Abstract
Cell adhesion molecules and neurotrophin receptors are crucial for the development and the function of the nervous system. Among downstream effectors of neurotrophin receptors and recognition molecules are ion channels. Here, we provide evidence that G protein-coupled inwardly rectifying K+ channel Kir3.3 directly binds to the neural cell adhesion molecule (NCAM) and neurotrophin receptor TrkB. We identified the binding sites for NCAM and TrkB at the C-terminal intracellular domain of Kir3.3. The interaction between NCAM, TrkB, and Kir3.3 was supported by immunocytochemical co-localization of Kir3.3, NCAM, and/or TrkB at the surface of hippocampal neurons. Co-expression of TrkB and Kir3.1/3.3 in Xenopus oocytes increased the K+ currents evoked by Kir3.1/3.3 channels. This current enhancement was reduced by the concomitant co-expression with NCAM. Both surface fluorescence measurements of microinjected oocytes and cell surface biotinylation of transfected CHO cells indicated that the cell membrane localization of Kir3.3 is regulated by TrkB and NCAM. Furthermore, the level of Kir3.3, but not of Kir3.2, at the plasma membranes was reduced in TrkB-deficient mice, supporting the notion that TrkB regulates the cell surface expression of Kir3.3. The premature expression of developmentally late appearing Kir3.1/3.3 in hippocampal neurons led to a reduction of NCAM-induced neurite outgrowth. Our observations indicate a decisive role for the neuronal K+ channel in regulating NCAM-dependent neurite outgrowth and attribute a physiologically meaningful role to the functional interplay of Kir3.3, NCAM, and TrkB in ontogeny.
Keywords: Adhesion, Cell Adhesion, Potassium Channels, Receptor Tyrosine Kinase, Trafficking, Girk3, Kir3.3, TrkB, Neural Cell Adhesion Molecule/NCAM, Neurite Outgrowth
Introduction
The neural cell adhesion molecule (NCAM),3 which is expressed as early as the developmental stage of neural tube closure (1), has been implicated in morphogenetic processes during nervous system development such as cell proliferation, migration, differentiation and survival, neuritogenesis, and synaptogenesis (2–6). NCAM is involved in neural cell interactions, including synaptic contact formation and modulation of synaptic activity (2–6).
Neurotrophic factors and their receptors regulate neuronal survival and neuritogenesis not only during development but also in the adult stage during synaptic plasticity (7). Similar to NCAM, the neurotrophin receptor TrkB and its ligand, brain-derived neurotrophic factor (BDNF), regulate neuronal cell survival, neurite outgrowth, synaptogenesis, and synaptic activity (8–14).
Interestingly, both NCAM and TrkB converge functionally to regulate ionic homeostasis via inwardly rectifying K+ channels of the Kir3 subfamily. Recombinantly expressed NCAM180 specifically reduces inward currents of neuron-specific Kir3.1/3.2 and Kir3.1/3.3 but not Kir3.1/3.4 channels in Xenopus oocytes and CHO cells (15). Activated TrkB determines the open state of Kir3.1/3.4 but not of Kir3.1/3.2 channels by phosphorylation; BDNF stimulation of TrkB results in tyrosine phosphorylation of the Kir3.4 and Kir3.1 subunits, but not the Kir3.2 subunit, and leads to an inhibition of the basal activity of these channels (16, 17). Thus, both NCAM and TrkB may target a common mechanism of membrane potential regulation that determines the excitability of neurons.
Here we show that NCAM, TrkB, and Kir3.3 interact directly with each other via their intracellular domains. Overexpression of the developmentally late appearing Kir3.3 subunit leads to a decrease in NCAM-mediated neurite outgrowth. Kir3.3 channel expression at the cell surface; thus activity is regulated by NCAM and TrkB independently of BDNF ligand binding. These observations indicate that the interplay of recognition molecules, neurotrophin receptors, and ion channels regulate neurite outgrowth.
EXPERIMENTAL PROCEDURES
Mice
Breeding and maintenance of mice are described in the accompanying article (37). TrkB-deficient mice were described elsewhere (18).
Antibodies
NCAM antibodies P61 and 5B8 reacting with the intracellular domain of NCAM, NCAM180-specific antibody D3, NCAM antibodies H28 and 1B2 against the extracellular domain of NCAM, panTrk antibody, TrkB-specific antibody H181, anti-penta His antibody, and secondary antibodies used in this study are described in the accompanying article (37). Kir3.3, Kir3.2, calreticulin, and rab11 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA).
DNA Constructs
Rat Kir3.1, human Kir3.2, rat Kir3.3, and human Kir3.4, the two NCAM isoforms, and full-length TrkB were subcloned into the pCDNA3 or pSGEM vector, which provides the 5′ and 3′ untranslated regions of the Xenopus β-globin gene for expression in Xenopus oocytes as described previously (15). The assembly of concatameric Kir3.1/3.2, Kir3.1/3.3, and Kir3.1/3.4 has also been described (19).
Production and Purification of Recombinant Intracellular Domains
Cloning of NCAM140, NCAM180, and the close homolog of L1 (CHL1) into the mammalian expression vector pcDNA3 is described in the accompanying article (37). Sequences encoding Kir3.3 intracellular domain (Kir3.3-ID, bp 469–1182) and Kir3.2 intracellular domain (Kir3.2-ID, bp 571–1278) were amplified from a mouse brain cDNA library and cloned into pQE30 plasmid. Protein expression and purification are described in the accompanying article (37). Native TrkB-ID prepared in a baculovirus expression system (20) was a kind gift of Shinichi Koizumi and Motohiko Kometani (Novartis Pharma K.K., Tsukuba Research Institute, Ibaraki, Japan).
Immunoprecipitation, ELISA, and Western Blot Analysis
These methods are described in the accompanying article (37). Detergent extracts of hippocampus or brain from 2–3-month-old C57BL/6 mice were centrifuged for 1 h at 100,000 × g and 4 °C before immunoprecipitation was performed.
Pulldown Assay
NCAM180-ID or CHL1-ID resuspended in 0.5 ml of PBS was incubated with brain detergent extracts from 2–3-month-old C57BL/6J mice for 30 min at room temperature. Nickel-nitrilotriacetic acid beads were added and incubated for 1 h at 4 °C. After washing the beads with PBS, bound protein was eluted from the beads by boiling them in SDS sample buffer (60 mm Tris/HCl, pH 6.8, 2% SDS, 1% β-mercaptoethanol, and 10% glycerol, 0.02% bromphenol blue).
Cell Surface Biotinylation, Transfection, and Microinjection of CHO Cells
CHO cells were grown in Glasgow modified Eagle's medium (GMEM) supplemented with 10% fetal calf serum (Invitrogen) in 6-well plates (BD Biosciences). After 4 h, the medium was replaced, and cultures were maintained at 37 °C in a humidified incubator gassed with 5% CO2. Cells were transiently transfected at 90% confluence with expression plasmids coding for concatameric Kir3.1/3.3, TrkB, and/or NCAM140 using a Lipofectamine Plus kit (Invitrogen) according to the manufacturer's protocol. For microinjections, CHO cells were grown on round 12-mm glass coverslips in GMEM. After 1–2 days, Kir3.1/3 expression plasmid alone (50 ng/μl) or in combination with either NCAM140 or TrkB expression plasmid (each at 50 ng/μl) were microinjected using a Transjector 5246 (Eppendorf, Hamburg, Germany). EGFP expression plasmid (20 ng/μl) was co-injected and served as a positive selection marker.
Neurite Outgrowth Measurements of Hippocampal Neurons and Immunocytochemistry
Preparation and transfection of hippocampal neurons and determination of neurite lengths were performed as described in the accompanying article (37).
Preparation of Subcellular Fractions
Mouse brains were homogenized in HOMO buffer (5 mm Tris-HCl, 0.32 m sucrose, 1 mm MgCl2, 1 mm CaCl2, 1 mm NaHCO3 plus CompleteTM EDTA-free protease inhibitor mixture (Roche Diagnostics), pH 7.5) and centrifuged at 1,000 × g for 10 min at 4 °C. The resulting supernatant was centrifuged at 17,000 × g for 20 min at 4 °C. For the isolation of plasma membranes, the 17,000 × g pellet was subjected to a hypotonic shock by adding 9 volumes ice-cold H2O plus CompleteTM EDTA-free protease inhibitor mixture and rapidly adjusted to 5 mm Tris-HCl by adding 1 m Tris-HCl (pH 7.5). After centrifugation at 25,000 × g for 20 min at 4 °C, the pellet fraction containing lysed membranes was resuspended in HOMO buffer and loaded onto a discontinuous sucrose gradient consisting of 0.8, 1.0, and 1.2 m sucrose. After centrifugation at 150,000 × g for 2 h at 4 °C, the interphase material between 1.0 and 1.2 m sucrose, which contained plasma membranes, was collected by centrifugation. Endosomes and endoplasmic reticulum were collected from the 17,000 × g supernatant by centrifugation at 100,000 × g for 1 h at 4 °C. The pellet was resuspended in a 2 m sucrose gradient solution, and a sucrose gradient containing 0.25, 0.8, 1.15, and 1.3 m sucrose was loaded on top of the samples. After centrifugation at 100,000 × g for 2 h at 4 °C, the interphase fraction between 0.8 and 1.15 m sucrose and between 1.3 and 2.0 m sucrose, which contained endosomes and endoplasmic reticulum, respectively, were collected, and proteins were precipitated by addition of methanol-chloroform.
Oocyte Electrophysiology and Cell Surface Expression
For recombinant protein expression in Xenopus oocytes, cDNAs of full-length NCAM, TrkA, TrkB, and Kir3 isoforms were subcloned into the polyadenylation vector pSGEM. N-terminal fusion of EGFP to rat Kir3.3 was generated by insertion of an EGFP-coding PCR product without a stop codon into the 5′ NotI site of Kir3.3 cDNA. The resulting fusion construct (EGFP-Kir3.3) separates the two entire ORFs by three in-frame alanine codons. Capped run-off poly(A)+ cRNA transcripts were synthesized from linearized cDNA and subsequently injected into defolliculated oocytes. Oocytes were incubated at 20 °C in ND96 solution (96 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 5 mm HEPES, pH 7.4) supplemented with 100 μg/ml gentamicin and 2.5 mm sodium pyruvate, and 48 h after injection either two electrode voltage clamp recordings or fluorescence measurements were performed. Currents were recorded with a Turbo Tec-10 amplifier (npi, Tamm, Germany) and sampled through an EPC9 (Heka Electronics, Lambrecht, Germany) interface using Pulse/Pulsefit software (Heka Electronics). For rapid exchange of external solution, oocytes were placed in a small perfusion chamber with a constant flow of ND96 or “96 K+” solution (96 mm KCl, 2 mm NaCl, 1 mm MgCl2, 1 mm CaCl2, and 5 mm HEPES, pH 7.4). Cell surface fluorescence in oocytes was measured with a Nikon C1 laser-scanning microscope equipped with a 10× water immersion objective. For quantification of fluorescence intensity, confocal images were taken under constant parameters from 10 different oocytes for each experiment. Current amplitudes and fluorescence intensity in arbitrary units are given as mean values ± standard deviations. Statistical analysis (Student's t test) was performed with Statview software.
Single Channel Recording and Analysis
For single channel recordings, CHO cells expressing Kir3.1/3 alone or in combination with either NCAM140 or TrkB were used 1–3 days after microinjection. The cells were bathed in a “135 K+” solution containing 135 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 5 mm HEPES and 20 mm sucrose with pH adjusted to 7.4 with KOH. Patch pipettes were pulled from thick-walled borosilicate glass and filled also with 135 K+ solution. Pipette-to-bath resistances were between 3.5 and 4 megohms. All single channel recordings were done in the cell-attached configuration with voltages applied to the patch membrane between +60 and −120 mV. Currents were recorded with an EPC9 patch clamp amplifier controlled by Pulse software (Heka Electronics). Signals were filtered at 3 kHz with 4-pole Bessel characteristics and acquired at a sampling interval of 40 μs. Only patches with no channel activity at +60 mV over 10 s were investigated further. Basal channel activity (no exogenous Gβγ or neurotrophin applied) at different negative voltages was monitored over periods of 10 to 260 s. Data analysis was performed with Fitmaster (Heka Electronics) and Kaleidagraph (Synergy Software, Reading, PA). Mean single channel current amplitudes (I) at different negative voltages (V) were obtained from all-points histograms, and the single channel conductance (γ) was determined for each patch by the slope of a linear fit to the I-V relationship. Single channel gating kinetics were studied at −60 mV in patches containing no more than 1 or 2 channels. Dwell time histograms were fitted with a single exponential function to obtain a mean open time (τopen) for each patch. The apparent open probability (Po) was obtained by dividing the cumulative open times by the entire recording time for each patch.
Statistical Analysis
All numerical data are presented as group mean values ± S.D. Parametric tests (t test or analysis of variance with subsequent Tukey's post hoc test) were used to compare group mean values as appropriate. Data conformed to the requirement for normal distribution (“normality” test, SigmaStat 2.0, SPSS, Chicago, IL). The threshold significance value for acceptance of differences between group mean values was 5%.
RESULTS
Identification of an Interaction between the Intracellular Domains of NCAM and Kir3.3
Because NCAM modulates the currents generated by the inwardly rectifying K+ channel subunits Kir3.3 and Kir3.2 (15), we investigated by ELISA whether NCAM-ID directly interacts with Kir3.3 or Kir3.2. NCAM140-ID (Fig. 1A) and NCAM180-ID (Fig. 1B) showed binding to substrate-coated Kir3.3-ID, whereas neither NCAM140-ID nor NCAM180-ID bound to Kir3.2-ID (Fig. 1, A and B). Because NCAM-ID binds to Kir3.3-ID but not Kir3.2-ID, and because Kir3.3-ID and Kir3.2-ID show significant sequence differences mainly over a stretch of ∼50 amino acids at the C terminus, overlapping peptides spanning the C-terminal stretch of Kir3.3 that is different from Kir3.2 were used in an ELISA to identify the binding site for NCAM-ID on Kir3.3-ID (Fig. 1C). Peptide A, comprising amino acids 344–364 of Kir3.3, bound to NCAM140-ID (Fig. 1D) and NCAM180-ID (Fig. 1E) but not to the more C-terminal peptides B and C (Fig. 1, D and E). To identify the site on NCAM for the binding of Kir3.3 a competition ELISA was performed using NCAM peptides matching the consecutive sequences of NCAM140-ID and NCAM180-ID (Fig. 2A). Peptide 1, comprising the membrane-proximal sequences of NCAM140-ID and NCAM180-ID but not the two other peptides deduced from the more distal intracellular sequences, reduced binding of NCAM180-ID to substrate-coated Kir3.3-ID (Fig. 2A). These observations indicate that NCAM140-ID and NCAM180-ID bind directly to the intracellular domain of the Kir3.3 but not to the Kir3.2 subunit of the inwardly rectifying K+ channel. This interaction is mediated by amino acids 730–764 of NCAM and amino acids 344–364 of Kir3.3.
FIGURE 1.
Identification of the binding site on Kir3.3-ID for interaction with NCAM-ID. A, B, D, and E, in ELISA experiments substrate-coated Kir3.3-ID, Kir3.2-ID (A and B), and Kir3.3 peptides A, B, and C (C, D, and E) were incubated with NCAM140-ID (A and D) and NCAM180-ID (B and E). C, sequences of Kir3.3-ID and Kir3.2-ID and the peptides A, B, and C used in ELISA experiments are shown. Dark gray bars feature the same amino acids and light gray bars similar amino acids. OH designates the C terminus of each protein, and numbers designate amino acid positions. A, B, D, and E, binding of the NCAM-IDs was detected by using the antibody P61. All experiments were performed at least three times. Error bars represent S.D.
FIGURE 2.
Identification of the binding site on NCAM-ID for interaction with Kir3.3-ID. A, for competition ELISA, substrate-coated Kir3.3-ID was preincubated with increasing amounts of NCAM140-ID sequence-derived peptides 1, 2, and 3 (upper panel) and then incubated with a constant amount of NCAM180-ID. Binding of the NCAM180-ID was detected by using the antibody P61. The experiment was performed at least three times. Error bars represent S.D. B, rabbit polyclonal NCAM antibody 1B2 or a nonspecific rabbit IgG (rIg) was used for immunoprecipitation (IP) from a detergent extract of hippocampal tissue. Western blot analysis (WB) of the immunoprecipitates was performed using the goat polyclonal Kir3.3 antibody or the monoclonal NCAM antibody 5B6 recognizing NCAM180 and NCAM140. The input lane and the lanes with the immunoprecipitates were derived from different exposure times of the same blot; the lanes were not adjacent to each other. The experiment was performed twice with identical results. C, NCAM180-ID (180-ID) or CHL1-ID (CHL-ID) was incubated with brain homogenate, and the bound proteins were isolated and subjected to Western blot analysis using a Kir3.3 antibody. The experiment was performed twice with identical results.
To obtain indications for an in vivo interaction of NCAM and Kir3.3, we performed co-immunoprecipitation and pulldown experiments. Co-immunoprecipitation of Kir3.3 from a detergent lysate of adult mouse hippocampus was seen with an antibody against NCAM but not with a non-immune IgG (Fig. 2B). The NCAM antibody, but not the non-immune IgG control, immunoprecipitated both NCAM140 and NCAM180 (Fig. 2B). In a pulldown assay using intracellular domains of NCAM180 and the close homolog of L1 (CHL1-ID), Kir3.3 was precipitated from brain detergent extracts with NCAM180-ID but not with CHL1-ID (Fig. 2C). The results confirm the interaction between NCAM and Kir3.3.
Effect of Kir3.3 Expression on Neurite Outgrowth of Cultured Hippocampal Neurons
In previous studies it was shown that substrate-coated NCAM enhances neurite outgrowth of dissociated early postnatal hippocampal neurons in comparison with substrate-coated poly-l-lysine (21). Therefore, we next investigated whether NCAM-dependent neurite outgrowth is affected by the interaction between the intracellular domains of NCAM and Kir3.3. Because Kir3.3 is not expressed in early postnatal neurons (22), we transfected hippocampal neurons with concatameric Kir3.1/3.3 or, as a negative control, Kir3.1/3.2 and measured neurite outgrowth on NCAM versus poly-l-lysine as a negative control or laminin as a positive control. When hippocampal neurons were plated on poly-l-lysine, expression of Kir3.1/3.2 or Kir3.1/3.3 did not change the mean length of neurites when compared with mock-transfected neurons (Fig. 3). Similarly, no difference in neurite outgrowth was seen with mock-, Kir3.1/3.2-, or Kir3.1/3.3-transfected neurons when plated on laminin, which enhanced neurite length relative to poly-l-lysine (Fig. 3). Interestingly, however, expression of Kir3.1/3.3 in neurons maintained on substrate-coated NCAM led to reduced neurite outgrowth when compared with mock- and Kir3.1/3.2-transfected neurons (Fig. 3). These observations indicate that the previously proven homophilic enhancement of neurite outgrowth via NCAM is negatively modified by Kir3.3. This is noteworthy in view of the fact that Kir3.3 only becomes expressed later in development of the mouse brain, namely within the second postnatal week, when neurite outgrowth ceases.
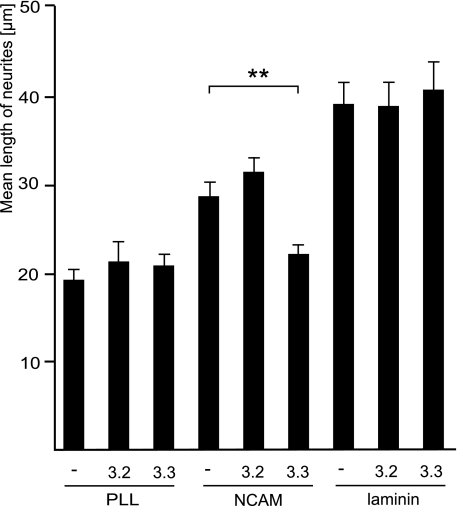
FIGURE 3.
Neurite outgrowth from hippocampal neurons transfected with Kir3.1/3.3 concatamers in vitro. Dissociated hippocampal neurons were transfected with either an empty vector or a vector encoding concatameric Kir3.1/3.2 or concatameric Kir3.1/3.3 and co-transfected with EGFP to identify the transfected neurons. The cells were plated on poly-l-lysine (PLL), NCAM-Fc (NCAM), or laminin as coated substrates and maintained for 24 h. The lengths of about 100 neurites of transfected neurons were measured. Bar graphs show a significant reduction of mean neurite length only when Kir3.1/3.3-transfected neurons are maintained on substrate-coated NCAM-Fc. The data represent the means from three independent experiments, with asterisks indicating statistically significant differences (Student's t test: **, p < 0.01). Error bars represent S.E.
Identification of an Interaction between the Intracellular Domains of TrkB and the Inwardly Rectifying K+ Channel Subunit, Kir3.3
Because we showed that NCAM interacts directly with Kir3.3 (this study) and TrkB (accompanying article (37)), we considered the possibility that Kir3.3 interacts with TrkB. Therefore, we performed co-immunoprecipitation experiments. Kir3.3 was co-immunoprecipitated with the panTrk antibody from synaptosomal fractions of wild-type and NCAM-deficient mice, whereas no co-immunoprecipitation of Kir3.3 was observed when a control non-immune IgG was used for immunoprecipitation (Fig. 4A). Western blot analysis with the TrkB-specific antibody showed that the panTrk antibody, which recognizes the intracellular domain of TrkB, immunoprecipitated only the full-length 155-kDa isoform. The panTrkB antibody did not immunoprecipitate the truncated 95-kDa TrkB isoform, which is devoid of most of the intracellular domain, whereas the control IgG did not precipitate TrkB at all (Fig. 4A). A similar experiment using a detergent extract from adult hippocampus and the antibody against TrkB revealed co-immunoprecipitation of Kir3.3, whereas no co-immunoprecipitation was observed when non-immune IgG was used (Fig. 4B). The TrkB antibody immunoprecipitated the full-length TrkB as well as the truncated isoform (Fig. 4B). These results indicate that TrkB interacts with Kir3.3 in adult brain tissue.
FIGURE 4.
Co-immunoprecipitation of TrkB and Kir3.3. A and B, detergent extracts of synaptosomal fractions from the brains of NCAM-deficient and wild-type littermate mice (A) or of hippocampi from wild-type mice (B) were incubated with the panTrk antibody (A), the TrkB-specific antibody H181 (B), or the corresponding non-immune IgG (Ig) (A and B) followed by incubation with protein A/G-agarose. Western blot analysis (WB) of the immunoprecipitates was performed using Kir3.3- or TrkB-specific antibodies. A, the arrowhead points to the IgG heavy chain, and the arrow indicates Kir3.3. B, the input lane and the lanes with the immunoprecipitates (IP) were from different parts of the same blot. The experiments were performed twice with identical results.
Next, to demonstrate direct binding of Kir3.3 to TrkB, the recombinant intracellular domain of Kir3.3 (Kir3.3-ID) and synthetic peptides of Kir3.3 spanning that C-terminal sequence stretch of Kir3.3 that is most different from Kir3.2 (see Fig. 1C) were used as substrate coat in an ELISA. Kir3.2 peptides and Kir3.2-ID served as controls. TrkB-ID bound to Kir3.3-ID and to Kir3.3 peptide C (Fig. 5A) but not to Kir3.3 peptide A or B (data not shown). TrkB did not bind to Kir3.2-ID or Kir3.2 peptide C (Fig. 5A). Using the biotinylated Kir3.3 peptide C as the soluble binding partner and TrkB-ID or the TrkB peptide that binds to NCAM-ID (accompanying article (37)) as substrate coat in a converse ELISA experiment, binding of peptide C to substrate-coated TrkB-ID and TrkB peptide (Fig. 5B) was observed. These results indicate a direct interaction between the intracellular domains of TrkB and Kir3.3 that is mediated by amino acids 377–393 of Kir3.3 and amino acids 669–680 of TrkB.
FIGURE 5.
Direct interaction between the intracellular domains of TrkB and Kir3.3. A and B, in ELISA experiments, substrate-coated Kir3.3-ID, Kir3.2-ID, Kir3.3 peptide C, and Kir3.2 peptide C (A) and TrkB-ID and TrkB peptide (B) were incubated with increasing amounts of TrkB-ID (A) or biotinylated Kir3.3 peptide C (B). Binding of TrkB-ID was detected by using the panTrk antibody (A), and binding of biotinylated Kir3.3 peptide C was determined using HRP-coupled streptavidin (B). All experiments were performed at least three times. Error bars represent S.D.
Competition of Kir3.3-ID and NCAM-ID for the Binding Site within TrkB-ID
Because both Kir3.3-ID and NCAM-ID bind to the TrkB peptide we analyzed by competition ELISA whether NCAM-ID and Kir3.3-ID compete for the binding to the TrkB peptide and thus bind to the same site on TrkB. NCAM peptide 1 and its derivatives peptide 1A and 1B (comprising amino acids 730–764, 730–750, and 748–764, respectively) and peptide 3 as negative control (Fig. 6A) were used for competition of the binding of biotinylated Kir3.3 peptide C to substrate-coated TrkB peptide. NCAM peptides 1 and 1A, but neither 1B nor control peptide 3, reduced the binding of the Kir3.3 peptide to the TrkB peptide (Fig. 6A). A competition ELISA using Kir3.3-ID or, as negative control, Kir3.2-ID for competition of the binding of NCAM140-ID and NCAM180-ID to the substrate-coated TrkB peptide revealed that binding of NCAM140-ID and NCAM180-ID to TrkB peptide was inhibited by Kir3.3-ID but not by Kir3.2-ID (Fig. 6B). These results show that the intracellular domains of NCAM and Kir3.3, but not Kir3.2, bind directly to the same binding site in the intracellular domain of TrkB comprising amino acids 669–680. These interactions are mediated by amino acids 377–393 of Kir3.3 and amino acids 730–750 of NCAM.
FIGURE 6.
Competition between the intracellular domains of NCAM and Kir3.3 for binding to the TrkB peptide. In a competition ELISA, substrate-coated TrkB peptide was preincubated with increasing amounts of NCAM-ID sequence-derived peptides 1, 1A, 1B, and 3 (A) and Kir3.3-ID and Kir3.2 (B) and then incubated with a constant amount of biotinylated Kir3.3 peptide C (A) or NCAM140-ID or NCAM180-ID (B). Binding of the biotinylated Kir3.3 peptide C was determined using HRP-coupled streptavidin (A), and binding of the NCAM-IDs (B) was detected by using NCAM antibody P61. All experiments were performed at least three times. Error bars represent S.D.
TrkB and NCAM Modulate Kir3.3-mediated K+ Currents
The binding experiments showed that Kir3.3-ID, but not Kir3.2-ID, interacts directly with TrkB-ID and NCAM-ID. Because it was shown in a previous study that NCAM influences Kir3.2- and Kir3.3-mediated currents (15), the influence of TrkB on Kir3.3-mediated macroscopic currents was investigated in Xenopus oocytes. Co-expression of TrkB and the concatamer of Kir3.1/Kir3.3 resulted in an increase in the Kir3.1/3.3-mediated currents in comparison with the currents observed when only Kir3.1/3.3 was expressed (Fig. 7, A and B), whereas Kir3.1/3.3-mediated currents were reduced by co-expression of NCAM180 (Fig. 5A) and NCAM140 (data not shown). Co-expression of Kir3.1/Kir3.3 with TrkA did not enhance the current (Fig. 7A). The increase of Kir3.1/3.3-evoked currents by TrkB was decreased by the concomitant expression of NCAM140 or NCAM180 (Fig. 7, A and B). The concatameric expression of Kir3.1/3.4 or Kir3.1/3.2 did not show enhanced currents when co-expressed with TrkB (Fig. 7C), indicating that the enhancement of K+ currents by TrkB is specific for Kir3.3. This specificity for Kir3.3 can be explained by differences in protein sequence between Kir3.3, on the one hand, and Kir3.2 and Kir3.4, on the other. To test whether the increase in K+ current was due to a higher cell surface expression, transcripts of EGFP-tagged Kir3.3 were co-injected with cRNA of Kir3.1 into oocytes. Co-expression of Kir3.1/3.3 with TrkB enhanced the cell surface expression of EGFP-tagged Kir3.1/Kir3.3 (Fig. 7, D and E); this enhancement in cell surface expression was abrogated when NCAM180 was co-expressed with Kir3.1/Kir3.3 and TrkB (Fig. 7, D and E). The presence or absence of BDNF had no effect on the K+ currents or the cell surface localization of the Kir3 channels for all subunits studied (data not shown). These observations indicate that surface localization of Kir3.1/Kir3.3, and thus the basic electric features of the plasma membrane, depends on levels of TrkB and NCAM expression.
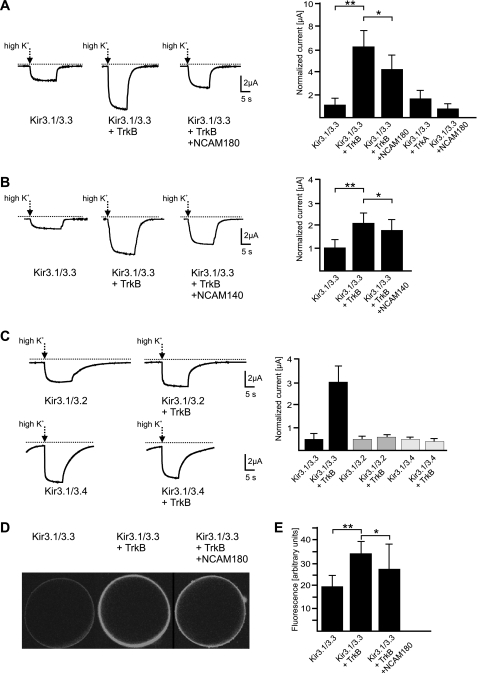
FIGURE 7.
Current recordings from Xenopus oocytes expressing Kir3 channels, TrkB, and NCAM and cell surface localization of Kir3.1/EGFP-Kir3.3 channels. A, oocytes were injected with concatameric Kir3.1/3.3 cRNA, concatameric Kir3.1/3.3 cRNA + TrkB cRNA (ratio 1:0.25), and concatameric Kir3.1/3.3 cRNA + TrkB cRNA + NCAM180 cRNA (ratio 1:0.25:1). Oocytes were clamped at −80 mV and perfused with high K+ (96 mm external K+) solution as indicated by arrows. The bar graph in the right panel shows current amplitude ratios of the measurements shown in the left panel. In addition, the current amplitude ratio is given for co-expression of Kir3.1/3.3 with TrkA and NCAM180, respectively. B, current recordings as shown in A, but triple injection is displayed for Kir3.1/3.3 + TrkB + NCAM140 instead of NCAM180 (cRNA ratio: 1:0.25:4). The bar graph in the right panel shows current amplitude ratios of the measurements shown in the left panel. C, current recordings from oocytes injected with concatameric Kir3.1/3.2 cRNA, concatameric Kir3.1/3.2 cRNA + TrkB cRNA (left panel, upper part), concatameric Kir3.1/3.4 cRNA, and concatameric Kir3.1/3.4 cRNA + TrkB cRNA (left panel, lower part). The bar graph in the right panel shows current amplitude ratios from measurements depicted in the left panel. The Kir3.1/3.3 data are from five independent experiments, and the Kir3.1/3.2 and Kir3.1/3.4 data are from two independent experiments each. D, representative confocal images are shown of Xenopus oocytes injected with cRNA of Kir3.1/3.3 (left), Kir3.1/3.3 with TrkB (middle), and Kir3.1/3.3 with TrkB and NCAM180 (right). Surface fluorescence of oocytes results from EGFP-tagged Kir3.3 subunits targeted to the plasma membrane. E, bar graph shows average fluorescence intensities of 10 oocytes injected with cRNAs as indicated in D. Data are shown as means with asterisks indicating statistically significant differences (Student's t test (A–C) or analysis of variance and Tukey's post hoc test (D and E): **, p < 0.01; *, p < 0.05). Error bars represent S.D.
Trk- and NCAM-dependent Cell Surface Localization of Kir3.3
To verify that the cell surface expression of Kir3.3 depends on the levels of TrkB and/or NCAM expression, cell surface biotinylation experiments were performed after transfection of CHO cells with Kir3.1/3.3 concatamer, TrkB, and/or NCAM. The cell surface level of Kir3.1/3.3 was increased ∼2-fold when CHO cells were co-transfected with Kir3.1/3.3 and TrkB compared with the level obtained after transfection with Kir3.1/3.3 alone (Fig. 8, A and B). Co-transfection of Kir3.1/3.3 with NCAM also resulted in a similar increase in the cell surface level of Kir3.1/3.3 (Fig. 8, A and B). Co-expression of Kir3.1/3.3 with both TrkB and NCAM led to cell surface levels of Kir3.1/3.3 similar to those obtained after transfection with Kir3.1/3.3 alone (Fig. 8, A and B), indicating that the interaction between TrkB and NCAM reduces the interaction of TrkB and NCAM to Kir3.3 and thus its surface expression. Because expression of Kir3.3 affects NCAM-mediated neurite outgrowth, we tested whether Kir3.1/3.3 expression also influences NCAM cell surface expression. Co-transfection of NCAM and Kir3.1/3.3 increased the cell surface levels of NCAM relative to the levels observed when NCAM alone was transfected, whereas the enhanced cell surface levels of NCAM were reduced when cells were co-transfected with NCAM, Kir3.1/3.3, and TrkB (Fig. 8, C and D). These data indicate that the interaction between TrkB and Kir3.3 reduces the interaction of NCAM and Kir3.3 and thus NCAM surface expression.
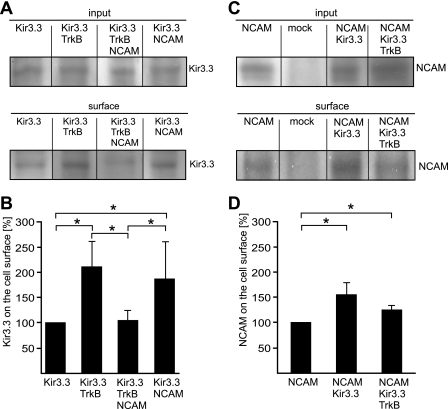
FIGURE 8.
Cell surface expression of Kir3.3 in CHO cells transfected with Kir3.3, TrkB, and NCAM. CHO cells were transfected with concatameric Kir3.1/3.3 (Kir3.3) or co-transfected with TrkB (Kir3.3+TrkB), NCAM140 (Kir3.3+NCAM), or NCAM140 and TrkB (Kir3.3+TrkB+NCAM) (A) or mock-transfected, transfected with NCAM140, or co-transfected with NCAM140 and Kir3.3 (NCAM+Kir3.3) or NCAM140, Kir3.3, and TrkB (NCAM+Kir3.3+TrkB) (C) and subjected to cell surface biotinylation. Cell lysates (Input) and biotinylated surface proteins isolated by streptavidin beads (surface) were subjected to Western blot analysis using the Kir3.3 antibody (A) or the NCAM antibody 5B8 (C). B and D, The bar graphs show the quantification of the surface levels of Kir3.3 (B) and NCAM (D) by densitometry. Cell surface levels of Kir3.3 and NCAM were determined by quantifying the amount of biotinylated Kir3.3 and NCAM; the total amount of Kir3.3 and NCAM was calculated from the amount in the input. The cell surface levels of Kir3.3 or NCAM relative to the total amount of Kir3.3 or NCAM after transfection of Kir3.3 alone or NCAM was set to 100%. Data are shown as means, with asterisks indicating statistically significant differences (Student's t test: *, p < 0.05). Error bars represent S.D.
To further support the notion that TrkB and NCAM regulate the cell surface expression of Kir3.3, the amounts of Kir3.3 in subcellular fractions isolated from the brains of TrkB-deficient, NCAM-deficient, and wild-type mice were determined by Western blot analysis. The total expression of Kir3.3 and TrkB, as well as their levels at the plasma membrane, which served as a marker protein for plasma membranes, was similar in NCAM-deficient and wild-type brains (Fig. 9A). However, relative to wild-type brains TrkB-deficient brains showed a nearly 2-fold higher total Kir3.3 expression and approximately two times more Kir3.3 in a fraction that is enriched in the endoplasmic reticulum marker calreticulin (Fig. 9B). In contrast, the levels of Kir3.3 in the plasma membrane fraction were reduced by more than 40% in TrkB-deficient brains in comparison with wild-type brains (Fig. 9B), whereas similar amounts were found in fractions enriched in the endosomal marker protein rab11 and deriving from wild-type and TrkB-deficient brains (Fig. 9B). In contrast to Kir3.3, the levels of Kir3.2 and of NCAM, which served as a marker for the plasma membranes, were not changed in brain extracts or in the plasma membrane fraction from brains of TrkB-deficient mice relative to those from wild-type mice (Fig. 9, B and C). The occurrence of double bands for Kir3.3 and 3.2 in some blots (Fig. 9, B and C) may be due to posttranslational modification, such as phosphorylation or proteolytic processing. These results confirm that the cell surface expression of Kir3.3, but not of Kir3.2, is regulated specifically by TrkB.
FIGURE 9.
Expression of Kir3.3 subcellular brain fractions from NCAM- and TrkB-deficient brains. A–C, detergent extracts of total brain and fractions enriched in plasma membranes were isolated from three NCAM-deficient (A), TrkB-deficient (B and C), or corresponding wild-type littermate mice (A–C). B, fractions enriched in endoplasmic reticulum (ER) or endosomes were prepared from four pools consisting of two TrkB-deficient or two wild-type littermate mice. A–C, samples were subjected to Western blot analysis using Kir3.3 antibody (A and B), NCAM antibody 1B2 (B and C), TrkB antibody H181 (A), calreticulin antibody (CRT), rab11 antibody (B), or Kir3.2 antibody (C). The bar graphs show the quantification of the relative levels of Kir3.3 (A and B), NCAM (A–C), calreticulin and rab11 (B), or Kir3.2 (C) by densitometry. The levels obtained for the wild-type brains were set to 100%. Data are shown as the means, with asterisks indicating statistically significant differences (Student's t test: *, p < 0.05). Error bars represent S.D.
The combined results suggest that the interaction between Kir3.3 and NCAM leads to the formation of a binary complex and an enhanced cell surface expression. Similarly, the interaction between Kir.3.3 and TrkB and between TrkB and NCAM may lead to the formation of binary complexes and enhanced cell surface expression. To test this idea, co-immunostaining of primary hippocampal neurons was performed after 14 days in culture when Kir3.3 was expressed. Co-immunostaining of Kir3.3 and TrkB, Kir3.3, and NCAM as well as NCAM and TrkB was frequently found in nearly every neuron and in particular along neurites (Fig. 10). Co-immunostaining of Kir3.3, NCAM, and TrkB was also observed occasionally in a few neurons and mainly in neurites (Fig. 10). This result suggests that NCAM·Kir3.3, TrkB·NCAM, and NCAM·TrkB binary complexes and also a ternary complex consisting of TrkB, NCAM, and Kir3.3 are expressed at the cell surface of neurites.
FIGURE 10.
Immunocytochemical localization of Kir3.3, TrkB, and NCAM in cultured hippocampal neurons. Hippocampal neurons were maintained for 14 days and stained with goat Kir3.3 antibody, rat NCAM antibody H28, and rabbit TrkB antibody H-181 followed by staining with secondary anti-goat, anti-rat, and anti-rabbit antibodies coupled to fluorescent dyes Cy3, Cy2, and Cy5, respectively. Co-localization is seen in the higher magnification of a neurite (indicated by the box). Co-localization of Kir3.3 (red) and NCAM (green) is shown by yellow staining, co-localization of Kir3.3 (red) and TrkB (blue) by turquoise staining, and co-localization of TrkB (blue) and NCAM (green) by violet staining; co-localization of all three proteins is shown by white staining.
Single Channel Properties of Kir3.1/3.3 Channels in the Absence and Presence of NCAM or TrkB
Because NCAM and TrkB interact with Kir3.3 and modulate Kir3.1/3.3-mediated macroscopic currents, we investigated whether the interaction of Kir3.1/3 with NCAM and/or TrkB might influence intrinsic channel properties including single channel conductance, mean open time, and open probability. To test whether NCAM or TrkB had any influence on these parameters, we took single channel recordings from CHO cells expressing the Kir3.1/3 concatamer alone (Fig. 11A) or in combination with either NCAM140 (Fig. 11D) or TrkB (Fig. 11E). The single channel conductance (γ) (Fig. 11B) and the mean open time (τopen) (Fig. 11C) for Kir3.1/3 channels expressed alone were not affected by co-expression of NCAM140 or TrkB (Fig. 11, F and G). To further characterize channel activity, we determined the apparent open probability (Po) during 2–4 min of recording. Basal channel activity, i.e. in the absence of exogenous Gβγ or BDNF, was reflected by low Po values (below 0.1) in all recordings (Fig. 11H). As for the other channel parameters, the observed reduction of Po in the presence of NCAM140 relative to its absence (p = 0.3614) as well as the increase of Po in the presence of TrkB relative to its absence (p = 0.1753) did not reach the set level of significance. Taken together, these results do not support a major impact of NCAM or TrkB interaction on the Kir3.1/3 channel permeation and gating properties under basal conditions.
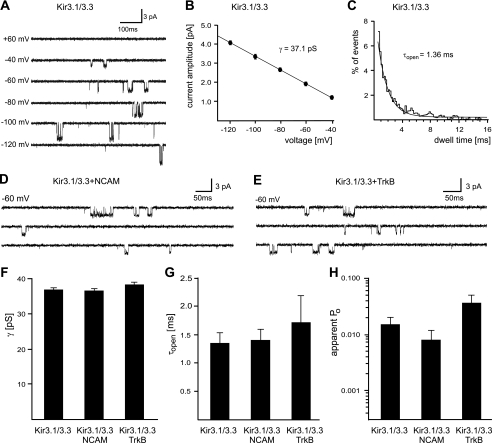
FIGURE 11.
Influence of NCAM and TrkB on single channel properties of Kir.3.1/3.3 in transfected CHO cells. Plasmids coding for concatameric Kir3.1/3.3, TrkB, or NCAM were microinjected into CHO cells. A, single channel recordings from microinjected CHO cells were performed in the cell-attached mode with 135 K+ solution in both the bath and the patch pipette. A, Kir3.1/3.3 basal activity at different voltages applied to the patch membrane. No channel activity was observed at +60 mV. B, single channel I-V relationship pooled from four patches containing Kir3.1/3.3 alone. Linear fitting of the data yields the single channel conductance, γ. C, dwell time histogram based on data pooled from four patches containing Kir3.1/3 alone. Fitting a single exponential function to the data yields the mean open time, τopen. D, consecutive cell-attached recordings at −60 mV from a cell expressing Kir3.1/3.3 together with NCAM140. E, consecutive recordings under the same conditions from a cell expressing Kir3.1/3.3 together with TrkB. F–H, bar graphs summarizing the results obtained for single channel conductance, γ (F), mean open time, τopen (G), and apparent open probability, Po (H), for Kir3.1/3 alone and in the presence of NCAM140 or TrkB. Note that γ and τopen for Kir3.1/3.3 alone (37.1 ± 0.35 pS, n = 8; 1.36 ± 0.18 ms, n = 4) were not significantly altered in the presence of NCAM140 (36.9 ± 0.74 pS, n = 5, p = 0.8167; 1.42 ± 0.20 ms, n = 5, p = 0.8309) or TrkB (38.4 ± 0.87 pS, n = 7, p = 0.2082; 1.72 ± 0.48 ms, n = 8, p = 0.5024). Po for Kir3.1/3.3 alone (0.0148 ± 0.0047, n = 4) was not significantly reduced in the presence of NCAM140 (0.0095 ± 0.0021, n = 5, p = 0.3614) and was not significantly increased in the presence of TrkB (0.0362 ± 0.0136, n = 8; p = 0.1753).
DISCUSSION
Our present study has revealed direct associations of three developmentally and functionally important molecules: the neural cell adhesion molecule NCAM, the tyrosine receptor kinase TrkB, and the inwardly rectifying K+ channel subunit Kir3.3. We provide evidence that the intracellular domain of Kir3.3, but not the structurally similar subunit Kir3.2, interacts directly with the intracellular domains of NCAM and TrkB, which also interact directly with each other (see accompanying article (37)). NCAM and TrkB bind to different sites in the intracellular C-terminal domain of Kir3.3. Co-expression of TrkB and Kir3.3 in CHO cells or oocytes leads to an increase in cell surface expression of Kir3.1/3.3 and Kir3.1/3.3-mediated K+ currents, indicating that the direct interaction of TrkB with Kir3.3 regulates cell surface expression and channel activity of Kir3.1/3.3. The analysis of Kir3.3 expression in TrkB-deficient mice revealed that ablation of TrkB leads to a reduction in Kir3.3 at the plasma membrane and to a concomitant accumulation of Kir3.3 in the endoplasmic reticulum, indicating that TrkB is involved in the regulation of Kir3.3 trafficking to the plasma membrane in vivo. On the other hand, TrkB does not trigger the cell surface expression of other Kir channels, namely Kir3.1/3.2 and Kir3.1/3.4. Furthermore, TrkA, which is structurally and functionally related to TrkB, does not affect the surface expression of Kir3.3. Both the TrkB-dependent increase in cell surface expression of Kir3.3 and the increase in Kir3.3-mediated K+ currents were reduced by co-expression of NCAM in CHO cells and oocytes. Because cell surface biotinylation of transfected CHO cells showed that co-expression of NCAM or TrkB increases the cell surface expression of Kir3.3 and because NCAM and TrkB interact with each other (accompanying article (37)), we concluded that the interaction of TrkB with NCAM reduces the levels of TrkB- and NCAM-mediated surface expression of Kir3.3. However, in contrast to TrkB, ablation of NCAM in NCAM-deficient mice does not affect the cell surface expression of Kir3.3. Moreover, because immunoprecipitation of TrkB from NCAM-deficient mice did not result in an enhanced co-immunoprecipitation of Kir3.3, we also concluded that interaction between TrkB and Kir3.3 was not enhanced in the absence of NCAM. These observations suggest that NCAM plays no or only a minor role in Kir3.3 trafficking to the plasma membrane in vivo.
Here, the observed single channel parameters of Kir3.1/3.3 concatamer transfected into CHO cells were similar to those obtained by others (23) for heteromeric Kir3 channels expressed in CHO cells. Our data convey the impression that the apparent open probability (Po) may differ depending on whether Kir3.1/3 channels associate with NCAM140 or with TrkB. This tendency, which proved not to be significant (p = 0.0935), would be in accordance with our macroscopic current measurements in Xenopus oocytes, leaving open the possibility that, in addition to the shown trafficking effects, small changes in intrinsic channel activity may also contribute to K+ current regulation by NCAM and TrkB. As we found no statistically significant influence of the co-expression of NCAM140 or TrkB on single channel properties, intrinsic channel modulation by NCAM or TrkB, however, would play a minor role in the regulation of macroscopic K+ currents.
Homophilic interaction of NCAM in a trans-position promotes neurite outgrowth from hippocampal neurons (21). Artificially premature expression of Kir3.1/3.3 in neonatal hippocampal neurons, which at this developmental stage do not express Kir3.3 channels, inhibits NCAM-dependent neurite outgrowth, suggesting that Kir3.3 disturbs the interaction between TrkB and NCAM that is required for TrkB-dependent NCAM-mediated neurite outgrowth (accompanying article (37)). Inhibition of NCAM-mediated neurite outgrowth by Kir3.1/3.3, which appears late during development (22), may be a physiologically meaningful mechanism during nervous system maturation.
Our observations reveal a novel role for the interplay between the functionally important molecules NCAM, TrkB, and Kir3.3 during different stages of neuronal development. First, the finding that TrkB binds directly to NCAM through its intracellular domain and is required for NCAM-mediated neurite outgrowth (accompanying article (37)) puts into focus the important role of TrkB for NCAM-mediated functions in early stages of development. That the interaction between TrkB and NCAM is counterproductive in the adult, when both molecules continue to be expressed after cessation of nervous system development, is evident, as strongly continued neurite outgrowth in the adult would lead to disturbances in the maintenance of once formed and viable neuronal networks. It is at this time of later neuronal development that expression of the Kir3.3 subunit of the inwardly rectifying K+ channel, which has important roles in the maintenance of membrane potential and excitability of neurons (23, 24), emerges. This channel disrupts the neurite outgrowth promoting interplay between TrkB and NCAM in that it binds via specific and distinct sites at the C-terminal intracellular domain to TrkB and NCAM, thereby inhibiting NCAM-mediated neurite outgrowth. Thus, the developmentally co-active pair of TrkB and NCAM, ensuring neurite outgrowth during development, becomes no longer functional, because the developmentally late appearing Kir3.3 subunit disturbs their relationship. In the adult, Kir3.1/3.3 channels generate, on the one hand, a constitutive basal K+ conductance to stabilize the membrane potential. On the other hand, they serve as a major target for Gi/Go-coupled neurotransmitter receptors, such as muscarinic M2, α2 adrenergic, dopamine D2, serotonin1A, adenosine, GABAB, and opioid (μ/κ/δ) receptors (25). Receptor activation induces a large K+ conductance that drives the membrane potential closer to the K+ equilibrium potential and thus reduces cellular excitability. TrkB may contribute to both stabilization of the membrane potential under basal conditions and enhancement of Kir3.3-mediated K+ currents upon receptor activation by increasing the number of Kir3.3 channels at the plasma membrane.
The functional interplay between Kir3.3 and NCAM and/or TrkB may play a crucial role in addictive and perhaps emotional behavior, as indicated by the following findings. Kir3.3-deficient mice are impaired in their ability to mediate the acute inhibitory effect of opioids on locus coeruleus neurons (26) and exhibit a less severe withdrawal from sedative hypnotics such as pentobarbital and ethanol (27), suggesting a role of Kir3.3 in addiction. NCAM-deficient mice show abnormal responses in fear conditioning and addictive responses (28–30), and TrkB has been associated with antisocial alcohol and nicotine dependence (31, 32) as well as anxiety-related behavior (33). Furthermore, NCAM- and TrkB-mediated signaling is dysregulated in the hippocampus and/or prefrontal cortex of patients with major depressive disorders (34, 35). However, whether there is a link between the interplay of these molecules with synaptic plasticity and behavior remains to be seen.
It would be interesting to investigate the ways by which Kir3.3 cooperates in vivo with NCAM and TrkB in synaptic plasticity, which not only regulates behavior but also learning and memory, using neuron-specific conditionally and doubly ablated TrkB- and NCAM-deficient mice. However, because the Kir3.3 expression pattern largely overlaps with that of Kir3.1 and Kir3.2 (19, 36) and because previous studies on K+ currents (15) indicated that Kir3.1/3.2 and Kir3.1/3.3 are co-expressed in hippocampal neurons, it will be difficult or even impossible to investigate the influence of NCAM or TrkB specifically on neuronal Kir3.1/3.3 currents.
Acknowledgments
We are grateful to Galina Dityateva for help with transfection of hippocampal neurons; Tanja Martini and Annett Hasse for technical assistance; Rüdiger Veh (Medizinische Fakultät Charité, Institut für Integrative Anatomie (CCM) Berlin) for the Kir cDNA constructs; Kathrin Hilke-Steen for typing the manuscript; Eva Kronberg for animal care; Fabio Morellini and Achim Dahlmann for breeding and genotyping of NCAM-deficient mice; Edgar Kramer for breeding and genotyping of TrkB−/− mice. Harold Cremer (Laboratoire de Genetique et Physiologie du Developpement Parc Scientifique de Luminy, Case 907, Marseille, France) for the NCAM−/− mice; Shinichi Koizumi and Motohiko Kometani (Novartis Institutes for BioMedical Research, Tsukuba, Japan) for the recombinantly expressed intracellular domain of TrkB; Jochen Heukeshoven (Heinrich-Pette-Institut, Hamburg, Germany) and Kai Schuh and Thomas Renné (Universität Würzburg, Würzburg, Germany) for access to their confocal microscope.
This work was supported by Deutsche Forschungsgemeinschaft Grant SCHA185/17-4.
- NCAM
- neural cell adhesion molecule
- BDNF
- brain-derived neurotrophic factor
- EGFP
- enhanced green fluorescent protein
- ID
- intracellular domain
- pS
- picosiemens
- TrkB
- receptor tyrosine kinase B.
REFERENCES
- 1.Edelman G. M., Gallin W. J., Delouvée A., Cunningham B. A., Thiery J. P. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 4384–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schachner M. (1997) Curr. Opin. Cell Biol. 9, 627–634 [DOI] [PubMed] [Google Scholar]
- 3.Lüthi A., McCormick D. A. (1998) Neuron 21, 9–12 [DOI] [PubMed] [Google Scholar]
- 4.Murase S., Schuman E. M. (1999) Curr. Opin. Cell Biol. 11, 549–553 [DOI] [PubMed] [Google Scholar]
- 5.Dityatev A., Dityateva G., Schachner M. (2000) Neuron. 26, 207–217 [DOI] [PubMed] [Google Scholar]
- 6.Dityatev A., Dityateva G., Sytnyk V., Delling M., Toni N., Nikonenko I., Muller D., Schachner M. (2004) J. Neurosci. 24, 9372–9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan D. R., Miller F. D. (2000) Curr. Opin. Neurobiol. 10, 381–391 [DOI] [PubMed] [Google Scholar]
- 8.Klein R., Parada L. F., Coulier F., Barbacid M. (1989) EMBO J. 8, 3701–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein R., Nanduri V., Jing S. A., Lamballe F., Tapley P., Bryant S., Cordon-Cardo C., Jones K. R., Reichardt L. F., Barbacid M. (1991) Cell 66, 395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Middlemas D. S., Lindberg R. A., Hunter T. (1991) Mol. Cell. Biol. 11, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soppet D., Escandon E., Maragos J., Middlemas D. S., Reid S. W., Blair J., Burton L. E., Stanton B. R., Kaplan D. R., Hunter T., Nikolics K., Parade L. F. (1991) Cell 65, 895–903 [DOI] [PubMed] [Google Scholar]
- 12.Squinto S. P., Stitt T. N., Aldrich T. H., Davis S., Bianco S. M., Radziejewski C., Glass D. J., Masiakowski P., Furth M. E., Valenzuela D. M. (1991) Cell 65, 885–893 [DOI] [PubMed] [Google Scholar]
- 13.Barbacid M. (1994) J. Neurobiol. 25, 1386–1403 [DOI] [PubMed] [Google Scholar]
- 14.Tannahill L., Klein R., Schachner M. (1995) Eur. J. Neurosci. 7, 1424–1428 [DOI] [PubMed] [Google Scholar]
- 15.Delling M., Wischmeyer E., Dityatev A., Sytnyk V., Veh R. W., Karschin A., Schachner M. (2002) J. Neurosci. 22, 7154–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogalski S. L., Appleyard S. M., Pattillo A., Terman G. W., Chavkin C. (2000) J. Biol. Chem. 275, 25082–25088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ippolito D. L., Temkin P. A., Rogalski S. L., Chavkin C. (2002) J. Biol. Chem. 277, 32692–32696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer E. R., Aron L., Ramakers G. M., Seitz S., Zhuang X., Beyer K., Smidt M. P., Klein R. (2007) PLoS Biol. 5, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wischmeyer E., Döring F., Wischmeyer E., Spauschus A., Thomzig A., Veh R., Karschin A. (1997) Mol. Cell. Neurosci. 9, 194–206 [DOI] [PubMed] [Google Scholar]
- 20.Iwasaki Y., Nishiyama H., Suzuki K., Koizumi S. (1997) Biochemistry 36, 2694–2700 [DOI] [PubMed] [Google Scholar]
- 21.Doherty P., Fruns M., Seaton P., Dickson G., Barton C. H., Sears T. A., Walsh F. S. (1990) Nature 343, 464–466 [DOI] [PubMed] [Google Scholar]
- 22.Grosse G., Eulitz D., Thiele T., Pahner I., Schröter S., Takamori S., Grosse J., Wickman K., Tapp R., Veh R. W., Ottersen O. P., Ahnert-Hilger G. (2003) Mol. Cell. Neurosci. 24, 709–724 [DOI] [PubMed] [Google Scholar]
- 23.Jelacic T. M., Kennedy M. E., Wickman K., Clapham D. E. (2000) J. Biol. Chem. 275, 36211–36216 [DOI] [PubMed] [Google Scholar]
- 24.Koyrakh L., Luján R., Colón J., Karschin C., Kurachi Y., Karschin A., Wickman K. (2005) J. Neurosci. 25, 11468–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isomoto S., Kondo C., Kurachi Y. (1997) Jpn. J. Physiol. 47, 11–39 [DOI] [PubMed] [Google Scholar]
- 26.Torrecilla M., Marker C. L., Cintora S. C., Stoffel M., Williams J. T., Wickman K. (2002) J. Neurosci. 22, 4328–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozell L. B., Walter N. A., Milner L. C., Wickman K., Buck K. J. (2009) J. Neurosci. 29, 11662–11673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stork O., Welzl H., Cremer H., Schachner M. (1997) Eur. J. Neurosci. 9, 1117–1125 [DOI] [PubMed] [Google Scholar]
- 29.Stork O., Welzl H., Wotjak C. T., Hoyer D., Delling M., Cremer H., Schachner M. (1999) J. Neurobiol. 40, 343–355 [DOI] [PubMed] [Google Scholar]
- 30.Maćkowiak M., Markowicz-Kula K., Fijał K., Wedzony K. (2005) Brain Res. 1055, 149–155 [DOI] [PubMed] [Google Scholar]
- 31.Xu K., Anderson T. R., Neyer K. M., Lamparella N., Jenkins G., Zhou Z., Yuan Q., Virkkunen M., Lipsky R. H. (2007) Pharmacogenomics J. 7, 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beuten J., Ma J. Z., Payne T. J., Dupont R. T., Lou X. Y., Crews K. M., Elston R. C., Li M. D. (2007) Biol. Psychiatry 61, 48–55 [DOI] [PubMed] [Google Scholar]
- 33.Bergami M., Berninger B., Canossa M. (2009) Commun. Integr. Biol. 2, 14–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brennaman L. H., Maness P. F. (2010) Adv. Exp. Med. Biol. 663, 299–317 [DOI] [PubMed] [Google Scholar]
- 35.Martinowich K., Manji H., Lu B. (2007) Nat. Neurosci. 10, 1089–1093 [DOI] [PubMed] [Google Scholar]
- 36.Karschin C., Dissmann E., Stühmer W., Karschin A. (1996) J. Neurosci. 16, 3559–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cassens C., Kleene R., Xiao M. F., Friedrich C., Dityateva G., Schafer-Nielsen C., Schachner M. (2010) J. Biol. Chem. 285, 28959–28967 [DOI] [PMC free article] [PubMed] [Google Scholar]