that GST-JN co-pelleted poorly with F-α-actin. Under the same experimental conditions and with the same amounts of starting materials, the amount of F-β-actin-bound GST-JN was ∼2-fold higher when compared with that bound to F-α-actin. α-Actinin was used as a control (Fig. 2C).
JN Has No Effect on G-actin Polymerization or F-actin Bundling in Vitro
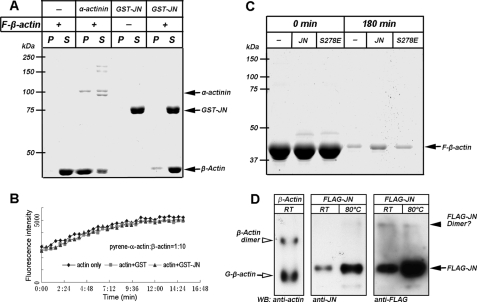
Actin polymerization assays were carried out to assess the role of JN in actin polymerization. Pyrene fluorescence increases when a pyrene-labeled actin subunit is incorporated into a polymerizing actin filament (21). Because juxtanodin was found to have isoform preference for β-actin binding, pyrene-conjugated α-actin was mixed with β-actin in a 1:10 ratio. To increase the sensitivity of the polymerization assay by reducing the starting background fluorescence, the samples were centrifuged at 160,000 × g for 2 h at 4 °C before induction of polymerization. As demonstrated by the standard curve, β-actin readily polymerized with pyrene-conjugated α-actin when KCl and MgCl2 were included (Fig. 3B). We did not detect any significant alteration in the rate and extent of actin polymerization in the presence of GST-JN in comparison with the GST control group. GST-tagged JN seemed to neither modulate the lag associated with the nucleation phase, nor influence the filament elongation phase. This is evidenced by the same rate of assembly during and after the initial lag phase among the GST-JN experimental, GST control, and actin only control groups (Fig. 3B).
FIGURE 3.
Effect of Juxtanodin on actin dynamics in vitro. A, JN did not show a significant effect on F-actin bundling. Pre-polymerized F-β-actin was allowed to react with GST-JN or α-actinin in F buffer for 30 min and then sedimented at 15,000 × g for 15 min. Under this condition, the linear, unbundled F-actin stays in the supernatant but bundled F-actin sediments. Both supernatant (S) and pellet (P) were separated on SDS-PAGE and stained with Coomassie Blue. α-Actinin was adopted as a positive control here. B, JN showed no clear effect on actin polymerization. Monomeric β-actin mixed with pyrene-labeled rabbit skeletal muscle actin at a ratio of 10:1 in general actin buffer was left to polymerize in the presence of GST-JN upon addition of actin polymerization buffer (triangles). Polymerization was monitored as an increase in fluorescence signal (see “Experimental Procedures”). Polymerization using the actins alone (diamonds) and polymerization of the actins in the presence of GST (squares) were used as controls. C, actin filament co-sedimentation assay showed F-actin stabilizing ability of polyhistidine-tagged JN, but not JN-S278E phosphomimetic mutant. Pre-polymerized F-β-actin alone (−) or together with polyhistidine-tagged JN or JN-S278E was incubated and ultracentrifuged. The pellets were then resuspended in G buffer to induce actin depolymerization. Three hours later, the samples were ultracentrifuged and the pellets were analyzed by SDS-PAGE and Coomassie Blue staining. D, native PAGE gel of FLAG-tagged JN (from lysate of transfected OLN-93 cells) and β-actin followed by Western blotting (WB) to assess the possibility of in vivo JN dimerization. The identity of the JN bands was cross-verified by anti-JN (lanes 2 and 3) and anti-FLAG tag antibodies (lanes 4 and 5). Cell lysates for lanes 3 and 5 were heated to 80 °C prior to being loaded for electrophoresis. β-Actin (lane 1 on the left) served as a control and its monomer and dimer bands as approximate markers of molecular mass. The various protein bands (A, C and D) are indicated by arrows or arrowheads. Molecular mass markers in kDa are labeled on the left (A and C). Other abbreviations: P, pellet; RT, room temperature; S, supernatant.
Actin filaments cross-link into closely packed parallel arrays of actin bundles (19). The formation of actin bundles requires the presence of actin-bundling proteins, which are small globular proteins such as α-actinin, fimbrin, and fascin (20). To determine whether JN acts as an actin-bundling protein, recombinant GST-JN was incubated together with F-β-actin. This was followed by low speed centrifugation to sediment bundled actin filaments. The result showed that GST-JN was ineffective in precipitating F-actin under low speed centrifugation even at excessively high concentrations (Fig. 3A). As a positive control, α-actinin was found together with actin in the low speed-sedimented pellet, indicating the induction of actin bundles by α-actinin. Actin filaments remained in the supernatant fraction in the absence of α-actinin (Fig. 3A).
One of the characteristics of certain actin filament-bundling proteins is dimer formation (19, 20). We tested by native PAGE gel and Western blotting if dimers could be formed between overexpressed FLAG-tagged JN molecules. As seen from Fig. 3D (filled arrow), FLAG-JN apparently existed mostly as monomers under the native PAGE gel condition. The possibility of JN dimerization in the cell could not be completely ruled out, but this accounted, if at all, for only a very small fraction of the overexpressed JN, as demonstrated by the very faint upper band of possible FLAG-JN dimers (filled arrowhead) in comparison with the prominent lower band of FLAG-JN monomers (filled arrow) in the FLAG-JN groups. Heating the cell lysate to 80 °C prior to loading on native PAGE gel increased JN antigenicity and detectability by both JN and FLAG antibodies, but did not significantly facilitate JN dimer formation. As a positive control, β-actin migrated as both monomers (open arrow) and dimers (open arrowhead) under the current experimental condition (Fig. 3D, left lane).
JN Inhibits Disassembly of Actin Filaments in Vitro
We next analyzed the possible influence of JN on F-actin disassembly. Because GST can form dimers and then have potentially undesirable effects on the activity of GST-JN, we employed polyhistidine-tagged JN for F-actin disassembly assays using a method previously described by Rybakova and Ervasti (22). F-actin disassembly was induced by dilution into low salt buffer conditions, and the effect of polyhistidine-tagged JN on actin disassembly was examined using the F-actin sedimentation assay. At 90 min into the depolymerization reaction, an average of 16.9 ± 2.3% (mean ± S.D., n = 2) F-actin remained in the F-actin alone control group, whereas 36.3 ± 2.2% (mean ± S.D., n = 2) F-actin was retained in the experimental group of actin together with polyhistidine-tagged JN. At 180 min after dilution with the F-actin depolymerization buffer, 3.6 ± 1.9% (mean ± S.D., n = 4) F-actin remained in the pellet in the actin alone group, whereas 8.3 ± 3.5% (mean ± S.D., n = 4) actin was pelleted in the experimental group added with polyhistidine-tagged JN (Fig. 3C). This indicates that JN significantly protected F-actin from depolymerization under the experimental condition.
Because a potential phosphorylation site of juxtanodin, serine 278, is located in the F-actin binding region, we examined whether phosphorylation of this site might influence the effect of JN on F-actin stability. JN phosphorylation status at this site was mimicked by site-directed mutation of serine 278 to glutamate. The polyhistidine-tagged JN mutant (His6-JN-S278E) was used to examine its effect on actin depolymerization by the above F-actin sedimentation assay. In the presence of His6-JN-S278E, an average of 3.8 ± 1.5% (mean ± S.D., n = 3) F-actin remained at 180 min into the depolymerization reaction. This was not significantly different from the 3.6 ± 1.9% retention rate in the F-actin alone control group (Fig. 3C). These results suggest JN could inhibit F-actin disassembly, and phosphorylation of JN at serine 278 directly disrupted the ability of the JN protein to stabilize F-actin. This inability of the His6-JN-S278E mutant to stabilize F-actin also agrees with the loss of F-actin binding ability of FLAG-tagged JN-S278E as seen from the above F-actin co-sedimentation assay (Fig. 2F).
Juxtanodin Inhibits F-actin Disassembly in Cultured Cells
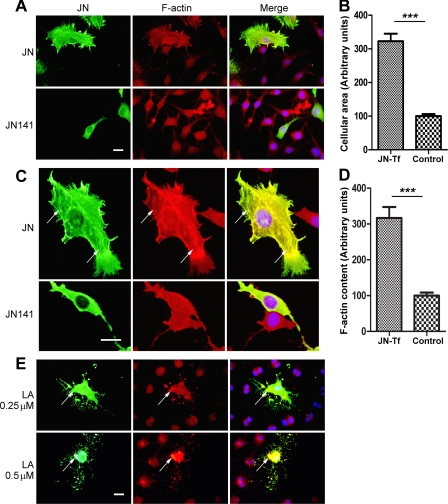
Given that JN was capable of binding and stabilizing actin filaments in vitro, we investigated whether JN can affect actin cytoskeletal architecture and dynamics in cultured cells. Because the F-actin binding of JN requires the last 14 amino acid residues of the C terminus of JN, JN141 was used as a negative control in this experiment. OLN-93 cells were transfected with constructs directing the expression of the FLAG epitope-tagged WT JN or truncated JN141 proteins. As observed under the fluorescence microscope, subcellular localization and effects of JN or JN141 protein on F-actin cytoskeleton (as revealed by binding to rhodamine-conjugated phalloidin) significantly differed. At low to medium cell densities, the majority of OLN-93 cells were bipolar in morphology and their cell bodies and processes were generally smooth. Staining with phalloidin revealed few F-actin-rich fibers in normal culture conditions. Overexpression of JN141 had little effect on morphology of the OLN-93 cells. Similar to neighboring untransfected cells, JN141 transfectants displayed only low amounts of thin actin filaments without particular orientations. JN141 itself showed a diffuse cytoplasmic distribution pattern in the transfected cells (Fig. 4C).
FIGURE 4.
Juxtanodin inhibited F-actin disassembly in OLN-93 cells. A, OLN-93 cells overexpressing exogenous JN, as compared with neighboring non-transfected or JN141 transfected control cells, were more spread out and exhibited more numerous and finer processes/filopodium-like prickles. B, quantification of the increase of the cellular area of JN-transfected OLN-93 cells (JN-Tf) as compared with neighboring untransfected cells (Control). ***, p < 0.001, unpaired Student's t test. Values are mean ± S.E. C, as visualized by rhodamine-conjugated phalloidin staining, JN-transfected cells showed thick filamentous structures (arrow) that resembled actin stress fibers and were associated with JN proteins (green). JN141-transfected cells, on the other hand, showed a pattern of diffuse distribution for both JN141 and F-actin. D, F-actin contents in JN-transfected OLN-93 cells in comparison with neighboring untransfected cells. ***, p < 0.001, unpaired Student's t test. Values are mean ± S.E. E, JN overexpression antagonized F-actin disassembly of OLN-93 cells induced by latrunculin A (LA). Forty-four hours after JN transfection of OLN-93 cells, 0.25 or 0.5 μm latrunculin A was added into the culture medium. OLN-93 cells were fixed 4 h later and then stained for immunofluorescence. LatA-resistant F-actin (red) clusters (arrow) were co-localized with JN-positive (green) patches in the transfected cells. For panels A, C, and E, transfections are indicated on the left, whereas fluorescent staining is indicated at the top. Scale bars, 20 μm.
Compared with neighboring untransfected cells or with JN141-transfected cells, JN-transfected cells were more spread out in morphology (Fig. 4A). Statistical analysis showed significant increases in the cellular area of cells overexpressing JN (on average 323 ± 22% of neighboring untransfected cells; n = 28 for JN transfectants) (Fig. 4B). Thick filamentous structures positive for rhodamine-conjugated phalloidin binding and reminiscent of actin stress fibers were observed in the JN transfectants, and JN was largely colocalized with the intense and widespread F-actin fibers (Fig. 4C).
The formation of actin fibers can result from bundling of pre-existing actin filaments or from de novo formation of actin filaments. To assay for an involvement of actin polymerization/depolymerization in JN-induced actin fiber formation, the fluorescence intensity of F-actin stains in JN overexpressors was compared with that of neighboring cells, and the ratio of the two values was used to plot possible changes in relative F-actin levels. The statistical result showed that the content of F-actin in JN overexpressors was also significantly augmented (average 317 ± 31% of neighboring untransfected cells; n = 28 for JN transfectants) (Fig. 4D). Yet, this increase in F-actin content seemed paralleled by the increase of the cellular area of JN transfectants, and did not result in significant alteration of F-actin intensity/level, as compared with neighboring untransfected cells. Together, these results indicate that overexpression of JN caused the formation of actin fibers and JN was subcellularly localized along these F-actin fibers.
Latrunculins promote F-actin depolymerization by binding to G-actin, preventing the incorporation of actin into filaments and depleting cellular F-actin over time (24). In the present study, LatA was used for disassembling the F-actin in OLN-93 cells. At 44 h after JN transfection, OLN-93 cells were further cultured in the presence of latrunculin A at concentrations of 0.25 or 0.5 μm for another 4 h to induce F-actin disassembly. Given that fetal bovine serum (FBS) could interfere with the function of LatA (25), FBS was reduced to a low concentration (from 10% to 1%) in the culture medium from the beginning of LatA addition. At the end of the experiment, the effect of LatA and JN was assessed by staining F-actin using rhodamine-conjugated phalloidin. As expected, F-actin in LatA-treated OLN-93 cells became reduced in levels (data not shown). Accompanying the LatA-induced reduction of F-actin content, the stress fiber appearance of F-actin in JN-transfected OLN-93 cells was also diminished (Fig. 4E). Overexpression of JN obviously counteracted the LatA-induced reduction of phalloidin-labeled F-actin, as LatA-resistant F-actin-rich clusters in the JN-transfected cells mostly overlapped the JN-intense patches (Fig. 4E). Except for its influence on cell F-actin, morphology (round-up of cell bodies) and division (note the double nuclei in many of the LatA-treated cells in Fig. 4E), no other overt toxic effect of LatA on OLN-93 cells was observed at the current concentrations and treatment durations and with 1% FBS in the culture medium.
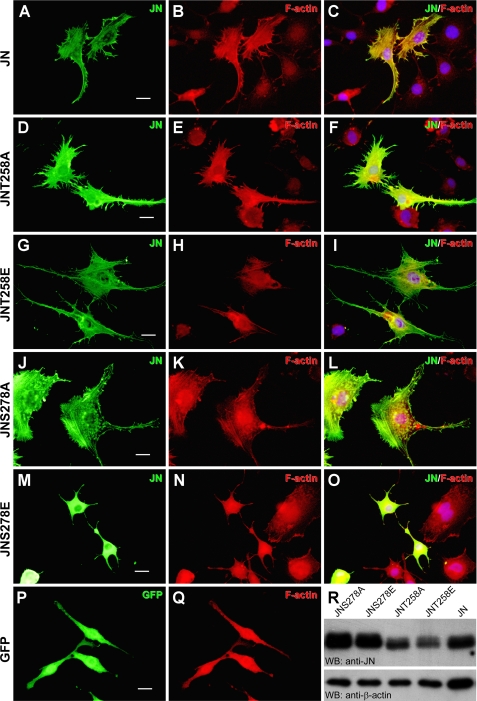
Phosphorylation at Serine 278 Abolishes JN Effects on Arborization and Actin Cytoskeleton of Cultured OLN-93 Cells
Bioinformatics search of possible phosphorylation sites revealed two conserved phosphorylation sites in JN. One is at threonine 258 and the other at serine 278. The positional context of Thr258 of JN is conserved in all ERM proteins and its phosphorylation is critical for releasing ERM functional domains (11), whereas the functional significance of the Ser278, of which phosphorylation has been reported in moesin, remains unclear (13). To determine the role of the two possible phosphorylation sites in JN, single point mutations of JN were generated using site-directed mutagenesis. The Thr to Ala or Ser to Ala mutation prevents phosphorylation, whereas the Thr to Glu or Ser to Glu mutation should mimic phosphorylation. The FLAG-tagged JN mutants were overexpressed in OLN-93 cells to examine their effects on the morphology and F-actin pattern in transfected cells. JN-transfected OLN-93 cells exhibited bigger sizes, more numerous and finer processes with many filopodium-like prickles (Fig. 5, A–C), in comparison to neighboring untransfected cells or cells transfected with green fluorescent protein (GFP, Fig. 5, P and Q). JN-S278E, the phosphorylation mimic of JN at Ser278, had lost much of the effects of wild-type JN on the size and arborization of OLN-93 cells. Additionally, in contrast to the colocalization of wild-type JN with F-actin fibers, JN-S278E showed a diffuse distribution in host cells (Fig. 5, M–O). JN-T258A, S278A (both being the dephosphorylation mimics of JN), or JN-T258E mutation (the other phosphorylation mimic of JN) caused no detectable alterations of JN effects on the morphology and F-actin organization of transfected OLN-93 cells (Fig. 5, D–L). As exemplified by the immunoblotting results of Fig. 5R, transfection efficiency and/or levels of expression of JN and the mutants were comparable although moderate variations were observed among the different plasmid constructs and across different repeats of transfection. Together, these results strongly suggest that specific phosphorylation at the site of Ser278 negatively interfered with the activity of JN in cells, consistent with the loss of JN-S278E in actin binding and inhibition of F-actin disassembly in vitro (see above).
FIGURE 5.
Phosphorylation of serine 278 abolished JN effects on OLN-93 cell arborization and F-actin organization. A–Q, overexpression of JN-S278E, the phosphorylation mimic of JN at Ser278, produced no significant effects on arborization and actin fiber organization of OLN-93 cells (M–O), in contrast to wild-type JN (A–C). However, JN-T258E (the phosphorylation mimic of JN at Thr258, G–I) T258A (D–F), and S278A (the dephosphorylation mimics of JN, J–L) mutations had no measurable effects on JN functions. GFP transfection was used as a negative control (P and Q). Transfections are indicated on the left, whereas fluorescent staining is indicated at the top right corner of each panel. Nuclei of the cells were counterstained with 4′,6-diamidino-2-phenylindole (blue) and shown for the right panels. Scale bars, 20 μm. R, Western blot showing comparable levels of expression of JN and its various point mutants in cultured OLN-93 cells. Immunoblotting of β-actin of the transfected cell lysates served as a loading control.
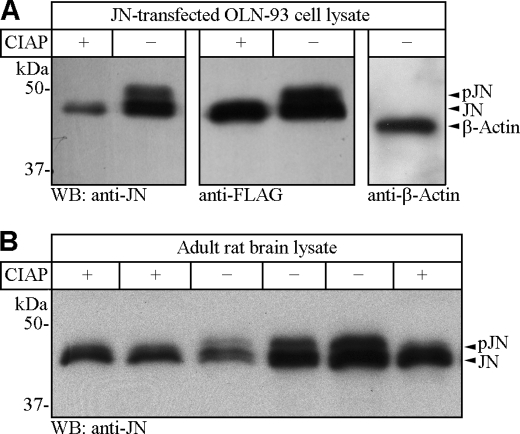
The extent of JN phosphorylation in vivo was assessed by dephosphorylation assays of brain and transfected cell lysates using CIAP. As reported previously, JN from normal brain lysate migrates as doublet bands on Western blot (10). Upon treatment of brain lysate with CIAP prior to SDS-PAGE, the upper band of the JN doublet disappeared on the Western blot, whereas the lower band of the doublet was not markedly affected. This indicates the existence of both phosphorylated and dephosphorylated forms of JN in vivo with the former migrating slightly slower than the latter during SDS-PAGE electrophoresis (Fig. 6B). Further dephosphorylation tests of lysates of JN-transfected OLN-93 cells also confirmed the presence of phosphorylated and dephosphorylated forms of overexpressed JN in the cultured cells (Fig. 6A). Judged according to the relative intensities of the doublet bands on Western blots of both native JN of the brain and overexpressed FLAG-tagged JN from transfected OLN-93 cells, the lower band of the JN doublet accounted for a majority of JN immunostaining, indicating that a larger portion of brain native JN and OLN-93-overexpressed JN remained dephosphorylated under physiological conditions (Fig. 6, A and B).
FIGURE 6.
Differential migration of phosphorylated and dephosphorylated JN on SDS-PAGE and in vitro JN dephosphorylation. A, Western blots showing differential migration of phosphorylated (pJN) and dephosphorylated FLAG-tagged JN (JN), and the disappearance of phosphorylated FLAG-JN (pJN, upper band of the FLAG-JN doublet) after transfected OLN-93 cell lysate was incubated with CIAP prior to being loaded for SDS-PAGE. Negative controls consisted of sham dephosphorylation reactions without CIAP. Identity of the FLAG-JN bands were verified by Western blots using both anti-JN (left) and anti-FLAG (middle) antibodies, respectively. Western blot of β-actin (right) served as a loading control. B, Western blots showing differential migration of phosphorylated and dephosphorylated brain native JN and the disappearance of phosphorylated JN (upper band of the JN doublet) after rat brain lysate was incubated with CIAP prior to being loaded for SDS-PAGE. Negative controls consisted of sham dephosphorylation reactions without CIAP (−). All lanes correspond to 2/2.5 μl of cell/brain lysates, except lanes 3 (1.3 μl) and 5 (5 μl) of panel B.
DISCUSSION
Binding of Juxtanodin to F-actin
The present co-sedimentation assays demonstrated that JN interaction with F-actin was direct between the two, and need not be mediated by other proteins. By deletion of different domains of JN, it was further demonstrated that the C-terminal 14 aa residues of JN were indispensable for its interaction with F-actin. JN268, which lacked the last 14 aa residues, was almost devoid of any binding ability with F-actin. These aa residues belong to the putative F-actin binding domain, which was first identified and mapped to the C-terminal 34 aa residues of ezrin, radixin, and moesin (26). The facts that JNc, but not JN141 or even JN268, retained the actin binding activity of JN support the importance of the C-terminal residues in JN actin binding. Although the N-terminal half of JN seemed nonessential for JN actin binding, it might not be entirely dispensable as evidenced by the seemingly higher affinity of JN to F-β-actin than the N terminus-truncated JNc (Fig. 2F). This is also consistent with our previous finding showing more prominent promotion of host cell arborization by JN than JNc (10).
Interestingly, JN exhibited stronger affinity to F-β-actin than to F-α-actin. Although actins are highly conserved proteins and cytoplasmic β-actin differs by less than 10% in terms of aa sequence from the muscle α-actin (27), they display several structural differences. These include the N-terminal conformation of β-actin bearing a turn rather than the helical structure found in F-α-actin, the distinct rotations within subdomains of the isoforms, and the different side chain orientations at residues 38–52 (28). Functional distinctions for the interaction of actin isoforms with actin-binding proteins have been reported. Profilin, for example, has different affinities for cytoplasmic β-actin and sarcomeric α-actin (17). For ERM proteins, the ability of their F-actin binding domain to discriminate between actin isoforms is still controversial. The interaction of ezrin with actin showed isoform preference. It co-localized with β-actin in gastric parietal cells. Purified parietal cell ezrin binds β-actin with a higher affinity than α-actin, and selectively promotes assembly of β-actin (18, 29). Moesin isolated from activated platelets, or ezrin activated in vitro by protein kinase C-θ (PKC-θ), however, did not show any difference in binding with actin isoforms (30, 31). In the present study, the high purity of FPLC-grade JN recombinant protein enabled us to investigate its isoform preference without contamination from lipids or other proteins.
Effect of Juxtanodin on Actin Dynamics
The ability of JN to induce cellular process extension and ramification is reminiscent of that of the C-terminal domain of ERM proteins (32, 33). The effects of JN and ERMs on cellular arborization seem to be dependent on their binding to F-actin, because: 1) deletion of its C-terminal 14-aa F-actin binding site abolished its influence on cellular arborization (10); 2) co-expression of an N-terminal fragment (residues 1–115) of ezrin, which could mask the F-actin binding site of the C-terminal domain of ERM proteins, antagonized its effect on process extension in some circumstances (32). These results suggested that the F-actin binding domains of JN and ERM proteins have similar effects on actin dynamics and is indispensable for the generation of cell process extensions caused by JN and ERM proteins.
A previous study showed that purified gastric ezrin could regulate actin dynamics by promoting the assembly of β-actin (18). However, as suggested by the present results, JN had no direct influence on actin polymerization. Although additional actin-binding sites (at aa residues 280–309, which bind F- and G-actin) have been identified in ezrin (34), which partly explained the ability of ezrin to promote actin assembly by providing nucleation sites, these extra sites were not detected by blot overlay assaying GST fusions or untagged N- and C-terminal domains of moesin, radixin, and merlin (35). As demonstrated by our study, JN also does not have other F- or G-actin binding sites except for the C-terminal F-actin binding domain.
JN was found to inhibit the disassembly of F-β-actin (Figs. 3C and 4E). Some F-actin-binding proteins stabilize actin cytoskeleton by bundling pre-existing actin filaments, such as Fascin and α-actinin (36, 37). However, as mentioned above, JN has no F-actin bundling activity. An F-actin-bundling protein usually harbors at least two F-actin binding sites or exhibits the ability to form dimers for its bundling activity. JN has only one F-actin binding site at its C terminus, and was not found to form dimers to any significant extent on native PAGE gel (Fig. 3D). These results are in line with the lack of F-actin bundling activity of JN. Instead, JN was found to concentrate along F-actin-rich fibers in transfected cells, implying lateral binding of JN to actin filaments in vivo, which presents a reasonable explanation for its inhibitory effect on F-actin depolymerization. Tropomyosin was first described to prevent actin filament disassembly by lateral binding to the actin filament (38). Recently, merlin has also been shown to inhibit F-actin disassembly through lateral association (14). In addition, the current study demonstrated no binding of JN to G-actin, neither does JN alter the rate of actin nucleation/elongation during polymerization. These observations indicate that the inhibitory effect of JN on F-actin depolymerization is not due to capping of the ends of actin filaments or to nucleating activity. Recently, Brockschnieder and co-workers (39) reported that endogenous Ermin, the ortholog of JN in mouse, was accumulated in spikes at the tip of F-actin-rich processes (termed “Ermin spikes”) in cultured primary oligodendrocytes. The concentration of JN at F-actin-rich spikes supports the notion that JN could directly bind and stabilize F-actin in vivo. This is also in agreement with the present observation that JN countered the F-actin depolymerizing effect of LatA in cultured OLN-93 cells (Fig. 4).
Regulation of JN Activity by Phosphorylation
The functions of ERM proteins are conformationally regulated, that is, the full-length dormant molecule has its activities masked because the N-terminal domain binds very tightly to its C-terminal domain (11). Activation of ERM proteins is predicted to require separation of the N- and C-terminal domains, thereby exposing the C-terminal F-actin binding site and the potential membrane association sites at the N-terminal domain (11). Phosphorylation of a conserved threonine residue in ERM proteins (Thr558 in moesin) results in activation of these proteins by disrupting the intramolecular and intermolecular self-associations. Although JN lacks the N-terminal FERM domain, which is conserved in ERM proteins, it shares an almost identical C terminus with ERM proteins and retains the conserved threonine site (Thr258). To our surprise, however, Thr258 modifications showed no clear influence to JN activities on host cell morphology and actin organization. This is probably because: 1) JN lacks the N-terminal FERM domain of ERMs for intramolecular or intermolecular self-association/masking, and 2) there might not be a FERM domain-containing protein in OLN-93 cells that interacts with and masks the JN C-terminal actin-binding domain in a JN-T258 phosphorylation-dependent way.
In contrast, the phosphomimetic Ser to Glu mutation of serine 278 of JN resulted in surprising alterations in JN molecular and cell biological activities. The corresponding serine residue in mouse moesin, Ser565, was identified in a proteome-wide analysis of murine WEHI-231 B lymphoma cells. The authors suggested that phosphorylation of this site might be involved in B-cell signaling (13). In the present study, phosphorylation-mimetic mutation of serine 278 obviously abolished JN binding to F-actin and JN effects on F-actin depolymerization and host cell arborization/actin fiber organization, whereas the Ser278 dephosphorylation-mimetic mutant of JN, JN-S278A, showed no difference from wild-type JN in its cell biological activity on cultured OLN-93 cell arborization and actin cytoskeleton. The mechanism of this serine 278 phosphorylation-dependent regulation of JN activity remains elusive. The functional importance of this phosphorylation-dependent regulation of JN activity, however, is obvious. It not only provided the possibility for regulating the activity of this regulatory protein for actin cytoskeleton, but also integrated the action of JN into the complex signaling pathways and functional context of the host cells.
Indeed, modification of the phosphorylation status is an important and general way for regulation of actin-binding proteins, such as calcium/calmodulin-dependent protein kinase IIβ (CaMKIIβ) (40), heLIM and SH3 domain protein (LASP), and Cortactin (41). Their binding to F-actin, important for in vivo maintenance of polymerized F-actin, was disrupted when highly phosphorylated. In OLs, the important actin-binding proteins, like myelin basic protein (MBP) and myosin II, are also regulated by phosphorylation for actin interaction. The results from Boggs and co-workers (42) showed that phosphorylation at two sites of MBP with mitogen-activated protein kinase (MAPK) could attenuate the ability of MBP to polymerize and bundle actin.
Both oligodendrocytes and myelin contain several kinases that can phosphorylate MBP on serine and threonine and phosphatases that can mediate rapid turnover of phosphate groups. Turnover is highest in the most mature myelin. Expectedly, the mechanism for the phosphorylation turnover of JN is present in myelin to regulate the interaction of JN with actin. In fact, both phosphorylated and dephosphorylated forms of brain native JN exist in vivo, as demonstrated by the dephosphorylation assays of the present study. The JN regulatory pathways/molecules also appear present and capable of modifying the phosphorylation status and activity of overexpressed JN in cultured OLN-93 cells (Fig. 6, A and B). JN has also been shown not to always colocalize with actin filaments (43).4 The dissociation of JN and actin filaments in vivo possibly involves the phosphorylation of serine 278 of JN. Further confirmation and functional elucidation of serine 278 phosphorylation of JN in vivo and identification of the related kinases and phosphatases call for future studies and the generation of specific antibodies against phosphorylated and dephosphorylated JN. It also remains to be clarified if JN is subject to phosphorylation or other modifications at other aa residues.
Taken together, our results point to Juxtanodin as an actin cytoskeleton-stabilizing protein that plays active roles in differentiation of oligodendrocytes and maintenance of the CNS myelin sheath. The results also suggest phosphorylation modification (most possibly at serine 278) as an important mechanism in the regulation of Juxtanodin functions.
This work was supported by Singapore Biomedical Research Council Research Grant BMRC/04/1/21/19/305 and National Medical Research Council Grant 0946/2005 (to F. L.).
J. Meng, W. Xia, J. Tang, and F. Liang, unpublished data.
- OL
- oligodendrocyte
- aa
- amino acid
- ERM
- ezrin/radixin/moesin
- JN
- Juxtanodin
- LatA
- latrunculin A
- CIAP
- calf intestinal alkaline phosphatase
- WASP
- Wiskott-Aldrich syndrome protein
- WAVE
- Wiskott-Aldrich syndrome protein family verprolin homologous protein
- MBP
- myelin basic protein.
REFERENCES
- 1.Kirby B. B., Takada N., Latimer A. J., Shin J., Carney T. J., Kelsh R. N., Appel B. (2006) Nat. Neurosci. 9, 1506–1511 [DOI] [PubMed] [Google Scholar]
- 2.Franklin R. J., Ffrench-Constant C. (2008) Nat. Rev. Neurosci. 9, 839–855 [DOI] [PubMed] [Google Scholar]
- 3.Bauer N. G., Richter-Landsberg C., Ffrench-Constant C. (2009) Glia 57, 1691–1705 [DOI] [PubMed] [Google Scholar]
- 4.Richter-Landsberg C. (2000) J. Neurosci. Res. 59, 11–18 [DOI] [PubMed] [Google Scholar]
- 5.Song J., Goetz B. D., Baas P. W., Duncan I. D. (2001) Mol. Cell. Neurosci. 17, 624–636 [DOI] [PubMed] [Google Scholar]
- 6.Rumsby M., Afsari F., Stark M., Hughson E. (2003) Glia 42, 118–129 [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Valle C., Gorman D., Gomez A. M., Bunge M. B. (1997) J. Neurosci. 17, 241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacon C., Lakics V., Machesky L., Rumsby M. (2007) Glia 55, 844–858 [DOI] [PubMed] [Google Scholar]
- 9.Kim H. J., DiBernardo A. B., Sloane J. A., Rasband M. N., Solomon D., Kosaras B., Kwak S. P., Vartanian T. K. (2006) J. Neurosci. 26, 5849–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B., Cao Q., Guo A., Chu H., Chan Y. G., Buschdorf J. P., Low B. C., Ling E. A., Liang F. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11527–11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramesh V. (2004) Nat. Rev. Neurosci. 5, 462–470 [DOI] [PubMed] [Google Scholar]
- 12.Bretscher A., Edwards K., Fehon R. G. (2002) Nat. Rev. Mol. Cell Biol. 3, 586–599 [DOI] [PubMed] [Google Scholar]
- 13.Shu H., Chen S., Bi Q., Mumby M., Brekken D. L. (2004) Mol. Cell. Proteomics 3, 279–286 [DOI] [PubMed] [Google Scholar]
- 14.James M. F., Manchanda N., Gonzalez-Agosti C., Hartwig J. H., Ramesh V. (2001) Biochem. J. 356, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher S. R. (2001) Curr. Protoc. Cell Biol. 6.5, 6.5.1–6.5.11 [DOI] [PubMed] [Google Scholar]
- 16.Attri A. K., Lewis M. S., Korn E. D. (1991) J. Biol. Chem. 266, 6815–6824 [PubMed] [Google Scholar]
- 17.Rozycki M., Schutt C. E., Lindberg U. (1991) Methods Enzymol. 196, 100–118 [DOI] [PubMed] [Google Scholar]
- 18.Yao X., Cheng L., Forte J. G. (1996) J. Biol. Chem. 271, 7224–7229 [DOI] [PubMed] [Google Scholar]
- 19.Winder S. J. (2003) Curr. Opin. Cell Biol. 15, 14–22 [DOI] [PubMed] [Google Scholar]
- 20.Otto J. J. (1994) Curr. Opin. Cell Biol. 6, 105–109 [DOI] [PubMed] [Google Scholar]
- 21.Cooper J. A., Walker S. B., Pollard T. D. (1983) J. Muscle Res. Cell Motil. 4, 253–262 [DOI] [PubMed] [Google Scholar]
- 22.Rybakova I. N., Ervasti J. M. (1997) J. Biol. Chem. 272, 28771–28778 [DOI] [PubMed] [Google Scholar]
- 23.Richter-Landsberg C., Heinrich M. (1996) J. Neurosci. Res. 45, 161–173 [DOI] [PubMed] [Google Scholar]
- 24.Coué M., Brenner S. L., Spector I., Korn E. D. (1987) FEBS Lett. 213, 316–318 [DOI] [PubMed] [Google Scholar]
- 25.Kuriu T., Inoue A., Bito H., Sobue K., Okabe S. (2006) J. Neurosci. 26, 7693–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turunen O., Wahlström T., Vaheri A. (1994) J. Cell Biol. 126, 1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang K. S., Zimmer W. E., Jr., Bergsma D. J., Dodgson J. B., Schwartz R. J. (1984) Mol. Cell. Biol. 4, 2498–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schutt C. E., Myslik J. C., Rozycki M. D., Goonesekere N. C., Lindberg U. (1993) Nature 365, 810–816 [DOI] [PubMed] [Google Scholar]
- 29.Yao X., Chaponnier C., Gabbiani G., Forte J. G. (1995) Mol. Biol. Cell 6, 541–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons P. C., Pietromonaco S. F., Reczek D., Bretscher A., Elias L. (1998) Biochem. Biophys. Res. Commun. 253, 561–565 [DOI] [PubMed] [Google Scholar]
- 31.Nakamura F., Huang L., Pestonjamasp K., Luna E. J., Furthmayr H. (1999) Mol. Biol. Cell 10, 2669–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin M., Roy C., Montcourrier P., Sahuquet A., Mangeat P. (1997) Mol. Biol. Cell 8, 1543–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litman P., Amieva M. R., Furthmayr H. (2000) BMC Cell Biol. 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy C., Martin M., Mangeat P. (1997) J. Biol. Chem. 272, 20088–20095 [DOI] [PubMed] [Google Scholar]
- 35.Huang L., Wong T. Y., Lin R. C., Furthmayr H. (1999) J. Biol. Chem. 274, 12803–12810 [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto Y., Parsons M., Adams J. C. (2007) Mol. Biol. Cell. 18, 4591–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coghill I. D., Brown S., Cottle D. L., McGrath M. J., Robinson P. A., Nandurkar H. H., Dyson J. M., Mitchell C. A. (2003) J. Biol. Chem. 278, 24139–24152 [DOI] [PubMed] [Google Scholar]
- 38.Broschat K. O. (1990) J. Biol. Chem. 265, 21323–21329 [PubMed] [Google Scholar]
- 39.Brockschnieder D., Sabanay H., Riethmacher D., Peles E. (2006) J. Neurosci. 26, 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Y. C., Redmond L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15791–15796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webb B. A., Zhou S., Eves R., Shen L., Jia L., Mak A. S. (2006) Arch. Biochem. Biophys. 456, 183–193 [DOI] [PubMed] [Google Scholar]
- 42.Boggs J. M., Rangaraj G., Gao W., Heng Y. M. (2006) Biochemistry 45, 391–401 [DOI] [PubMed] [Google Scholar]
- 43.Tang J., Tang J., Ling E. A., Wu Y., Liang F. (2009) Neuroscience 161, 249–258 [DOI] [PubMed] [Google Scholar]