Abstract
The β1-adrenergic receptor (β1AR) is the predominant βAR in the heart, mediating the catecholamine-stimulated increase in cardiac rate and force of contraction. Regulation of this important G protein-coupled receptor is nevertheless poorly understood. We describe here the biosynthetic profile of the human β1AR and reveal novel features relevant to its regulation using an inducible heterologous expression system in HEK293i cells. Metabolic pulse-chase labeling and cell surface biotinylation assays showed that the synthesized receptors are efficiently and rapidly transported to the cell surface. The N terminus of the mature receptor is extensively modified by sialylated mucin-type O-glycosylation in addition to one N-glycan attached to Asn15. Furthermore, the N terminus was found to be subject to limited proteolysis, resulting in two membrane-bound C-terminal fragments. N-terminal sequencing of the fragments identified two cleavage sites between Arg31 and Leu32 and Pro52 and Leu53, which were confirmed by cleavage site and truncation mutants. Metalloproteinase inhibitors were able to inhibit the cleavage, suggesting that it is mediated by a matrix metalloproteinase or a disintegrin and metalloproteinase (ADAM) family member. Most importantly, the N-terminal cleavage was found to occur not only in vitro but also in vivo. Receptor activation mediated by the βAR agonist isoproterenol enhanced the cleavage in a concentration- and time-dependent manner, and it was also enhanced by direct stimulation of protein kinase C and adenylyl cyclase. Mutation of the Arg31–Leu32 cleavage site stabilized the mature receptor. We hypothesize that the N-terminal cleavage represents a novel regulatory mechanism of cell surface β1ARs.
Keywords: Adrenergic Receptor, G Protein-coupled Receptors (GPCR), Glycosylation, Metalloprotease, Post-translational Modification, Protein Processing, Receptor Regulation
Introduction
The β1-adrenergic receptor (β1AR)3 is one of the three βAR subtypes that are activated by the endogenous catecholamines adrenaline and noradrenaline (1). These receptors belong to the G protein-coupled receptor (GPCR) family, one of the largest membrane protein families involved in cellular signaling (2, 3). The β1AR is the predominant βAR subtype in the heart, mediating the increase in cardiac rate and force of contraction (4, 5). This makes it the most important target receptor for the β-blockers that are used to treat common cardiac diseases such as chronic heart failure, coronary artery disease, hypertension, and arrhythmias. The mechanisms that regulate human β1AR (hβ1AR) are therefore of considerable interest.
The hβ2AR is one of the most extensively studied GPCRs, but much less is known about hβ1AR. The suggestion that it may be more resistant to agonist-mediated desensitization (6), internalization (7–12), and down-regulation (7, 10, 13, 14) could indicate that the two receptors are regulated by distinct mechanisms. Their ligand-binding sites are well conserved, but the overall homology of the two βARs is only 54% (15). The most diverse regions are the intervening loops that connect the transmembrane domains, the extracellular N terminus and the intracellular C terminus. The third intracellular loops and the C-terminal tails of β1AR and β2AR have been implicated in the mediation of interactions with distinct intracellular proteins, including a number of PSD-95/Discs-large/ZO-1-homology (PDZ) domain-containing proteins that have divergent roles in receptor signaling and trafficking (reviewed in Refs. 16, 17). In contrast, the extracellular domains of the two βARs have aroused only limited interest and have been thought to have a negligible role in receptor activation and regulation. This is probably related to the relatively short length of the N termini and the absence of noticeable functional domains with conserved motifs as opposed to GPCRs, e.g. in the adhesion receptor family (2, 3). Recent evidence nevertheless suggests that the extracellular βAR domains may have a more important role than had been anticipated. For example, the crystal structure of turkey β1AR revealed that the second extracellular loop participates in ligand binding, defining the entrance to the ligand binding pocket within the 7-transmembrane domain bundle (18). On the other hand, ligand binding was found to lead to conformational changes in the β2AR N terminus and extracellular loops, as was revealed by conformation-specific antibodies and NMR spectroscopy, respectively (19, 20).
To initiate investigations into the mechanisms that regulate hβ1AR, we set out to characterize the specific steps that take place during the biogenesis of this GPCR. We demonstrate here that the receptor N terminus is extensively modified by mucin-type O-glycosylation during the rapid transport to the cell surface in addition to one N-glycan. Furthermore, the N terminus of the mature receptor with fully processed oligosaccharides was found to be subject to proteolytic cleavage by metalloproteinases, a cleavage that occurred both in vitro and in vivo. This cleavage was enhanced by agonist-mediated receptor activation and by direct activation of protein kinase C and adenylyl cyclase. On the other hand, mutation of the primary cleavage site was found to lead to stabilization of mature receptors. These results allow us to hypothesize that the N-terminal cleavage represents a novel way of controlling the number of functionally active cell surface β1ARs.
EXPERIMENTAL PROCEDURES
Materials
Endo-β-N-acetylglucosaminidase H (Endo H), peptide-N-glycosidase F (PNGase F), O-glycosidase, and neuraminidase were obtained from Roche Applied Science. Anti-FLAG M2 and anti-HA monoclonal antibodies, anti-FLAG M2 and anti-c-Myc antibody affinity resins, tunicamycin, phorbol 12-myristate 13-acetate (PMA), and forskolin were products from Sigma. The anti-hβ1AR polyclonal antibody and anti-c-Myc (9E10) monoclonal antibody were from Santa Cruz Biotechnology. EZ-linked sulfo-N-hydroxysuccinimide (NHS) biotin and immobilized streptavidin were from Pierce. FLAG and c-Myc peptides were obtained from Sigma or were synthesized at the Biocenter Oulu Protein Analysis Core Facility. The ligands propranolol, isoproterenol (ISO), dobutamine, and 1-[2-((3-carbamoyl-4-hydroxy)phenoxy)ethylamino]-3-[4-(1-methyl-4-trifluoromethyl-2-imidazolyl)phenoxy]-2-propanol dihydrochloride (CGP-20712) were purchased from Tocris, and cell culture reagents were from BioWhittaker, Invitrogen, or Sigma. TAPI-1 (tumor necrosis factor-α protease inhibitor-1: N-(R)-[2-(hydroxyaminocarbonyl)methyl]-4-methylpentanoyl-l-naphthylalanyl-l-alanine,2-aminoethyl amide), GM6001, and its inactive form (negative control) were obtained from Calbiochem, and bisindolylmaleimide I was from Alexis. All the other reagents were of analytical grade and purchased from various commercial suppliers.
DNA Constructs
A DNA construct encoding the hβ1AR with a cleavable influenza hemagglutinin signal peptide (KTIIALSYIFCLVFA), N-terminal Myc tag (EQKLISEEDL), and C-terminal FLAG tag (DYKDDDDK) was created. Briefly, cDNA for the hβ1AR (GenBankTM accession number P08588) (a generous gift from Professor M. Bouvier, Montreal, Canada) was amplified by PCR using the oligonucleotides 5′-TCGCCCGCTAGCATGGGCGCGGGGGTGCTC-3′ and 5′-CGCCGGCCTAGGCACCTTGGATTCCGAGGC-3′, digested with NheI and AvrII (New England Biolabs), ligated into the pFT-SMMF vector, and transformed into Escherichia coli JM109. The hβ1AR construct with a mutated cleavage site (R31H,L32A) was created by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene) and 5′-GGCCACCGCGGCGCATGCGCTGGTGCCCGC-3′ and the complementary oligonucleotide. The N-terminally truncated hβ1AR constructs were cloned by PCR amplification from cDNAs encoding Met32–447 and Met53–447, using the primer pairs 5′-GTCGAAGCTTATGCTGCTGGTGCCCGCG-3′/5′-GTCGAAGCTTATGCTGTCTCAGCAGTGGACAG-3′, and 5′-CGCCGGCCTAGGCACCTTGGATTCCGAGGC-3′, followed by digestion with HindIII and AvrII (New England Biolabs), and ligation into the pFT-SMMF vector digested with the same enzymes. The pFT-SMMF vector was modified from the pcDNA5/FRT/TO vector (Invitrogen) as described previously (21). The DNA construct for the N-terminally HA-tagged hβ1AR in pcDNA3 (Invitrogen) was obtained from Professor M. Bouvier and has been described elsewhere (22).
Cell Culture and Transfections
Cells were cultured at 37 °C in a humidified atmosphere of 5% CO2. Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% (w/w) fetal bovine serum, 100 units/ml penicillin, 0.1 mg/ml streptomycin (complete DMEM), and the appropriate selection antibiotics, was used for the human embryonic kidney (HEK)293-derived cell lines. The medium was supplemented with Zeocin (100 μg/ml, Invitrogen or InvivoGen) and blasticidin S (4 μg/ml, InvivoGen) for the maintenance of the HEK293i cells that express the Tet repressor (23), and with Zeocin (100 μg/ml) for the Flp-In-293 cells (Invitrogen). Flp-In-CHO (Chinese hamster ovary) cells (Invitrogen) were cultured in Ham's F-12 nutrient mixture (Sigma), supplemented with 10% (w/w) fetal bovine serum, 2 mm glutamine, 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 100 μg/ml Zeocin. Stable cell lines with inducible hβ1AR expression were established by co-transfecting the receptor construct and pOG44 plasmid (Invitrogen) into HEK293i cells with the Lipofectamine 2000 transfection reagent (Invitrogen) under blasticidin S (4 μg/ml) and hygromycin (400 μg/ml) selection. For maintenance of the isolated clones, the hygromycin concentration was lowered to 100 μg/ml. The selected clones were sensitive to Zeocin, lacked β-galactosidase activity, and showed very low basal but highly inducible hβ1AR expression (see Fig. 1A). After a 24-h induction, the maximal binding capacity (Bmax) for the clone that was routinely used for the experiments reached 160 pmol/mg membrane protein, as determined by saturation binding assays with [3H]alprenolol. The calculated binding affinity for [3H]alprenolol (kd) was 2.9 nm. An independent clone with lower receptor expression was also isolated and was used to verify the key findings.
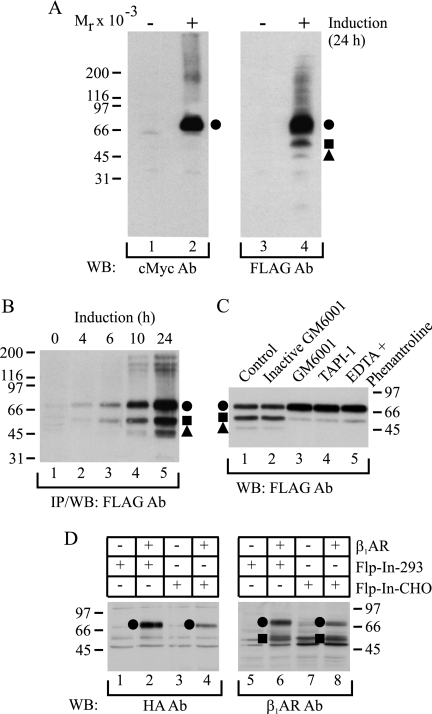
FIGURE 1.
hβ1AR is susceptible to N-terminal cleavage. A, receptor species expressed after long term induction. HEK293i cells stably transfected with the N-terminally Myc-tagged and C-terminally FLAG-tagged hβ1AR were induced with 0.5 μg/ml tetracycline for 24 h (lanes 2 and 4) or not (lanes 1 and 3). Isolated membranes were solubilized in SDS-sample buffer and subjected to Western blotting, using either anti-c-Myc (lanes 1 and 2) or anti-FLAG M2 (lanes 3 and 4) antibodies (Ab). B, time-dependent induction of receptor expression. Stably transfected HEK293i cells were induced for 0, 4, 6, 10, or 24 h, and the receptors were solubilized in DDM buffer, subjected to anti-FLAG M2 antibody immunoprecipitation, and analyzed by Western blotting with the same antibody. C, in vitro proteolysis. Stably transfected HEK293i cells were induced for 24 h, and cellular membranes were isolated using 25 mm Tris-HCl, pH 7.4, containing 2 μg/ml aprotinin, 0.5 mm phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 5 μg/ml trypsin inhibitor, and 10 μg/ml benzamidine (lane 1). The buffer was supplemented with 10 μm inactive GM6001 (lane 2), 10 μm GM6001 (lane 3), 10 μm TAPI-1 (lane 4), or 2 mm EDTA and 2 mm 1,10-phenanthroline (lane 5). The DDM-solubilized receptors were subjected to anti-FLAG M2 antibody immunoprecipitation in the presence of the respective reagents, and the purified samples were analyzed by Western blotting with the same antibody. D, receptor expression in transiently transfected cells. The N-terminally HA-tagged hβ1AR was transiently transfected into Flp-In-293 (lanes 2 and 6) or Flp-In-CHO (lanes 4 and 8) cells. Control cells (lanes 1, 3, 5, and 7) were transfected with vector DNA. The receptors were solubilized in DDM buffer and subjected to Western blotting, using either the anti-HA antibody (lanes 1–4) or the anti-hβ1AR antibody (lanes 5–8), which is directed against the C terminus of the receptor. The full-length receptor is indicated with a closed circle and the proteolytic fragments with a closed square and a triangle. Molecular weight markers are indicated. IP, immunoprecipitation; WB, Western blotting.
The cells for the experiments were plated onto culture flasks/plates (4.5 × 106 cells/75-cm2 flask or 100-mm plate or 1.5 × 106 cells/25-cm2 flask) and cultured for 3 days. Receptor expression was induced by adding tetracycline (0.5 μg/ml unless otherwise indicated) (Invitrogen) into the medium for different periods of time, as specified in the figures. Metalloproteinase inhibitors were added to the culture medium 60 min before tetracycline, except in the metabolic labeling experiments (see below). Transient transfections were performed as described earlier (24). Cells were harvested on ice in cold phosphate-buffered saline, quick frozen in liquid nitrogen, and stored thereafter at −70 °C.
Radioligand Binding Assays
Cellular membranes were prepared, and saturation ligand binding assays were performed as described previously (25), using 1–4 μg of membrane protein and increasing concentrations of [3H]dihydroalprenolol (PerkinElmer Life Sciences, 117.8 Ci/mmol; 0.1–15 nm). The nonspecific binding was determined in the presence of 10 μm propranolol.
Metabolic Labeling with [35S]Methionine/Cysteine
Prior to pulse-chase labeling, the stably transfected cells were treated with 0.5 μg/ml tetracycline for 16 h and then incubated in methionine- and cysteine-free DMEM for 60 min (depletion) before labeling in fresh medium containing 75 μCi/ml [35S]methionine/cysteine (EasyTagTM Express 35S-protein labeling mix, 1175 Ci/mmol; PerkinElmer Life Sciences) for 15 min. After washing twice with the chase medium (complete DMEM supplemented with 5 mm methionine), the cells were chased for various periods of time before harvesting, as specified in the figures. The transiently transfected cells were labeled in a similar manner 20 h after transfection. In the case of the brefeldin A (BFA, Alexis)-treated cells, the drug was added to the depletion medium to a final concentration of 5 μg/ml and was maintained thereafter. The βAR agonists ISO (0.001–10 μm) and dobutamine (10 μm), the protein kinase C activator PMA (0.5 μm), and the adenylyl cyclase activator forskolin (15 μm) were added to the chase medium 45 min after the pulse, and the βAR antagonist CGP-20712 (10 μm), the metalloproteinase inhibitor GM6001 (10 μm), and the protein kinase C inhibitor bisindolylmaleimide I (10 μm) 30 min after the pulse. The induction time for the tunicamycin-treated cells was 14 h, and the drug was added simultaneously with tetracycline to a concentration of 5 μg/ml, after which it was increased to 25 μg/ml during depletion, which was extended to 150 min. The chase was performed in the absence of the drug.
Preparation and Solubilization of Membranes and Whole Cell Extracts
Membranes and whole cell extracts were prepared and solubilized as described before (25). The homogenization buffers used for membrane preparation (25 mm Tris-HCl, pH 7.4, 20 mm N-ethylmaleimide) contained the protease inhibitors EDTA (2 mm), 1,10-phenanthroline (2 mm), aprotinin (2 μg/ml), phenylmethylsulfonyl fluoride (0.5 mm), leupeptin (5 μg/ml), soybean trypsin inhibitor (5 μg/ml), and benzamidine (10 μg/ml), although for some experiments the metalloproteinase inhibitors EDTA and 1,10-phenanthroline were omitted or replaced with inactive or active GM6001 (10 μm) or TAPI-1 (10 μm), as indicated in the figures. The buffer for membrane solubilization and for preparation of cellular lysates was supplemented with 0.5% (w/v) n-dodecyl β-d-maltoside (DDM) (Alexis) and 140 mm NaCl, without N-ethylmaleimide. Alternatively, the membranes were solubilized directly in SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol, 0.001% (w/v) bromphenol blue).
Immunoprecipitation of Solubilized Membranes
Solubilized membranes were supplemented with 0.1% (w/v) bovine serum albumin, subjected to immunoprecipitation using either immobilized anti-c-Myc or anti-FLAG M2 antibodies, and eluted with 200 μg/ml c-Myc or FLAG peptide, respectively, as described previously (26, 27).
Deglycosylation of Immunoprecipitated Receptors
Enzymatic deglycosylation was performed as described previously (23, 28). Prior to the digestions, the immunoprecipitates were diluted either 7.5- (Endo H and PNGase F) or 5-fold (neuraminidase and O-glycosidase) in the corresponding digestion buffers containing 1% (v/v) β-mercaptoethanol.
Cell Surface Biotinylation
Cell surface biotinylation was performed as described previously (26), and the receptors were subjected to one-step purification with the anti-c-Myc antibody or immobilized streptavidin (Fig. 2C) or two-step purification using either immobilized streptavidin followed by the anti-FLAG M2 antibody or two consecutive steps with the antibody (Fig. 7C).
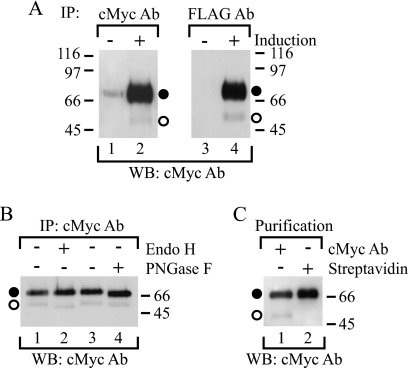
FIGURE 2.
Identification of full-length precursor and mature hβ1AR species. A, identification of the two full-length receptor species. Stably transfected HEK293i cells were treated with (lanes 2 and 4) or without (lanes 1 and 3) tetracycline for 6 h, and DDM-solubilized receptors were subjected to immunoprecipitation (IP) with anti-c-Myc (lanes 1 and 2) or anti-FLAG M2 (lanes 3 and 4) antibodies (Ab) and to Western blotting (WB) with the former antibody. B, deglycosylation of purified receptors. Stably transfected HEK293i cells were induced for 6 h, and anti-c-Myc antibody-immunoprecipitated receptors were digested with Endo H (50 milliunits/ml; lane 2) or PNGase F (50 units/ml; lane 4) for 16 h at 30 °C. The control samples (lanes 1 and 3) contained buffer only. The samples were analyzed by Western blotting using the anti-c-Myc antibody. C, cell surface biotinylation. Stably transfected HEK293i cells were induced for 6 h, and the cell surface proteins were labeled with sulfo-NHS-biotin (0.5 mg/ml) for 30 min on ice. DDM-solubilized receptors were purified with the anti-c-Myc antibody (lane 1) or immobilized streptavidin (lane 2) and subjected to Western blotting with the anti-c-Myc antibody. Symbols are as in Fig. 1.
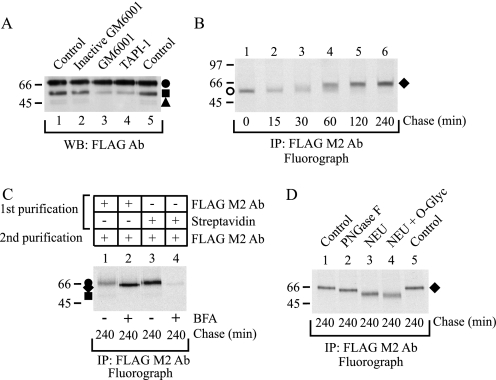
FIGURE 7.
hβ1AR N-terminal cleavage takes place after receptors leave the ER. A, inhibition of in vivo cleavage by metalloproteinase inhibitors. Stably transfected HEK293i cells were induced to express hβ1ARs for 6 h in the absence or presence of various protease inhibitors, as indicated. DDM-solubilized receptors were analyzed by Western blotting (WB) with the anti-FLAG M2 antibody (Ab). B, pulse-chase labeling of BFA-treated cells. Stably transfected HEK293i cells were induced, labeled with [35S]methionine/cysteine, and chased as described in the legend to Fig. 3 in the presence of 5 μg/ml BFA, which was added to the culture medium 60 min before labeling. DDM-solubilized receptors were purified with the anti-FLAG M2 antibody and analyzed by SDS-PAGE and fluorography. C, cell surface biotinylation. Stably transfected control and BFA-treated HEK293i cells were labeled as described for B and chased for 240 min. Cell surface proteins were biotinylated with sulfo-NHS biotin before harvesting. DDM-solubilized receptors were subjected to a two-step purification with the anti-FLAG M2 antibody (lanes 1 and 2; one-fifth of the samples) or immobilized streptavidin, followed by immunoprecipitation (IP) with the anti-FLAG M2 antibody (lanes 3 and 4; four-fifths of the samples). The samples were analyzed by SDS-PAGE and fluorography. D, enzymatic deglycosylation. Stably transfected BFA-treated HEK293i cells were labeled as described for B and chased for 240 min. DDM-solubilized receptors were purified with the anti-FLAG M2 antibody and digested with glycosidases for 16 h at 30 °C or not before analysis by SDS-PAGE and fluorography. Lanes 1 and 5, untreated controls. Lane 2, PNGase F (50 units/ml). Lane 3, neuraminidase (NEU; 50 milliunits/ml). Lane 4, neuraminidase (50 milliunits/ml) and O-glycosidase (O-glyc; 100 milliunits/ml). The receptor precursor in the BFA-treated cells is indicated with an open circle and the receptor species carrying partially processed oligosaccharides with a closed diamond. Symbols are as in Fig. 1.
SDS-PAGE, Western Blotting, and Fluorography
Samples were reduced by heating at 95 °C for 2 min in the presence of 50 mm dithiothreitol, and SDS-PAGE was run on a Bio-Rad Mini-PROTEAN 3 cell apparatus (10% SDS-polyacrylamide gels) using reagents from Bio-Rad or Amresco (NextGel system). Broad Range molecular weight standards from Bio-Rad were used as markers and stained with Ponceau S (Sigma) after blotting. Proteins were electroblotted onto Immobilon P (Millipore) or ProBlott (Applied Biosystems) membranes using a Bio-Rad mini trans-blot cell apparatus at 35–50 V for 16 h or 100 V for 60 min at 4 °C. The blots were probed with anti-FLAG M2 (0.5 μg/ml), anti-c-Myc 9E10 (1:10,000), anti-hβ1AR (1:100), or anti-HA (1:1,000) antibodies, followed by horseradish peroxidase-conjugated goat anti-mouse (1:15,000, Invitrogen), donkey anti-mouse F(ab)2, or donkey anti-rabbit F(ab)2 antibodies (Jackson Immunochemicals; 1:15,000 for ECL and 1:100,000 for ECL Plus, respectively). ECL or ECL Plus detection reagents (GE Healthcare) were used to reveal the blots. Gels containing radiolabeled samples were treated for fluorography as described previously (21). Exposed films were scanned with the Umax PowerLook 1120 color scanner and Image Master 2D Platinum 6.0 software, and the data were quantified and analyzed as described previously (21).
N-terminal Sequencing of Cleaved Receptor Fragments
Purified receptor fragments on ProBlott membranes were stained with Coomassie Blue as instructed by the manufacturer. The fragments were excised and subjected to automated Edman degradation on the protein sequencer ProciseTM 492 (Applied Biosystems). The amino acid residues sequentially removed from the N terminus were identified by reverse-phase high performance liquid chromatography.
Sequence Alignment
The sequence alignment was performed as described previously (24).
Data Analysis
The data were analyzed using the GraphPad Prism 4.01 software.
RESULTS
hβ1AR Is Subject to Metalloproteinase-mediated N-terminal Cleavage in Vitro
The hβ1AR was expressed in stably transfected tetracycline-inducible HEK293i cells with N-terminal Myc and C-terminal FLAG epitope tags. Western blot analysis of solubilized membranes from 24-h induced cells revealed that only one major specific band with an apparent molecular weight of 69,000 was detected with the anti-c-Myc antibody (Fig. 1A, lane 2). This species was also recognized with the anti-FLAG M2 antibody (Fig. 1A, lane 4), indicating that it represents the full-length receptor. Interestingly, the latter antibody also detected smaller specific species of Mr 54,000 and 47,000 (Fig. 1A, lane 4), the intensity of which varied from one experiment to another. The relative abundance of the three receptor forms remained unaltered in cells that were induced for various periods of time (Fig. 1B), indicating that their appearance was not dependent on the receptor expression level. Thus, the smaller molecular weight species are likely to represent proteolytic products of the full-length receptor, missing a part of their N terminus. This conclusion is consistent with previous reports on N-terminal proteolysis in the turkey β1AR (29).
Because various protease inhibitors, including those that affect metalloproteinases, have been shown to inhibit the proteolysis of βARs in vitro (29–31), we tested this possibility on the hβ1AR by omitting EDTA and 1,10-phenanthroline routinely used for membrane preparation from the homogenization buffer. As seen in Fig. 1C, the cleaved receptor species were substantially less abundant in the presence of the two inhibitors than in their absence (lanes 5 and 1, respectively). The cleavage was also inhibited by GM6001, a broad spectrum hydroxamate inhibitor of metalloproteinases (Fig. 1C, lane 3), but not by its inactive form (Fig. 1C, lane 2), and likewise by TAPI-1, another hydroxamate inhibitor showing some specificity for tumor necrosis factor-α-converting enzyme (TACE; also known as a disintegrin and metalloproteinase (ADAM)-17)) (Fig. 1C, lane 4). These results indicate that the hβ1AR is very susceptible to in vitro proteolysis, resulting in two cleaved fragments. The cleavage is mediated by a matrix metalloproteinase or a protease belonging to the ADAM family of metalloproteinases.
To verify that the observed susceptibility of the hβ1AR to N-terminal cleavage was not dependent on the Myc and FLAG epitope tags added to the receptor N and C termini, an N-terminally HA-tagged receptor construct was transiently transfected into Flp-In-293 cells, and the receptor species expressed were identified by Western blotting using either HA antibody or an antibody directed against the receptor C terminus. As expected, the former antibody recognized only one receptor species, whereas the C-terminal antibody identified two specific receptor forms (Fig. 1D, lanes 2 and 6, respectively). Similar results were obtained with transiently transfected Flp-In-CHO cells (Fig. 1D, lane 4 and 8, respectively), confirming that the N-terminal cleavage of the receptor is not a cell-specific phenomenon.
hβ1AR Is Expressed as Two Full-length Receptor Forms
The predicted molecular weight of the hβ1AR as calculated from the amino acid sequence (477 amino acids) is 51,323 (15). With the added Myc and FLAG epitope tags, the calculated molecular weight for the construct used for preparing the stable cell line was 53,815. Thus, the apparent Mr 69,000 detected for the full-length receptor by Western blotting suggests that this species represents a post-translationally modified cell surface receptor rather than an intracellular biosynthetic intermediate. To characterize the Mr 69,000 hβ1AR species further and to find out whether a biosynthetic intermediate can be identified, the receptors were immunoprecipitated with either anti-c-Myc or anti-FLAG M2 antibodies and subjected to Western blotting with the former antibody. This technique visualizes only the full-length receptor. As expected, the Mr 69,000 receptor species was detected in both immunoprecipitates (Fig. 2A, lanes 2 and 4), but in addition, one smaller band became apparent (Fig. 2A, lanes 2 and 4). This barely detectable Mr 54,100 species migrated slightly more slowly in SDS-PAGE than the more abundant Mr 54,000 N-terminally cleaved receptor form that was detected in the anti-FLAG M2 antibody immunoprecipitates probed with the same antibody (see Fig. 1B; see also Fig. 3B).
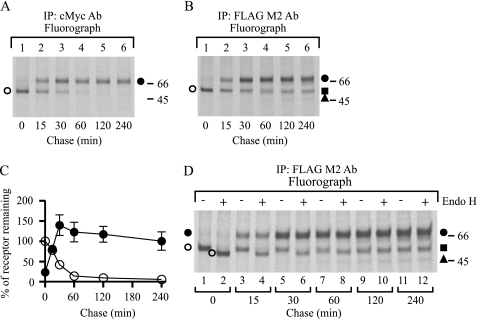
FIGURE 3.
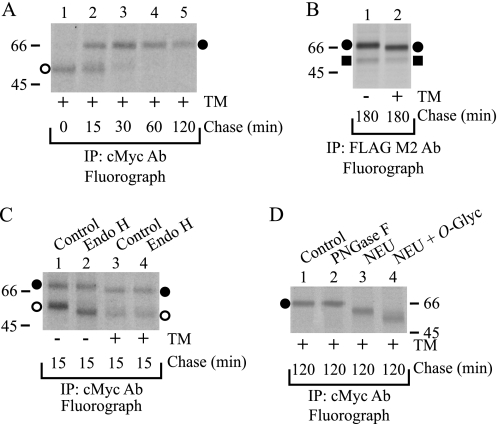
Pulse-chase analysis of hβ1AR synthesis and processing. Stably transfected HEK293i cells were treated with tetracycline for 16 h, labeled with 75 μCi/ml [35S]methionine/cysteine for 15 min, and chased for the times indicated. Cellular membranes were isolated, and the DDM-solubilized extracts were divided into 2 equal aliquots and subjected to immunoprecipitation (IP) with immobilized anti-c-Myc (A) or anti-FLAG M2 antibodies (Ab) (B). The latter samples were further subjected to Endo H digestion (50 milliunits/ml, 16 h, 30 °C), or not, as indicated (D). Aliquots were analyzed by SDS-PAGE and fluorography. C shows the time course of the appearance and disappearance of mature and precursor receptor forms, respectively, when purified with the anti-c-Myc antibody. Symbols refer to those used to identify the different receptor forms. The intensities of labeled receptor species were obtained by densitometric scanning, the values being normalized to the labeling of the receptor precursors at the end of the pulse. The values given are means ± S.E. from seven independent experiments. Symbols are as in Fig. 1.
The identities of the two full-length Mr 54,100 and 69,000 receptor species were studied by enzymatic deglycosylation. As the former species was sensitive to Endo H (Fig. 2B, lane 2), it must represent a receptor carrying mannose-type N-glycans, as is typical for intracellular biosynthetic intermediates. The shift in its electrophoretic mobility was small (∼3,000), yet detectable, correlating with the removal of one N-linked glycan attached to Asn15, the only putative N-glycosylation site on the hβ1AR (15). In contrast, the Mr 69,000 species was sensitive only to PNGase F and was digested to a species of Mr 67,000 (Fig. 2B, lane 4). This indicates that the N-glycan of this receptor form has been processed in the Golgi to a hybrid or complex type. Its electrophoretic mobility was not increased any further when the PNGase F concentration was increased (data not shown), indicating that the enzyme reaction was complete. Thus the larger full-length receptor form apparently carries other post-translational modifications in addition to the one N-glycan.
The cellular localization of the Mr 69,000 and 54,100 receptor forms was then investigated by cell surface biotinylation. Induced cells were treated with membrane-impermeable sulfo-NHS-biotin, and the receptors, which were purified with immobilized anti-c-Myc antibody or streptavidin, were identified by Western blotting using the anti-c-Myc antibody. As expected, both receptor forms were purified by the antibody, but only the Mr 69,000 form was purified by streptavidin (Fig. 2C, lanes 1 and 2, respectively). These results confirm that the Mr 69,000 receptor form represents the mature cell surface species and the Mr 54,100 one the intracellular precursor.
Maturation of the hβ1AR Is Efficient and Displays Relatively Fast Kinetics
The low relative amount of hβ1AR precursors in HEK293i cells suggests that synthesized receptors mature efficiently, a finding that is in contrast to those obtained in the same cellular background for two other GPCRs in the rhodopsin family, namely the human δ-opioid (24, 26) and the rat-luteinizing hormone receptors (21). Thus, to investigate hβ1AR processing and maturation in more detail, cells were subjected to metabolic labeling. Induced cells were pulse-labeled with [35S]methionine/cysteine for 15 min, chased for 0, 15, 30, 60, 120, or 240 min, and subjected to immunoprecipitation with the anti-c-Myc antibody. As seen in Fig. 3A, the major receptor form detected at the end of the pulse was the Mr 54,100 precursor (lane 1). This had already disappeared after the 60-min chase, however, and its half-life was calculated to be about 23 min (Fig. 3C). In line with the fast disappearance of the precursor, the mature Mr 69,000 receptor form was already detected at the end of the 15-min pulse (Fig. 3A, lane 1), and the calculated half-time for its maturation was only 26 min (Fig. 3C). Thus, as predicted from the high mature receptor to precursor ratio seen in Fig. 2A, hβ1AR maturation appears to be both efficient and rapid.
The labeled samples were then subjected to purification with the anti-FLAG M2 antibody to examine the appearance of the cleaved receptor forms. As seen in Fig. 3B, the fragments were clearly observable after the 60-min chase (lane 4), at a time when the receptor precursors had almost disappeared (compare Fig. 3A, lane 4). This suggests that only mature receptors are susceptible to cleavage. To confirm this notion, the purified samples were digested with Endo H. As expected (Fig. 3D), only the precursors were sensitive to the enzyme. The more slowly migrating Endo H-resistant receptor fragment first became detectable after the 15-min chase (Fig. 3D, lanes 3–4), was the predominant form after 60 min, and remained relatively stable until the end of the chase (lanes 9–12).
hβ1AR Is O-Glycosylated
Analysis of the amino acid sequence of the hβ1AR N-terminal domain revealed that it contains numerous Ser and Thr residues that may be O-glycosylation sites (Fig. 4). This post-translational modification was suggested by the observation that precursor and mature receptor forms showed a considerable difference in migration in the enzymatic N-deglycosylation experiments (see Fig. 2B). To investigate O-glycosylation of the hβ1AR, N-glycosylation was first inhibited with tunicamycin, and the effectiveness of the drug treatment was verified in pulse-chase labeling experiments. As seen in Fig. 5A, lane 1, a single receptor species of Mr 51,000 was purified from the treated cells with the anti-c-Myc antibody at the end of the pulse. This species was resistant to Endo H and co-migrated with Endo H-digested precursors from the nontreated control cells (Fig. 5C, compare lanes 2 and 4). During the chase, the non-N-glycosylated precursor was converted to a species of Mr 67,000 (Fig. 5A). This confirms that the receptor acquires additional modifications after leaving the endoplasmic reticulum (ER). Interestingly, when chase samples from the tunicamycin-treated cells were subjected to anti-FLAG M2 antibody purification, a smaller receptor fragment was identified, which co-migrated with the Mr 54,000 fragment of the nontreated control cells (Fig. 5B). This indicates that the N-glycan at Asn15 does not appear to have any effect on receptor cleavage.
FIGURE 4.
Sequence analysis of the N terminus of vertebrate β1ARs. The sequence alignment was performed as described under “Experimental Procedures.” The last N-terminal amino acid preceding the first transmembrane domain was predicted according to the crystal structure of the turkey β1AR (18). The O-glycosylated Ser/Thr residues predicted by the NetOGlyc 3.1 Server (52) are indicated in boldface. The conserved consensus sequence for N-glycosylation is framed, and the hβ1AR proteolytic cleavage sites identified are indicated with arrows. Dashes show gaps introduced in the sequence to optimize the alignment. The accession numbers are as follows: P08588 (Homo sapiens), P47899 (Macaca mulatta), Q9TT96 (Bos taurus), P79148 (Canis familiaris), Q9TST6 (Felis silvestris catus), Q28998 (Sus scrofa), B0FL73 (Cavia porcellus), P34971 (Mus musculus), P18090 (Rattus norvegicus), Q28927 (Ovis aries), P07700 (Meleagris gallopavo), O42574 (Xenopus laevis), and B3DHM6 (Danio rerio).
FIGURE 5.
hβ1AR carries O-linked glycans. Stably transfected HEK293i cells were labeled with [35S]methionine/cysteine as described in the legend to Fig. 3, in the presence or absence of 25 μg/ml tunicamycin (TM), as indicated. DDM-solubilized receptors were subjected to immunoprecipitation (IP) with anti-c-Myc (A, C, and D) or anti-FLAG M2 (B) antibodies (Ab) and analyzed by SDS-PAGE and fluorography. Some samples were subjected to enzymatic deglycosylation for 16 h at 30 °C or not before SDS-PAGE, as indicated (C and D). C, lanes 1 and 3, untreated control. Lanes 2 and 4, Endo H (50 milliunits/ml). D, lane 1, untreated control. Lane 2, PNGase F (50 units/ml). Lane 3, neuraminidase (NEU; 50 milliunits/ml). Lane 4, neuraminidase (50 milliunits/ml) and O-glycosidase (O-glyc; 100 milliunits/ml). Symbols are as in Fig. 1.
To find out whether the Mr 67,000 receptor form in the tunicamycin-treated cells carries O-glycans, samples that had been chased for 2 h were subjected to enzymatic deglycosylation. The Mr 67,000 species was resistant to PNGase F, as was expected, but was sensitive to both neuraminidase and O-glycosidase, resulting in a species of Mr 59,000 (Fig. 5D). Thus, the receptor carries sialylated O-glycans. In view of the specificity of O-glycosidase (32), it can be concluded that the O-glycans are of the mucin-type and consist of the disaccharide galactose-N-acetylgalactosamine attached to Ser/Thr.
Identification of N-terminal Cleavage Sites of the hβ1AR
The finding of a cleaved receptor fragment of Mr 54,000 in both tunicamycin-treated and nontreated control cells (Fig. 5B) suggests that the cleavage site resulting in the formation of this species lies in the C-terminal direction relative to Asn15. To confirm this finding, we subjected anti-FLAG M2 antibody immunoprecipitated samples of nontreated control cells to deglycosylation with PNGase F and analyzed the samples by Western blotting. EDTA and 1,10-phenanthroline were omitted from the homogenization buffer to maximize the amount of the cleaved receptor species. PNGase F reduced the molecular weight of the full-length receptor to about Mr 67,000 in a concentration-dependent manner, but it had no effect on the electrophoretic mobility of the Mr 54,000 or 47,000 species (Fig. 6A, lanes 1–5). Thus, the hβ1AR appears to be cleaved in the N terminus at sites that reside in a C-terminal direction relative to Asn15. This is consistent with findings obtained for the turkey β1AR, which has been shown to be cleaved between amino acids 15 and 28, after the consensus site for N-linked glycosylation at Asn14 (29, 33).
FIGURE 6.
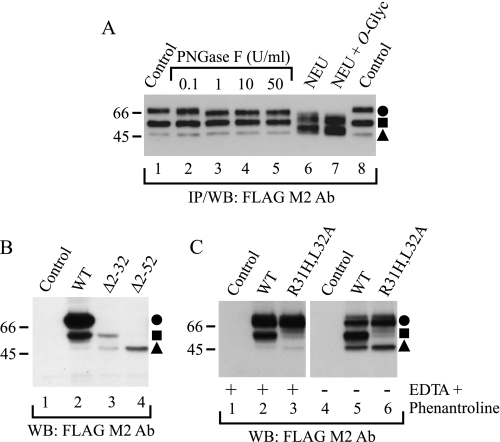
Identification of hβ1AR cleavage sites. A, localization of cleavage sites in relation to N- and O-glycans. Stably transfected HEK293i cells were induced for 24 h, and DDM-solubilized receptors were purified with the anti-FLAG M2 antibody (Ab). EDTA and 1,10-phenanthroline were omitted from the homogenization buffer. Aliquots of the eluates were subjected to digestion with 0.1, 1, 10, or 50 units/ml PNGase F (lanes 2–5, respectively) or, alternatively, with 50 milliunits/ml neuraminidase (NEU; lane 6), or 50 milliunits/ml neuraminidase and 100 milliunits/ml O-glycosidase (O-glyc; lane 7) for 16 h at 30 °C. The control samples (lanes 1 and 8) contained buffer only. B, identification of the cleavage sites by means of receptor truncation mutants. C, site-directed mutagenesis of the major cleavage site at Arg31. Constructs for the wild-type hβ1AR, the truncation mutants hβ1ARΔ2–31 and hβ1ARΔ2–52, and the R31H,L32A mutant were transiently expressed for 24 h in Flp-In-293 cell, the control cells being transfected with vector DNA. Cells were homogenized in the absence or presence of EDTA and 1,10-phenanthroline, as indicated, and DDM-solubilized receptors were subjected to Western blotting (WB) using the anti-FLAG M2 antibody. The truncation mutants were expressed at a substantially lower level than the wild-type receptor (C), most likely because they lacked the HA signal sequence. WT, wild type; IP, immunoprecipitation. Symbols are as in Fig. 1.
The hβ1AR N-terminal domain carries several Ser/Thr residues that are located C-terminally relative to Asn15 (Fig. 4). We therefore tested whether the proteolytic fragments carry O-glycans by subjecting the immunoprecipitated receptors to digestion with neuraminidase and O-glycosidase. As seen in Fig. 6A, lanes 6 and 7, the two enzymes were able to reduce the molecular weight of the full-length and the cleaved Mr 54,000 receptor down to about Mr 61,000 and 48,000, respectively. However, the enzymes were not able to digest the cleaved Mr 47,000 species. These results are consistent with the notion that the cleavage site resulting in the formation of the larger fragment lies C-terminally relative to at least one of the O-glycosylated Ser/Thr residues that are located after the N-glycosylated Asn15. On the other hand, the second cleavage site resulting in the formation of the smaller receptor fragment lies C-terminally relative to all the O-glycosylation sites (Fig. 4).
To identify the cleavage sites more specifically, a larger scale receptor purification was performed using cellular membranes that were prepared in a buffer devoid of EDTA and 1,10-phenanthroline. After a two-step immunoprecipitation with the anti-FLAG M2 antibody, the purified samples were subjected to SDS-PAGE and blotted onto PVDF membrane, and the Coomassie Blue-stained bands of Mr 54,000 and 47,000 were excised and subjected to N-terminal sequencing. The sequence obtained for the larger cleaved fragment, 32LLVPA36, was found to reside C-terminally relative to Asn15 and one of the predicted O-glycosylation sites, Thr28 (Fig. 4). Thus, the first cleavage site was identified as being between Arg31 and Leu32. The Mr 47,000 fragment gave the sequence 53LSQQXT58 (Trp at position 57 was not identified), which is located close to the first transmembrane domain of the receptor, showing the second cleavage site to lie between Pro52 and Leu53 (Fig. 4).
To verify the cleavage sites determined by N-terminal sequencing, alternative strategies were employed. First, the receptor constructs hβ1ARΔ2–31 and hβ1ARΔ2–52 were prepared, which were truncated at the first and second proposed cleavage sites, respectively. These truncated receptors lacking the N-terminal Myc tag were then expressed in transiently transfected Flp-In-293 cells and analyzed by Western blotting using the anti-FLAG M2 antibody. As seen in Fig. 6B, the larger truncated receptor, hβ1ARΔ2–31, was expressed as two species that co-migrated with the wild-type receptor fragments of Mr 54,000 and 47,000, whereas the second truncated receptor, hβ1ARΔ2–52, was expressed as a single species that co-migrated with the smaller cleaved form.
As another way of confirming the cleavage sites, a mutant receptor construct was prepared in which the Arg31 and Leu32 that flank the first proposed cleavage site were replaced by His and Ala, respectively. Analysis of the expressed N-terminally Myc-tagged and C-terminally FLAG-tagged protein in transiently transfected Flp-In-293 cells (Fig. 6C) revealed that these mutations inhibited the appearance of the larger receptor fragment but not that of the smaller fragment. Taken together, these results confirm the two cleavage sites identified at the receptor N terminus and suggest further that the two cleavage steps do not occur sequentially but are independent events. Furthermore, the cleavage at Arg31↓Leu32 appears to be highly sequence-specific.
N-terminal Cleavage of the hβ1AR Also Occurs in Vivo
As only the mature hβ1AR carrying fully processed N-glycans appears to be subject to proteolytic cleavage, and as the pulse-chase labeling experiments showed that the cleaved receptor fragments appeared with somewhat slower kinetics than the full-length receptor, cleavage of the hβ1AR is not likely to be entirely an in vitro event. We thus tested whether the cleavage can be inhibited in vivo. For this purpose GM6001 and TAPI-1 were added to the culture medium 60 min after tetracycline, and receptor expression was induced for 5 h. As seen by Western blotting (Fig. 7A), both inhibitors, but not the inactive GM6001, were able to reduce the amount of the Mr 54,000 cleaved product, in line with the view that at least the cleavage between Arg31 and Leu32 takes place in vivo.
To find out whether the cleavage requires receptor delivery to the cell surface, the appearance of receptor fragments was studied in metabolically labeled cells treated with the transport blocker BFA, which causes disassembly of the Golgi and intracellular accumulation of newly synthesized proteins (25, 34). Purification of labeled receptors from BFA-treated cells with the anti-FLAG M2 antibody revealed that the hβ1AR precursors were converted to the Mr 66,000 species (Fig. 7B), and notably, no smaller receptor species were identified. This was in contrast to the situation in the nontreated controls (compare lane 6 in Fig. 7B with Fig. 3B) or the tunicamycin-treated cells (Fig. 5B). The Mr 66,000 receptor remained intracellular, as was verified by cell surface biotinylation (Fig. 7C), and carried both N- and O-glycans (Fig. 7D). This was expected, because Golgi enzymes are retrotranslocated to the ER in BFA-treated cells, causing partial processing of N- and O-glycans in the ER-retained proteins (26). Taken together, these results indicate that the N-terminal cleavage of the receptor occurs in vivo after the newly synthesized receptors have been transported through the trans-Golgi network to the plasma membrane.
We also tested whether the cleaved hβ1AR N terminus is extracted into the culture medium and made several attempts to immunoprecipitate the cleaved fragment from the medium with the anti-c-Myc antibody. The attempts were unsuccessful, however. This negative finding suggests that the fragment (or the N-terminal epitope) is either degraded soon after cleavage or that the fragment somehow remains attached to cellular membranes despite the cleavage. Alternative explanations would be that the Myc epitope of the cleaved fragment is masked as a consequence of a conformational change in the peptide structure or that the cleavage occurs in endosomes after internalization.
In Vivo N-terminal Cleavage of the hβ1AR Is a Regulated Event
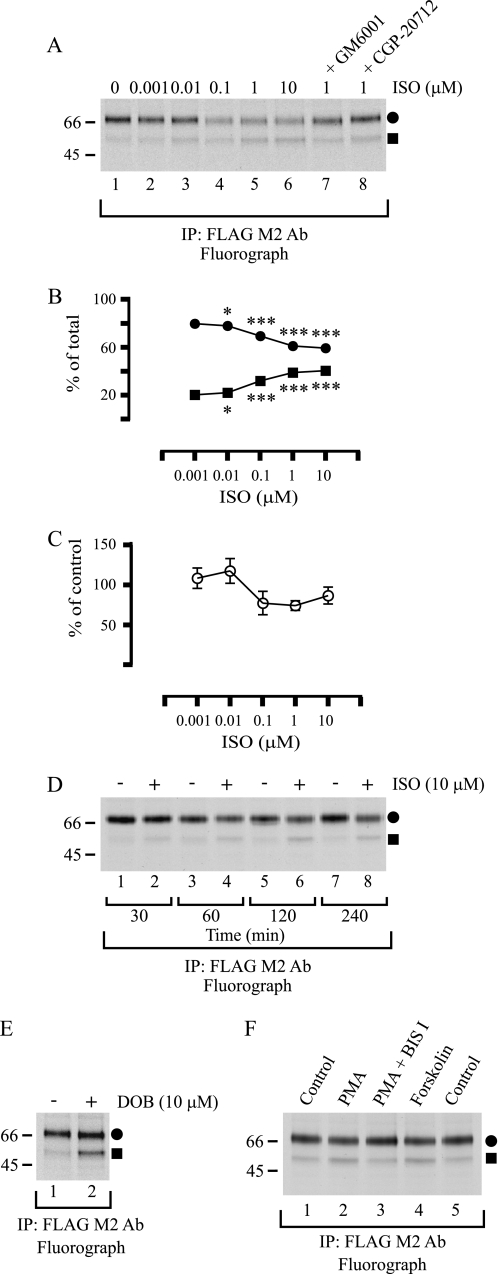
We next wanted to find out whether there is any regulation of hβ1AR N-terminal cleavage. For this purpose, cells were subjected to metabolic labeling, and the appearance of cleaved receptor fragments was studied during the chase in the presence of the βAR agonist ISO. When the ligand was added to the medium 45 min after the beginning of the chase and cells were harvested 195 min later, there was a significant concentration-dependent increase in the proportion of the Mr 54,000 species relative to the full-length receptor (Fig. 8, A and B). At the same time, the total amount of receptor decreased, but only slightly (Fig. 8, A and C). The changes in the relative amounts of the full-length and cleaved receptor fragments were already apparent after 30 min of incubation and were enhanced with time (Fig. 8D), but they could be inhibited by adding 10 μm GM6001 to the medium 15 min before ISO (Fig 8A, compare lane 5 with 7). Another βAR agonist, dobutamine, was also able to enhance receptor cleavage (Fig. 8E), whereas the enhanced cleavage induced by 1 μm ISO was inhibited with 10 μm CGP-20712, a β1AR-specific antagonist (Fig. 8A, compare lane 5 with 8). These results are in line with the notion that the agonist-induced β1AR cleavage most likely results from direct activation of the receptor.
FIGURE 8.
hβ1AR N-terminal cleavage is a regulated process. A, receptor activation by increasing concentrations of the βAR-agonist ISO. Stably transfected HEK293i cells were induced, labeled with [35S]methionine/cysteine, and chased for 240 min, as described in the legend to Fig, 3. ISO at the concentrations indicated was added to the chase medium 45 min after the beginning of the chase. GM6001 (10 μm) and CGP-20712 (10 μm) were added 15 min prior to ISO. Cellular membranes were prepared using a buffer containing EDTA and 1,10-phenanthroline, and DDM-solubilized receptors were purified with the anti-FLAG M2 antibody (Ab) and analyzed by SDS-PAGE and fluorography. Changes in the relative amounts of the full-length mature receptor (a closed circle) and the larger cleaved fragment (a closed square), as revealed by densitometric scanning of fluorograms, are shown in B. The data are expressed as (Fl/(Fl + Fr)) × 100 and (Fr/(Fl + Fr)) × 100 for the full-length receptor and cleaved fragment, respectively, where Fl = full-length receptor, and Fr = receptor fragment. C shows relative changes in the total amount of receptor. The values were normalized to that in the nontreated cells, which was set to 100%. The data shown represent the means ± S.E. from three independent experiments and were analyzed by GraphPad Prism using the repeated measures two-way analysis of variance followed by Bonferroni's post-test to compare the ISO-treated samples with the nontreated controls. The post-test did not reveal any significant differences for the data in C, although the analysis of variance performed before normalization revealed significant differences between the samples (p = 0.0081). D, time-dependent receptor activation by ISO. E, receptor activation by dobutamine. F, activation of protein kinase C and adenylyl cyclase by PMA and forskolin, respectively. Stably transfected HEK293i cells were induced and labeled with [35S]methionine/cysteine as described for A. ISO (10 μm) was added to the chase medium 45 min after the beginning of the chase or not, and the cells were harvested 30, 60, 120, or 240 min later, as indicated (D). Dobutamine (DOB) (10 μm) was added 45 min after the beginning of the chase, and the cells were harvested 195 min later (E), Bisindolylmaleimide I (Bis I; 10 μm) was added 30 min after the beginning of the chase, and PMA (0.5 μm) and forskolin (15 μm) 15 min later, and the cells were harvested after a further 195 min (F). DDM-solubilized receptors were analyzed as described for A. Symbols are as in Fig. 1. ***, p < 0.001; *, p < 0.05. IP, immunoprecipitation.
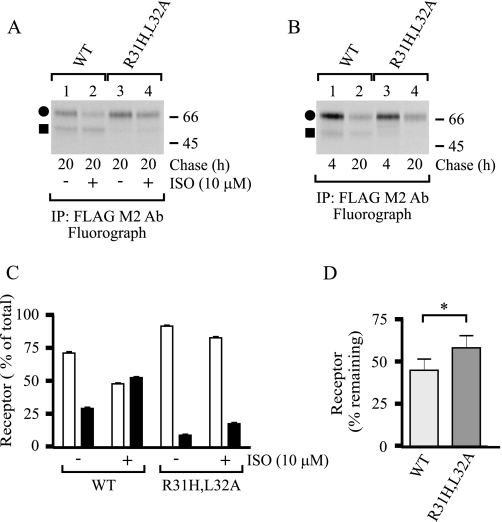
As the N-terminal cleavage of the hβ1AR appears to be a regulated event involving agonist-mediated activation, it can be speculated that it may have a role in receptor down-regulation. To test this possibility, the wild-type receptor and the R31H,L32A cleavage site mutant were transiently transfected into Flp-In-293 cells, and ISO-mediated long term changes in receptor numbers were monitored in the pulse-chase labeling assay. A 20-h treatment with ISO enhanced cleavage of the wild-type receptor (Fig. 9, A and C), and the amount of total receptors was decreased by 26 ± 6% when compared with the nontreated control cells. The cleavage of the mutant receptor was significantly attenuated, as expected, but was enhanced in ISO-treated cells (Fig. 9, A and C). The agonist-mediated decrease in the amount of total mutant receptors was similar than in cells expressing the wild-type receptor (26 ± 8%). Agonist-induced hβ1AR down-regulation thus does not appear to be dependent on receptor cleavage at the Arg31↓Leu32 cleavage site. It can be argued that the residual cleavage that occurs at, or very close to, the mutated site was adequate enough to lead to similar long term down-regulation of the mutant receptor. Interestingly, the total amount of the mutant receptor was consistently higher than that of the wild-type receptor in the absence of ISO (Fig. 9A, compare lane 1 with 3), suggesting that the N-terminal cleavage might have a role in the general turnover of mature receptors. Consistent with this idea, the total amount of wild-type and R31H,L32A mutant receptors decreased by 55 and 42% within 16 h, respectively (Fig. 9, B and D).
FIGURE 9.
Inhibition of the N-terminal cleavage stabilizes the mature hβ1AR. The N-terminally Myc-tagged and C-terminally FLAG-tagged wild-type (WT) and R31H,L32A mutant hβ1ARs were transiently transfected into Flp-In-293 cells. The cells were labeled with [35S]methionine/cysteine after 20 h and chased for 4 or 20 h, as described in the legend to Fig. 3. ISO (10 μm) was added to the chase medium or not 45 min after the beginning of the chase. Cellular membranes were prepared using a buffer containing EDTA and 1,10-phenanthroline, and DDM-solubilized receptors were purified with the anti-FLAG M2 antibody (Ab) and analyzed by SDS-PAGE and fluorography (A and B). Receptor bands were quantified by densitometric scanning of the fluorograms. Agonist-mediated changes in the relative amounts of the full-length mature receptors (closed circles and white bars) and the larger cleaved fragments (closed squares and black bars), are shown in C. The total amounts of receptors were set to 100%. D shows the decreases in the total receptor amount within 16 h in cells not treated with ISO. The data represent the means ± S.E. from five (C) and four (D) independent experiments, and were analyzed by GraphPad Prism using the paired t test (D). Symbols are as in Fig. 1. *, p < 0.05. IP, immunoprecipitation.
Finally, we wanted to assess whether the hβ1AR N-terminal cleavage might be regulated in a heterologous manner. As seen in Fig. 8F, addition of PMA to the cell culture medium in the pulse-chase labeling assay increased the proportion of the Mr 54,000 receptor species, whereas the enhanced cleavage was inhibited with 10 μm bisindolylmaleimide I, a protein kinase C inhibitor (lanes 2 and 3). The cleavage was also enhanced by forskolin (Fig. 8F, lane 4). These results suggest that hβ1AR cleavage maybe under heterologous regulation, involving protein kinase C as well as adenylyl cyclase activation.
DISCUSSION
The present description of the biosynthetic profile of the hβ1AR in a heterologous expression system reveals several distinct features of the behavior of the receptor that are relevant to its regulation. We show that the hβ1AR is a GPCR that is efficiently expressed at the cell surface and is extensively glycosylated during transport to the plasma membrane. One N-glycan is added to the conserved Asn at position 15, and several mucin-type sialylated O-glycans are added to the N-terminal Ser/Thr residues. However, despite these modifications, the N terminus appears to be very susceptible to cleavage by a metalloproteinase of either the matrix metalloproteinase or the ADAM families, resulting in the formation of at least two smaller membrane-bound C-terminal receptor fragments. The cleavage was found to take place constitutively in vivo and, importantly, was enhanced after agonist-mediated receptor activation or direct stimulation of protein kinase C and adenylyl cyclase. Furthermore, inhibition of the cleavage by mutation of the major Arg31↓Leu32 cleavage site of the receptor was found to stabilize the mature receptors. These results suggest that the extracellular N terminus of the β1AR has a distinct functional role in receptor regulation. This N-terminal cleavage may be a way of altering the number of β1ARs at the cell surface, thus regulating cellular responsiveness to βAR ligands.
Metalloproteinase-mediated N-terminal cleavage of the β1AR was first reported for the turkey receptor expressed in erythrocytes (29). Various mammalian species, including rats, rabbits, dogs, pigs, and humans, have also been shown to express multiple β1AR forms in endogenous tissues (31), and the same phenomenon has been described for a number of heterologous expression systems expressing hβ1ARs (10, 13, 14). Thus, susceptibility to limited proteolysis appears to be an evolutionally conserved property of this GPCR. Nevertheless, the general notion has been that the cleavage is entirely an in vitro event that takes place during sample preparation. To our knowledge, this study represents the first demonstration that the receptor is also cleaved in a regulated manner in vivo. Several lines of direct and indirect evidence support this notion. (i) Only the mature hβ1AR with fully processed N- and O-glycans was found to be cleaved, whereas the receptor precursor existed only as a full-length polypeptide. In pulse-chase labeling experiments, the cleaved receptor fragments appeared after the receptor precursors had been converted to the mature form, and furthermore, if receptor transport to the cell surface was blocked with BFA, no cleavage occurred. These results suggest that the receptor is cleaved constitutively after the newly synthesized receptors have reached the cell surface or are being transported through the trans-Golgi network. (ii) The appearance of the cleaved receptor fragments was partially inhibited by adding the metalloproteinase inhibitors GM6001 and TAPI-1 to the culture medium, whereas the inactive GM6001 had no apparent effect. (iii) Inhibition of the N-terminal cleavage by mutation of the major Arg31↓Leu32 cleavage site stabilized mature receptors. (iv) The cleavage was enhanced by PMA and forskolin, indicating that the event can be modulated by protein kinase C and adenylyl cyclase activation. (v) Furthermore, the βAR agonists ISO and dobutamine were found to enhance the cleavage. As the N-terminal sequences of the Mr 54,000 fragments purified from cells treated with 10 μm ISO for 3 h or not were identical (data not shown), the cleavage induced by the agonists appears to take place at the same site(s) as the cleavage occurring in vitro during membrane preparation. The agonist-mediated enhancement of receptor cleavage was both concentration- and time-dependent and was inhibited by the β1AR-specific antagonist CGP-20712. There is thus compelling evidence that mature hβ1ARs are subject to both homologous and heterologous regulation via limited proteolysis of the N terminus.
Metalloproteinase-mediated N-terminal cleavage has been described previously for a limited number of GPCRs in the rhodopsin family, i.e. for endothelin B (35, 36) and thyrotropin (37–39) receptors, and for the protease-activated receptor-1 (PAR-1) (40, 41), which is the main GPCR for thrombin. The V2 vasopressin receptor, on the other hand, has been shown to be cleaved at the interface between the second transmembrane domain and the first extracellular loop (42, 43). Consistent with the findings obtained for the hβ1AR, the endothelin B, V2 vasopressin, and thyrotropin receptors are cleaved in an activation-dependent manner following agonist binding (36–38, 42), although conflicting reports have been published for the thyrotropin receptor (44). On the other hand, the thrombin-mediated cleavage of PAR-1 has been shown to take place following protein kinase C activation (40), and furthermore, it has been shown that matrix metalloproteinase I can mediate the cleavage and activation of PAR-1, which leads to invasion and metastasis of tumor cells (41). Otherwise, the functional consequences of metalloproteinase-mediated cleavage have not been fully revealed for any other family A GPCRs. The existence of this phenomenon in distinct subfamilies of the rhodopsin-type GPCRs suggests, however, that regulated cleavage may be a more common property than has previously been anticipated.
What is the functional significance of the hβ1AR N-terminal cleavage? Several other cell surface transmembrane proteins are known to be cleaved in the extracellular domain by metalloproteinases. All the epidermal growth factor ligand family members, for example, are synthesized as inactive transmembrane precursors that undergo ectodomain proteolytic cleavage. The growth factors released this way then act in an endocrine, paracrine, or autocrine manner via binding to their cognate receptors (45). A similar functional role for the cleaved hβ1AR N terminus is very unlikely, however, as it lacks any apparent functional protein domains. It is more likely that the cleavage has a role in attenuating receptor activity and activation-mediated signaling in cells. This notion was supported by the observation that the inhibition of receptor cleavage by mutation of the Arg31↓Leu32 cleavage site stabilized mature receptors, suggesting that the proteolytic cleavage may have a role in the turnover of cell surface receptors. Removal of the receptor N terminus may represent the first step in a pathway that directs the receptor protein toward degradation. It remains to be demonstrated in future studies whether cleavage of the receptor requires internalization or whether it occurs at the cell surface. Internalization and targeting to lysosomal degradation may be the primary way of disposing of cell surface GPCRs (46), but it has also been suggested, e.g. for the β2AR (47), that receptor degradation may occur without internalization at the cell surface.
The β1AR ligand-binding site resides within the 7-transmembrane helix bundle and involves amino acids in transmembrane helices 3 and 5–7 and in the second extracellular loop (18). Thus, it is not immediately obvious why agonist binding and receptor activation should lead to enhanced cleavage of the receptor in the N-terminal domain. It can be hypothesized, however, that activation-induced conformational changes in the transmembrane α-helices may be reflected in the N terminus. This may in turn increase susceptibility to enzymatic cleavage. This possibility is in line with previous findings that have suggested that metalloproteinase-mediated cleavage is not necessarily dependent on the primary structure of the substrate protein but rather on conformational attributes (48). Interestingly, recent studies by Gupta et al. (19, 49) have demonstrated that activation-induced conformational alterations do indeed occur in the N terminus of several GPCRs in the rhodopsin family, including the β2AR. Furthermore, it was also found that an N-terminally directed antibody of the δ-opioid receptor was able to recognize post-activation-mediated changes in the receptor C terminus, an ability that was impaired after mutations at the C-terminal phosphorylation sites (49). If changes in the phosphorylation of β1AR intracellular domains are converted to conformational changes in the receptor N terminus, it can be hypothesized that the observed increase in the cleavage of the hβ1AR by PMA and forskolin may be mediated by the same mechanism as binding of the agonist to the receptor. An alternative hypothesis is that the enhanced cleavage of the receptor results from increased functional activity of the cleaving enzyme itself. This could occur via phosphorylation of the metalloprotease through a protein kinase that is activated by the second messengers. Extracellular signal-regulated kinase (ERK), for example, is known to phosphorylate ADAM-17 in response to PMA at Thr735 (50), a modification that is indispensable for maturation of the protein and its inducible trafficking to the cell surface (51). These highly speculative mechanisms that may be responsible for regulated hβ1AR cleavage need to be verified in future studies.
The hβ1AR was found to be modified by both N- and O-glycosylation. The latter modification was first suggested by the fact that removal of the single N-glycan at Asn15 in the mature receptor resulted in a receptor form that migrated more slowly than the corresponding de-N-glycosylated receptor precursor. Treatment of the mature receptor with neuraminidase and O-glycosidase revealed that it carries mucin-type sialylated O-glycans. Because of a lack of the consensus sequence for O-glycosylation, we used the NetOGlyc 3.1 Server (52) to predict the mucin-type O-glycosylation sites in the receptor N terminus. This program predicts five potential O-glycosylation sites for the hβ1AR, namely Thr28, Ser37, Ser41, Ser47, and Ser49, which are well conserved in mammalian β1ARs (Fig. 4).
Apart from the hβ1AR, an increasing number of other GPCRs in the rhodopsin family have been reported to be modified by O-linked carbohydrates. These include the human δ-opioid (26, 28), κ-opioid (53), V2 vasopressin (54), CCR5 chemokine (55, 56), and bradykinin B2 (57) receptors, as well as octopus rhodopsin and chicken iodopsin (58). The functional significance of the modification has nevertheless remained elusive. Bannert et al. (56) found that O-glycosylation contributes to the binding of chemokines to the CCR5 chemokine receptor by providing an array of negative charges that allow electrostatic interactions. On the other hand, Sadeghi and Birnbaumer (54) showed that neither the level of expression of the V2 vasopressin receptor nor its function is altered by eliminating the putatively O-glycosylated Ser/Thr residues. We cannot at the moment deduce the functional role of the O-glycosylation of the hβ1AR, and further studies will have to be performed before any definite conclusions are drawn. One intriguing possibility is that the extensive O-glycosylation might modulate the susceptibility of the N-terminal domain to metalloproteinases, possibly by restricting the conformational flexibility of the N terminus or by introducing negative charges. By comparison, O-glycans of several other cell surface proteins, including the transferrin receptor, have been shown to protect the protein from excessive proteolytic cleavage (59). The clustered Ser residues in the hβ1AR N terminus (Ser37, Ser41, Ser47, and Ser49) are flanked by two proteolytic cleavage sites identified here, Arg31↓Leu32 and Pro52↓Leu53 (Fig. 4), which speaks for the possibility that O-glycans may indeed have a protective role.
Acknowledgments
We thank Dr. Hongmin Tu (Biocenter Oulu Protein Sequencing and Amino Acid Analysis Core Service) for assistance with the N-terminal sequencing and Paula Salmela for technical assistance. We are grateful to other members of the GPCR team for discussions during this work and to Prof. Hannu Rajaniemi for fruitful ideas. We also thank Prof. Michel Bouvier for the cDNAs encoding the untagged and HA-tagged hβ1AR constructs.
This work was supported in part by the Sigrid Juselius Foundation and Grants 107922 and 127199 from the Academy of Finland.
- β1AR
- β1-adrenergic receptor
- h
- human
- ADAM
- a disintegrin and metalloproteinase
- CGP-20712
- 1-[2-((3-carbamoyl-4-hydroxy)phenoxy)ethylamino]-3-[4-(1-methyl-4-trifluoromethyl-2-imidazolyl)phenoxy]-2-propanol dihydrochloride
- DDM
- n-dodecyl-β-d-maltoside
- Endo H
- endo-β-N-acetylglucosaminidase H
- ER
- endoplasmic reticulum
- GPCR
- G protein-coupled receptor
- ISO
- isoproterenol
- NHS
- N-hydroxysuccinimide
- PMA
- phorbol 12-myristate 13-acetate
- PNGase F
- peptide-N-glycosidase F
- BFA
- brefeldin A.
REFERENCES
- 1.Bylund D. B., Eikenberg D. C., Hieble J. P., Langer S. Z., Lefkowitz R. J., Minneman K. P., Molinoff P. B., Ruffolo R. R., Jr., Trendelenburg U. (1994) Pharmacol. Rev. 46, 121–136 [PubMed] [Google Scholar]
- 2.Fredriksson R., Lagerström M. C., Lundin L. G., Schiöth H. B. (2003) Mol. Pharmacol. 63, 1256–1272 [DOI] [PubMed] [Google Scholar]
- 3.Lagerström M. C., Schiöth H. B. (2008) Nat. Rev. Drug Discov. 7, 339–357 [DOI] [PubMed] [Google Scholar]
- 4.Rockman H. A., Koch W. J., Lefkowitz R. J. (2002) Nature 415, 206–212 [DOI] [PubMed] [Google Scholar]
- 5.Brodde O. E., Bruck H., Leineweber K. (2006) J. Pharmacol. Sci. 100, 323–337 [DOI] [PubMed] [Google Scholar]
- 6.Rousseau G., Nantel F., Bouvier M. (1996) Mol. Pharmacol. 49, 752–760 [PubMed] [Google Scholar]
- 7.Suzuki T., Nguyen C. T., Nantel F., Bonin H., Valiquette M., Frielle T., Bouvier M. (1992) Mol. Pharmacol. 41, 542–548 [PubMed] [Google Scholar]
- 8.Green S. A., Liggett S. B. (1994) J. Biol. Chem. 269, 26215–26219 [PubMed] [Google Scholar]
- 9.Shiina T., Kawasaki A., Nagao T., Kurose H. (2000) J. Biol. Chem. 275, 29082–29090 [DOI] [PubMed] [Google Scholar]
- 10.McLean A. J., Milligan G. (2000) Br. J. Pharmacol. 130, 1825–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Y., Devic E., Kobilka B. (2002) J. Biol. Chem. 277, 33783–33790 [DOI] [PubMed] [Google Scholar]
- 12.Liang W., Curran P. K., Hoang Q., Moreland R. T., Fishman P. H. (2004) J. Cell Sci. 117, 723–734 [DOI] [PubMed] [Google Scholar]
- 13.Dunigan C. D., Hoang Q., Curran P. K., Fishman P. H. (2002) Biochemistry 41, 8019–8030 [DOI] [PubMed] [Google Scholar]
- 14.Liang W., Austin S., Hoang Q., Fishman P. H. (2003) J. Biol. Chem. 278, 39773–39781 [DOI] [PubMed] [Google Scholar]
- 15.Frielle T., Collins S., Daniel K. W., Caron M. G., Lefkowitz R. J., Kobilka B. K. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 7920–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall R. A. (2004) Semin. Cell Dev. Biol. 15, 281–288 [DOI] [PubMed] [Google Scholar]
- 17.Marchese A., Paing M. M., Temple B. R., Trejo J. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 601–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta A., Décaillot F. M., Gomes I., Tkalych O., Heimann A. S., Ferro E. S., Devi L. A. (2007) J. Biol. Chem. 282, 5116–5124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokoch M. P., Zou Y., Rasmussen S. G., Liu C. W., Nygaard R., Rosenbaum D. M., Fung J. J., Choi H. J., Thian F. S., Kobilka T. S., Puglisi J. D., Weis W. I., Pardo L., Prosser R. S., Mueller L., Kobilka B. K. (2010) Nature 463, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietilä E. M., Tuusa J. T., Apaja P. M., Aatsinki J. T., Hakalahti A. E., Rajaniemi H. J., Petäjä-Repo U. E. (2005) J. Biol. Chem. 280, 26622–26629 [DOI] [PubMed] [Google Scholar]
- 22.Lavoie C., Mercier J. F., Salahpour A., Umapathy D., Breit A., Villeneuve L. R., Zhu W. Z., Xiao R. P., Lakatta E. G., Bouvier M., Hébert T. E. (2002) J. Biol. Chem. 277, 35402–35410 [DOI] [PubMed] [Google Scholar]
- 23.Apaja P. M., Tuusa J. T., Pietilä E. M., Rajaniemi H. J., Petäjä-Repo U. E. (2006) Mol. Biol. Cell 17, 2243–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leskelä T. T., Markkanen P. M., Alahuhta I. A., Tuusa J. T., Petäjä-Repo U. E. (2009) Traffic 10, 116–129 [DOI] [PubMed] [Google Scholar]
- 25.Leskelä T. T., Markkanen P. M., Pietilä E. M., Tuusa J. T., Petäjä-Repo U. E. (2007) J. Biol. Chem. 282, 23171–23183 [DOI] [PubMed] [Google Scholar]
- 26.Petäjä-Repo U. E., Hogue M., Laperriere A., Walker P., Bouvier M. (2000) J. Biol. Chem. 275, 13727–13736 [DOI] [PubMed] [Google Scholar]
- 27.Petäjä-Repo U. E., Hogue M., Laperriere A., Bhalla S., Walker P., Bouvier M. (2001) J. Biol. Chem. 276, 4416–4423 [DOI] [PubMed] [Google Scholar]
- 28.Markkanen P. M., Petäjä-Repo U. E. (2008) J. Biol. Chem. 283, 29086–29098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jürss R., Hekman M., Helmreich E. J. (1985) Biochemistry 24, 3349–3354 [DOI] [PubMed] [Google Scholar]
- 30.Benovic J. L., Stiles G. L., Lefkowitz R. J., Caron M. G. (1983) Biochem. Biophys. Res. Commun. 110, 504–511 [DOI] [PubMed] [Google Scholar]
- 31.Stiles G. L., Strasser R. H., Lavin T. N., Jones L. R., Caron M. G., Lefkowitz R. J. (1983) J. Biol. Chem. 258, 8443–8449 [PubMed] [Google Scholar]
- 32.Umemoto J., Bhavanandan V. P., Davidson E. A. (1977) J. Biol. Chem. 252, 8609–8614 [PubMed] [Google Scholar]
- 33.Yarden Y., Rodriguez H., Wong S. K., Brandt D. R., May D. C., Burnier J., Harkins R. N., Chen E. Y., Ramachandran J., Ullrich A., Ross E. M. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 6795–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petäjä-Repo U. E., Hogue M., Bhalla S., Laperrière A., Morello J. P., Bouvier M. (2002) EMBO J. 21, 1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozuka M., Ito T., Hirose S., Lodhi K. M., Hagiwara H. (1991) J. Biol. Chem. 266, 16892–16896 [PubMed] [Google Scholar]
- 36.Grantcharova E., Furkert J., Reusch H. P., Krell H. W., Papsdorf G., Beyermann M., Schulein R., Rosenthal W., Oksche A. (2002) J. Biol. Chem. 277, 43933–43941 [DOI] [PubMed] [Google Scholar]
- 37.Couet J., Sar S., Jolivet A., Hai M. T., Milgrom E., Misrahi M. (1996) J. Biol. Chem. 271, 4545–4552 [DOI] [PubMed] [Google Scholar]
- 38.Ando T., Latif R., Pritsker A., Moran T., Nagayama Y., Davies T. F. (2002) J. Clin. Invest. 110, 1667–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaczur V., Puskas L. G., Nagy Z. U., Miled N., Rebai A., Juhasz F., Kupihar Z., Zvara A., Hackler L., Jr., Farid N. R. (2007) J. Mol. Recognit. 20, 392–404 [DOI] [PubMed] [Google Scholar]
- 40.Ludeman M. J., Zheng Y. W., Ishii K., Coughlin S. R. (2004) J. Biol. Chem. 279, 18592–18599 [DOI] [PubMed] [Google Scholar]
- 41.Boire A., Covic L., Agarwal A., Jacques S., Sherifi S., Kuliopulos A. (2005) Cell 120, 303–313 [DOI] [PubMed] [Google Scholar]
- 42.Kojro E., Fahrenholz F. (1995) J. Biol. Chem. 270, 6476–6481 [DOI] [PubMed] [Google Scholar]
- 43.Kojro E., Postina R., Gilbert S., Bender F., Krause G., Fahrenholz F. (1999) Eur. J. Biochem. 266, 538–548 [DOI] [PubMed] [Google Scholar]
- 44.Chazenbalk G. D., Chen C. R., McLachlan S. M., Rapoport B. (2004) Endocrinology 145, 4–10 [DOI] [PubMed] [Google Scholar]
- 45.Blobel C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 46.Hanyaloglu A. C., von Zastrow M. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 47.Jockers R., Angers S., Da Silva A., Benaroch P., Strosberg A. D., Bouvier M., Marullo S. (1999) J. Biol. Chem. 274, 28900–28908 [DOI] [PubMed] [Google Scholar]
- 48.Huovila A. P., Turner A. J., Pelto-Huikko M., Kärkkäinen I., Ortiz R. M. (2005) Trends Biochem. Sci. 30, 413–422 [DOI] [PubMed] [Google Scholar]
- 49.Gupta A., Rozenfeld R., Gomes I., Raehal K. M., Décaillot F. M., Bohn L. M., Devi L. A. (2008) J. Biol. Chem. 283, 10735–10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Díaz-Rodríguez E., Montero J. C., Esparís-Ogando A., Yuste L., Pandiella A. (2002) Mol. Biol. Cell 13, 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soond S. M., Everson B., Riches D. W., Murphy G. (2005) J. Cell Sci. 118, 2371–2380 [DOI] [PubMed] [Google Scholar]
- 52.Julenius K., Mølgaard A., Gupta R., Brunak S. (2005) Glycobiology 15, 153–164 [DOI] [PubMed] [Google Scholar]
- 53.Li J. G., Chen C., Liu-Chen L. Y. (2007) Biochemistry 46, 10960–10970 [DOI] [PubMed] [Google Scholar]
- 54.Sadeghi H., Birnbaumer M. (1999) Glycobiology 9, 731–737 [DOI] [PubMed] [Google Scholar]
- 55.Farzan M., Mirzabekov T., Kolchinsky P., Wyatt R., Cayabyab M., Gerard N. P., Gerard C., Sodroski J., Choe H. (1999) Cell 96, 667–676 [DOI] [PubMed] [Google Scholar]
- 56.Bannert N., Craig S., Farzan M., Sogah D., Santo N. V., Choe H., Sodroski J. (2001) J. Exp. Med. 194, 1661–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michineau S., Alhenc-Gelas F., Rajerison R. M. (2006) Biochemistry 45, 2699–2707 [DOI] [PubMed] [Google Scholar]
- 58.Nakagawa M., Miyamoto T., Kusakabe R., Takasaki S., Takao T., Shichida Y., Tsuda M. (2001) FEBS Lett. 496, 19–24 [DOI] [PubMed] [Google Scholar]
- 59.Rutledge E. A., Root B. J., Lucas J. J., Enns C. A. (1994) Blood 83, 580–586 [PubMed] [Google Scholar]