Abstract
During apoptosis, the pro-apoptotic Bcl-2 family proteins BAK and BAX form large oligomeric pores in the mitochondrial outer membrane. Apoptotic factors, including cytochrome c, are released through these pores from the mitochondrial intermembrane space into the cytoplasm where they initiate the cascade of events leading to cell death. To better understand this pivotal step toward apoptosis, a method was developed to induce membrane permeabilization by BAK in the membrane without using the full-length protein. Using a soluble form of BAK with a hexahistidine tag at the C terminus and a liposomal system containing the Ni2+-nitrilotriacetic acid lipid analog that can bind hexahistidine-tagged proteins, BAK oligomers were formed in the presence of the activator protein p7/p15Bid. In this system, we determined the conformational changes in BAK upon membrane insertion by applying the site-directed spin labeling method of EPR to 13 different amino acid locations. Upon membrane insertion, the BH3 domains were reorganized, and the α5-α6 helical hairpin structure was partially exposed to the membrane environment. The monomer-monomer interface in the oligomeric structure was also mapped by measuring the distance-dependent spin-spin interactions for each residue location. Spin labels attached in the BH3 domain were juxtaposed within 5–10 Å distance in the oligomeric form in the membrane. These results are consistent with the current hypothesis that BAK or BAX forms homodimers, and these homodimers assemble into a higher order oligomeric pore. Detailed analyses of the data provide new insights into the structure of the BAX or BAK homodimer.
Keywords: Electron Paramagnetic Resonance (EPR), Liposomes, Membrane Proteins, Mitochondrial Apoptosis, Protein Structure, BAX and BAK, BCL-2, Homodimer, Oligomeric Pore, Site-directed Spin Labeling
Introduction
In mitochondrial apoptosis, the mitochondrial outer membrane is permeabilized by either BAX (Bcl-2-associated X protein) or its homolog BAK (Bcl-2 antagonist/killer) (Fig. 1) (1). Upon arrival of cell death signals, BAX and BAK are activated and form large pores in the outer membrane of mitochondria with an estimated diameter of ∼30–60 Å (Fig. 1B) (2, 3). As a result of the pore formation, many apoptosis-promoting proteins, including cytochrome c, escape from the mitochondrial intermembrane space into the cytoplasm of the cell through the BAX (or BAK) pore (2–5). Released cytochrome c then triggers a cascade of protease activation in the cytoplasm of the cell, leading to apoptotic cell death (6). Thus, BAX or BAK serves as a critical gateway in the mitochondrial cell death pathways (7). The ischemic brain damage after stroke or in neurodegenerative diseases is caused by this mechanism (8).
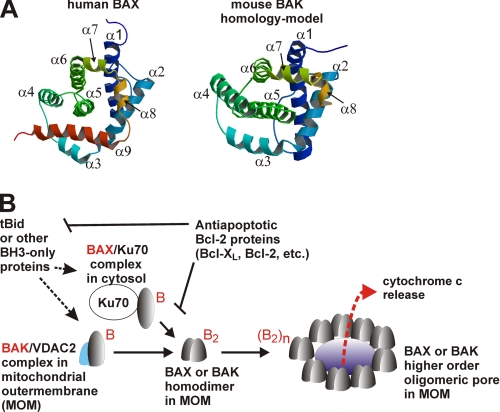
FIGURE 1.
Regulation of the mitochondrial membrane permeabilization by the Bcl-2 family proteins. A, BAX and BAK structure in solution state. The ribbon diagrams for the human BAX (Protein Data Bank code 2F16) (68) and the mouse BAK homology model were generated in Molscript (69). The homology model depicts the structure for residues 16–179 of mouse BAK, which was modeled after the human BAK structure (Protein Data Bank code 2IMS) (49). B, mechanism of BAX or BAK oligomeric pore formation in the mitochondrial outer membrane. The multidomain proapoptotic Bcl-2 proteins BAX and BAK that are held in check by Ku70 protein (70) and VDAC2 protein (71, 72), respectively, are activated by the proapoptotic BH3-only proteins such as truncated Bid (37, 73–75), which can be inhibited by the antiapoptotic Bcl-2 proteins such as Bcl-XL. Currently, the activated BAX or BAK monomer, B, is hypothesized to form a homodimer, B2, that in turn further oligomerizes to form higher order pore structures, (B2)n (14–16). The large pore serves as a conduit for the release of the apoptotic factors, including cytochrome c into the cytosol from the intermembrane space of the mitochondria, leading to cell death (1).
BAX and BAK are the members of the Bcl-2 (B cell lymphoma-2) family of proteins (1, 9, 10) (Fig. 1). The Bcl-2 family proteins carry conserved sequences known as the Bcl-2 homology (BH)2 domains (1). The antiapoptotic Bcl-2 proteins such as Bcl-2 or Bcl-XL share up to four BH domains (BH1–4). The proapoptotic multidomain Bcl-2 proteins such as BAX or BAK also contain four BH domains (BH1–4) according to a recent report (11). The “BH3-only” proteins constitute a subset of the proapoptotic proteins that display sequence conservation only in the BH3 domain (1). BH3-only proteins antagonize the antiapoptotic proteins through direct interactions via the BH3 domain (1). A discrete subset of the BH3-only members, including Bid, also activate BAX and BAK directly (Fig. 1) (1).
Currently, it is hypothesized that BAX and BAK form homodimers, and they assemble to form a higher order oligomeric pore structure (Fig. 1B) (12–16). It is also hypothesized that the BH3 domain in BAK forms a monomer-monomer interface in the homodimer (13–15). Understanding the nature of these homodimers or the oligomeric pore has been hampered due to difficulties in applying conventional structural biological methods such as x-ray crystallography or NMR. Only recently has limited structural information on these molecules in the membrane-inserted state begun to emerge from chemical cross-linking studies (14–16) and other approaches, including the site-directed spin labeling (SDSL) method of electron paramagnetic resonance (EPR) spectroscopy (12, 17, 18). The SDSL method of EPR is a powerful structural method that can be applied to proteins or biopolymers that are not easily amenable to x-ray crystallography or NMR. In the SDSL experiment (19–24), a residue of interest in a protein is substituted with a cysteine by site-directed mutagenesis, and a spin label is specifically attached to the thiol group in the cysteine residue. The EPR spectrum of the spin-labeled residue provides information regarding the dynamic nature of the labeled site (25, 26). The accessibility parameters for a spin-labeled residue measured by the power saturation method can be used to determine the topological location of the residue either in a soluble protein or in a membrane protein (27, 28). In a membrane protein, the membrane-immersion depth of a spin-labeled residue can also be determined (28). Furthermore, using the spin-spin interactions between two labels in close proximity, distance can be readily determined (29–31).
Inspired by our earlier observation that targeting of a Bid BH3 peptide to the membrane surface significantly enhances its potency in BAX activation (32, 33), in this study we have developed a proteoliposomal system that markedly enhances the membrane permeabilizing activity of a soluble form of BAK. Using a Ni2+-nitrilotriacetic acid (Ni-NTA) liposomal system that can conjugate hexahistidine-tagged proteins to the surface of the simulated outer mitochondrial membrane, we demonstrate that nanomolar concentrations of the soluble form of BAK when targeted to the membrane surface can efficiently permeabilize the membrane. Furthermore, tBid further enhances the membrane permeabilization by the membrane-targeted BAK, whereas Bcl-XL inhibits it, demonstrating the biological relevance of this system.
Utilizing the Ni-NTA liposomal system above, we also carried out the SDSL experiment to determine the conformational changes that occur in BAK upon membrane insertion. For SDSL experiments, 13 different residue locations were selected in BAK that represent various domains or structural elements. They include the following: (i) the residues in and around the BH3 domain, (ii) the residues in the α5-α6 helical hairpin structure, and (iii) the residues in other loops and α-helices. By comparing the EPR spectra and the accessibility parameters for the 13 spin-labeled protein samples both in the solution and in the membrane-inserted state, we determined the conformational changes occurring at various locations in BAK upon membrane insertion. The topological locations of these residues in the membrane-inserted state were determined. Furthermore, the monomer-monomer interface in the oligomeric structure was also mapped by measuring the spin-spin interactions for each residue location. The data provide new insights into the structure of the BAK homodimer and advance our knowledge of the mechanism of membrane permeabilization by BAK or BAX.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Proteins
The mouse BAK protein containing amino acids 16–184 was expressed as a fusion protein with an N-terminal bacterial hemoglobin and a C-terminal hexahistidine tag (designated as Hb-sBAK-ΔC-His) in the cytoplasm of Escherichia coli BL21(DE3) using plasmid pPosKJ-sBAK-CHis (see supplemental Fig. S1 for details). The C-terminally hexahistidine-tagged soluble mouse BAK protein (sBAK-ΔC-His; Fig. 2) was purified as described in detail in supplemental Fig. S2 after digesting Hb-sBAK-ΔC-His with glutathione S-transferase-tagged tobacco etch virus protease (GST-TEV 219V) (34). This protein construct sBAK-ΔC-His contains residues 16–184 of the mouse BAK and additional sequences of “GHM” and “AAALGHHHHHH” at the N and C terminus, respectively. For spin labeling, a cysteine to serine substitution mutation was first introduced in the codon of the BAK gene corresponding to residue location 154 in pPosKJ-sBAK-CHis plasmid using the QuickChange® site-directed mutagenesis kit (Stratagene). This plasmid was designated as pPosKJ-sBAKC154S-CHis. All the cysteine mutants were prepared using this plasmid coding for a cysteine-less backbone (Figs. 2 and 3). TEV 219V protease (residues 1–246 with 219V substitution) was expressed and purified as a glutathione S-transferase (GST) fusion protein (GST-TEV 219V) using the pGex-4T-1 vector (GE Healthcare). GST-BCL-XLΔC-His, a truncated BCL-XL (residues 3–212) fused to the GST and a His6 tag at the N and C termini, respectively, was similarly prepared using the pGex-4T-1 vector (GE Healthcare). A mutant form of GST-BCL-XLΔC-His containing G138E/R139L/I140N triple amino acid substitution mutations, designated as GST-BCL-XLΔC-His (m8), was also similarly prepared after site-directed mutagenesis. N-terminally His6-tagged Bid (p22 Bid) was purified as described previously (35). Caspase-8 was purified as described previously (36). Using caspase-8 in the presence of 10 mm dithiothreitol (DTT), His6-tagged p22Bid was cleaved into the N-terminal 7-kDa and C-terminal 15-kDa fragments that form a tight complex designated as p7/p15Bid (37). DTT was removed by dialysis. The isolated p15Bid fragment, also known as the truncated Bid (tBid), was prepared from p7/p15Bid as described previously (35). All the proteins above were treated with 18% (v/v) glycerol for storage at −80 °C.
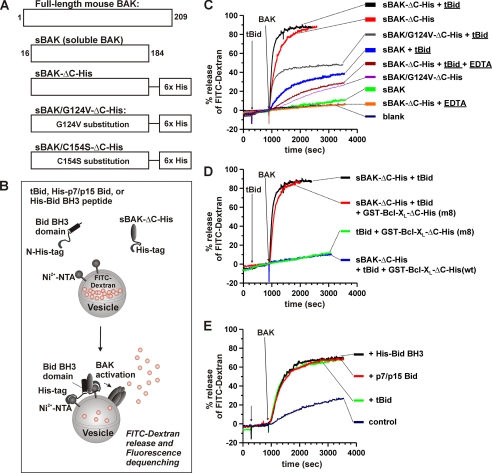
FIGURE 2.
Permeabilization of membrane by membrane-targeted BAK and Bid. A, schematic representation of mouse BAK protein constructs. Soluble form of mouse BAK protein (sBAK, residues 16–184) was prepared with (sBAK-ΔC-His) or without a C-terminal hexahistidine tag as described under “Experimental Procedures.” sBAK/G124V-ΔC-His and sBAK/C154S-ΔC-His represent sBAK-ΔC-His, in which residues Gly-124 and Cys-154 are substituted to valine and serine, respectively. B, schematic representation of the liposome dye release assay. The C-terminally His6-tagged sBAK (sBAK-ΔC-His) is targeted to the membrane via the binding of the His6 tag to the Ni2+-NTA moiety on the surface of the LUVs containing 5% (w/w) DOGS-NTA-Ni and other lipids mimicking the mitochondrial outer membrane contact sites (Ni-Nta-MCS LUVs) (see “Experimental Procedures”) (39). tBid (p15Bid without His6 tag), His6 tagged p7/p15 Bid, or His6-tagged Bid BH3 also targets to the membrane and exposes its BH3 domain, promoting BAK to form oligomeric pores, the conduit for the encapsulated FITC-dextran (10 kDa) release into the medium, resulting in fluorescence dequenching. C, effect of the hexahistidine tag, tBid, metal chelator, and the G124V amino acid substitution mutation on the membrane permeabilization by soluble BAK (sBAK). In the presence or absence of 5 mm EDTA, tBid (5 nm) and/or soluble BAK (10 nm) with (sBAK-ΔC-His) or without the C-terminal His6 tag (sBAK) were added at the indicated times to the Ni-Nta-MCS LUVs (10 μg/ml, or 0.125 nm (32)) encapsulating FITC-dextran (10 kDa). D, effect of Bcl-XL, an antiapoptotic Bcl-2 family protein on the tBid-catalyzed membrane permeabilization by soluble BAK. In the presence or absence of wild type or mutant form (m8) of 200 nm Bcl-XL fusion protein (GST-Bcl-XL-ΔC-His (wt) or (m8)), tBid (5 nm) and sBAK-ΔC-His (10 nm) were added at the indicated times to the Ni-Nta-MCS LUVs (10 μg/ml) encapsulating FITC-dextran (10 kDa). Bcl-XL (m8) mutant protein contains G138E/R139L/I140N triple amino acid substitution mutations. E, effect of tBid, His6-tagged Bid BH3 peptide, and the N-terminally His6-tagged p7/p15 Bid on the membrane permeabilization by the membrane-targeted BAK. tBid, His-Bid BH3 peptide, or His-p7/p15 Bid (all at 50 nm) and/or sBAK-ΔC-His (2 nm) were added at the indicated times to the liposomes (10 μg/ml) encapsulating FITC-dextran (10 kDa). C–E, all the experiments were carried out at 37 °C with the Ni-Nta-MCS LUVs containing 5% (wt) DOGS-NTA-Ni.
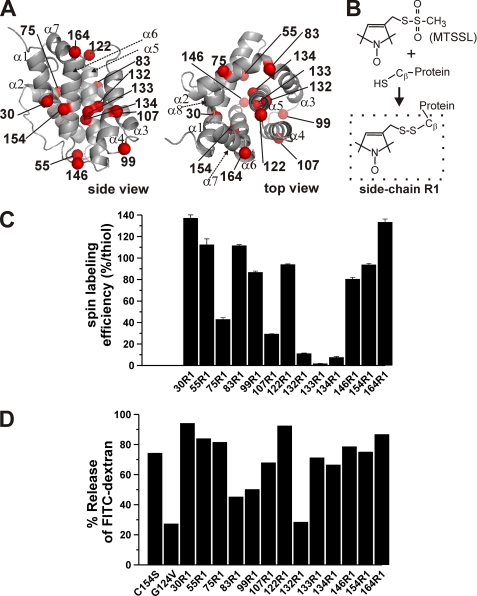
FIGURE 3.
Membrane permeabilization activity of spin-labeled soluble BAK measured by liposome dye release assay. A, sites of spin labeling in mouse BAK. The α-carbon atoms of the 13 residues selected for single cysteine substitution mutagenesis and spin labeling reaction in sBAK/C154S-ΔC-His are shown in red spheres in a ribbon diagram generated in PyMOL (76) in two views. The α-helices are denoted as α1–α8. B, SDSL reaction. The MTSSL reacts with the thiol group from a cysteine residue to form a spin-labeled side chain designated as R1. C, spin labeling efficiency. The percentage of spin labeling for cysteine residues at the indicated locations was determined as described under “Experimental Procedures.” Experiments were done in duplicate. D, membrane permeabilization activity of the spin-labeled sBAK-ΔC-His proteins. Liposome dye release assays were carried out with the indicated spin-labeled sBAK-ΔC-His proteins (5 nm) in the presence of 25 nm N-terminally His6-tagged p7/p15 Bid. The reported percent release values represent the values of the percent dye release averaged over the last 60 s after 25 min of incubation of the liposomes with the indicated sBAK-ΔC-His proteins (5 nm) in the presence of N-terminally His6-tagged p7/p15 Bid (25 nm) at 37 °C. All the experiments were carried out with Ni-Nta-MCS LUVs (10 μg/ml) encapsulating FITC-dextran (10 kDa). The error range was typically ±5% (not shown). The percent release values for sBAK/C154S-ΔC-His and sBAK/G124V-ΔC-His are shown as a positive and a negative control, respectively.
Peptide Synthesis
A peptide corresponding to the BH3 domain of Bid with an N-terminal hexahistidine sequence was synthesized at the Tufts Peptide Synthesis Core facility. The peptide was purified by reverse-phase high pressure liquid chromatography, and its mass was confirmed by mass spectrometry. A stock solution of the peptide was prepared in dimethyl sulfoxide for the liposomal release assay. The amino acid sequence of the synthesized peptide is as follows: HHHHHHESQEEIIHNIARHLAQIGDEMDHN.
Preparation of Large Unilamellar Vesicles (LUVs) Mimicking the Mitochondrial Contact Site
LUVs mimicking the lipid composition of the mitochondrial outer membrane contact sites (MCS) (38, 39) and containing a metal chelator lipid analog were made with a mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, beef liver phosphatidylinositol, beef heart cardiolipin, cholesterol, and 1,2-dioleoyl-sn-glycero-3-{[N-5-amino-1-carboxylpentyl)iminodiacetic acid]succinyl} (nickel salt) (DOGS-NTA-Ni) in 20 mm Hepes, 150 mm KCl (pH 7.0) (buffer A) as described previously (32). Briefly, the lipid emulsion was freeze-thawed 20 times and extruded through the polycarbonate membranes with a pore size of 100 nm as described previously (32). These vesicles were designated as Ni-Nta-MCS LUVs. The weight ratio of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, phosphatidylinositol, cholesterol, cardiolipin, and DOGS-NTA-Ni was 36:22:9:8:20:5 (32). DOGS-NTA-Ni was included to capture His6-tagged proteins on the liposomal membrane surface as described previously (32). Ni-Nta-MCS LUVs encapsulating fluorescein isothiocyanate-dextran 10 (FITC-dextran, 10 kDa, Invitrogen) were also prepared as described previously (35, 40–42). The phosphate contents of the vesicles were determined as described previously (43). The resulting LUVs were treated with glycerol (18% (v/v)), divided in 50-μl aliquots, and flash-frozen in liquid nitrogen for prolonged storage at −80 °C until they were used for the liposomal release assays. All the lipids were purchased from Avanti Polar Lipids, Inc. The molar concentration of LUVs was estimated as described previously (32).
Liposomal Release Assay
Liposomal release assays were performed as described previously (32) by adapting the fluorescence dequenching assay described by Terrones et al. (40) and Kuwana et al. (41, 42) to assess the biological activity of the BAK protein constructs. The release of the FITC-labeled dextran 10 from the LUVs was monitored by fluorometry using a SLM 8000C fluorometer (Olis Instruments, Inc.) in a thermostated 1-cm path length quartz cuvette with constant stirring at 37 °C. Excitation and emission wavelengths were 488 and 525 nm, respectively (slits, 4 nm). The extent of marker release was quantified on a percentage basis as described previously (32). In all assays, LUV concentration was 10 μg/ml lipids or 0.125 nm as described previously (32).
EPR Spectroscopy
To investigate the conformational changes occurring in BAK upon membrane insertion, we used the SDSL approach of the EPR spectroscopy. A few milligrams of single cysteine mutants of sBAK/C154S-ΔC-His were prepared as described above and spin-labeled with 10–20-fold molar excess (0.25–1.5 mm) of (1-oxyl-2,2,5,5,-tetramethylpyrroline-3-methyl)methanethiosulfonate spin label (MTSSL, Toronto Research Chemicals, Toronto, Canada) in buffer A at room temperature for 16 h. Unreacted MTSSL was removed by gel filtration using the Superdex 75 10/30 column (GE Healthcare) with buffer A as an eluant. If necessary, spin-labeled proteins were further concentrated using centrifugal concentrators (molecular mass cutoff of 3.5 kDa, Millipore). The spin labeling efficiency was determined as described using 3-carboxyproxyl (Sigma) as a standard (32). To generate BAK oligomers in the membrane, the experimental conditions for the liposomal release assay were adapted in a smaller reaction volume with minor modifications in the protein quantity ensuring appropriate signal strength for the EPR experiments. Typically, a total of 2 mg of Ni-Nta-MCS LUVs in a total reaction volume of 150 μl was used to form BAK oligomers with a total of 3.5 nmol of BAK in the presence of equimolar p7/p15Bid. Briefly, a mixture of 1.5 nmol of spin-labeled sBAK-ΔC-His and 2.0 nmol of sBAK/C154S-ΔC-His (unlabeled) (3:4 mixture) was added dropwise to the Ni-Nta-MCS LUVs while vortexing. Immediately after this, equimolar (3.5 nmol) p7/p15Bid was added to the mixture while vortexing it. The reaction mixture was incubated at 37 °C for 45 min. To stop the reaction and remove Ni2+ ions from the liposomes, EDTA was added to a final concentration of 20 mm, and the vesicles were pelleted by centrifugation at ∼110,000 × g for 15–20 min using an Airfuge (Beckman). The vesicle pellets were resuspended in 100 μl of buffer A containing 5 mm EDTA and spun down again. The vesicles were further washed with 100 μl of buffer A by two successive resuspension-centrifugation cycles. The resulting pellets were resuspended in a few microliters of buffer A for EPR experiments. To investigate possible monomer-monomer interaction in the membrane, we also reconstituted the spin-labeled BAK in the membrane in the absence of unlabeled BAK, i.e. sBAK/C154S-ΔC-His (7:0 mixture), following the same procedures described above. EPR spectra were obtained on a Bruker ElexSys 580 spectrometer using a Bruker High Sensitivity resonator or a loop gap resonator (JAGMAR, Krakow, Poland) at 2-milliwatt incident microwave power using a field modulation of 1.0–1.5 gauss at 100 kHz at room temperature. All the spectra were normalized to the same central peak height or to the same number of spins. For the latter, the double integration method was used making base-line corrections at each integration step using the Xepr program provided by Bruker. To measure the accessibility parameters, Π, of O2 and NiEDDA (at 5 mm), power saturation experiments were carried out with the above loop-gap resonator (44), and the data were analyzed using the R program (version 2.5.1) (45) as described previously (46–48). The immersion depths of membrane-inserted R1 residues were determined by the power saturation method using air O2 and 50 mm NiEDDA as collision reagents (35). Depth calibration curve was determined as described in supplemental Fig. S4.
Homology Modeling of the Mouse BAK Protein
Because the structure of mouse BAK in solution state is not available, a homology model of the mouse BAK solution structure was constructed using the crystal structure of the human BAK (Protein Data Bank code 2IMS (49)) with 77% sequence identity. The calculations were performed in a Swiss-Model as described by Schwede et al. (50). Briefly, the backbone of the human BAK structure was modified in the loop region (residues 48–57) by constrained space programming. The side chains were reconstructed starting with the conserved residues using a backbone-dependent rotamer library. The model was energy-minimized using the GROMOS96 empirical force fields (51).
RESULTS
Reconstitution of BAK Apoptotic Machinery in an Artificial Membrane as a Platform for Structure/Function Studies on BAK
Because of the technical difficulty in expressing full-length BAK protein in E. coli, soluble protein constructs of mouse BAK (sBAK, residues 16–184) were generated as shown in Fig. 2A. These sBAK constructs contain all the α-helices except the C-terminal helix α9 that is believed to be a transmembrane helix keeping the BAK protein tethered to the mitochondrial outer membrane. We reasoned that the tethering role of the helix α9 could be replaced with a C-terminal hexahistidine (His6) tag that can bind to the Ni-NTA moiety present on the membrane surface. Thus, we tested the membrane permeabilizing activity of these sBAK constructs with or without the C-terminal His6 tag in the Ni-NTA liposomal system containing 5% DOGS-NTA-Ni and other lipids mimicking the composition of the mitochondrial outer membrane contact sites (Fig. 2B) (32). The sBAK alone at 10 nm did not permeabilize the membrane efficiently (Fig. 2C, green trace). In the presence of the truncated Bid (tBid, 50 nm), which is known to target to the membrane and activate BAK and BAX via its BH3 domain (Fig. 2B), there was a modest increase in the membrane permeabilization by sBAK (Fig. 2C, blue trace). This indicates that in sBAK the binding site for the Bid BH3 domain is still fully or partially functional despite the lack of the N- and the C-terminal sequences.
To test whether the activity of sBAK can be further enhanced by membrane targeting, we tested sBAK with a C-terminal His6 tag (sBAK-ΔC-His) (Fig. 2A). When sBAK-ΔC-His (10 nm) was added to the Ni-NTA-MCS LUVs, there was a marked enhancement in the release of the 10-kDa FITC-dextran molecules both in the initial rate and the final extent of release (Fig. 2C, red trace). Note that FITC-dextran 10 (10 kDa) is comparable with cytochrome c (13 kDa) in its size. Thus, this result indicates that sBAK-ΔC-His efficiently permeabilized the membrane. Furthermore, the membrane permeabilization by sBAK-ΔC-His was dose-dependent over the concentration range of 0.1–20 nm (see supplemental Fig. S4B). These data indicate that sBAK can readily permeabilize the membrane once it is targeted to the membrane at nanomolar concentrations. Additionally, tBid further enhanced the membrane permeabilization by sBAK-ΔC-His (Fig. 2C, black trace). The effect of tBid was observed over a wide range of concentrations of sBAK-ΔC-His (0.1–20 nm; see supplemental Fig S4D). These results demonstrate that tBid interacts with the membrane-targeted sBAK and further promotes the membrane permeabilization by sBAK-ΔC-His.
To test whether the membrane targeting of sBAK-ΔC-His was specifically mediated by the interaction between the His6 tag and the Ni-NTA moieties on the liposomal surface, we measured the effect of the metal chelator EDTA on the membrane permeabilizing activity. In the presence of 5 mm EDTA that was used to abstract the Ni2+ ions from the Ni-NTA moieties, the membrane permeabilization activity of sBAK-ΔC-His was completely abolished in the absence of tBid (Fig. 2C, orange trace). This indicates that sBAK-ΔC-His was indeed targeted to the Ni-NTA liposomal membrane surface by the specific interactions between the C-terminal His6 tag and the Ni-NTA groups present on the liposomal surface. In the presence of tBid, however, EDTA significantly reduced the membrane permeabilization by sBAK-ΔC-His but not completely (Fig. 2C, dark red). This is expected because tBid could recruit sBAK-ΔC-His to the membrane as if it were sBAK (Fig. 2C, blue trace) and because EDTA would not affect the activity of tBid in activating sBAK significantly (supplemental Fig. S4E).
To test whether the membrane permeabilization by sBAK-ΔC-His is indeed mediated by the BAK protein itself, rather than by other contaminants, a mutant sBAK-ΔC-His with a G124V amino acid substitution mutation (sBAK/G124V-ΔC-His) was tested. This mutation is known to abrogate the activity of the human BAK in vivo (15). The activity of the sBAK/G124V-ΔC-His was significantly compromised in the presence (Fig. 2C, gray trace) or absence (Fig. 2C, violet trace) of tBid. These results clearly showed that the membrane permeabilization by sBAK-ΔC-His was indeed mediated by the functional activity of the sBAK protein and not by artifacts. The results taken together clearly demonstrated that the tethering role of the transmembrane helix α9 could be substituted with a chemical (His6 tag/Ni-NTA) and/or a biological (tBid) means. Furthermore, these results also suggest that sBAK retains the membrane-permeabilizing functions and thus the truncated sequences of BAK, i.e. the N-terminal 13 amino acid residues and the C-terminal TM helix α9 are not absolutely necessary for the membrane permeabilization.
Additionally, we investigated the effect of the antiapoptotic protein, Bcl-XL, on the activity of sBAK-ΔC-His in the presence or absence of tBid. Bcl-XL is known to inhibit the functions of the proapoptotic proteins such as tBid, BAX, and BAK, by heterodimerizing with them, thus enhancing the cell survival (52–54). As shown in Fig. 2D (blue trace), the Bcl-XL expressed as a fusion protein (GST-Bcl-XL-ΔC-His, C-terminally His6-tagged wild type Bcl-XL fused to a glutathione S-transferase) completely abrogated the membrane permeabilization mediated by sBAK-ΔC-His and tBid. The same result was observed in the absence of tBid (data not shown). In contrast, a mutant form of Bcl-XL with G138E/R139L/I140N triple amino acid substitutions (GST-Bcl-XL-ΔC-His (m8)), which lost its heterodimerizing activity (55), did not block the membrane permeabilization by sBAK-ΔC-His and tBid (Fig. 2D, red trace). These results show that Bcl-XL can specifically inhibit the membrane permeabilization by sBAK-ΔC-His, thus displaying its antiapoptotic effect against sBAK-ΔC-His and/or tBid.
Finally, to find the most active form of Bid in promoting the membrane permeabilization by sBAK-ΔC-His for SDSL experiments, we compared the activity of the His6-tagged Bid BH3 domain (32) and two different forms of Bid, i.e. tBid and the N-terminally His6-tagged p7/p15Bid (p7/p15Bid). As shown in Fig. 2E, all these reagents were almost equally effective in enhancing the membrane permeabilization by sBAK-ΔC-His at 2 nm. Therefore, p7/p15 Bid was chosen for further studies because it could be conveniently prepared by digesting full-length Bid (p22 Bid) with caspase-8 (37).
The above results showed that the functions of the representative members of the Bcl-2 proteins, i.e. the proapoptotic multidomain protein BAK, the proapoptotic “BH3 only protein” Bid, and the antiapoptotic protein Bcl-XL, can be recapitulated in the artificial system we used, reconstituting the BAK apoptotic machinery in a cell-free system. Thus, we have generated a convenient, biologically relevant platform with which the membrane permeabilization process by BAK can be studied by various means, including the SDSL method of EPR.
Functional Integrity of the Spin-labeled BAK Proteins and the Adaptation of the Liposomal Release Assay Conditions to the SDSL Experiments
To study the conformational changes in BAK upon membrane insertion and oligomerization, we chose 13 different residue locations in BAK for an SDSL study (Fig. 3A). They are as follows: (i) residues 75 and 83 from the α2-α3 helix-loop-helix structure in the solution state, which encompasses the BH3 domain (residues 76–85) that is known to be essential in the homodimerization of BAK (15); (ii) residues 122, 132–134, 146, 154, and 164 all of which are located in the α5-α6 helical hairpin structure that has been hypothesized to insert into the membrane upon membrane insertion of BAK (56, 57); and (iii) residues 30, 55, 99, and 107 that are located at helix α1, the α1-α2 loop, the α3-α4 loop, and the helix 4, respectively, which partially sandwich the helical hairpin structure.
After labeling the single cysteine mutant proteins with the spin label MTSSL, the labeling efficiency was determined for each of the spin-labeled side chain R1 residues (Fig. 3B). Residues 30R1, 55R1, 83R1, 99R1, 122R1, 146R1, 154R1, and 164R1 had labeling efficiency higher than 80%. Residues 75R1 and 107R1 had lower efficiency than the former group. Residues 132–134 that are expected to be located in the interior of the protein in solution state displayed very low labeling efficiency in contrast to other more-exposed residues mentioned (Fig. 3, A and C). To test if all the spin-labeled proteins were functionally intact, we performed the liposomal release assay (Fig. 3D). Most of the proteins retained membrane permeabilizing activity comparable with or slightly less than the parent molecule sBAK/C154S-ΔC-His. The activity of BAK/132R1 (i.e. sBAK/C154S/132R1-ΔC-His) was reduced to the level of the functionally defective G124V mutant (Fig. 2, A and C), indicating that cysteine mutation and/or spin labeling at this location compromised its function.
EPR Data for the Spin-labeled Soluble BAK Proteins Are Consistent with the Structural Model for the Soluble Mouse BAK
The EPR spectra for the R1 residues attached to the solution state of mouse BAK show location-dependent variations in their mobility (Fig. 4A, black traces). Residues such as 55R1, 132R1, and 134R1, predicted to be located in the interior of the protein in solution state, show a spectral feature representing a spin population that is severely restricted in motion, which is denoted in Fig. 4A by the dotted vertical line marked with “i” (immobile). Other residues show EPR spectra that indicate more rapidly tumbling nitroxide side chains (R1s), denoted in Fig. 4A by the dotted line marked with “m” (mobile). The accessibility parameters, Π(O2) and Π(NiEDDA), for the solution state also showed variations reflecting their predicted topological locations (Fig. 4B). The accessibility parameters Π(O2) and Π(NiEDDA) are proportional to the frequency of collision between the R1 residue and each of the added collision reagents, i.e. the molecular oxygen or the hydrophilic nickel complex NiEDDA (28). In solution state, these parameters measure the solvent accessibility of a given R1 residue. As expected, residues 55R1, 132R1, and 134R1 that are predicted to be located in the protein interior by the homology model showed small Π(O2) values and very small Π(NiEDDA) values (Fig. 4B). Other residues had relatively large values of Π(O2) and Π(NiEDDA) (Fig. 4B). These observations are consistent with the mouse homology model.
FIGURE 4.
EPR spectra and accessibility parameters of the spin label side chain R1s. A, EPR spectra of the spin-labeled soluble BAK in solution and in membrane-inserted state. The EPR spectra for the R1 residues in BAK in solution state (black trace) and in membrane-inserted state (red trace) are superimposed. The spectral features representing the immobile and mobile spin populations are indicated by i and m along with the vertical dotted lines in the CW spectra of the 100 G scan that are normalized to the same height for the central line. The solution samples contained 18% glycerol (v/v). (Note that in these oligomeric BAK samples in membrane, the spin-labeled BAK proteins were mixed with unlabeled BAK at a 3:4 ratio to suppress the spin-spin interactions between neighboring BAK molecules.) Briefly, the oligomeric BAK was prepared using 1.5 nmol of spin-labeled sBAK-ΔC-His, 2.0 nmol of sBAK/C154S-ΔC-His, and 3.5 nmol of p7/p15 Bid per mg of Ni-Nta-MCS LUVs. B, accessibility parameters for R1s of air oxygen, Π(O2), and 5 mm NiEDDA, Π(NiEDDA). Power saturation method was used to determine the accessibility parameters of molecular oxygen and NiEDDA for each R1 in BAK protein in solution and in membrane-inserted state. C, two-dimensional plot of Π(O2) and Π(NiEDDA). The accessibility parameters from B for the indicated R1s for solution (black diamonds) and for the oligomeric BAK in membrane (red dots) are shown in two-dimensional plot with the residue numbers indicated (black for solution; red italic for membrane). The topological locations of the R1s are categorized in three groups as follows: protein surface, protein interior/tertiary contact sites, or membrane interior according to the following: (i) the prediction by the mouse BAK homology modeled structure in solution and (ii) from the accessibility parameters for oligomeric samples. The changes in accessibility parameters from the solution state to the membrane-inserted state are indicated with thin dotted arrows for certain residues. D, topological locations of the R1s in the oligomeric state in the membrane. The topological locations of the R1s in BAK in the membrane-inserted state are indicated for the corresponding residues in the mouse BAK homology model. For residues 146R1, 154R1, and 164R1, the immersion depths from the membrane surface are indicated without error ranges for clarity (see supplemental Fig. S3 for details). Residue 134R1 is in a tertiary contact site in the membrane. The topological locations for 132 and 133 are unknown.
The accessibility parameters are plotted in a two-dimensional map as shown in Fig. 4C (black diamonds), made to aid in recognizing the overall pattern of the accessibility of the R1 residues. Residues predicted to be located in the protein interior such as 55R1, 132R1, and 134R1 have small values of Π(O2) and Π(NiEDDA), and thus their positions in the two-dimensional map cluster near the origin of the plot. Other residues that are predicted to be located in the flexible loops or on the helical surface have much larger values of Π(O2) and Π(NiEDDA), and their positions are scattered over a wider area far away from the origin. Thus the area near the origin was designated as the “protein interior/tertiary contact sites” and the other wider region for the surface exposed residues as the “protein surface.”
Residue 154R1, predicted to be located on the surface of helix α6, displayed a somewhat low Π(NiEDDA) value while showing a Π(O2) value slightly below average among the protein surface residues, indicating that the R1 chain is perhaps involved in a tertiary contact. This could be due either to the structural perturbation caused by spin labeling or due to the discrepancy between the actual structure of the mouse BAK protein and the homology model built after the human BAK (49). In addition, despite its predicted location within the tightly folded protein interior, 133R1 displayed a mobile component (m) in addition to the predicted immobile (i) component in the EPR spectrum. It also has relatively high Π(O2) and Π(NiEDDA) values, placing it in the protein surface region between residues 99 and 107 in Fig. 4C. These observations indicate that the cysteine substitution and/or spin labeling reactions caused a major perturbation in the structure, resulting in an anomaly. Despite this, residue 133R1 retained membrane permeabilizing activity that was almost comparable with its parent molecule sBAK/C154S-ΔC-His as shown in Fig. 3D. Taken together, except at two locations, i.e. residue 133 and 154, the EPR data of the tested spin-labeled mutants are consistent with the predictions based on the mouse BAK homology model, validating the homology model of mouse BAK used in this study.
EPR Data for the Membrane-associated BAK Protein Indicate That Conformational Changes Occur at Various Locations in BAK Structure upon Membrane Insertion
Structural Reorganization Involving the BH3 Domain
The EPR spectra for the R1 residues in BAK that formed oligomers in the Ni-Nta-MCS liposomes indicate that at almost all the locations tested, the mobilities of the R1 residues were altered (Fig. 4A, red trace). Among them, the changes occurring in 83R1 is very surprising. Residue 83R1 is located within the BH3 domain in a distorted helical turn, interconnecting α2 and α3 in solution structure (Fig. 3A, top view). In solution state, 83R1 showed a relatively mobile line shape that is typically observed on a helical surface (Fig. 4A) (58–60). Consistent with this, the accessibility parameter Π(NiEDDA) for 83R1 in solution was relatively large, and the Π(O2) was close to an average value (Fig. 4, B and C). Upon membrane insertion of BAK, this residue became entirely immobile (Fig. 4A), and its accessibility parameters Π(O2) and Π(NiEDDA) were markedly reduced, locating it in the region of “protein interior/tertiary contact sites” in the Π(O2) versus Π(NiEDDA) two-dimensional plot (Fig. 4C). The accessibility parameters were in fact the least among all the tested residues both in solution and membrane-associated states of BAK (Fig. 4, B and C). These results indicate that this flexible, water-exposed residue 83R1 in the BH3 domain in solution state becomes completely buried in the protein interior, virtually inaccessible to either oxygen or NiEDDA upon membrane insertion of BAK (Fig. 4D). Similarly, residue 75R1 that is on helix α2 and two helical turns away from 83R1 also showed similar changes in its EPR spectra and accessibility parameters. Residue 75R1 had slightly larger accessibility parameters than 83R1, indicating that 75R1 is slightly more accessible to these collision reagents than 83R1. These data indicate that residue 75R1 is located at a tertiary contact site rather than in the highly inaccessible protein interior in the membrane-associated form of BAK (Fig. 4D). Noting that residue 83R1 is within the BH3 domain of BAK and residue 75R1 is located at the immediate upstream of the beginning of the BH3 domain in amino acid sequence (supplemental Fig. S5), these results show that there is a large conformational change with the BH3 domain and its vicinity.
α1-α2 Loop Interconnecting Helices α1 and α2 Loses Tertiary Interactions and Becomes Exposed to Water
Residue 55R1 exists in a long loop formed by the 20 amino acids that interconnect helices α1 and α2 in the mouse BAK homology model in solution state (Fig. 4D). This loop is not entirely flexible and is partially folded making tertiary contacts with nearby domains. The homology model predicts that 55R1 would be located in such a region in the loop with its side chain surrounded by residues from α1, α5, and α1-α2 loop, well shielded from the aqueous milieu. As described above (Fig. 4, A–C), this residue displayed EPR line shape and accessibility parameters consistent with its predicted location in the solution state. In stark contrast, upon membrane insertion of BAK, this residue became extremely mobile (Fig. 4A, red trace). Its accessibility parameters Π(O2) and Π(NiEDDA) also increased greatly, both of which were the largest among all the tested residues in membrane-associated BAK (Fig. 4, B, red bars, and C, red dots and also see the thin dotted arrow to 55), indicating that this residue becomes highly exposed to water losing its tertiary interactions upon membrane insertion of BAK (Fig. 4D). Taken together, these data clearly show that α1-α2 loop is freed from the interacting domains and exposed to the water phase upon membrane insertion of BAK.
Helices α5-α6 Appear to Be Partially Exposed to the Membrane
In Fig. 4C, certain residues such as 133R1, 134R1, 146R1, 154R1, and 164R1 were clustered in the region near the axis of Π(O2) away from the origin, which does not overlap with either the protein surface or the protein interior/tertiary contact regions. It is well known that across the membrane the polar collision reagent NiEDDA forms a concentration gradient with decreasing concentration toward the hydrophobic core of the membrane (61). On the other hand, the oxygen has an opposite trend in its gradient with increasing concentration toward the hydrophobic core (61). Therefore, residues residing in the membrane interior are expected to display small accessibility to NiEDDA but relatively large accessibility to oxygen, which would be located near the axis of Π(O2) away from the origin in the Π(O2) versus Π(NiEDDA) two-dimensional map (e.g. 134R1 denoted by the dotted arrow in Fig. 4C). For this reason, the area for these newly clustering residues in the membrane-inserted state of BAK was designated as the “membrane interior” (Fig. 4C).
Among the residues that belong to membrane interior group, residues 133R1 and 134R1 on helix α5 displayed EPR spectra with a dominant immobile component (i in Fig. 4A, red traces), indicating that these residues are involved in tertiary contact even in the membrane interior. In contrast, the EPR spectra of residues 146R1, 154R1, and 164R1, located in the α5-α6 loop, α6 helix, and the α6-α7 loop, respectively, did not indicate the presence of a single predominantly immobile population (Fig. 4, red traces). Thus, we assumed that these residues are not at the tertiary contact sites in the membrane-inserted state and their immersion depths from the membrane surface were estimated as described under “Experimental Procedures” (see also supplemental Fig. S3). The estimated immersion depths for 146R1, 154R1, and 164R1 were 3.0 Å (error range −3.5 and 6.5 Å), 6.0 Å (error range 3.4 and 7.5 Å), and 7.0 Å (error range 5.8 and 8.1 Å), respectively, which are reported in Fig. 4D without the error ranges (the asymmetric error ranges are explained in supplemental Fig. S3 legend). Assuming that the thickness of the phosphate headgroup region is a minimum of 5 Å (62–64), residue 146R1 appears to be immersed in the headgroup region, and residues 154R1 and 164R1 are inserted deeper into the headgroup region or the acyl chain compartment of the lipid bilayer.
Of note, the functionally defective 132R1 (Fig. 3D) displayed accessibility parameters corresponding to the protein interior/tertiary contact region even after membrane targeting (Fig. 4, B and C), indicating that this mutant is still in its solution form without unfolding. Consistent with this, the EPR spectrum displayed a line shape for a very “immobile” spin population (Fig. 4A). Essentially, this mutant served as a negative control for this SDSL study.
Residues 30R1, 99R1, 107R1, and 122R1 Remain Exposed to Water in the Membrane-inserted BAK
Among the water-exposed residues on the protein surface in solution, residues 30R1, 99R1, 107R1, and 122R1 still maintain high accessibility values to oxygen and NiEDDA in the membrane-inserted state (Fig. 4, B and C) that are comparable with those residues mapped in the protein surface region in Fig. 4C. The EPR spectra for these residues in the membrane-inserted state are also consistent with this interpretation, displaying line shapes consisting of relatively mobile spin populations (Fig. 4A). These data indicate that these residues are still exposed to the water phase in the membrane-inserted state (Fig. 4D).
EPR Data Directly Show That BH3 Domains Are Brought Close to Each Other in the Oligomeric State in the Membrane
Proximity of the BH3 Domains in the Membrane-inserted State of BAK
Dewson et al. (15) proposed that BAK dimerization occurs by juxtaposition of the two BH3 domains (residues 76–87 in mouse BAK within the α2-α3 helices). We attempted to test this by determining if spin labels attached to a single residue within the BH3 domain or other domains of BAK are brought close to each other in the membrane-inserted state. For this, we carried out the so-called “spin-dilution experiment” (65) using BAK oligomers prepared by mixing the spin-labeled BAK proteins with an unlabeled one at 7:0 or 3:4 ratios, respectively. In the absence of the unlabeled BAK (Fig. 5A, top left), all the BAK proteins will have R1 chains, and if a spin label in one monomer is in close proximity (within 25 Å) to another from the neighbor (29, 30), the continuous wave (CW) EPR spectrum (normalized to the same number of spins) will be broadened, and the intensity will decrease in a distance-dependent way, i.e. the closer the distance, the bigger the effect. In the presence of excess unlabeled BAK in the oligomers (Fig. 5A, top right), the chances for a spin-labeled BAK to have another spin-labeled BAK as a neighbor will sharply decrease assuming that the BAK monomers associate with each other randomly regardless of the spin label. This will effectively reduce the spin-spin interactions between neighboring BAK monomers and thus increase the signal intensity of the normalized EPR spectrum. If the spin-spin interactions between two labels are primarily mediated by the dipolar interaction, the line broadening and amplitude reduction in the CW EPR spectrum will be detectable up to ∼25 Å (29, 30).
FIGURE 5.
Spin dilution experiment. A, rationale for the spin dilution experiment and the result for sBAK/C154S/83R1-ΔC-His. Top panel, diagrams explain how the distance between the nearest spin-labeled proteins is affected by the presence or absence of the excess unlabeled protein. The BAK oligomers were prepared using the spin-labeled sBAK-ΔC-His protein (BAK/154S/83R1, red disks) in the presence (right panel) and the absence (left panel) of excess unlabeled protein sBAK/C154S-ΔC-His (BAK/154S, black disks), respectively, at the indicated ratio using Ni-Nta-MCS LUVs. The two partially overlapping disks represent BAK dimers. Bottom panel, EPR spectra of BAK/154S/83R1 in the presence and the absence of spin dilution. EPR spectra were taken from each of the samples as described above, normalized to the same number of spins by double integration, and superimposed. The red trace represents the EPR spectrum of 83R1 without spin dilution (7:0 mixture) and the black trace with spin dilution (3:4 mixture) with its vertical scale expanded for clarity. The red arrows indicate EPR line splitting in the EPR spectrum in the absence of the spin dilution caused by strong spin-spin interactions between the two 83R1 residues. The estimated distance between the two 83R1 residues existing at the monomer-monomer interface in the membrane-inserted state is 5–10 Å. B, EPR spectra for R1 residues in the presence or absence of spin dilution. EPR spectra were measured with the membrane-inserted form of the indicated singly spin-labeled BAK proteins in the presence (black trace) and the absence (red trace) of excess unlabeled BAK (BAK/C154S), respectively, at the indicated molar ratios. All the EPR spectra were normalized to the same number of spins by double integration. The h0 and hd represent the maximum peak height values of the central lines of the normalized EPR spectra in the absence and presence of the excess unlabeled BAK, respectively. The hd/h0 ratio is shown for each R1 on the right. Spectra were obtained by 150 G scans.
In Fig. 5, the results are summarized for selected residues in BAK (see supplemental Fig. S6 for details). For these, the line-broadening effect due to spin-spin interactions was quantitated by the ratio of the maximum intensity of the EPR spectra that were obtained in the presence and absence of the excess unlabeled BAK and normalized to the same number of spins (hd/h0 ratio in Fig. 5B). Very surprisingly, among the residues tested, only residue 83R1, located within the BH3 domain of BAK (Fig. 3A and supplemental Fig. S5), showed a marked increase in the intensity of the EPR spectrum upon spin dilution, resulting in the hd/h0 ratio of 2.42. Furthermore, the EPR spectrum of 83R1 in the absence of the additional unlabeled protein showed fine splittings (black arrows in Fig. 5A), which can only be observed in a spin label pair in which the spin labels are separated within 5–10 Å distance (29, 30). This is direct evidence that two molecules of BAK are almost in contact with each other at this residue location in the oligomeric structure in the membrane.
Proximity of Other Domains in the BAK Oligomeric Structure
In contrast to 83R1, residue 122R1 located in a loop N-terminal to α5 helix showed practically no change in the intensity of the EPR spectra upon spin dilution (Fig. 5C), indicating that residue 122R1 is separated from one another at a distance greater than 25 Å in the oligomeric state in the membrane. Similarly, residue 154R1 located in helix α6 did not show any significant spin dilution effect, indicating the same conclusion. In contrast, residues 146R1 and 164R1 showed signs of spin-spin interaction with hd/h0 ratios of 1.34 and 1.17, respectively, indicating that these residues are within 25 Å when they exist in the oligomers. This will be further discussed below.
It should be noted that in spin dilution experiments, the tested proteins must be almost 100% labeled with the spin label (Fig. 3C). Otherwise, because of the contaminating unlabeled single cysteine mutant proteins in the “spin-labeled” protein sample, the spin dilution effect would already occur in the “undiluted sample.” Thus, the effect of real dilution by the unlabeled protein would be diminished, obscuring the interpretation of the results. For this reason, the BH3 residue 75R1 and residues in others domains such as 107R1 (on α4) and 132R1–134R1 (on α5) were excluded due to their low spin labeling efficiency (less than 50%, Fig. 3C). Additionally, the nonspecific spin-spin interactions caused by the overcrowded spin-labeled proteins should be avoided for a successful spin dilution experiment. In our experiments, a total of 3.5 nmol of the BAK proteins bind to a total of 1 mg of the LUVs that have the average diameter of 100 nm. Under these conditions, the average distance between the closest BAK neighbors is estimated to be greater than 100 Å if the BAK monomers are evenly distributed over the surface of the LUVs (32). This distance far exceeds the detection limit of the dipolar broadening in the CW EPR spectrum. Thus, the spin-spin interactions observed above must originate from the neighboring BAK molecules in the oligomeric structure (i.e. from the neighboring BAK molecules in the same homodimer or those at the homodimer-homodimer interface).
DISCUSSION
In this study, the BAK apoptotic machinery was reconstituted in an artificial membrane system. The VDAC2 protein was not included, and other protein or lipid factors might be missing in the system. It was noticed that the soluble form of BAK was very active even in the absence of tBid upon membrane targeting. This is perhaps due to the following: (i) the lack of VDAC2 in the liposomal system, (ii) the missing N-terminal and/or the C-terminal sequences that could be involved in the regulation of the BAK activity, or (iii) other missing factors in the experimental system. Despite these, this system proved useful for the purpose of studying the conformational changes occurring in BAK upon membrane insertion as demonstrated here.
Recently, Kim et al. (17) have shown that the helix α1 in BAX becomes exposed upon activation by tBid, making it available for the recognition by the 6A7 antibody that is specific to the N-terminal sequence of BAX. Similarly, helix α1 in BAK that corresponds to the α1 helix in BAX (Fig. 1A and supplemental Fig. S5) appears to be exposed upon activation by tBid, making it available for binding to specific antibodies, according to Dewson et al. (14). Relatedly, Bleicken and Zeth (66) reported that in detergent-treated BAX, the α1-α2 loop becomes susceptible to protease cleavage. Our current observations are consistent with these reports and extend them. For example, residue 55R1, which is hidden within a tightly folded structure of the α1-α2 loop in the solution state, became highly flexible (Fig. 4A) and was exposed to the water phase upon membrane insertion of BAK (Fig. 4, B–D). Furthermore, residue 134R1 on α5 that is shielded from the water phase by the helix α1 and other residues nearby also became exposed to the membrane (Fig. 4), indicating that the interface between helices α1 and α5 was disrupted. Additionally, the accessibility parameters for 30R1 that is located on the surface of helix α1 in solution state also increased upon membrane insertion of BAK, indicating that it is more exposed to the aqueous environment (Fig. 4). These results suggest that helix α1 is liberated from the remaining structure in BAK along with the loop that interconnects α1 and α2, consistent with the above findings by others (17, 66). Thus, these results show that the experimental system developed in this study is biologically relevant both structurally and functionally, lending credence to the new findings in this study.
One of the most interesting observations in this study is that in the membrane-inserted state BAK molecules form a monomer-monomer interface via the BH3:BH3 domain interactions as evidenced by the results from the spin-dilution experiment (Fig. 5). This is consistent with the BAK homodimer model proposed by Dewson et al. (15). Dewson et al. (15) reported that in the oligomers formed in the cells undergoing apoptosis, two neighboring BAK molecules can be chemically cross-linked between certain residues on the helix α2 in one BAK monomer and other residues on the helices α3 and α4 in another BAK molecule. Based on these results, they proposed that BAK homodimerizes via the interaction between the BH3 domain (existing within the α2/α3 helices, see supplemental Fig. S5) of one BAK monomer and a groove formed by the helices α2, α3, and α4 from another monomer (Fig. 6A) (15). In this BAK homodimer model, helices α2 and α3 form a single extended helix (α2/α3), and two of these extended helices (α2/α3:α2′/α3′) are juxtaposed in an anti-parallel manner (Fig. 6A) (15). The EPR data from the spin dilution experiment (Fig. 5) indicate that in the membrane-inserted state of BAK residue 83R1 located within the BH3 domain is brought close to each other within a 10-Å distance, which almost warrants molecular contact between the domains. Although these data, per se, do not prove the anti-parallel arrangement of the two BH3 domains, this is strong evidence for the existence of a direct contact between the two BH3 domains in BAK oligomeric structure in the membrane, consistent with the BAK homodimer model proposed by Dewson et al. (15).
FIGURE 6.
Current models of BAX/BAK homodimers. A, BAK homodimer model. The α2 helix (red box) in one BAK binds to the binding pocket formed by the helices α3 and α4 (green boxes) from another molecule, forming a dimerization interface between the α2/α3 extended helices. The two overlapping deep yellow ovals represent BAK molecules forming a homodimer. Adapted from Dewson et al. (15). B, BAX homodimer models. Two α2/α3 extended helices are juxtaposed to form a symmetric BAX homodimer on the surface of the membrane (12). The distance between the two symmetry related cysteine residues (C62) is estimated to be ∼24 Å. The α2/α3 helices can be either in the non-salt bridged conformation (left) or in the salt bridged conformation (right) indicated with the locations of the two residues Cys-62 and Leu-70 shown (small red and yellow ovals). Corresponding BAK residues are indicated. Adapted from Bleicken et al. (12).
Because of the homology in both the amino acid sequence and the structure between BAK and BAX, it is highly likely that they would form higher order oligomers by the same mechanism. Because of this reason, the BAX homodimer model recently proposed by Bleicken et al. (12) is insightful in understanding the possible organization of the proposed BAK homodimer. In this model, the two α2/α3 extended helices (i.e. α2/α3:α2′/α3′) can exist in two different conformations, with or without the salt bridges formed between α2/α3 and α2′/α3′ (12), which will be called the “salt bridged conformation” and the “non-salt bridged conformation,” respectively (Fig. 6B). In the non-salt bridged conformation of the BAX homodimer (Fig. 6B, left), a cysteine residue at position 62 (Cys-62) is exposed to the water phase, with its side chain pointing to a direction almost perpendicular to the membrane surface without making any tertiary contacts (12). In the salt bridged conformation of the BAX homodimer (Fig. 6B, right), the two extended helices are rotated through their respective helical axes relative to the non-salt bridged conformation to allow the salt bridge formation between the charged residues on the helical surface, placing the side chains of the residue 62 at the interface between the two helices (Fig. 6B, right) (12).
Note that BAX residues Cys-62 and Leu-70 correspond to BAK residues Gln-75 and Ile-83, respectively, according to the sequence alignment (supplemental Fig. S5). Therefore, BAK molecules can be fit into these BAX homodimer models. Thus, assuming that BAK homodimers are in such proposed conformations of the BAX homodimer, the predictions by these models for the inter-residue distances, spin label mobility states, and topological locations for 75R1 and 83R1 in BAK were tabulated and compared with the experimental results from this study (Table 1).
TABLE 1.
Comparison of the experimental data and the predictions by the BAX homodimer models
The EPR data for spin-labeled residues 75R1 and 83R1 in BAK in the membrane-inserted state are compared with the predictions by the two BAX homodimer models (12) assuming that BAK takes the conformations indicated.
| Items compared | Residue Q75R1 (=Cys-62 in BAX) |
Residue I83R1 (=Leu-70 in BAX) |
||||
|---|---|---|---|---|---|---|
| 75R1–75R1′ distance | R1 mobility by EPR line shape | Topological location | 83R1–83R′ distance | R1 mobility by EPR line shape | Topological location | |
| Experimental data | NA | Immobile | Tertiary contact site | 5–10 Å | Very immobile | Protein interior |
| Predictions from BAK assuming salt bridged conformation | ∼24 Å | Immobile | Tertiary contact site | ∼10 Å (Cβ–Cβ distance) | Mobile | Exposed to membrane |
| Predictions from BAK assuming non-salt bridged conformation | >24 Å | Mobile | Exposed to water | ∼15 Å (Cβ−-Cβ distance) | Immobile | Tertiary contact site |
Regarding the distance between the two symmetry-related Ile-83 residues in the BAK homodimer, both conformations in the BAX homodimer model appear to be compatible with the data (Table 1). Regarding the mobility and the accessibility parameters for residues 75R1 and 83R1 in BAK, however, none of the above two conformations of the BAX homodimer appear to be fully compatible with the experimental data (Table 1). Residue 75R1 in BAK, corresponding to BAX residue 62, would be highly exposed to the water phase in the non-salt bridged conformation of the BAX homodimer. Contrary to this prediction, BAK residue 75R1 on helix α2 appears to be located at a tertiary contact site in the membrane-inserted state of BAK (Fig. 4). This is supported by the EPR line shape that corresponds to a spin label side chain restricted motion (Fig. 4A) and further by the very limited accessibility of the water-soluble collision reagent NiEDDA for the residue (Fig. 4, B and C). In the salt bridged conformation, isoleucine 83 in BAK, corresponding to leucine 70 in BAX (supplemental Fig. S5), would be pointing to the membrane phase removed from the aqueous environment. Thus oxygen, but not NiEDDA, would be readily accessible to this residue. The R1 chain would be relatively mobile due to its exposure to the fluidic membrane environment without any tertiary contact interactions. Contrary to these predictions, residue 83R1 is located in a very tightly sealed protein interior virtually inaccessible to oxygen and NiEDDA, and the side chain motion is severely restricted as shown in Fig. 4. Therefore, this conformation is not consistent with the data for 83R1.
These discrepancies are not likely due to functional or structural defects of these two spin-labeled mutant proteins because they both retained membrane-permeabilizing activity as demonstrated in Fig. 3D. The BAK protein with 75R1 was as active as its parent molecule, i.e. sBAK/C154S-ΔC-His. The BAK protein with 83R1 still retained a certain level of activity, although at a considerably lower level than C154S mutant. Nevertheless, it is likely that the less active 83R1 mutant BAK inserts into the membrane, dimerizes, and further oligomerizes after binding to the membrane. If a monomer, the residue will display high accessibility to both oxygen and NiEDDA, and the EPR line shape will be that for a flexible nitroxide label due to its highly exposed location in the α2-α3 loop. The EPR line shape might be explainable as aggregated protein, but it is highly unlikely that it will result in such strong spin-spin interactions in randomly aggregated BAK molecules.
The above discrepancies can be explained. The EPR data from BAK residue 75R1 was compatible with the salt bridged conformation of the BAX homodimer (Table 1), and in this conformation the BAX residue 70 (equivalent to BAK 83R1) was pointing to the membrane (Fig. 6B). Thus, if the two extended helices (i.e. α2/α3:α2′/α3′) were in tight contact at or around the residue 83 with an unknown domain in BAK within the membrane as schematically shown in Fig. 7A, 83R1 will be located in a protein interior-like environment. Thus, we posit that the lack of this unknown domain in the salt bridged BAX homodimer model by Bleicken et al. (12) caused the discrepancies between the EPR data and the model when BAK assumed this conformation. In a recent review article, Dewson and Kluck (13) proposed that α5 helix might be in a nontransmembrane orientation associated with the α2/α3 extended helix. This is called the “TM-only” model for the BAK homodimer structure. In this model, without experimental evidence, only the C-terminal TM domain (α9) of BAK was assumed to take the transmembrane orientation (13). Thus, the “unknown domain” above could be the helix α5, which needs to be tested experimentally. Relatedly, residue 134R1, a residue on helix α5 located away from the α5-α6 contact interface, became exposed to the membrane but was still involved in tertiary contact interactions in the membrane-inserted state of BAK as described under “Results” (Fig. 4). Furthermore, residues on or near the N and C termini of helix α6 such as residues 146R1, 154R1, and 164R1 had relatively shallow immersion depths in the membrane (Fig. 4D). All these results are not easily reconciled with the current BAX homodimer model(s) in which the α5-α6 helical bundle exists in an isolated transmembrane conformation (12). It remains to be seen whether this helix constitutes a transmembrane aqueous channel as observed in other amphipathic helices (67).
FIGURE 7.
Modified BAK homodimer model. A, topological locations of the spin-labeled residues in the proposed BAK homodimer model in the membrane. Portions of the two BAK monomers are schematically depicted in blue and green, respectively. The cylinders and thick lines represent the α-helices and the interconnecting loops, respectively. Blue dots represent water-exposed residues. The two sets of anti-parallel cylinders in the center represent the extended α2/α3 helices juxtaposed in an anti-parallel orientation (α2/α3, α2′/α3′) to form the monomer-monomer contact interface. The dotted parallelogram on α2/α3, α2′/α3′, represents the approximate positions of the two BH3 domains from each of the α2-α3 sequences. The red and yellow ovals on the α2/α3 helices represent 2-fold symmetry-related 75R1s and 83R1s. The distance between the two 83R1s is estimated to be 5–10 Å. The large gray oval under the helices represents an unknown domain of BAK in tight contact with α2/α3, α2′/α3′. The large flat box drawn in thin lines represents the membrane. The relative orientations of the two juxtaposed α2/α3 helices are the same as in the salt bridged conformation model for BAX homodimer (Fig. 6B). The water-membrane interface for α2/α3, α2′/α3′ is unknown. B, topological locations of the residues in the α5-α6 helical hairpin structure. Topological locations of residues 134R1, 146R1, 154R1, and 164R1 in the membrane-inserted BAK are indicated in the mouse homology model (solution structure) corresponding to α5–α7. The membrane-immersion depth for residues 146R1, 154R1, and 164R1 are shown in Å. The topological locations of residues 132 and 133 are unknown. Side chains in green and blue represent hydrophobic and hydrophilic residues, respectively. The ribbon diagram was generated in PyMOL (76).
Regarding the formation of the higher order oligomers, based on the chemical cross-linking experiments, Dewson et al. (14) recently reported that the α6 helix in BAK forms an interface between the BAK homodimers. Consistent with this, residue 164R1 located at the kink between the α6 and α7 helices showed a modest spin-dilution effect (Fig. 5B), indicating that 164R1 is near the monomer-monomer interface within the distance of 25 Å from each other. This spin-spin interaction might originate from the two α6 helices in close proximity at the interface between the two neighboring BAK homodimers as proposed by Dewson et al. (14). However, depending on the three-dimensional arrangement of the α6 helices in the homodimer in the oligomer, the spin-spin interactions between the 164R1 side chains could originate from the two BAK monomers constituting the same homodimer. Contrary to 164R1, residue 154R1 despite its location on the α6 helix, did not show any spin dilution effect (Fig. 5), indicating that this residue is not close to the α6-α6 interface proposed by Dewson et al. (14). Similar to residue 164R1, residue 146R1 located in the α5-α6 loop also showed the spin dilution effect (Fig. 5). Despite the lower spin labeling efficiency than that of 164R1 (Fig. 3C), this residue showed a bigger spin dilution effect (Fig. 5B). This indicates that the intra- or inter-dimeric distance(s) between the 146R1 residues are shorter than for the residue 164R1. However, whether 146R1 is near the α6-α6 interface is uncertain. Further studies are needed to resolve these issues. As for the number of BAK proteins in the higher order oligomer or the size of the pores formed in the system used in this study, further studies are needed (supplemental Fig. S7). Martinez-Caballero et al. (3) estimated that there could be as many as nine monomers of BAX in a pore if the helical hairpin structure of the BAX were arranged like staves in a barrel to form a transmembrane aqueous pore.
In conclusion, as summarized in Fig. 7, the EPR data suggest the following: (i) large conformational changes occur in BAK upon formation of oligomers in the membrane; (ii) in the symmetric BAK homodimer model proposed earlier by Dewson et al. (15), the two juxtaposed α2/α3 extended helices (i.e. α2-α3:α2′-α3′ complex) should be in an additional tertiary contact with other domain(s) of BAK, perhaps with the α5 helix as suggested in the TM-only model (13). It is likely that the same would be true for the BAX homodimer structural models by others (12, 16, 17). Further studies are necessary to test these possibilities.
Supplementary Material
Acknowledgments
We thank Prof. Byung-ha Oh (Pohang University of Science and Technology, Pohang City, South Korea) for providing us the pPosKJ-BAK vector; Dr. Ronald Kaplan for the helpful discussions and support; Drs. Jun-Yong Choe and Christina Iancu for their helpful comments, preparing figures, and sharing equipments for protein purification; Dr. David Mueller for sharing equipments for EPR sample preparation; Drs. Carl Correll and Binal Shah for their help in fluorometry; Drs. Kenneth Neet, John Keller, Marc Glucksman, and Carl Correll for their comments on the manuscript; Drs. Wayne Hubbell, Christian Altenbach, Piotr Fajer, Likai Song, and Yeon-Kyun Shin for useful programs for the EPR data analysis and/or helpful discussions; and Sunaina Yadav and Dr. Kalpesh Bhesaniya for their help in this work. We thank the support of the EPR Center at the Rosalind Franklin University of Medicine and Science.
This work was supported in part by the Scientist Development Grant from the American Heart Association (0835026G) and the Start-up fund from the Rosalind Franklin University of Medicine and Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and additional references.
- BH
- Bcl-2 homology
- Ni-NTA
- Ni2+-nitrilotriacetic acid
- sBAK
- soluble BAK
- His
- hexahistidine
- LUV
- large unilamellar vesicle
- MCS
- mitochondrial outer membrane contact site
- MOM
- mitochondrial outer membrane
- DOGS-NTA-Ni
- 1,2-dioleoyl-sn-glycero-3-{[N-5-amino-1-carboxylpentyl)iminodiacetic acid]succinyl} (nickel salt)
- MTSSL
- (1-oxyl-2,2,5,5,-tetramethylpyrroline-3-methyl)methanethiosulfonate spin label
- SDSL
- site-directed spin labeling
- CW
- continuous wave
- Bid
- truncated Bid
- NiEDDA
- nickel(II)-ethylenediaminediacetate
- TM
- transmembrane.
REFERENCES
- 1.Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., Green D. R. (2010) Mol. Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito M., Korsmeyer S. J., Schlesinger P. H. (2000) Nat. Cell Biol. 2, 553–555 [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Caballero S., Dejean L. M., Kinnally M. S., Oh K. J., Mannella C. A., Kinnally K. W. (2009) J. Biol. Chem. 284, 12235–12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang X., Wang X. (2004) Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 5.Korsmeyer S. J., Wei M. C., Saito M., Weiler S., Oh K. J., Schlesinger P. H. (2000) Cell Death Differ. 7, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 6.Kinnally K. W., Antonsson B. (2007) Apoptosis 12, 857–868 [DOI] [PubMed] [Google Scholar]
- 7.Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culmsee C., Krieglstein J. (2007) EMBO Rep. 8, 129–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danial N. N., Korsmeyer S. J. (2004) Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 10.Adams J. M., Cory S. (2007) Curr. Opin. Immunol. 19, 488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kvansakul M., Yang H., Fairlie W. D., Czabotar P. E., Fischer S. F., Perugini M. A., Huang D. C., Colman P. M. (2008) Cell Death Differ. 15, 1564–1571 [DOI] [PubMed] [Google Scholar]
- 12.Bleicken S., Classen M., Padmavathi P. V., Ishikawa T., Zeth K., Steinhoff H. J., Bordignon E. (2010) J. Biol. Chem. 285, 6636–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewson G., Kluck R. M. (2009) J. Cell Sci. 122, 2801–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewson G., Kratina T., Czabotar P., Day C. L., Adams J. M., Kluck R. M. (2009) Mol. Cell 36, 696–703 [DOI] [PubMed] [Google Scholar]
- 15.Dewson G., Kratina T., Sim H. W., Puthalakath H., Adams J. M., Colman P. M., Kluck R. M. (2008) Mol. Cell 30, 369–380 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z., Zhu W., Lapolla S. M., Miao Y., Shao Y., Falcone M., Boreham D., McFarlane N., Ding J., Johnson A. E., Zhang X. C., Andrews D. W., Lin J. (2010) J. Biol. Chem. 285, 17614–17627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2009) Mol. Cell 36, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annis M. G., Soucie E. L., Dlugosz P. J., Cruz-Aguado J. A., Penn L. Z., Leber B., Andrews D. W. (2005) EMBO J. 24, 2096–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbell W. L., Gross A., Langen R., Lietzow M. A. (1998) Curr. Opin. Struct. Biol. 8, 649–656 [DOI] [PubMed] [Google Scholar]
- 20.Hubbell W. L., Cafiso D. S., Altenbach C. (2000) Nat. Struct. Biol. 7, 735–739 [DOI] [PubMed] [Google Scholar]
- 21.Feix J. B., Klug C. S. (1998) in Biological Magnetic Resonance (Berliner L. J. ed) Vol. 14, pp. 251–281, Plenum Publishing Corp., New York [Google Scholar]
- 22.Columbus L., Hubbell W. L. (2002) Trends Biochem. Sci. 27, 288–295 [DOI] [PubMed] [Google Scholar]
- 23.Klug C. S., Feix J. B. (2008) Methods Cell Biol. 84, 617–658 [DOI] [PubMed] [Google Scholar]
- 24.Klare J. P., Steinhoff H. J. (2009) Photosynth. Res. 102, 377–390 [DOI] [PubMed] [Google Scholar]
- 25.Barnes J. P., Liang Z., Mchaourab H. S., Freed J. H., Hubbell W. L. (1999) Biophys. J. 76, 3298–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeSensi S. C., Rangel D. P., Beth A. H., Lybrand T. P., Hustedt E. J. (2008) Biophys. J. 94, 3798–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altenbach C., Marti T., Khorana H. G., Hubbell W. L. (1990) Science 248, 1088–1092 [DOI] [PubMed] [Google Scholar]
- 28.Altenbach C., Froncisz W., Hemker R., McHaourab H., Hubbell W. L. (2005) Biophys. J. 89, 2103–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabenstein M. D., Shin Y. K. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8239–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altenbach C., Oh K. J., Trabanino R. J., Hideg K., Hubbell W. L. (2001) Biochemistry 40, 15471–15482 [DOI] [PubMed] [Google Scholar]
- 31.Jeschke G. (2002) Chemphyschem 3, 927–932 [DOI] [PubMed] [Google Scholar]
- 32.Oh K. J., Barbuto S., Pitter K., Morash J., Walensky L. D., Korsmeyer S. J. (2006) J. Biol. Chem. 281, 36999–37008 [DOI] [PubMed] [Google Scholar]
- 33.Walensky L. D., Pitter K., Morash J., Oh K. J., Barbuto S., Fisher J., Smith E., Verdine G. L., Korsmeyer S. J. (2006) Mol. Cell 24, 199–210 [DOI] [PubMed] [Google Scholar]
- 34.Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 35.Oh K. J., Barbuto S., Meyer N., Kim R. S., Collier R. J., Korsmeyer S. J. (2005) J. Biol. Chem. 280, 753–767 [DOI] [PubMed] [Google Scholar]
- 36.Liu X., Zou H., Slaughter C., Wang X. (1997) Cell 89, 175–184 [DOI] [PubMed] [Google Scholar]
- 37.Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. (1998) Cell 94, 481–490 [DOI] [PubMed] [Google Scholar]
- 38.Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. (2000) Nat. Cell Biol. 2, 754–761 [DOI] [PubMed] [Google Scholar]
- 39.Ardail D., Privat J. P., Egret-Charlier M., Levrat C., Lerme F., Louisot P. (1990) J. Biol. Chem. 265, 18797–18802 [PubMed] [Google Scholar]
- 40.Terrones O., Antonsson B., Yamaguchi H., Wang H. G., Liu J., Lee R. M., Herrmann A., Basañez G. (2004) J. Biol. Chem. 279, 30081–30091 [DOI] [PubMed] [Google Scholar]
- 41.Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. (2005) Mol. Cell 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 42.Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 43.Böttcher C. J., van Gent C. M., Pries C. (1961) Anal. Chim. Acta 24, 203–204 [Google Scholar]
- 44.Hubbell W. L., Froncisz W., Hyde J. S. (1987) Rev. Sci. Instrum. 58, 1879–1886 [Google Scholar]
- 45.Ihaka R., Gentleman R. (1996) J. Comput. Graph. Stat. 5, 229–314 [Google Scholar]
- 46.Hubbell W. L., Altenbach C. (1994) Membrane Protein Structure: Experimental Approaches (Holst I. ed) pp. 244–248, Oxford University Press, London [Google Scholar]
- 47.Farahbakhsh Z. T., Altenbach C., Hubbell W. L. (1992) Photochem. Photobiol. 56, 1019–1033 [DOI] [PubMed] [Google Scholar]
- 48.Oh K. J., Altenbach C., Collier R. J., Hubbell W. L. (2000) Methods Mol. Biol. 145, 147–169 [DOI] [PubMed] [Google Scholar]
- 49.Moldoveanu T., Liu Q., Tocilj A., Watson M., Shore G., Gehring K. (2006) Mol. Cell 24, 677–688 [DOI] [PubMed] [Google Scholar]
- 50.Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Gunsteren W. F., Billeter S. R., Eising A., Hunenberger P. H., Kruger P., Mark A. E., Scott W. R., Tironi I. G. (1996) The GROMOS96 Manual and User Guide, VdF Hochschulverlag ETHZ, Zurich [Google Scholar]
- 52.Sedlak T. W., Oltvai Z. N., Yang E., Wang K., Boise L. H., Thompson C. B., Korsmeyer S. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7834–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., Fesik S. W. (1997) Science 275, 983–986 [DOI] [PubMed] [Google Scholar]
- 54.Minn A. J., Kettlun C. S., Liang H., Kelekar A., Vander Heiden M. G., Chang B. S., Fesik S. W., Fill M., Thompson C. B. (1999) EMBO J. 18, 632–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng E. H., Wei M. C., Weiler S., Flavell R. A., Mak T. W., Lindsten T., Korsmeyer S. J. (2001) Mol. Cell 8, 705–711 [DOI] [PubMed] [Google Scholar]
- 56.Muchmore S. W., Sattler M., Liang H., Meadows R. P., Harlan J. E., Yoon H. S., Nettesheim D., Chang B. S., Thompson C. B., Wong S. L., Ng S. L., Fesik S. W. (1996) Nature 381, 335–341 [DOI] [PubMed] [Google Scholar]
- 57.Antignani A., Youle R. J. (2006) Curr. Opin. Cell Biol. 18, 685–689 [DOI] [PubMed] [Google Scholar]
- 58.Mchaourab H. S., Lietzow M. A., Hideg K., Hubbell W. L. (1996) Biochemistry 35, 7692–7704 [DOI] [PubMed] [Google Scholar]
- 59.Mchaourab H. S., Kálai T., Hideg K., Hubbell W. L. (1999) Biochemistry 38, 2947–2955 [DOI] [PubMed] [Google Scholar]
- 60.Langen R., Oh K. J., Cascio D., Hubbell W. L. (2000) Biochemistry 39, 8396–8405 [DOI] [PubMed] [Google Scholar]
- 61.Altenbach C., Greenhalgh D. A., Khorana H. G., Hubbell W. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1667–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagle J. F., Tristram-Nagle S. (2000) Biochim. Biophys. Acta 1469, 159–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tristram-Nagle S., Nagle J. F. (2004) Chem. Phys. Lipids 127, 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kucerka N., Nagle J. F., Sachs J. N., Feller S. E., Pencer J., Jackson A., Katsaras J. (2008) Biophys. J. 95, 2356–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gross A., Columbus L., Hideg K., Altenbach C., Hubbell W. L. (1999) Biochemistry 38, 10324–10335 [DOI] [PubMed] [Google Scholar]
- 66.Bleicken S., Zeth K. (2009) J. Bioenerg. Biomembr. 41, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh K. J., Zhan H., Cui C., Hideg K., Collier R. J., Hubbell W. L. (1996) Science 273, 810–812 [DOI] [PubMed] [Google Scholar]
- 68.Suzuki M., Youle R. J., Tjandra N. (2000) Cell 103, 645–654 [DOI] [PubMed] [Google Scholar]
- 69.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 1–1042 [Google Scholar]
- 70.Sawada M., Sun W., Hayes P., Leskov K., Boothman D. A., Matsuyama S. (2003) Nat. Cell Biol. 5, 320–329 [DOI] [PubMed] [Google Scholar]
- 71.Cheng E. H., Sheiko T. V., Fisher J. K., Craigen W. J., Korsmeyer S. J. (2003) Science 301, 513–517 [DOI] [PubMed] [Google Scholar]
- 72.Roy S. S., Ehrlich A. M., Craigen W. J., Hajnóczky G. (2009) EMBO Rep. 10, 1341–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. (2008) Cell 135, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 74.Zha J., Weiler S., Oh K. J., Wei M. C., Korsmeyer S. J. (2000) Science 290, 1761–1765 [DOI] [PubMed] [Google Scholar]
- 75.Wei M. C., Lindsten T., Mootha V. K., Weiler S., Gross A., Ashiya M., Thompson C. B., Korsmeyer S. J. (2000) Genes Dev. 14, 2060–2071 [PMC free article] [PubMed] [Google Scholar]
- 76.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.