Abstract
Internal ribosome entry sites (IRESs) are specialized mRNA elements that allow recruitment of eukaryotic ribosomes to naturally uncapped mRNAs or to capped mRNAs under conditions in which cap-dependent translation is inhibited. Putative cellular IRESs have been proposed to play crucial roles in stress responses, development, apoptosis, cell cycle control, and neuronal function. However, most of the evidence for cellular IRES activity rests on bicistronic reporter assays, the reliability of which has been questioned. Here, the mechanisms underlying cap-independent translation of cellular mRNAs and the contributions of such translation to cellular protein synthesis are discussed. I suggest that the division of cellular mRNAs into mutually exclusive categories of “cap-dependent” and “IRES-dependent” should be reconsidered and that the implications of cellular IRES activity need to be incorporated into our models of cap-dependent initiation.
Keywords: Gene Regulation, mRNA, RNA-binding Protein, Translation Control, Translation Initiation Factors, Internal Ribosome Entry Site (IRES)
Introduction
Eukaryotic mRNAs are modified by the addition of an m7GpppN cap structure to their 5′-ends. The m7G cap is thought to stimulate translation of most mRNAs by enhancing binding of a 43 S preinitiation complex (containing 40 S ribosomal subunits, methionine initiator tRNA, and initiation factors eIF2 and eIF3) to 5′-UTRs of mRNAs through recognition of the m7G cap by a complex of the cap-binding protein, eIF4E; a large scaffold protein, eIF4G; and an ATP-dependent RNA helicase, eIF4A. Subsequent movement of the 43 S complex in a 5′- to 3′-direction (scanning) locates the initiating AUG through recognition by the anticodon of the initiator tRNA. The discovery that naturally uncapped picornaviral mRNAs can efficiently recruit the host cell translation machinery via internal ribosome entry sites (IRESs)2 raised the possibility that certain cellular mRNAs might have a similar capability (1, 2).
The cellular IRES hypothesis offered an attractive solution to two problems. First, a number of cellular stress responses involve inhibition of one or more general translation initiation factors, yet the adaptive responses to stress require new protein synthesis. Cellular IRES elements could allow mRNAs encoding key regulatory proteins to escape the general inhibition of translation. The observation that some cellular mRNAs continue to be translated in poliovirus-infected cells after the inhibition of cap-dependent initiation (through cleavage of eIF4G by a virally encoded protease) is consistent with this hypothesis (3). Second, the existence of cellular IRESs could explain how mRNAs with very long 5′-UTRs or containing numerous predicted stem-loop structures or upstream AUG codons within their 5′-UTRs could be translated with reasonable efficiency, despite evidence that such features can significantly reduce translation of model mRNAs (4–6).
Both arguments in favor of the cellular IRES hypothesis rest on certain assumptions about the predominant mechanism of cap- and scanning-dependent initiation that this minireview will re-examine in light of recent publications that (i) demonstrate that long GC-rich 5′-UTRs can be efficiently translated by a cap-dependent mechanism, in contrast to the prevailing view, and (ii) reveal the surprising range of translational efficiencies displayed by cellular mRNAs under conditions in which cap-dependent initiation is presumed to be the predominant mechanism for translation.
To be clear, it is not my intention to deny the existence of cellular IRESs or to discourage newcomers to the translational control field from testing their favorite genes for IRES activity. Rather, one purpose of this discussion is to ensure that such newcomers do not fall victim to either the logical or experimental pitfalls that have plagued the cellular IRES field. The ultimate goals of this field are to understand both the molecular mechanisms underlying cap-independent initiation and the physiological function(s) of cellular IRES-dependent translation. Because neither of these purposes is served when 5′-UTRs are erroneously claimed to promote IRES-dependent initiation, I begin by discussing the source of most such errors.
Pitfalls of Bicistronic Reporter Assays
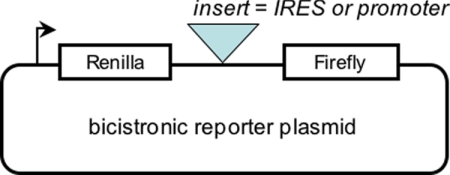
Eukaryotic ribosomes do not efficiently reinitiate translation of a downstream ORF after translating a full-length ORF located upstream in the same mRNA. If, however, the downstream ORF is preceded by an IRES, internal ribosome recruitment can result in high levels of translation of the second cistron. Based on this property, bicistronic reporters have been used to investigate the cis-acting elements required for viral IRES activity (7). A typical bicistronic reporter construct is illustrated in Fig. 1. Such reporters have also been employed to test the capacity of cellular 5′-UTRs for IRES-dependent initiation. It will be immediately clear to most readers that the insertion of a promoter sequence between the two cistrons will also promote reporter activity from the downstream cistron, by activating transcription of a capped monocistronic mRNA. In principle, it seems straightforward to distinguish between these two mechanisms by determining whether or not the inserted sequence leads to the production of monocistronic messages. In practice, despite abundant evidence that cryptic promoter artifacts are widespread in the cellular IRES literature (8–16), most publications claiming cellular IRES activity do not include controls that are adequate to determine whether a candidate sequence really is an IRES and not a cryptic promoter. Why is this the case?
FIGURE 1.
Typical bicistronic reporter plasmid used for IRES assays. A strong promoter such as SV40 drives expression of a bicistronic mRNA. Renilla luciferase activity reports the level of cap-dependent initiation in the experiment. Firefly luciferase activity is very low unless the intercistronic region contains an IRES or a promoter.
Most putative cellular IRESs are much less active than their viral counterparts when tested in assays that reliably measure translational activity (in vitro translation or in vivo translation of transfected in vitro transcribed mRNAs) (8, 9, 16). Although it is true that the presumed raison d'être of most cellular IRESs, to permit expression of key regulatory proteins, often of low abundance, under conditions of global inhibition of cap-dependent translation, does not require that cellular IRES-dependent initiation be nearly as efficient as viral IRES-dependent translation (which typically drives unregulated high-level expression of viral proteins), the very low level of activity of most putative cellular IRESs makes it far more difficult to rule out alternative explanations for activity in bicistronic reporter assays, mechanisms that do not involve translation at all, such as cryptic promoter activity. The use of extremely sensitive reporters like firefly luciferase permits detection of protein produced from almost undetectably small quantities of m7G-capped monocistronic mRNA. A typical Northern blot exposure is simply not adequate to rule out the possibility that 1% of the total mRNA encoding the 3′-cistron is monocistronic. This is the appropriate level of detection to consider for most putative cellular IRESs, whose demonstrably IRES-dependent translation is only ∼1% as efficient as cap-dependent translation of a control reporter mRNA (8, 9, 16). It is important to note that the popular Renilla/firefly luciferase reporter is not the only bicistronic reporter system vulnerable to cryptic promoter artifacts. The β-gal/chloramphenicol acetyltransferase reporter system has been shown to produce similar results (14).
No matter what reporter is used, the induction of 3′-cistron expression through non-translational mechanisms must be rigorously excluded before cellular IRES activity is concluded. A number of appropriate controls have been proposed, including showing very overexposed Northern blots, using siRNAs targeting the 5′-cistron of bicistronic mRNAs, and testing candidate IRESs for promoter activity in vectors that retain the SV40 enhancer element (but omit the SV40 promoter). Each of these control experiments behaves as expected for true IRES-dependent initiation when tested with viral UTR sequences such as the encephalomyocarditis virus (EMCV) IRES. Notably, most cellular IRES publications do not undertake such tests, and when tested, many putative cellular IRESs fail (8–16).
The DNA-based bicistronic reporter assay is not hopelessly flawed. The use of tightly regulated inducible promoters to drive expression of bicistronic mRNAs in vivo can permit discrimination between translational activation of the downstream cistron by an IRES, in which case activity from the downstream reporter will disappear in parallel with the upstream reporter when the 5′-promoter is repressed, and transcriptional activation via the insertion of a promoter, in which case activity of the two reporters will be uncoupled (17, 18). Note that the presence of promoter activity, cryptic or otherwise, does not rule out the possibility that a sequence may normally function as an IRES (the hepatitis C virus (HCV) IRES is an example of such a sequence), but it necessitates the use of some assay other than DNA transfection of bicistronic reporters to study the activity of putative IRESs.
Cellular IRESs, Weak and Strong
So, are there cellular 5′-UTRs with unambiguous IRES activity? Yes. A number of cellular 5′-UTRs stimulate translation of uncapped mRNA and/or promote translation of 3′-cistrons in RNA-based reporter assays that eliminate the possibility of cryptic promoter activity (9, 16, 19–21). The question is, how much does IRES-dependent initiation contribute to the overall level of protein synthesis for IRES-containing genes? As noted above, most cellular IRESs promote translation that is very inefficient (<2%) compared with cap-dependent translation of control reporters. Not all cellular IRESs are so weak. Internal initiation at AUG94 of the URE2 gene of Saccharomyces cerevisiae occurs ∼22% as often as cap-dependent initiation at the first AUG under normal growth conditions and ∼50% as often under conditions in which cap-dependent initiation is reduced by a mutation in the cap-binding protein (22). This level of IRES activity is comparable with that of viral IRESs. Importantly, the truncated protein produced by internal initiation at AUG94 has distinct functional properties, suggesting mechanisms whereby IRES-dependent initiation could affect cellular physiology. The authors ruled out IRES-independent mechanisms for the efficient production of the C-terminal fragment of Ure2p, including proteolysis of full-length protein and leaky scanning past the first AUG initiation codon. Heavily exposed Northern blots showed no signs of a smaller mRNA species (22), and sequencing of full-length cDNAs revealed transcription start sites between −216 and −208 exclusively (23).
URE2 is not the only cellular IRES with activity comparable with viral IRESs. Other yeast cellular IRESs show similarly high activity in in vitro translation assays (19). Furthermore, strong cellular IRES activity is not restricted to yeast. The 5′-UTR of mammalian c-src contains a potent IRES, having >80% of the activity of the poliovirus IRES in in vitro translation assays and >100% of the activity of the HCV IRES in in vivo RNA transfection experiments. In a bicistronic m7G-Renilla-IRES-firefly reporter mRNA, translation initiation by the c-src IRES produced a firefly/Renilla ratio of >1 (24).
Given that some cellular IRESs show activity comparable with viral IRESs and within the same order of magnitude as efficiently translated m7G-capped mRNAs, what are we to make of cellular IRESs that are <2% as efficient as cap-dependent controls? It has been argued that this is not the relevant comparison. Control m7G-capped mRNAs usually have short 5′-UTRs that are thought to mediate very efficient initiation. In contrast, most reported cellular IRESs are found in mRNAs with unusually long 5′-UTRs predicted to form extensive RNA secondary structures and therefore presumed incapable of mediating efficient cap-dependent initiation. Furthermore, cellular IRESs are generally proposed to function under conditions in which cap-dependent translation is inhibited.
Presumption of Inefficient Cap-dependent Initiation
Predicted 5′-UTR RNA secondary structure is frequently invoked as a reason a gene might require an IRES for efficient translation. The evidence for an inhibitory effect of RNA secondary structure seems clear from experiments using artificial 5′-UTRs with hairpins of defined stability and placement within the 5′-UTR (4–6). It was therefore quite surprising when the long GC-rich 5′-UTRs of several putative cellular IRES-dependent genes (including HIF-1α, c-myc, and Apaf-1) were found to mediate cap-dependent translation nearly as efficiently as the 5′-UTR from β-globin or a short unstructured control 5′-UTR. Even somewhat less efficiently translated 5′-UTRs were translated almost exclusively by a cap-dependent mechanism (8, 9). Clearly, our current understanding of what makes an mRNA amenable to cap-dependent translation is insufficient to allow accurate predictions. To avoid this pitfall in the future, researchers should directly compare the efficiencies of cap-dependent and IRES-dependent translation mediated by a given 5′-UTR. One cannot conclude that any 5′-UTR, no matter how long or burdened with “inhibitory” features like predicted stem-loops or AUG codons, is poorly translated via cap-dependent initiation without direct experimental evidence.
How strongly is cap-dependent translation inhibited under conditions in which cap-independent initiation is proposed to predominate? Glucose withdrawal causes a 10–20-fold reduction in global protein synthesis by a mechanism that requires the decapping machinery (25, 26). A cap-independent mechanism of translation that was only 10% as efficient as cap-dependent initiation under normal growth conditions could nevertheless be responsible for the majority of new protein synthesis in starved cells, if the mechanism of “global” inhibition were specific to cap-dependent initiation. Hypoxia is another cellular stress that causes global down-regulation of translation and for which cellular IRES-dependent translation has been suggested to be important for the adaptive response. Might a mechanism that is 2% as efficient as cap-dependent initiation contribute significantly to overall levels of protein synthesis? Given recent work showing that global translation decreases by 20–70% during oxygen deprivation, it seems unlikely (16). One should carefully consider both the extent of inhibition of cap-dependent initiation and the relative efficiencies of cap-dependent and IRES-dependent translation of a given gene when trying to determine the likely biological role of a 5′-UTR with IRES activity.
There may be dedicated cellular IRESs, 5′-UTRs that are incapable of cap-dependent initiation, as well as local cellular environments where cap-dependent initiation is so strongly inhibited as to render even inefficient cellular IRESs physiologically relevant, but this needs to be demonstrated experimentally. Something is wrong when a study revealing that a putative cellular IRES is actually a cryptic promoter is followed by a subsequent study investigating the regulation of said “cellular IRES” activity using transfection of bicistronic DNA reporters.
Role of RNA Tertiary Structure in Cellular IRES Activity: Is There One?
The nature and function of RNA structures in a variety of viral IRESs that use different mechanisms to initiate have been reviewed recently (27). There is reason to suspect that cellular IRESs may also use a variety of mechanisms. Here, I consider the evidence that cellular IRESs rely on the formation of defined RNA structures that functionally substitute for one or more translation factors to recruit ribosomes. Mammalian c-src is a good candidate for a dedicated cellular IRES gene, as addition of an m7G-cap does not stimulate translation of mRNA containing the c-Src 5′-UTR (24). This observation is also consistent with the hypothesis that the c-Src 5′-UTR is extensively folded, as predicted in silico. Although it is tempting to speculate that the cellular IRES activity of c-Src depends on the formation of a defined RNA tertiary architecture, similar to structured viral IRESs, this need not be the case. The IRES-containing YMR181c 5′-UTR is extensively folded, at least in vitro, yet deletion of the structured 5′-portion of the 5′-UTR has no effect on IRES activity (19). In contrast, the URE2 IRES does appear to require RNA structure for full activity, but the minimal IRES is both smaller and less structured than well characterized viral IRESs (18, 28).
Hints that virus-like structured cellular IRESs may exist can be found in studies of mutants with reduced translation from the structured cricket paralysis virus (CrPV) IRES (17, 29). In yeast lacking the nonessential ribosomal protein Rps25, CrPV IRES activity is reduced by 97%. Cellular protein synthesis, measured by [35S]methionine incorporation, is reduced by 19%. Polysome analysis showed a similarly modest decrease in the polysome/monosome ratio in the rps25Δ strain, consistent with a mild defect in translation initiation (17). The identities of the affected cellular mRNAs were not determined, but it is tempting to speculate that at least some of them might require a specific interaction between their 5′-UTRs and the 40 S subunit of the ribosome for efficient translation initiation. It seems unlikely that 19% of normal yeast protein synthesis proceeds via a CrPV IRES-like initiation mechanism, but this possibility cannot yet be ruled out. Alternatively, our models for cap-dependent initiation must be altered to explain why certain mRNAs require a specific nonessential ribosomal protein for their translation.
In summary, the jury is still out on whether virus-like structured cellular IRESs exist. Even if they exist, they may not be the norm for cellular IRESs. In other aspects of mRNA metabolism, such as mRNA export, eukaryotic host cells employ diverse RNA-binding proteins (RBPs) to do a job that is performed by structured viral RNA elements (30). This may reflect a trade-off between constitutive efficiency in viral gene expression and a need for regulation in cellular gene expression.
Dedicated IRES or Translational Enhancer?
Does the capacity of a 5′-UTR to promote IRES-dependent initiation, even in cases in which the IRES-dependent mechanism is quite efficient, necessarily mean that a particular protein is synthesized by a cap-independent mechanism? In the literature, “has an IRES” is often taken to mean “is normally translated via internal ribosome entry.” This is a dangerous assumption. Unlike some viral mRNAs, cellular IRES-containing mRNAs are generally capped. A 5′-UTR element that is capable of promoting internal ribosome entry might also function as an enhancer of cap-dependent initiation, depending on its mechanism of action. For example, some yeast cellular IRESs enhance the recruitment of eIF4G via binding of the poly(A)-binding protein to A-rich elements within 5′-UTRs (19). Unless tightly folded intervening RNA elements are present to preclude a productive interaction between eIF4E bound to the cap and eIF4G recruited more internally, there is no reason a priori to assume that internal 5′-UTR elements could not act synergistically with the m7G cap to increase the overall efficiency of initiation. Such a capability could permit some mRNAs to be preferentially translated under conditions in which global cap-dependent initiation is reduced but not abolished.
The purpose of this discussion is not to dwell on possible errors of interpretation in the cellular IRES literature, but to suggest that mechanistic studies of cellular IRES-dependent initiation ought to be reconsidered in light of what they may be telling us about the mechanism(s) of eukaryotic translation. Whether or not a particular gene is likely to rely on a cap-independent mechanism of protein synthesis, the fact that some 5′-UTRs, but not others, are capable of recruiting the eukaryotic translation machinery internally is interesting. The prevailing model for cap-dependent initiation treats 5′-UTRs as passive substrates that contribute nothing to the recruitment of ribosomes. According to this model, most mRNAs are translated by a constitutive mechanism whose efficiency is largely determined by the global availability of active initiation factors. Deviations from the “typical” (idealized) 5′-UTR are presumed to affect translation negatively. Consistent with this view, most examples of 5′-UTR-mediated translational control involve inhibition of cap-dependent initiation (reviewed in Ref. 31).
Why should there not also be enhancers of cap-dependent translation? The core eukaryotic translation initiation machinery includes several proteins known to have direct RNA-binding capacity, which viruses exploit. The IRESs of poliovirus and EMCV bind specifically to eIF4G; the HCV IRES binds eIF3 as well as the 40 S subunit; and the CrPV IRES binds ribosomes directly (27). It is unlikely to be true that all cellular 5′-UTRs have identical affinities for all factors. Indeed, genome-wide studies investigating the consequences of reducing the activity of the “core” cap-dependent initiation factor eIF4G reveal striking gene-specific consequences rather than uniform reduction of translation of most mRNAs (32, 33). This is analogous to recent studies in the splicing field, in which reductions in different core components of the splicing machinery were shown to cause intron-specific effects in both yeast and metazoans (34, 35). One could argue that certain mRNA transcripts are inherently poor substrates for the splicing or translation machineries and are therefore sensitized to partial loss-of-function conditions. This “sensitive substrate” model does not adequately explain why genes would respond differentially to reductions of some but not other core components, if one envisions a single pathway taken by all genes. An alternative model proposes that specific interactions between individual 5′-UTRs and RNA-binding translation factors contribute significantly to the efficiency of the translation of some genes.
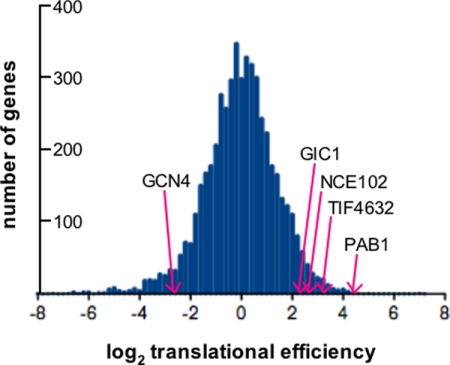
Even under growth conditions in which “standard” cap-dependent translation is presumed to predominate, the translational efficiencies of yeast genes vary by 2 orders of magnitude (36). This surprising conclusion was reached using an elegant new method developed by Ingolia, Weissman, and colleagues. Their method, ribosome footprint profiling, permits quantitative measurement in vivo of translational efficiency genome-wide and is capable of confidently distinguishing small changes in translation over a large dynamic range. We have repeated these experiments in our laboratory and seen similar results.3 Even if one allows for 5-fold differences in translational efficiency to be ignored, which is generous given that the replicate error of measurement is <1.5-fold, there is still a lot of variation to be explained, as illustrated in Fig. 2. For the ∼200 genes that are translated with very low efficiency (lower than GCN4, which is known to be poorly translated in rich media), one could argue that each of these mRNAs is burdened with a “bad” 5′-UTR. But how shall we explain the hundreds of genes that are translated much more efficiently than the average yeast gene? Although it remains to be seen how much of this variability is due to differences in 5′-UTR features, translation studies of putative cellular IRESs may be informative.
FIGURE 2.
Genome-wide measurements reveal large differences in translational efficiency under conditions in which the canonical cap-dependent initiation mechanism is presumed to predominate. Translational efficiency is determined by comparing the amount of ribosome-associated mRNA with the total pool of mRNA for each gene. The data are normalized such that the median translational efficiency is equal to 1 (log2 = 0). Some genes with unusual translational efficiencies are highlighted and discussed in the text. The data are for wild-type yeast grown in rich media (36).
Whereas most mechanistic studies of cap-dependent initiation have focused on a very limited collection of mRNA substrates, the cellular IRES field has investigated the molecular requirements for translation of a more diverse group of 5′-UTR constructs. Assays for cellular IRES activity artificially force 5′-UTRs to rely exclusively on internal ribosome entry, thereby revealing contributions to translation that do not require recognition of the m7G cap by eIF4E. Most yeast cellular IRESs characterized to date show activity that is strongly affected by the level of eIF4G (19). Some of these eIF4G-dependent IRES-containing genes are among the yeast genes that are very efficiently translated in rapidly dividing cells: PAB1, TIF4632, NCE102, and GIC1 (Fig. 2). One attractive hypothesis to explain this observation is that 5′-UTR sequences that are capable of recruiting eIF4G in the absence of eIF4E greatly enhance the efficiency of translation in the presence of eIF4E and an m7G cap. Several mammalian 5′-UTRs that are capable of (relatively inefficient) IRES-dependent initiation are translated with surprising efficiency in a cap-dependent context, given that these 5′-UTRs are quite long and GC-rich (8, 9). The in vitro IRES activity of these 5′-UTRs is strongly dependent on the level of eIF4G (20). eIF4A may also act as an mRNA-specific translational enhancer. Cap-independent translation of mRNAs containing the c-myc or BiP 5′-UTRs was strongly stimulated by increased levels of eIF4A compared with a control m7G-capped reporter containing an artificial 56-nucleotide 5′-UTR (21). Selective recruitment of multiple molecules of eIF4A to certain 5′-UTRs could explain why eIF4A is severalfold more abundant than eIF4E in yeast (37, 38). These results are consistent with the model that specific recruitment of a general initiation factor can lead to enhanced cap-dependent initiation and permit some level of cap-independent initiation, as depicted in Fig. 3. The m7G cap may also cooperate with the cellular IRES element to facilitate initiation, e.g. by increasing the local concentration of eIF4F. Thus, a stimulatory effect of the m7G cap on translation does not necessarily reflect the use of a canonical cap-dependent initiation mechanism.
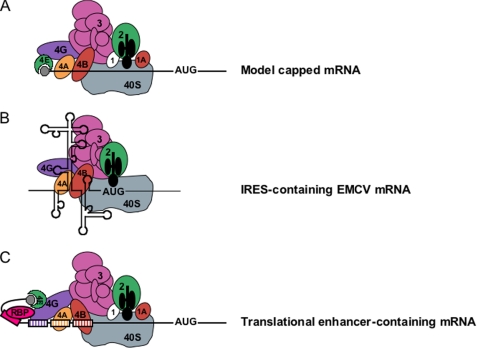
FIGURE 3.
Mechanisms of 5′-UTR-mediated translational enhancement. A, according to the canonical model of cap-dependent eukaryotic ribosome recruitment, the only specific point of contact between the 5′-UTR and the translation machinery is the m7G cap, which is bound by eIF4E. B, other eIFs, including eIF4G, eIF4A, and eIF4B, have RNA-binding activity, which viral IRESs such as EMCV exploit for efficient cap-independent ribosome recruitment. C, cellular 5′-UTRs may also use translational enhancer elements (shown as color-coded boxes along the RNA) to recruit the translation machinery via either cap-stimulated or cap-independent pathways. Sequence-specific RBPs can bridge interactions between 5′-UTR elements and general translation factors.
Cellular 5′-UTRs may also contain translational enhancer elements that recruit dedicated RBPs. Such enhancers of cap-dependent initiation need not be large RNA elements. In the case of splicing enhancers, 6-nucleotide elements are sufficient to stimulate splicing severalfold (39). This likely involves the recognition of the enhancer element by specific RBPs that subsequently bind to and stabilize the association of one or more spliceosome components with the pre-mRNA. Although it is not yet clear how many RBPs might similarly bridge interactions between cellular 5′-UTRs and the translation machinery, even the relatively small yeast genome is predicted to encode >300 RBPs (exclusive of ribosomal proteins), each with a specific set of RNA targets (40). Precedent for this mode of translational enhancement by specific RBPs exists in the form of 5′-UTRs that specifically bind Pab1 to enhance recruitment of eIF4G in yeast and neuronal mRNAs that specifically bind HuD to enhance recruitment of eIF4A in mammals (19, 41). Some of the RBPs proposed to act as IRES trans-activating factors may function similarly. Of course, RBPs might also regulate translation by antagonizing the activity of a translational enhancer element.
Concluding Remarks
Most 5′-UTRs with cellular IRES activity can be efficiently translated by a cap-dependent mechanism, despite the presence of features (unusual length, GC richness, predicted RNA structure, upstream AUG codons) long presumed to inhibit cap-dependent initiation. The significance of this fact is only beginning to be appreciated. In future work, researchers investigating the mechanisms of eukaryotic translation initiation, whether cap-dependent or IRES-dependent, will need to account for the surprising range of in vivo translational efficiencies revealed by new high-throughput methods. It seems likely that many molecular connections linking specific 5′-UTR sequences, RBPs, and the translation machinery remain to be discovered.
Acknowledgments
I thank members of the Gilbert laboratory for critical reading of the manuscript and helpful comments, as well as Dr. Chris Burge for thoughtful discussions regarding the parallels between splicing and translation.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R00GM081399-03. This is the fifth article in the Thematic Minireview Series on Protein Synthesis. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
M. K. Thompson and W. Gilbert, unpublished data.
- IRES
- internal ribosome entry site
- EMCV
- encephalomyocarditis virus
- HCV
- hepatitis C virus
- CrPV
- cricket paralysis virus
- RBP
- RNA-binding protein.
REFERENCES
- 1.Jang S. K., Kräusslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., Wimmer E. (1988) J. Virol. 62, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelletier J., Kaplan G., Racaniello V. R., Sonenberg N. (1988) Mol. Cell. Biol. 8, 1103–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johannes G., Sarnow P. (1998) RNA 4, 1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozak M. (1988) Mol. Cell. Biol. 8, 2737–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M. (1989) J. Cell Biol. 108, 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sagliocco F. A., Vega Laso M. R., Zhu D., Tuite M. F., McCarthy J. E., Brown A. J. (1993) J. Biol. Chem. 268, 26522–26530 [PubMed] [Google Scholar]
- 7.Doudna J. A., Sarnow P. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. B. eds.) pp. 129–153, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 8.Andreev D. E., Dmitriev S. E., Terenin I. M., Prassolov V. S., Merrick W. C., Shatsky I. N. (2009) Nucleic Acids Res. 37, 6135–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bert A. G., Grépin R., Vadas M. A., Goodall G. J. (2006) RNA 12, 1074–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuesta R., Martínez-Sánchez A., Gebauer F. (2009) Mol. Cell. Biol. 29, 2841–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elango N., Li Y., Shivshankar P., Katz M. S. (2006) J. Cell. Biochem. 99, 1108–1121 [DOI] [PubMed] [Google Scholar]
- 12.Han B., Zhang J. T. (2002) Mol. Cell. Biol. 22, 7372–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang B. L., Dornbach L. M., Lyons K. M. (2007) J. Cell Commun. Signal. 1, 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saffran H. A., Smiley J. R. (2009) RNA 15, 1980–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Weaver M., Magnuson N. S. (2005) Nucleic Acids Res. 33, 2248–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young R. M., Wang S. J., Gordan J. D., Ji X., Liebhaber S. A., Simon M. C. (2008) J. Biol. Chem. 283, 16309–16319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landry D. M., Hertz M. I., Thompson S. R. (2009) Genes Dev. 23, 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reineke L. C., Komar A. A., Caprara M. G., Merrick W. C. (2008) J. Biol. Chem. 283, 19011–19025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert W. V., Zhou K., Butler T. K., Doudna J. A. (2007) Science 317, 1224–1227 [DOI] [PubMed] [Google Scholar]
- 20.Hundsdoerfer P., Thoma C., Hentze M. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thoma C., Bergamini G., Galy B., Hundsdoerfer P., Hentze M. W. (2004) Mol. Cell 15, 925–935 [DOI] [PubMed] [Google Scholar]
- 22.Komar A. A., Lesnik T., Cullin C., Merrick W. C., Trachsel H., Altmann M. (2003) EMBO J. 22, 1199–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., Morishita S., Ito T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17846–17851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allam H., Ali N. (2010) J. Biol. Chem. 285, 5713–5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashe M. P., De Long S. K., Sachs A. B. (2000) Mol. Biol. Cell 11, 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes L. E., Campbell S. G., De Long S. K., Sachs A. B., Ashe M. P. (2004) Mol. Cell. Biol. 24, 2998–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filbin M. E., Kieft J. S. (2009) Curr. Opin. Struct. Biol. 19, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reineke L. C., Merrick W. C. (2009) RNA 15, 2264–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon A., Peng G., Brandenburger Y., Brandenburg Y., Zollo O., Xu W., Rego E., Ruggero D. (2006) Science 312, 902–906 [DOI] [PubMed] [Google Scholar]
- 30.Fontoura B. M., Faria P. A., Nussenzveig D. R. (2005) IUBMB Life 57, 65–72 [DOI] [PubMed] [Google Scholar]
- 31.Hentze M. W., Gebauer F., Preiss T. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. B. eds.) pp. 269–295, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32.Clarkson B. K., Gilbert W. V., Doudna J. A. (2010) PLoS ONE 5, e9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramírez-Valle F., Braunstein S., Zavadil J., Formenti S. C., Schneider R. J. (2008) J. Cell Biol. 181, 293–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park J. W., Parisky K., Celotto A. M., Reenan R. A., Graveley B. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15974–15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pleiss J. A., Whitworth G. B., Bergkessel M., Guthrie C. (2007) PLoS Biol. 5, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009) Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 38.von der Haar T., McCarthy J. E. (2002) Mol. Microbiol. 46, 531–544 [DOI] [PubMed] [Google Scholar]
- 39.Fairbrother W. G., Yeh R. F., Sharp P. A., Burge C. B. (2002) Science 297, 1007–1013 [DOI] [PubMed] [Google Scholar]
- 40.Hogan D. J., Riordan D. P., Gerber A. P., Herschlag D., Brown P. O. (2008) PLoS Biol. 6, e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukao A., Sasano Y., Imataka H., Inoue K., Sakamoto H., Sonenberg N., Thoma C., Fujiwara T. (2009) Mol. Cell 36, 1007–1017 [DOI] [PubMed] [Google Scholar]