Abstract
The feline leukemia virus subgroup C receptor (FLVCR) is a heme export protein that is required for proerythroblast survival and facilitates macrophage heme iron recycling. However, its mechanism of heme export and substrate specificity are uncharacterized. Using [55Fe]heme and the fluorescent heme analog zinc mesoporphyrin, we investigated whether export by FLVCR depends on the availability and avidity of extracellular heme-binding proteins. Export was 100-fold more efficient when the medium contained hemopexin (Kd < 1 pm) compared with albumin (Kd = 5 nm) at the same concentration and was not detectable when the medium lacked heme-binding proteins. Besides heme, FLVCR could export other cyclic planar porphyrins, such as protoporphyrin IX and coproporphyrin. However, FLVCR has a narrow substrate range because unconjugated bilirubin, the primary breakdown product of heme, was not transported. As neither protoporphyrin IX nor coproporphyrin export improved with extracellular hemopexin (versus albumin), our observations further suggest that hemopexin, an abundant protein with a serum concentration (6.7–25 μm) equivalent to that of the iron transport protein transferrin (22–31 μm), by accepting heme from FLVCR and targeting it to the liver, might regulate macrophage heme export and heme iron recycling in vivo. Final studies show that hemopexin directly interacts with FLVCR, which also helps explain why FLVCR, in contrast to some major facilitator superfamily members, does not function as a bidirectional gradient-dependent transporter. Together, these data argue that hemopexin has a role in assuring systemic iron balance during homeostasis in addition to its established role as a scavenger during internal bleeding or hemolysis.
Keywords: Heme, Hemopexin, Membrane Proteins, Reactive Oxygen Species (ROS), Receptor Regulation, Heme Export, Heme Homeostasis, Iron Recycling
Introduction
The biological roles of heme are diverse and encompass most areas of cell metabolism and gene regulation. Heme serves as a prosthetic group in the hemeproteins, which are involved in crucial biological functions, including oxygen binding (hemoglobin and myoglobin), oxygen metabolism (oxidases, peroxidases, catalases, and hydroxylases), electron transfer (cytochromes) (4), and signal transduction (nitric-oxide synthases) (5). In addition, heme serves as a signaling molecule in erythropoiesis and other physiological systems. Because heme generally signals by binding and inactivating a transcriptional repressor (e.g. Bach1) or translational inhibitor (e.g. heme-regulated inhibitor and thus eIFα2 kinase), its impact is immediate (6–8). Perhaps for this reason, heme is utilized in time-sensitive processes, such as the regulation of circadian rhythm (9), N-end rule pathway/protein ubiquitination (10), and globin synthesis. Heme is also required for microRNA processing (11). However, excess free or non-protein-bound heme damages lipid, protein, and DNA through the generation of reactive oxygen species, resulting in cellular injury and death (12). Therefore, free cellular heme levels must be tightly regulated to provide an adequate supply yet avoid heme toxicities (4, 13–15).
Most studies of heme homeostasis focus on its biosynthesis and degradation. The rate of heme biosynthesis varies significantly among various cells and tissues and is especially high in hepatocytes and erythroid cells, where large amounts of heme are required for cytochrome P450 and hemoglobin, respectively (4). The addition of aminolevulinic acid (ALA)3 bypasses the initial rate-limiting step in the heme synthesis pathway and allows the synthesis of intermediates up to and including protoporphyrin IX (PPIX). If sufficient iron is available, ferrochelatase inserts the iron into PPIX, completing heme synthesis. Heme is degraded by heme oxygenases to biliverdin, which is then converted to bilirubin by biliverdin reductase (16). Excess heme further induces heme oxygenase-1 levels by increasing the transcription of heme oxygenase-1.
Recently, we demonstrated (1) that intracellular heme can also be exported by a cell-surface membrane protein termed FLVCR, initially identified as the feline leukemia virus subgroup C receptor (17, 18). Inhibition of FLVCR function in the human erythroid cell line K562 decreases heme export, impairs erythroid maturation, and leads to apoptosis (1). FLVCR-null mice lack definitive erythropoiesis, and mice with FLVCR that is deleted neonatally develop severe macrocytic anemia with proerythroblast maturation arrest (2), arguing that erythroid precursors export excess heme to ensure survival. Interestingly, FLVCR-deleted mice develop iron overload in macrophages, liver, and duodenum. These data suggest that the iron homeostatic cycle includes heme export at these sites. This hypothesis is supported by direct observations that FLVCR-deficient macrophages accumulate more ferritin than wild-type macrophages when ingesting opsonized red cells (2).
The goal of this study was to determine how FLVCR-mediated heme export occurs and whether FLVCR has limited substrate specificity or can also transport structurally and metabolically related compounds. We demonstrate that the substrate specificity of FLVCR export involves heme and proximal planar porphyrins, but not bilirubin; that the export of heme is preferentially facilitated by hemopexin; and that hemopexin binds to FLVCR at a site thought to be important for heme export. A physiological role for hemopexin in heme iron recycling may explain the very high serum concentrations of hemopexin, 0.55–1.25 mg/ml (9.2–20.8 μm) in mice (19) and 0.4–1.5 mg/ml (6.7–25 μm) in humans (20).
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
Normal rat kidney cells transduced with human FLVCR (NRK/FLVCR) or empty vector alone (NRK/Neo) (1) were cultured in high glucose DMEM (Invitrogen) with 10% FBS (HyClone, Logan, UT) and supplemented with nonessential amino acids, l-glutamine, sodium pyruvate, and penicillin/streptomycin/Fungizone (all from Invitrogen). Wild-type dynamin-GFP and dominant-negative K44A dynamin-GFP (21) were introduced by retroviral gene transfer into NRK/FLVCR and NRK/Neo cells. FLVCR-null and littermate control mouse embryonic fibroblasts (MEFs) isolated from day 12.5 embryos (2) were cultured in DMEM with 10% FBS as described above. The MTT assay (American Type Culture Collection, Manassas, VA) was performed as described previously (22).
Preparation of Human Macrophages
Human peripheral blood was obtained from normal donors with an approved protocol from the University of Washington. Mononuclear cells were isolated from blood with lymphocyte separation medium (Mediatech, Inc., Herndon, VA) as described previously (23). Monocytes/macrophages were subsequently isolated by plastic adherence for 18 h in Iscove's modified Dulbecco's medium (Invitrogen) supplemented with 20% FBS, 1% BSA, penicillin/streptomycin/Fungizone, and 10 ng/ml macrophage colony-stimulating factor (PeproTech, Rocky Hill, NJ). Adherent cells were cultured for an additional 5 days to generate mature macrophages.
Isolation and Purification of Rabbit Hemopexin
Intact apohemopexin was isolated from rabbit serum as described previously (24). This protein is free of contaminants, binds heme as characterized and quantitated following published procedures and extinction coefficients (25), and is not toxic for cells (26).
Zinc Mesoporphyrin (ZnMP) and [55Fe]Heme Transport
NRK/FLVCR and NRK/Neo cell lines were grown on 8-well chamber slides (BD Biosciences) in complete medium until near confluency. ZnMP export studies were performed, as described previously (27), by preloading for 30 min at 37 °C using 5 μm ZnMP (Frontier Scientific, Logan, UT) dissolved in cell culture-grade Me2SO at a concentration of 4 mm and then diluted to 5 μm in working buffer (25 mm HEPES (pH 7.4), 130 mm NaCl, 10 mm KCl, 1 mm CaCl2, and 1 mm MgSO4) containing 2.5 μm BSA (Sigma). Next, the cells were washed three times and incubated in working buffer with either albumin or hemopexin at the desired concentrations for up to 90 min at 37 °C. Export was determined by comparison of residual intracellular fluorescence on microscopic images using a Nikon upright Eclipse E600 microscope and Texas Red filter with excitation at 540–580 nm and emission at 600–680 nm.
For quantitative studies, ZnMP export was also performed using flow cytometry. NRK/FLVCR and NRK/Neo cell lines were trypsinized, washed, and resuspended in working buffer contain 2.5 μm BSA. The cells were then processed as described above. Export was quantitated by comparing residual intracellular fluorescence at channel FL2 using a BD FACScan instrument (BD Biosciences).
For [55Fe]heme uptake and export studies, cells were seeded in 12-well dishes and allowed to attach overnight in culture medium. They were incubated in working buffer with 5 μm ZnMP (included only for export studies) and 2.5 μm BSA for 15 min, and [55Fe]hemin (RI Consultants LLC, Hudson, NH) was added to a final concentration of 0.9 μm for an additional 30 min. The cells were washed three times and then harvested as described below for uptake studies or incubated with either albumin or hemopexin at the desired concentrations for 90 min at 37 °C for export studies. To measure cellular radioactivity, the cells were rinsed twice with working buffer, detached from the plates by a 15-min incubation with 0.1 n NaOH, mixed with liquid scintillation mixture (MP Biomedicals, Solon, OH), and placed in an LF 5000 CE liquid scintillation counter (Beckman Coulter, Brea, CA). Radioactive counts (cpm) were normalized by total cell protein content and are presented as the mean ± S.D. of three replicate samples. To combine multiple experiments for statistical analysis, NRK/Neo control cells were considered to have 100% uptake, and all other samples within each experiment were normalized to the NRK/Neo data.
Preparation of FLVCR Peptide-GST Fusion Proteins
Two FLVCR peptide-GST fusion proteins were generated as immunogens in an attempt to elicit anti-FLVCR antibody. The first fusion peptide contained amino acids 132–201 (and thus His145 and His198), and the second fusion peptide contained amino acids 262–369 (and no histidine residue). During the terminal purification steps, the first (but not second) peptide was red in color and had Soret absorption at ∼425 nm compared with GST alone, and HPLC confirmed heme binding (data not shown). This fragment was subsequently used as an immunogen to generate antibody to FLVCR.4
FLVCR and Hemopexin Binding Assay
FLVCR peptide-GST fusion protein and GST alone in a 1:2 dilution series (72.5–580 nm) were coated onto 96-well Pro-BindTM microplates (Becton Dickinson Labware, Franklin Lakes, NJ) at 100 μl/well at 4 °C overnight. After incubation with 1% BSA in PBS for 2 h, hemopexin (290 nm) was added and allowed to bind to the FLVCR peptide-GST fusion (or control) protein for 2 h. Next, rabbit anti-hemopexin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at 1 μg/ml and horseradish peroxidase-labeled goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) at 0.5 μg/ml were added sequentially. The microplate was washed with PBS three times for 5 min after each incubation with hemopexin and antibodies. The color-developing reaction was carried out by incubation with 3,3′,5,5′-tetramethylbenzidine chromogenic solution (Sigma) for 10 min and stopped by the addition of 2 m H2SO4 according to the manufacturer's instructions. The known concentration of hemopexin in a 1:2 dilution series (36.25–290 nm) was used to standardize the assay results. For the competitive binding study, the FLVCR peptide-GST fusion protein was incubated for 1 h at room temperature with or without heme at a 1:1 molar ratio before coating onto the microplate. Competitive binding studies were also performed by adding the FLVCR peptide-GST fusion protein at a 1:1 molar ratio to hemopexin prior to its binding to the FLVCR peptide-GST fusion protein-coated microplate.
Unconjugated Bilirubin (UCB) Uptake and Export Studies
NRK/FLVCR and NRK/Neo cells were cultured in 12-well dishes as described above and incubated in working buffer containing 10 μm BSA and 10 μm highly purified [3H]UCB (specific activity of 10.8 mCi/mmol) biosynthetically labeled in dogs (28, 29), corresponding to a free bilirubin concentration of 100 nm (30). After 1, 10, and 30 min, the cells were washed three times with working buffer, and bilirubin uptake was then determined from cell-associated radioactivity. Bilirubin export was studied using cells preloaded for 30 min as described above, washed three times with working buffer, and incubated for 0, 30, or 90 min in working buffer containing 151.5 μm (1%) BSA. Exported [3H]UCB was washed off with working buffer, and retained [3H]UCB was quantitated and normalized to total cell protein content.
Induction of PPIX Synthesis by Treatment with ALA
NRK cells in cell culture complete medium were supplemented with 1 mm ALA for up to 24 h to induce the synthesis of PPIX. At time points of 12 and 24 h, respectively, cell culture supernatants were collected, and cell pellets were harvested and briefly rinsed with PBS. Both supernatants and cell pellets were stored at −80 °C until ready for process of analysis.
Measurement of Heme and Porphyrin Content for Export Studies
The extraction of heme and porphyrins was adapted from previously described methods (31). Cell culture media (10-ml fractions) were extracted in parallel three times with 10 ml of ethyl acetate/acetic acid (2:1, v/v) and then washed twice with 0.5 volume of saturated sodium acetate. The sodium acetate washings were also extracted with fresh ethyl acetate and combined with the original extract, which was then washed once with 0.1 volume of 3% (w/v) sodium acetate and dried down with argon. Samples were dissolved in buffer, and the heme and porphyrins were separated by HPLC as described (32). Standards of heme, porphyrins, and ZnMP (all from Frontier Scientific) were used for peak identification and quantification. The flow cytometric analysis of protoporphyrin export by NRK/FLVCR and NRK/Neo cells was performed using methods similar to those described for the ZnMP export study described above except that PPIX (Frontier Scientific) was substituted for ZnMP, and residual intracellular fluorescence was analyzed at channel FL3.
RESULTS
Heme Export Depends on the Avidity and Concentration of Extracellular Heme-binding Proteins
To understand the mechanism of heme export by FLVCR, we first studied the export of ZnMP, a fluorescent heme analog. ZnMP traffics similarly to heme and also inhibits heme degradation by competitively blocking heme oxygenase activity (27). Fig. 1 shows the effects of various concentrations of two heme-binding proteins, BSA (albumin) and hemopexin, on the export of ZnMP from preloaded NRK/FLVCR or NRK/Neo cells. In the absence of BSA, no detectable export occurred. However, ZnMP export was nearly complete in the presence of 151.5 μm (1%) BSA, less extensive with 15.15 μm BSA, and almost nil with 1.515 μm BSA. Interestingly, residual cellular fluorescence in the presence of 1.515 μm hemopexin was similar to that with 151.5 μm BSA, so hemopexin appears to be 100-fold more efficacious than BSA in inducing ZnMP export. When quantitated with flow cytometry, 55.9 ± 3.6% and 45.9 ± 3.8% of ZnMP was retained intracellularly when the export studies were performed in the presence of 151.5 μm BSA and 1.515 μm hemopexin, respectively. NRK/Neo cells failed to export ZnMP (data not shown) in all circumstances; this is consistent with our prior data (1) and documents the requirement for FLVCR for heme export in these cells.
FIGURE 1.
ZnMP export by FLVCR is carrier protein-dependent. NRK/FLVCR cells were loaded with 5 μm ZnMP for 30 min (upper panel) and then washed and cultured for 90 min in the absence or presence of either bovine serum albumin (middle panels) or hemopexin (lower panels) at the indicated concentrations. The retained intracellular ZnMP was visualized by fluorescence microscopy. Export was comparable with 1.515 μm hemopexin and 151.5 μm albumin. No export occurred when a heme-binding protein was absent (0 μm albumin). NRK/Neo cells failed to export ZnMP (data not shown). Scale bar = 200 μm.
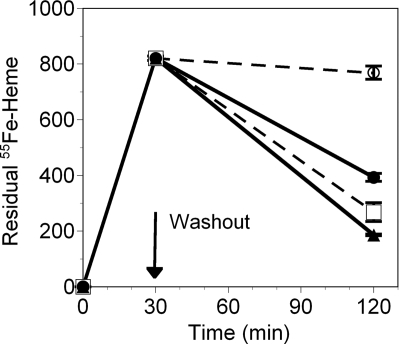
In additional similar studies, the export of [55Fe]heme by human peripheral macrophages was quantified. As shown in Fig. 2, only 6% of the [55Fe]heme load was exported in the presence of 1.515 μm albumin, whereas 52% was exported in the presence of 1.515 μm hemopexin in the same time period. When the albumin concentration in the medium was increased by 100-fold to 151.5 μm, 67% of the [55Fe]heme was exported. These studies confirm that heme export is ∼100 times more efficient in the presence of hemopexin than albumin on a molar basis. An equally important observation of Fig. 2 is that export is completely dependent on the extracellular presence of a heme-binding protein.
FIGURE 2.
[55Fe]Heme export by FLVCR is carrier protein-dependent. Human mononuclear cells were isolated from peripheral blood, and macrophages were generated by in vitro culture. The macrophages were preincubated with 5 μm ZnMP, and [55Fe]heme was added to a final concentration of 0.9 μm for an additional 30 minutes. The cells were washed and cultured for 90 min more in the presence of 151.5 μm (□) or 1.515 μm (○) albumin (dashed lines) or 15.15 μm (▴) or 1.515 μm (●) hemopexin (solid lines). The retained intracellular [55Fe]heme was quantitated in a liquid scintillation counter over the washout interval. Radioactive counts were normalized to the total cellular protein concentration and demonstrate that hemopexin facilitates heme export. The data show the mean ± S.D. of triplicate samples.
FLVCR Is Not a Bidirectional Heme Transporter
Heme uptake in NRK cells was not increased when the cells expressed FLVCR (100% in NRK/Neo cells versus 101.6 ± 6.2% in NRK/FLVCR cells; p = 0.6; n = 6). In additional studies, we used dominant-negative K44A dynamin (33) to block endocytosis-mediated heme uptake. Heme uptake decreased by 15–30%. yet the heme uptake by NRK/Neo/K44A dynamin and NRK/FLVCR/K44A dynamin cells remained comparable (74.8 ± 4.0% versus 72.9 ± 9.9% of NRK/Neo/dynamin uptake, respectively; p > 0.3; n = 3).
FLVCR Can Export Endogenously Synthesized Heme
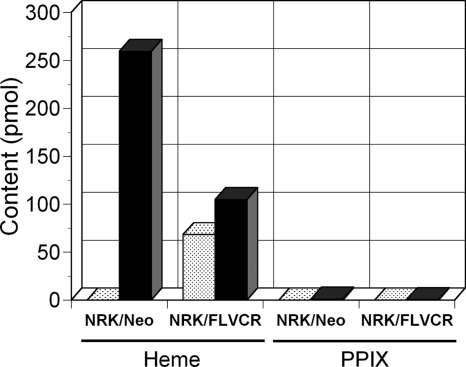
To determine whether cells can export endogenously synthesized heme via FLVCR and not only heme supplied exogenously, we performed studies in NRK/FLVCR and NRK/Neo cells (Fig. 3). Using HPLC, 68.4 pmol of heme was detected in a 10-ml sample of the cell culture supernatant from 8 × 106 NRK/FLVCR cells in this representative study (n = 4). However, heme was not detectable in the cell culture supernatant from NRK/Neo cells. Conversely, heme contents were higher in the cell pellet from NRK/Neo cells (259.4 pmol/sample) than in that from NRK/FLVCR cells (104.8 pmol/sample). PPIX was not detected in the 10-ml sample of the cell culture supernatant from either NRK/Neo or NRK/FLVCR cells and was barely detected in the cell pellet at 1.0 and 0.5 pmol/sample from 8 × 106 NRK/Neo and NRK/FLVCR cells, respectively, implying that iron is not limiting and that heme is the primary substrate for FLVCR export under basal conditions.
FIGURE 3.
FLVCR exports endogenously synthesized heme. NRK/FLVCR and NRK/Neo cells were maintained under normal cell culture conditions as described under “Experimental Procedures.” The cultured medium (stippled bars) and cell pellets (solid bars) were collected, and the heme and porphyrins were extracted and separated by HPLC. Standards of heme and porphyrins were used for peak identification and quantification. The amount of heme and PPIX from each sample is presented.
FLVCR Can Also Export Non-metallo-planar Porphyrins
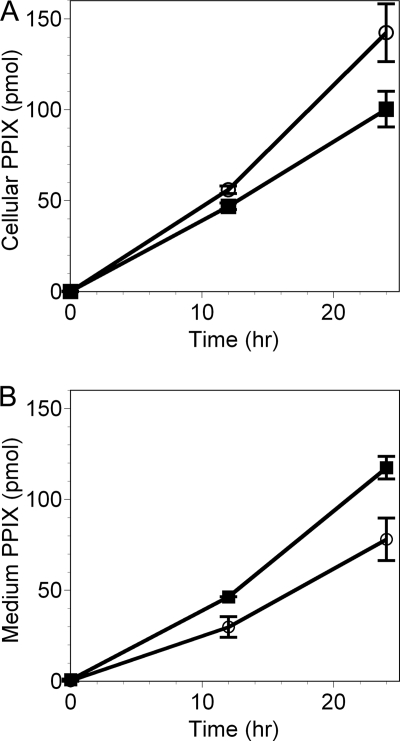
Similar studies were done to determine whether coproporphyrin and PPIX, which are both intermediate products in the heme biosynthetic pathway (34), can be exported via FLVCR should their intracellular concentrations be sufficiently high. When cells were supplemented with ALA to bypass the rate-limiting step in heme biosynthesis, PPIX steadily accumulated in the cells, but more extensively in NRK/Neo cells than in NRK/FLVCR cells (nearly 50% more at 24 h) (Fig. 4A). To determine whether this lower net accumulation reflected lower synthesis or greater export of PPIX by the NRK/FLVCR cells, PPIX content in the medium was measured at the same time points. After 24 h, the total PPIX content (cells + medium) was similar for both cell types (217 versus 220 pmol), but of this, 53.9 ± 2.8% of total PPIX was present in the culture medium of NRK/FLVCR cells versus 35.4 ± 2.7% in the culture medium of NRK/Neo cells (Fig. 4B). Thus, the lower accumulation of PPIX in the NRK/FLVCR cells was due to enhanced export provided by FLVCR transgene expression in these cells.
FIGURE 4.
FLVCR is able to export PPIX. NRK/FLVCR (■) and NRK/Neo (○) cells were maintained under normal cell culture conditions and then supplemented with 1 mm ALA for up to 24 h to induce the synthesis of PPIX. The cell pellets (A) and culture medium (B) were collected at 0, 12, and 24 h after ALA addition, and tetrapyrroles were extracted and separated by HPLC. Standards of heme and porphyrins were used for peak identification and quantification. The mean ± S.D. of three measurements is shown. The export of PPIX from NRK/FLVCR (and also FLVCR/Neo) cells required an extracellular protein, but in contrast to the results from the heme export studies, hemopexin and albumin functioned equivalently (data not shown), as anticipated by their similar but low affinities for PPIX (Kd = 1.75 and 0.75 μm, respectively) (40, 41).
Quantification of coproporphyrin export yielded similar results. In cultures of both NRK cell lines, coproporphyrin was not detectable in either the culture medium or cell pellet under base-line conditions but was detected after ALA supplementation (n = 3; representative result) (Fig. 5). NRK/FLVCR cells accumulated less coproporphyrin in the cell pellet (Fig. 5A) and exported more into the culture supernatant (Fig. 5B) than NRK/Neo control cells. After 24 h, the total coproporphyrin contents of the two cell lines were similar, but more was exported by cells expressing FLVCR (89.4% compared with 55.4% by NRK/Neo cells).
FIGURE 5.
FLVCR is able to export coproporphyrin. NRK/FLVCR (solid bars) and NRK/Neo (stippled bars) cells were maintained under normal cell culture conditions supplemented with no addition (Control) or with 1 mm ALA with or without 20 μm ferric citrate (Fe) as indicated for 24 h to induce heme synthesis. The cell pellets (A) and cultured medium (B) were collected, and the heme and porphyrins were extracted and separated by HPLC. Standards of heme and porphyrins were used for peak identification and quantification.
These data suggest that FLVCR can export PPIX and coproporphyrin but predict that other transporters, presumably ABCG2 (35), can do so also. The extracellular presence of hemopexin, which binds these porphyrins with relatively low affinity (Kd ∼ 1 μm) (36, 37), did not improve the export efficiency compared with albumin (data not shown), which binds these porphyrins with similarly low affinity (Kd ∼ 1 μm) (37).
FLVCR Does Not Export UCB
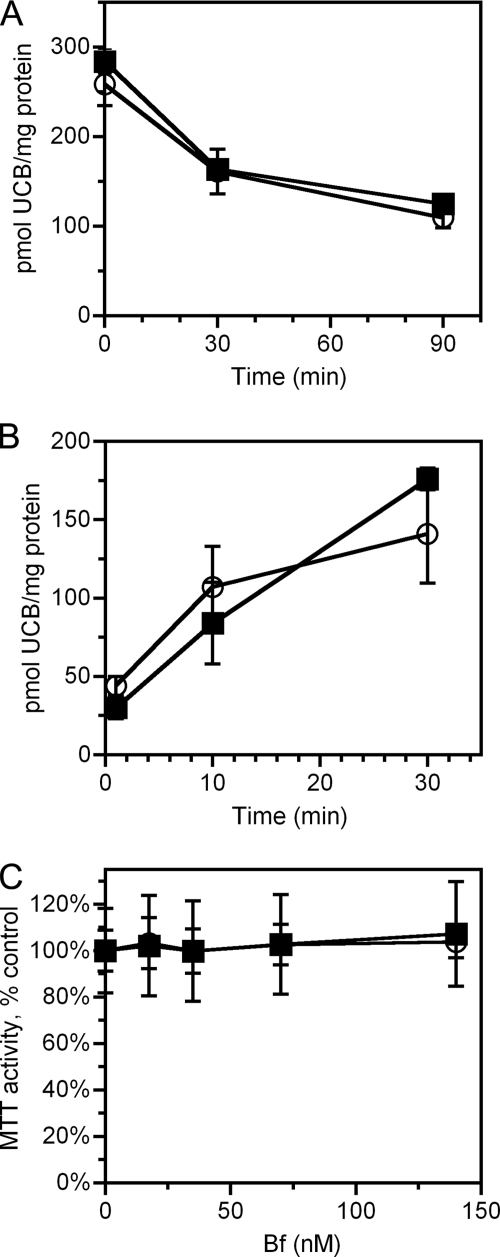
Heme metabolism by heme oxygenase-1 leads to the production of biliverdin, free ferrous iron, and carbon monoxide. Biliverdin is further reduced into UCB (reviewed in Ref. 16). In most neonates, immaturity of the conjugation and biliary secretion of bilirubin leads to retention of UCB, which, if severe, results in neurotoxicity from accumulation of UCB in the central nervous system (38). The impaired hepatic elimination is partially compensated by enhanced excretion of UCB across the intestinal wall (38), which might theoretically be mediated by FLVCR, prompting these studies. As shown in Fig. 6, both NRK/Neo and NRK/FLVCR cells exhibited similar rates of uptake and rapidly exported UCB, implying that export occurs via a mechanism distinct from FLVCR (39). Because UCB is toxic to cells that lack the ability to export it (22), we used an indirect approach as a secondary method to determine whether FLVCR is capable of exporting bilirubin from cells. We compared the toxicity of UCB in FLVCR-containing versus FLVCR-null MEFs. As assessed by MTT activity (a test of mitochondrial function), there was no significant difference in sensitivity to UCB toxicity between MEFs that contain or lack FLVCR (Fig. 6C). Together, these data indicate that FLVCR does not export bilirubin.
FIGURE 6.
FLVCR does not export bilirubin. A, NRK/FLVCR (■) and NRK/Neo (○) cells were loaded with 10 μm UCB for 30 min; washed and cultured for 0, 30, and 90 min; and assayed for retention of UCB. B, NRK/FLVCR (■) and NRK/Neo (○) cells were loaded with 10 μm UCB for 1, 10, and 30 min and then assayed for uptake of UCB. C, FLVCR-positive (■) and FLVCR-negative (○) MEFs were treated with UCB to achieve the indicated free bilirubin (Bf) concentration for 4 h and then assayed for MTT activity. MTT activity is presented as the percent of MTT activity present in the untreated samples. Data are presented as the mean ± S.D. of triplicate samples (A and B) or of two independent MEF lines assayed in triplicate (six samples; C). There were no significant differences between FLVCR-containing and FLVCR-lacking cells for UCB uptake, export, or toxicity.
Hemopexin Binds to FLVCR to Facilitate Heme Export
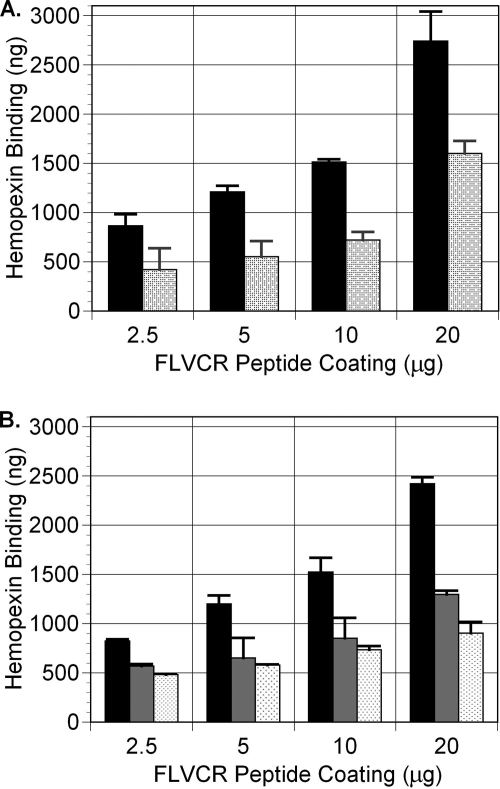
To examine whether hemopexin directly interacts with FLVCR, we performed a competitive binding ELISA (Fig. 7) using a FLVCR peptide (amino acids 132–201)-GST fusion protein previously shown to bind heme (see “Experimental Procedures”). When the FLVCR peptide-GST fusion protein was coated onto 96-well microplates at 2.5, 5, 10, and 20 μg/well, the amount of hemopexin that bound was 865 ± 118, 1210 ± 63, 1515 ± 27, and 2740 ± 301 ng, respectively. However, when the FLVCR peptide-GST fusion protein was preincubated with heme prior to coating the microplates, the amount of hemopexin that bound was reduced to 420 ± 217, 550 ± 158, 720 ± 83, and 1600 ± 129 ng, respectively (Fig. 7A). Together, these data suggest that apohemopexin attaches to FLVCR, binds heme, and then leaves, so less residual bound hemopexin is detected in the ELISA. To confirm this, several additional studies were performed. First, we showed that the preincubation of hemopexin with heme decreased its ability to bind to the FLVCR peptide-GST fusion protein (Fig. 7B), implying that apohemopexin preferentially binds FLVCR. Second, we confirmed that the binding of hemopexin to the FLVCR peptide-GST fusion protein was a property of the FLVCR peptide and not GST, as hemopexin did not bind to GST alone as the control in our studies (data not shown). Third, we observed that exogenous FLVCR peptide-GST fusion protein could facilitate the export of heme (ZnMP) by FLVCR but only when its binding sites for heme were open and not when they were saturated. By flow cytometry, 54.1% of the preloaded ZnMP was exported in the presence of 15.15 μm hemopexin. When both the FLVCR peptide-GST fusion protein and hemopexin were present extracellularly, 60.1% of the preloaded ZnMP was exported (p < 0.05). However, when the FLVCR peptide-GST fusion protein was preincubated with heme (and excess heme was removed by dialysis), the export of ZnMP was not improved (52.7%; data representative of three experiments), data that also confirm binding specificity.
FIGURE 7.
FLVCR directly interacts with hemopexin. A, the FLVCR peptide-GST fusion protein preincubated with heme (stippled bars) or without heme (solid bars) was coated onto a 96-well microplate in a 1:2 series dilution as indicated. Hemopexin (290 nm) binding to the FLVCR peptide-GST fusion protein was detected by rabbit anti-hemopexin antibody and horseradish peroxidase-labeled goat anti-rabbit antibody, followed by incubation with 3,3′,5,5′-tetramethylbenzidine chromogenic solution. Hemopexin bound to the FLVCR peptide-GST fusion protein in a dose-dependent manner. B, preincubation of hemopexin with the FLVCR peptide (gray bars) or heme (stippled bars), in comparison with the untreated control (black bars), decreased its ability to bind to the FLVCR peptide-GST fusion protein. Data from A and B together show that apohemopexin bound to FLVCR more avidly than heme-hemopexin.
DISCUSSION
Here, we have demonstrated that the export of heme via FLVCR is tightly regulated by the presence and avidity of the extracellular heme-binding proteins hemopexin and albumin. Export via FLVCR is most efficient in the presence of 1.515 μm hemopexin. A comparable efficiency of export requires a 100-fold higher concentration of BSA, which reflects the much higher affinity of hemopexin for heme (Kd < 1 pm) compared with albumin (Kd = 5 nm) (40, 41). Because hemopexin is present at a concentration in human serum (6.7–25 μm) similar to transferrin (22–31 μm), the protein responsible for elemental iron transport (42), we suspect that it functions physiologically to facilitate the recycling and trafficking of heme iron, in addition to its role as a scavenger protein.
We would anticipate that albumin, the most abundant serum protein (522–776 μm, 21–122-fold more prevalent than hemopexin), could substitute for hemopexin under steady-state conditions, despite its much lower affinity for heme. This would explain the mild base-line phenotype of hemopexin-null mice. Although there is no obvious tissue lesion caused by oxidative damage or abnormal iron deposition in hemopexin-null mice, these animals have severe renal damage and defective recovery after acute hemolysis (43) and increased iron deposition and oxidative stress markers in brain (44).
There are two physiological reasons that cells might export heme via FLVCR: 1) to avoid cellular toxicity when heme synthesis exceeds use (as hemoproteins) and metabolism (via heme oxygenase-1) and 2) to facilitate systemic iron homeostasis. Although these roles have been best characterized in erythroid marrow (2) and macrophages (Ref. 2 and this report), respectively, it appears that FLVCR has importance for other tissues where it is highly expressed, such as liver, duodenum, kidney, placenta, and brain (2). Because FLVCR-deleted mice develop iron overload in liver and duodenum, as well as tissue macrophages, by 12 weeks (2) and accumulate iron in renal tubular cells by 9 months,5 heme trafficking via FLVCR at these sites likely contributes to iron homeostasis (3). Heme export via FLVCR may also provide protection from heme or ferrous iron toxicities in liver and brain, as our observations support emerging hypotheses concerning hepatic (45) and neuronal (46, 47) injury.
Hemopexin is a plasma glycoprotein produced primarily by liver parenchymal cells (48). However, it is also expressed in human neurons (49) and several types of mouse brain cells (26, 44, 48), implying that this protein is synthesized locally in brain. Interestingly, hemopexin protects the brain of adult mice against ischemic stroke-related damage in murine models (26). In addition to hemopexin, FLVCR is also present in developing mouse neural tissues and human brain tissues and cell lines (1, 2), thus providing a mechanism to remove excess heme iron from this protected site and prevent damaging iron deposits.
Heme-mediated cellular toxicity can also be caused by oxidized products of heme and heme precursors such as porphyrins (50). The ATP-binding cassette transporter ABCG2 is widely expressed in a variety of drug-resistant cancer cells and in numerous normal tissues and stem cells (51) and is a promiscuous transporter of many substrates, including drugs. ABCG2 was also identified as the major exporter for porphyrins, as PPIX accumulates in tissues of ABCG2-deleted mice (35). It was shown that ABCG2 also specifically interacts with heme, as it was isolated by binding hemin-agarose in a dose-dependent manner (52). Although heme appears to be the prime substrate for FLVCR (Fig. 3), our study reveals that FLVCR can export endogenously synthesized PPIX and coproporphyrin, induced by treatment with ALA alone or with iron. The fact that PPIX was present in the supernatant of NRK/Neo cells, although at a lower level than in NRK/FLVCR cells, indicates that another porphyrin exporter, probably ABCG2, is functioning in NRK cells to maintain homeostasis of intracellular porphyrins.
Our data show that UCB, the critical end product of heme catabolism, is not exported by FLVCR, despite its dicarboxylic tetrapyrrole structure. Unlike PPIX, coproporphyrin, and heme, which have been shown here to be substrates for FLVCR, the actual structure of UCB resembles a ridge tile or folded book, with extensive internal hydrogen bonding between each -COOH side chain and the lactam group of the opposite dipyrrinone half of the molecule (38). Apparently, the uniplanar configuration of heme and the porphyrins and/or the freedom of their carboxyl groups is an essential feature for transport of a tetrapyrrole by FLVCR, but this requires further study.
FLVCR has not yet been crystallized. However, the three-dimensional structures of three related major facilitator superfamily (MFS) transporters (oxalate transporter, lactose permease (LacY), and glycerol 3-phosphate transporter) have been established (53–55) and are remarkably similar (56), despite divergent transport function and only 21% sequence similarity. Also, modeling other distantly related MFS members (e.g. TetAB and vesicular monoamine transporter) on this MFS structure predicts tertiary configurations consistent with their known biochemical and structural properties (56). When we model FLVCR on this structure, regions ECL1 and ECL6 of the FLVC-binding site (57) are adjacent to each other, despite being separated by ∼330 amino acids in the sequence (data not shown), implying that the model can provide functional information about FLVCR.
To transport substrates, MFS transporters generally use an open-in then open-out mechanism termed a rocker-switch mechanism (55). Based on the functional data for LacY, which are most complete (58–60), the cytoplasmic substrate enters the transporter channel, and the channel then closes to the inside and opens to the outside, permitting the passage of substrate and thus export. Conceptually, a heme-binding protein like hemopexin or albumin might rapidly remove heme from the channel at the cell surface, allowing FLVCR to change conformation, reopen in, and accept a second heme molecule for export. Thus, the concentration and type of heme-binding protein that is present outside the cell regulate the export efficiency.
Across all mammalian species, there are two invariant histidine residues in FLVCR at positions 145 and 198, located adjacent to the outside opening of the exporting channel. These two histidine residues are not conserved in related paralogs that do not transport heme. It is possible that these two histidine residues each form a salt bridge with the -COO- groups of heme, which, in coordination with other functional domains either within FLVCR itself or on an adjacent membrane protein, serve as a transient binding site for heme, facilitating its transfer to hemopexin. When occupied, FLVCR may be maintained in an open-out configuration. Our data showing that heme (see “Experimental Procedures”) and hemopexin bind to a FLVCR peptide (amino acids 132–301) that contains these residues, as well as the results of our competitive binding and ZnMP export studies, support this concept and the rocker-switch mechanism of export. Of note, the internal H-bonding of the -COOH groups of UCB may render them unavailable to form salt bridges with the histidines in FLVCR and may contribute to the inability of FLVCR to transport bilirubin.
As an additional approach to link FLVCR structure with its heme export function, we surveyed the structures of prokaryotic heme transporters. Recently, it was reported that CcsBA, an integral membrane protein of Helicobacter hepaticus, is involved in heme transport for cytochrome c biogenesis (61). CcsBA has a highly conserved tryptophan-rich WWD domain that orients the heme to be translocated in concert with histidine residues. Although FLVCR does not have a WWD domain, it does have amino acid sequence WLLD in intracellular loop 1 surrounding the transport channel (17, 18). We propose that this similar sequence, or other unidentified functional domains in FLVCR, can serve as a docking site or orient heme entering the transport channel. These structural findings and the presence of hemopexin outside (but not inside) of cells may explain why FLVCR exports (but does not import) heme (1), whereas many MFS family members mediate bidirectional concentration-dependent transport and support its physiological role.
This study and our previous observations suggest that heme export by FLVCR is a mechanism for heme iron trafficking to prevent cellular heme and iron toxicity and to maintain heme and iron homeostasis. Hemopexin as an available and effective heme-binding protein facilitates this function by targeting heme to liver (3). It seems possible that hemopexin or other heme-binding molecules might facilitate removal of excess heme iron from tissues, ameliorate the anemia of chronic inflammation (which results from impaired export of elemental iron via ferroportin (62, 63)) or the systemic iron overload of hemochromatosis or other disorders, and provide a therapeutic benefit.
Acknowledgments
We thank K. R. Rish and T. Davis for hemopexin isolation and characterization and J. Kaplan for the dynamin-GFP constructs.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL31823, R01 DK020503, P30 DK072437, and DK085146. This work was also supported by Telethon Grant GGP05062 and the University of Missouri-Kansas City Research Incentive Funds.
Z. Yang, J. D. Philips, R. T. Doty, and J. L. Abkowitz, unpublished data.
S. B. Keel, R. T. Doty, and J. L. Abkowitz, unpublished data.
- ALA
- aminolevulinic acid
- PPIX
- protoporphyrin IX
- FLVCR
- feline leukemia virus subgroup C receptor
- NRK
- normal rat kidney
- MEF
- mouse embryonic fibroblast
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ZnMP
- zinc mesoporphyrin
- UCB
- unconjugated bilirubin
- MFS
- major facilitator superfamily.
REFERENCES
- 1.Quigley J. G., Yang Z., Worthington M. T., Phillips J. D., Sabo K. M., Sabath D. E., Berg C. L., Sassa S., Wood B. L., Abkowitz J. L. (2004) Cell 118, 757–766 [DOI] [PubMed] [Google Scholar]
- 2.Keel S. B., Doty R. T., Yang Z., Quigley J. G., Chen J., Knoblaugh S., Kingsley P. D., De Domenico I., Vaughn M. B., Kaplan J., Palis J., Abkowitz J. L. (2008) Science 319, 825–828 [DOI] [PubMed] [Google Scholar]
- 3.Smith A., Morgan W. T. (1979) Biochem. J. 182, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponka P. (1997) Blood 89, 1–25 [PubMed] [Google Scholar]
- 5.Stuehr D. J. (1997) Annu. Rev. Pharmacol. Toxicol. 37, 339–359 [DOI] [PubMed] [Google Scholar]
- 6.Suzuki H., Tashiro S., Hira S., Sun J., Yamazaki C., Zenke Y., Ikeda-Saito M., Yoshida M., Igarashi K. (2004) EMBO J. 23, 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han A. P., Yu C., Lu L., Fujiwara Y., Browne C., Chin G., Fleming M., Leboulch P., Orkin S. H., Chen J. J. (2001) EMBO J. 20, 6909–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hintze K. J., Katoh Y., Igarashi K., Theil E. C. (2007) J. Biol. Chem. 282, 34365–34371 [DOI] [PubMed] [Google Scholar]
- 9.Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 10.Hu R. G., Wang H., Xia Z., Varshavsky A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faller M., Matsunaga M., Yin S., Loo J. A., Guo F. (2007) Nat. Struct. Mol. Biol. 14, 23–29 [DOI] [PubMed] [Google Scholar]
- 12.Kumar S., Bandyopadhyay U. (2005) Toxicol. Lett. 157, 175–188 [DOI] [PubMed] [Google Scholar]
- 13.Tsiftsoglou A. S., Tsamadou A. I., Papadopoulou L. C. (2006) Pharmacol. Ther. 111, 327–345 [DOI] [PubMed] [Google Scholar]
- 14.Sassa S. (2006) J. Clin. Biochem. Nutr. 38, 138–155 [Google Scholar]
- 15.Furuyama K., Kaneko K., Vargas P. D. (2007) Tohoku J. Exp. Med. 213, 1–16 [DOI] [PubMed] [Google Scholar]
- 16.Ryter S. W., Tyrrell R. M. (2000) Free Radic. Biol. Med. 28, 289–309 [DOI] [PubMed] [Google Scholar]
- 17.Quigley J. G., Burns C. C., Anderson M. M., Lynch E. D., Sabo K. M., Overbaugh J., Abkowitz J. L. (2000) Blood 95, 1093–1099 [PubMed] [Google Scholar]
- 18.Tailor C. S., Willett B. J., Kabat D. (1999) J. Virol. 73, 6500–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang X., Lin T., Sun G., Beasley-Topliffe L., Cavaillon J. M., Warren H. S. (2009) J. Leukocyte Biol. 86, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delanghe J. R., Langlois M. R. (2001) Clin. Chim. Acta 312, 13–23 [DOI] [PubMed] [Google Scholar]
- 21.De Domenico I., Ward D. M., Langelier C., Vaughn M. B., Nemeth E., Sundquist W. I., Ganz T., Musci G., Kaplan J. (2007) Mol. Biol. Cell 18, 2569–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calligaris S., Cekic D., Roca-Burgos L., Gerin F., Mazzone G., Ostrow J. D., Tiribelli C. (2006) FEBS Lett. 580, 1355–1359 [DOI] [PubMed] [Google Scholar]
- 23.Abkowitz J. L., Taboada M. R., Sabo K. M., Shelton G. H. (1998) Stem Cells 16, 288–293 [DOI] [PubMed] [Google Scholar]
- 24.Smith A., Morgan W. T. (1984) J. Biol. Chem. 259, 12049–12053 [PubMed] [Google Scholar]
- 25.Eskew J. D., Vanacore R. M., Sung L., Morales P. J., Smith A. (1999) J. Biol. Chem. 274, 638–648 [DOI] [PubMed] [Google Scholar]
- 26.Li R. C., Saleem S., Zhen G., Cao W., Zhuang H., Lee J., Smith A., Altruda F., Tolosano E., Doré S. (2009) J. Cereb. Blood Flow Metab. 29, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Worthington M. T., Cohn S. M., Miller S. K., Luo R. Q., Berg C. L. (2001) Am. J. Physiol. Gastrointest. Liver Physiol. 280, G1172–G1177 [DOI] [PubMed] [Google Scholar]
- 28.Bayón J. E., Pascolo L., Gonzalo-Orden J. M., Altonaga J. R., González-Gallego J., Webster C., Haigh W. G., Stelzner M., Pekow C., Tiribelli C., Ostrow J. D. (2001) J. Lab. Clin. Med. 138, 313–321 [DOI] [PubMed] [Google Scholar]
- 29.Ostrow J. D., Mukerjee P. (2007) Transl. Res. 149, 37–45 [DOI] [PubMed] [Google Scholar]
- 30.Roca L., Calligaris S., Wennberg R. P., Ahlfors C. E., Malik S. G., Ostrow J. D., Tiribelli C. (2006) Pediatr. Res. 60, 724–728 [DOI] [PubMed] [Google Scholar]
- 31.Smith K. M. (1975) Porphyrins and Metalloporphyrins, Elsevier Scientific Publishing Co., Amsterdam [Google Scholar]
- 32.Lübben M., Morand K. (1994) J. Biol. Chem. 269, 21473–21479 [PubMed] [Google Scholar]
- 33.Damke H., Baba T., Warnock D. E., Schmid S. L. (1994) J. Cell Biol. 127, 915–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ajioka R. S., Phillips J. D., Kushner J. P. (2006) Biochim. Biophys. Acta 1763, 723–736 [DOI] [PubMed] [Google Scholar]
- 35.Jonker J. W., Buitelaar M., Wagenaar E., Van Der Valk M. A., Scheffer G. L., Scheper R. J., Plosch T., Kuipers F., Elferink R. P., Rosing H., Beijnen J. H., Schinkel A. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15649–15654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seery V. L., Muller-Eberhard U. (1973) J. Biol. Chem. 248, 3796–3800 [PubMed] [Google Scholar]
- 37.Morgan W. T., Smith A., Koskelo P. (1980) Biochim. Biophys. Acta 624, 271–285 [DOI] [PubMed] [Google Scholar]
- 38.Vítek L., Ostrow J. D. (2009) Curr. Pharm. Des. 15, 2869–2883 [DOI] [PubMed] [Google Scholar]
- 39.Rigato I., Pascolo L., Fernetti C., Ostrow J. D., Tiribelli C. (2004) Biochem. J. 383, 335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hrkal Z., Vodrázka Z., Kalousek I. (1974) Eur. J. Biochem. 43, 73–78 [DOI] [PubMed] [Google Scholar]
- 41.Hargrove M. S., Barrick D., Olson J. S. (1996) Biochemistry 35, 11293–11299 [DOI] [PubMed] [Google Scholar]
- 42.Fletcher J., Huehns E. R. (1968) Nature 218, 1211–1214 [DOI] [PubMed] [Google Scholar]
- 43.Tolosano E., Hirsch E., Patrucco E., Camaschella C., Navone R., Silengo L., Altruda F. (1999) Blood 94, 3906–3914 [PubMed] [Google Scholar]
- 44.Morello N., Tonoli E., Logrand F., Fiorito V., Fagoonee S., Turco E., Silengo L., Vercelli A., Altruda F., Tolosano E. (2009) J. Cell. Mol. Med. 13, 4192–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duvigneau J. C., Piskernik C., Haindl S., Kloesch B., Hartl R. T., Hüttemann M., Lee I., Ebel T., Moldzio R., Gemeiner M., Redl H., Kozlov A. V. (2008) Lab. Invest. 88, 70–77 [DOI] [PubMed] [Google Scholar]
- 46.Zukor H., Song W., Liberman A., Mui J., Vali H., Fillebeen C., Pantopoulos K., Wu T. D., Guerquin-Kern J. L., Schipper H. M. (2009) J. Neurochem. 109, 776–791 [DOI] [PubMed] [Google Scholar]
- 47.Benvenisti-Zarom L., Chen-Roetling J., Regan R. F. (2006) Neurosci. Lett. 398, 230–234 [DOI] [PubMed] [Google Scholar]
- 48.Tolosano E., Cutufia M. A., Hirsch E., Silengo L., Altruda F. (1996) Biochem. Biophys. Res. Commun. 218, 694–703 [DOI] [PubMed] [Google Scholar]
- 49.Morris C. M., Candy J. M., Edwardson J. A., Bloxham C. A., Smith A. (1993) Neurosci. Lett. 149, 141–144 [DOI] [PubMed] [Google Scholar]
- 50.Krishnamurthy P., Xie T., Schuetz J. D. (2007) Pharmacol. Ther. 114, 345–358 [DOI] [PubMed] [Google Scholar]
- 51.Zhou S., Schuetz J. D., Bunting K. D., Colapietro A. M., Sampath J., Morris J. J., Lagutina I., Grosveld G. C., Osawa M., Nakauchi H., Sorrentino B. P. (2001) Nat. Med. 7, 1028–1034 [DOI] [PubMed] [Google Scholar]
- 52.Krishnamurthy P., Ross D. D., Nakanishi T., Bailey-Dell K., Zhou S., Mercer K. E., Sarkadi B., Sorrentino B. P., Schuetz J. D. (2004) J. Biol. Chem. 279, 24218–24225 [DOI] [PubMed] [Google Scholar]
- 53.Hirai T., Heymann J. A., Shi D., Sarker R., Maloney P. C., Subramaniam S. (2002) Nat. Struct. Biol. 9, 597–600 [DOI] [PubMed] [Google Scholar]
- 54.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. (2003) Science 301, 610–615 [DOI] [PubMed] [Google Scholar]
- 55.Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 56.Vardy E., Arkin I. T., Gottschalk K. E., Kaback H. R., Schuldiner S. (2004) Protein Sci. 13, 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown J. K., Fung C., Tailor C. S. (2006) J. Virol. 80, 1742–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abramson J., Iwata S., Kaback H. R. (2004) Mol. Membr. Biol. 21, 227–236 [DOI] [PubMed] [Google Scholar]
- 59.Smirnova I., Kasho V., Choe J. Y., Altenbach C., Hubbell W. L., Kaback H. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16504–16509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guan L., Mirza O., Verner G., Iwata S., Kaback H. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15294–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frawley E. R., Kranz R. G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10201–10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nemeth E., Tuttle M. S., Powelson J., Vaughn M. B., Donovan A., Ward D. M., Ganz T., Kaplan J. (2004) Science 306, 2090–2093 [DOI] [PubMed] [Google Scholar]
- 63.Donovan A., Lima C. A., Pinkus J. L., Pinkus G. S., Zon L. I., Robine S., Andrews N. C. (2005) Cell Metab. 1, 191–200 [DOI] [PubMed] [Google Scholar]