Abstract
Abscisic acid (ABA) is one of the most important phytohormones in plant. PYL proteins were identified to be ABA receptors in Arabidopsis thaliana. Despite the remarkably high degree of sequence similarity, PYL1 and PYL2 exhibit distinct responses toward pyrabactin, an ABA agonist. PYL1 inhibits protein phosphatase type 2C upon binding of pyrabactin. In contrast, PYL2 appears relatively insensitive to this compound. The crystal structure of pyrabactin-bound PYL1 revealed that most of the PYL1 residues involved in pyrabactin binding are conserved, hence failing to explain the selectivity of pyrabactin for PYL1 over PYL2. To understand the molecular basis of pyrabactin selectivity, we determined the crystal structure of PYL2 in complex with pyrabactin at 1.64 Å resolution. Structural comparison and biochemical analyses demonstrated that one single amino acid alteration between a corresponding valine and isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2. These characterizations provide an important clue to dissecting the redundancy of PYL proteins.
Keywords: Amino Acid, Crystal Structure, Ligand-binding Protein, Plant, Receptor Regulation, ABA Receptor, ABA Signaling, PYL Proteins, Abscisic Acid, Pyrabactin
Introduction
The PYR1/PYL/RCAR family of proteins (hereafter referred to as PYLs for simplicity) in Arabidopsis thaliana contains 14 members that share a remarkably high degree of sequence similarity. Except PYL13, all of the other PYLs are ABA3 receptors (1–3). Recent biochemical and structural investigations revealed the molecular mechanisms by which PYLs perceive ABA (4–9).
An elegant linear ABA signaling pathway mediated by PYLs was successfully recapitulated in vitro (9), where PYLs, upon binding to ABA, inhibit the downstream protein phosphatases type 2C (PP2Cs), such as ABI1, ABI2, HAB2, for example. PP2Cs dephosphorylate a family of protein kinases SnRK2, whose autophosphorylation is required for the phosphorylation and activation of transcription factors that regulate the expression of ABA-responsive genes. Structural and biochemical studies further revealed the molecular basis of ABA-PYL-mediated inhibition of PP2Cs (4–8). Structures of apo- or ligand-bound PYR1, PYL1, and PYL2 were determined (4–8, 10). In all of these structures, PYLs are homodimers, with each protomer comprising a conserved ligand-accommodating pocket surrounded by four conserved loops CL1–CL4 (4). Upon binding of one ABA molecule by each protomer, the switch loop CL2 undergoes pronounced conformational rearrangement, creates a novel binding surface for PP2C (4, 5, 7, 8), and leads to the weakened dimer interface (4). Each PYL protomer can be recognized by PP2C via the newly formed surface upon ABA binding (4–6). Formation of a PYL/PP2C heterodimer blocks the substrate entry to PP2Cs, thus releasing PP2C-mediated inhibition of SnRK2 (4–6).
Despite the rapid progress in understanding PYL-mediated ABA signaling (11, 12), a number of questions remain unanswered. In particular, pyrabactin (13), an ABA agonist, selectively inhibits seed germination, but not seedling (2). In planta analyses revealed redundant roles for PYR1, PYL1, PYL2, and PYL4 in response to ABA (2); but these PYLs displayed distinct responses to pyrabactin. Thus, pyrabactin may represent a more sensitive ligand for functional dissection of PYLs. What is the structural basis for the selectivity for or against pyrabactin? Preliminary analysis revealed that pyrabactin binds to PYL1 and PYL2 with similar affinity. The crystal structure of pyrabactin-bound PYL1 revealed that, despite lack of chemical similarity, pyrabactin and ABA share a similar set of interactions with PYL1. Unfortunately, most of the PYL1 residues involved in pyrabactin binding are conserved (10), failing to explain the selectivity of pyrabactin for PYL1 over other PYLs such as PYL2.
To understand the molecular basis of pyrabactin selectivity, we determined the crystal structure of PYL2 in complex with pyrabactin at 1.64 Å resolution. Structural comparison and biochemical analysis revealed that, unexpectedly, a single amino acid alteration between valine and isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2. This study provides an important clue to understanding the redundancy of PYLs and suggests a complex regulation mechanism of ABA signaling by PYLs.
EXPERIMENTAL PROCEDURES
Protein Preparation and Crystallization
All PYR/PYLs homologs and ABI1 (AT4G26080) were subcloned from the A. thaliana cDNA library using standard PCR-based protocol. All mutants of PYLs were generated with two-step PCR, verified by plasmid sequencing. All proteins were purified according to the protocol described before (4). All of the PYLs were expressed in Escherichia coli strain BL21(DE3) using vector pET-15b induced at 22 °C for 12 h. The individual protein was purified with Ni-nitrilotriacetic acid resin (Qiagen) followed by anion exchange chromatography (Source-15Q; GE Healthcare) and size exclusion chromatography (Superdex-200; GE Healthcare). Before crystallization, a His6 tag was removed from PYL2 by thrombin. The protein was incubated with pyrabactin at a molecular ratio of 1:2. Crystals were grown at 18 °C using the hanging-drop vapor diffusion method. Diamond-shaped crystals appeared overnight in the well buffer containing 1.8 m (NH4)2SO4, 0.1 m Li2SO4, 100 mm BisTris, pH 6.5, 67 mm NDSB-256 (Hampton Research), and grew to full size in 2 days.
Data Collection, Structure Determination, and Refinement
The diffracting data of PYL2/pyrabactin were collected at the Shanghai Synchrotron Radiation Facility beamline BL17U and integrated with MOSFLM (14). Further processing was carried out using programs from the CCP4 suite (15). Data collection statistics are summarized in Table 1. The structure of PYL2/PYB was determined by molecular replacement with the program PHASER (16). The PYL2 model (Protein Data Bank code 3KDH) was translated into the PYL2/pyrabactin cell. Manually model-iterative rebuilding and refinement were performed with COOT (17) and PHENIX (18). The pyrabactin molecules were built into the cavity of the host molecules.
TABLE 1.
Data collection and refinement statistics
One crystal was used for each structure.
| Parameter | PYL2/pyrabactin |
|---|---|
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 62.52, 104.60, 184.30 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 1.28020 |
| Resolution (Å) | 50∼1.64 (1.70∼1.64) |
| Rmerge (%) | 6.8 (47.6) |
| I/σI | 27.16 (3.23) |
| Completeness (%) | 99.8 (99.3) |
| Redundancy | 7.4 |
| Resolution (Å) | 50∼1.64 |
| No. reflections | 72,397 |
| Rwork/Rfree (%) | 21.23/22.28 |
| No. atoms | |
| Protein | 4,405 |
| Ligand/ion | 86 |
| Water | 378 |
| B-factors | |
| Protein | 36.89 |
| Ligand/ion | 60.74 |
| Water | 46.70 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.229 |
| Ramachandran plot statistics (%) | |
| Most favored | 90.8 |
| Additional allowed | 8.8 |
| Generously allowed | 0.2 |
| Disallowed | 0.2 |
Phosphatase Activity Assay
The phosphatase activity was measured by the Ser/Thr phosphatase assay system (Promega). The protocol of phosphatase activity assay was described previously with some modifications (4). For the systematic examination of the effects of pyrabactin on PYLs, each reaction was performed in a 50-μl reaction volume containing 0.4 μm ABI1 and 4 μm PYLs. Pyrabactin was applied at indicated concentrations. For IC50 measurement, each reaction was performed in a 100-μl reaction volume containing 0.4 μm ABI1, 0.4 μm PYL2, and pyrabactin at the indicated concentrations. In each reaction, after incubation with peptide substrate in 50 mm imidazole, pH 7.2, 5 mm MgCl2, 0.2 mm EGTA, and 0.1 mg/ml BSA at 30 °C for 20 min, the reaction was stopped by the addition of 50 μl of molybdate dye and incubated for another 15 min at room temperature. Absorbance at 630 nm was measured. All data are the means ± S.D. from three independent experiments.
RESULTS
Recognition of Pyrabactin by PYL2
PYL2 appeared to be insensitive to pyrabactin (2). Nonetheless, PYL2 exhibited a weak inhibition of PP2C in response to pyrabactin in the in vitro biochemical assay. We tried to measure the IC50 of pyrabactin on PYL2-mediated inhibition of ABI1. The concentrations of PYL2 and ABI1 were both set at 0.4 μm in the experiments. However, ABI1 retained up to 70% of the phosphatase activity even in the presence of 100 μm pyrabactin (Fig. 1A). By contrast, PYL1 can effectively inhibit the phosphatase activity of ABI1 upon binding to pyrabactin (10). Interestingly, pyrabactin displayed a similar or even higher binding affinity to PYL2 than to PYL1 as observed in the preliminary surface plasmon resonance analysis. We sought to determine the structure of pyrabactin-bound PYL2 to understand the molecular mechanism underlying the pyrabactin selectivity for PYL1 over PYL2.
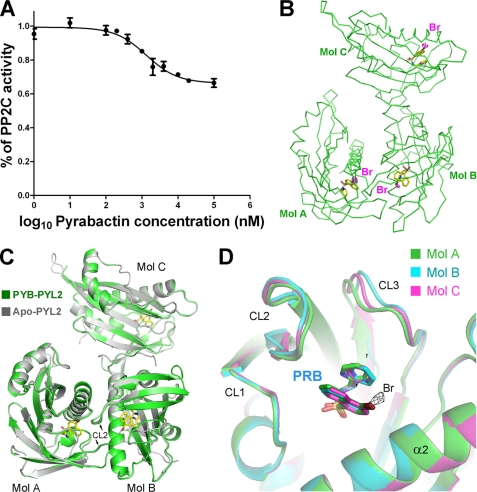
FIGURE 1.
Structure of pyrabactin-bound PYL2. A, PYL2 inhibits ABI1 in response to pyrabactin. The concentration of ABI1 was 0.4 μm for all of the assays throughout the paper. The details of the experiments are described under “Experimental Procedures.” B, there are three pyrabactin-bound PYL2 molecules in each asymmetric unit in the crystal structure. The positions of the pyrabactin molecules are confirmed by the anomalous signal of bromide, which, shown as a magenta dash, is contoured at 3σ. C, overall structure of pyrabactin-bound PYL2 is identical to that of apo-PYL2, with an open conformation despite pyrabactin binding. The structure of three pyrabactin-bound PYL2 molecules (named Mol A, Mol B, and Mol C, respectively) from each asymmetric unit is superimposed on that of apo-PYL2. Pyrabactin molecules are shown as yellow sticks. D, Mol A (green), Mol B (cyan), and Mol C (magenta) from each asymmetric unit of PYL2 are superimposed. The anomalous signal of bromide (black mesh) in Mol A is contoured at 3σ. All structure figures were prepared with PyMOL (19).
We crystallized PYL2 in the presence of pyrabactin and determined its structure at 1.64 Å resolution (Table 1). There are three PYL2 molecules in each asymmetric unit (Fig. 1B). The assignment of pyrabactin was confirmed by the anomalous signal of bromide. There is one pyrabactin molecule bound within the conserved pocket of each PYL2 protomer (Fig. 1B). The pyrabactin-bound PYL2 adopts almost the same conformation as that of apo-PYL2 (Fig. 1C), including the switch loop CL2. The three protein molecules within each asymmetric subunit of pyrabactin-bound PYL2 can be superimposed on that of the three corresponding apo-PYL2 with a root mean square deviation of 0.48 Å over a total of 529 Cα atoms. This observation suggests that the PYL2 dimer is likely to adopt an “open” conformation even in the presence of pyrabactin. The three PYL2 molecules in each asymmetric unit adopt similar conformation (Fig. 1D).
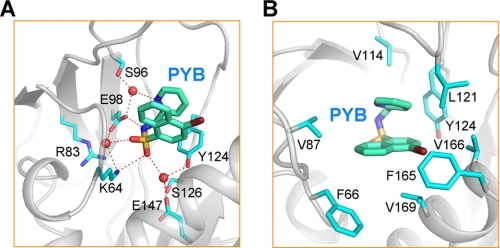
Similar to previous examples of ligand-PYL interactions (10), coordination of the “U”-shaped pyrabactin by PYL2 is mediated by both polar and van der Waals interactions (Fig. 2). Four conserved, charged residues, Lys64, Arg83, Glu98, and Glu147 anchor the polar module of pyrabactin through direct or water-mediated H-bonds (Fig. 2A). In addition to Lys64, which coordinates pyrabactin through both direct and water-mediated H-bonds with the sulfone group, Glu98 also appears to play an important role, interacting directly with the amine group and coordinating the sulfone and the pyridyl nitrogen through water-mediated H-bonds. Three other polar residues, Ser96, Tyr124, and Ser126, also contribute to pyrabactin binding through water-mediated H-bonds (Fig. 2A). The hydrophobic arms of the U-shaped pyrabactin are surrounded by eight hydrophobic residues, Phe66 from CL1, Val87 from CL2, Val114 from CL3, Leu121/Tyr124 from CL4, and Phe165/Val166/Val169 from helix α2 (Fig. 2B). Notably, CL2, which only contributes one hydrophobic residue, Val87, for van der Waals contact with pyrabactin, appears to play a less important role in the recognition of pyrabactin in PYL2 compared with that in PYL1, likely due to its open conformation. This observation again confirms the suggestion that ligand binding precedes closure of CL2 (10).
FIGURE 2.
Recognition of pyrabactin by PYL2. A, coordination of pyrabactin by PYL2 through polar interactions. B, coordination of pyrabactin by PYL2 through van der Waals contacts.
Molecular Basis of the Pyrabactin Selectivity
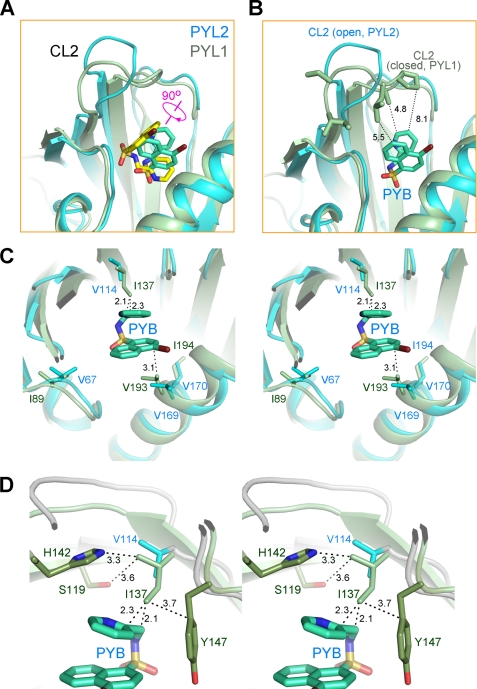
To understand the pyrabactin selectivity, we superimposed the structures of pyrabactin-bound PYL1 (10) and PYL2. Interestingly, the pyrabactin is rotated for 90 degrees in PYL2 relative to that in PYL1 (Fig. 3A). Consequently, an important hydrophobic module, the naphthalene ring of pyrabactin that attracts CL2 in PYL1, is now perpendicular to CL2 in PYL2. As analyzed previously (10), a bulky, hydrophobic portion of the ligand, either ABA or pyrabactin, should be placed approximately in parallel with the geometry plate of CL2 to stabilize a closed conformation. The orientation of pyrabactin in PYL2 may not be able to provide sufficient contacts to stabilize a closed conformation of CL2 (Fig. 3B). Consequently, CL2 remains open even in the presence of pyrabactin, which explains the poor response of PYL2 to pyrabactin.
FIGURE 3.
Pyrabactin adopts distinct orientations in PYL1 and PYL2. A, pyrabactin recognition by PYL1 and PYL2 is compared. The structures of pyrabactin-bound PYL1 and PYL2 protomers are superimposed. PYL1 protein is shown in pale green, with the bound pyrabactin shown as yellow sticks. PYL2 is shown in cyan, with the bound pyrabactin shown as cyan sticks. Note that the bound pyrabactin is rotated for ∼90 degrees in PYL2 relative to that in PYL1. B, orientation of pyrabactin in PYL2 cannot stabilize a closed conformation of CL2. Structures of pyrabactin-bound PYL1 (pale green) and PYL2 (cyan) are superimposed as in A, and the pyrabactin in PYL2 is shown as cyan sticks. CL2 is unlikely to form enough contacts with pyrabactin in PYL2 even if it is closed as in PYL1. C, in PYL1, the side chain of Ile137 is too close to pyrabactin if the ligand were localized as in PYL2. Structures of pyrabactin-bound PYL1 and PYL2 are superimposed as in A. The pyrabactin in PYL2 is shown as cyan sticks. D, surrounding residues may prevent Ile137 from adopting other rotamers. Rotation of the side chain of Ile137 for 90, 180, or 270 degrees may cause potential clash with the side chains of Ser119, His142, and Tyr147.
To understand why pyrabactin adopts two distinct orientations in the highly conserved pocket of PYL1 and PYL2, we took a pairwise comparison of the residues involved in pyrabactin coordination in both PYLs. Most of the residues surrounding pyrabactin are invariants in PYL1 and PYL2 except for three valines, Val67, Val114, and Val170 in PYL2, all of which are replaced with Ile in PYL1 (Ile89, Ile137, and Ile194, respectively). The substitution of Val67 (Ile89) and Val170 (Ile194) should have no effect on pyrabactin coordination because they are not in direct contact with pyrabactin (Fig. 3C). However, if Val114 were to be replaced by Ile in PYL2, the distance between the side chain of this residue and the naphthalene ring of pyrabactin would be too close to allow the observed orientation of pyrabactin (Fig. 3C). Notably, this potential steric clash would have been avoided if the side chain of Ile137 were rotated for 90, 180, or 270 degrees; nonetheless, the surrounding residues, Ser119, His142, and Tyr147, which are in close proximity to Ile137, may prevent Ile137 from adopting other rotamers (Fig. 3D).
Single Amino Acid Alteration Determines Pyrabactin Selectivity
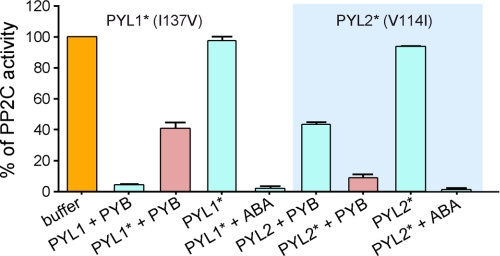
To validate the structural analysis, we generated swapped mutants of PYL1 and PYL2, with Ile137 replaced by Val in PYL1, and Val114 of PYL2 replaced by Ile. Then we examined their reception of pyrabactin. These two mutants by themselves cannot inhibit the phosphatase activity of ABI1, but they exhibit complete inhibition upon addition of ABA, indicating that both are functional proteins (Fig. 4). When pyrabactin was applied, PYL1 (I137V) could no longer completely inhibit ABI1 as wild type (WT) PYL1 did. ABI1 retained up to 40% of phosphatase activity in the presence of excess PYL1 (I137V) and pyrabactin, an observation reminiscent of WT PYL2. By contrast, PYL2 (V114I) is able to inhibit 90% of the phosphatase activity of ABI1 in the presence of pyrabactin, a result reminiscent of WT PYL1. This observation indicates that single-residue alteration between isoleucine and valine does account for the distinct pyrabactin selectivity by PYL1 over PYL2.
FIGURE 4.
Single amino acid alteration between valine and isoleucine determines pyrabactin selectivity. Swap of the isoleucine and valine residues switched pyrabactin selectivity of PYL1 and PYL2. PYL1* contains the single missense mutation I137V, and PYL2* contains V114I.
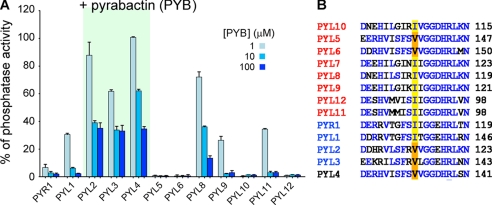
To see whether this conclusion can be generalized to other PYL proteins, we performed systematic biochemical characterizations for all the ABA-responsive PYLs, except PYL7, which defied recombinant expression. We expressed and purified all of the other 12 PYL proteins from A. thaliana and examined their dependence on pyrabactin. ABI1 and PYLs were used at 0.4 μm and 4 μm in each assay. Three concentrations of pyrabactin, 1, 10, and 100 μm, were applied. Although PYR1, PYL1, and PYL5, 6, 9–12 almost completely inhibited ABI1 in the presence of 10 μm pyrabactin, PYL2–4 only inhibited 60% of the phosphatase activity of ABI1 with the addition of 100 μm pyrabactin. PYL8 was able to inhibit >80% of the phosphatase activity of ABI1 in the presence of 100 μm pyrabactin, and it responded to pyrabactin in a concentration-dependent manner. Therefore, PYL2–4 are the only pyrabactin-insensitive PYLs (Fig. 5A).
FIGURE 5.
Systematic examination of the inhibitory effect on ABI1 by PYLs in response to pyrabactin revealed a common determinant of pyrabactin selectivity. A, inhibition of ABI1 by 12 PYLs in response to pyrabactin. The concentration of all PYLs was set at 4 μm, and three concentrations of pyrabactin (1, 10, and 100 μm) were tested for each PYL. The details of the experiments are described under “Experimental Procedures.” B, sequence alignment reveals that the single-residue alteration between valine and isoleucine underlies the distinct pyrabactin selectivity by PYLs. PYL5 and PYL6, which contain valine at the corresponding position, are sensitive to pyrabactin due to a distinct mechanism which will be discussed in a separate manuscript.
With this result, we reexamined the sequence alignment of PYLs. Indeed, PYL3 and PYL4 both contain a Val, whereas all other PYLs, except PYL5 and PYL6, have Ile at this key position (Fig. 5B). As will be seen in a separate manuscript, PYL5 and PYL6 represent a unique family of PYLs, whose selectivity of pyrabactin was accounted for by a distinct mechanism.
DISCUSSION
In this study, we attempted to understand the molecular basis for the pyrabactin selectivity by different PYLs. The structure of pyrabactin-bound PYL2 showed that pyrabactin is still accommodated into the ligand-binding pocket of PYL2. However, the orientation of pyrabactin in PYL2 is rotated by 90 degrees compared with that in PYL1. Such an accommodation of pyrabactin fails to pull over the CL2 due to the lack of hydrophobic module of the ligand in proximity to CL2 (10).
The determinant of pyrabactin selectivity turned out to be a surprise. It would be almost impossible to predict that a single residue variation between Val and Ile defines the pyrabactin selectivity of PYL1, PYL2, and other PYLs. Val and Ile differ from each other by only one methyl group. If it were not by structural comparison, it would be almost impossible to realize that this subtle alteration between two hydrophobic residues completely changed the ligand selectivity of a protein. This discovery showcased the beauty and power of structural biology, and called for caution in the sequence-based functional prediction.
The experimental evidence presented here suggested that the function of PYLs may be distinguished from each other even with subtle alterations in the primary sequence. Although PYL proteins are highly conservative based on sequence alignment and structural comparison, subtle variations can play a dominant role in determining their distinct functions. Therefore, the existence of 13 similar but distinct PYLs may provide a very complex regulation system for ABA signaling. Despite the rapid progress in the elucidation of PYL-mediated ABA signaling, as the domain name of PYLs suggested, it may be just a START (star-related lipid transfer) to understand PYLs. In particular, the physiological roles of these PYLs remain to be investigated. All 13 PYLs are capable of ABA binding and subsequent inhibition of downstream PP2Cs. However, are there any variations in their downstream signaling? Are they involved in different processes? To dissect the redundancy of PYLs further, selective agonists of ABA, such as pyrabactin, may provide a powerful tool. On the other hand, selective ABA antagonists may also be developed. These compounds may selectively activate or inhibit a few or even only one PYL protein. Therefore, such small molecule probes may help characterize the specific physiological role of a specific PYL protein in vivo. On the other hand, the observation that a single missense mutation of PYL1 and PYL2 resulted in altered ligand sensitivity provided a tantalizing clue to potential protein engineering which may lead to enhanced stress tolerance for plants.
Acknowledgments
We thank J. He and S. Huang at Shanghai Synchrotron Radiation Facility.
This work was supported by Ministry of Science and Technology Grant 2009CB918802, Tsinghua University 985 Phase II funds, Yuyuan Foundation, and the Li Foundation.
The atomic coordinates and structure factors (code 3NR4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ABA
- abscisic acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PP2C
- protein phosphatase type 2C.
REFERENCES
- 1.Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009) Science 324, 1064–1068 [DOI] [PubMed] [Google Scholar]
- 2.Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T. F., Alfred S. E., Bonetta D., Finkelstein R., Provart N. J., Desveaux D., Rodriguez P. L., McCourt P., Zhu J. K., Schroeder J. I., Volkman B. F., Cutler S. R. (2009) Science 324, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., Park S. Y., Márquez J. A., Cutler S. R., Rodriguez P. L. (2009) Plant J. 60, 575–588 [DOI] [PubMed] [Google Scholar]
- 4.Yin P., Fan H., Hao Q., Yuan X., Wu D., Pang Y., Yan C., Li W., Wang J., Yan N. (2009) Nat. Struct. Mol. Biol. 16, 1230–1236 [DOI] [PubMed] [Google Scholar]
- 5.Melcher K., Ng L. M., Zhou X. E., Soon F. F., Xu Y., Suino-Powell K. M., Park S. Y., Weiner J. J., Fujii H., Chinnusamy V., Kovach A., Li J., Wang Y., Li J., Peterson F. C., Jensen D. R., Yong E. L., Volkman B. F., Cutler S. R., Zhu J. K., Xu H. E. (2009) Nature 462, 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyazono K., Miyakawa T., Sawano Y., Kubota K., Kang H. J., Asano A., Miyauchi Y., Takahashi M., Zhi Y., Fujita Y., Yoshida T., Kodaira K. S., Yamaguchi-Shinozaki K., Tanokura M. (2009) Nature 462, 609–614 [DOI] [PubMed] [Google Scholar]
- 7.Nishimura N., Hitomi K., Arvai A. S., Rambo R. P., Hitomi C., Cutler S. R., Schroeder J. I., Getzoff E. D. (2009) Science 326, 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santiago J., Dupeux F., Round A., Antoni R., Park S. Y., Jamin M., Cutler S. R., Rodriguez P. L., Márquez J. A. (2009) Nature 462, 665–668 [DOI] [PubMed] [Google Scholar]
- 9.Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S. Y., Cutler S. R., Sheen J., Rodriguez P. L., Zhu J. K. (2009) Nature 462, 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao Q., Yin P., Yan C., Yuan X., Li W., Zhang Z., Liu L., Wang J., Yan N. (2010) J. Biol. Chem. 285, 28946–28952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klingler J. P., Batelli G., Zhu J. K. (2010) J. Exp. Bot. 61, 3199–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010) Annu. Rev. Plant Biol. 61, 26.21–26.29 [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y., Chow T. F., Puckrin R. S., Alfred S. E., Korir A. K., Larive C. K., Cutler S. R. (2007) Nat. Chem. Biol. 3, 716–721 [DOI] [PubMed] [Google Scholar]
- 14.Leslie A. G. (2006) Acta Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 15.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 16.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emsley P., Cowtan K. (2004) Acta Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 19.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos CA [Google Scholar]