Abstract
The DnaX complex (DnaX3δδ′χψ) within the Escherichia coli DNA polymerase III holoenzyme serves to load the dimeric sliding clamp processivity factor, β2, onto DNA. The complex contains three DnaX subunits, which occur in two forms: τ and the shorter γ, produced by translational frameshifting. Ten forms of E. coli DnaX complex containing all possible combinations of wild-type or a Walker A motif K51E variant τ or γ have been reconstituted and rigorously purified. DnaX complexes containing three DnaX K51E subunits do not bind ATP. Comparison of their ability to support formation of initiation complexes, as measured by processive replication by the DNA polymerase III holoenzyme, indicates a minimal requirement for one ATP-binding DnaX subunit. DnaX complexes containing two mutant DnaX subunits support DNA synthesis at about two-thirds the level of their wild-type counterparts. β2 binding (determined functionally) is diminished 12–30-fold for DnaX complexes containing two K51E subunits, suggesting that multiple ATPs must be bound to place the DnaX complex into a conformation with maximal affinity for β2. DNA synthesis activity can be restored by increased concentrations of β2. In contrast, severe defects in ATP hydrolysis are observed upon introduction of a single K51E DnaX subunit. Thus, ATP binding, hydrolysis, and the ability to form initiation complexes are not tightly coupled. These results suggest that although ATP hydrolysis likely enhances β2 loading, it is not absolutely required in a mechanistic sense for formation of functional initiation complexes.
Keywords: ATPases, DNA Polymerase, DNA Replication, Enzyme Mechanisms, Nucleic Acid Enzymology, DNA Polymerase III Holoenzyme, DnaX Complex, Replicase, Walker A Motif, Clamp Loader
Introduction
DNA polymerase III holoenzyme (pol III HE)2 exhibits features common to all other cellular replicases. It is a tripartite assembly composed of a replicative polymerase (pol III), a sliding clamp processivity factor (β2), and an AAA+ ATPase that assembles the β2 clamp around DNA (DnaX complex) (1, 2). The DnaX complex contains three DnaX subunits and one each of δ, δ′, χ, and ψ. The DnaX subunit is either the full-length dnaX translation product τ or a shorter version, γ, which is generated by translational frameshifting. γ lacks two C-terminal domains that bind the replicative helicase and pol III.
Current models for clamp loading, largely derived from γ-only DnaX complexes in the absence of pol III, propose that ATP binds to all three DnaX subunits, increasing the affinity of the clamp loader for β2 and primed DNA (1, 3). Once a β2-primed DNA complex is formed, hydrolysis of the three ATPs places the DnaX complex into a conformation with decreased affinity for DNA, leading to its dissociation from DNA-bound β2 (3). pol III then associates with β2 in a downstream step, leading to formation of an initiation complex for processive replication.
We have shown that some of the characteristics of initiation complex formation catalyzed by τ-containing DnaX complexes are different from γ-only complexes. For example, τ complex, when bound to pol III, can form initiation complexes in the leading strand half of a dimeric replicase in the presence of the nonhydrolyzed analog, ATPγS (4). In addition, τ-containing DnaX complexes chaperone pol III onto the newly loaded β2, reducing the required levels of pol III and enabling initiation rates sufficient to support Okazaki fragment synthesis under physiological conditions (5). This chaperoning reaction is required for ATPγS-dependent initiation complex formation. A model was presented in which DnaX complex-ATPγS-β2 binds primed DNA, with a small percentage of β2 closing around DNA spontaneously in equilibrium with a mostly open β2 population. pol III was proposed to attach to the closed β2 and displace DnaX from β2 and the DNA in a reaction that does not necessarily require nucleotide hydrolysis. These observations suggest that initiation complex formation reactions catalyzed by τ-containing DnaX complex might have different ATP binding and hydrolysis requirements than a β2 loading reaction that occurs in the absence of pol III. To investigate this possibility, we constructed a series of DnaX complexes containing varying numbers of DnaX subunits competent to bind and hydrolyze ATP. Although the fully wild-type complexes are the most active, we demonstrate that only one DnaX protomer with an active ATP-binding site is required to support initiation complex formation.
EXPERIMENTAL PROCEDURES
Proteins
Wild-type γ and τ were purified as described previously (6). Specific activity was 2.1 × 108 and 5.2 × 106 units/mg, respectively. δ and δ′ were purified (7) at the specific activity of 2.2 × 108 and 2.3 × 108 units/mg, respectively. χ and ψ were purified as described (8). The τγ2 and τ2γ wild-type complexes were purified (9) with a specific activity of 1.1 × 107 and 8.4 × 106 units/mg, respectively. pol III (specific activity 7.6 × 106 units/mg) was purified as described (10). A DNA synthesis unit was defined as 1 pmol of dNTP incorporation/min assayed as described previously (8).
Buffers
Buffer A is 50 mm Tris-HCl (pH 7.5), 20% glycerol, 1 mm EDTA, 0.1 m NaCl, 5 mm DTT added fresh. Buffer S is 25 mm HEPES (pH 7.5), 100 mm NaCl, 10% glycerol, 0.5 mm EDTA. Buffer T/20 is 20 mm Tris-HCl (pH 7.5), 20% glycerol, 5 mm DTT. ATPase reaction buffer is 50 mm HEPES (pH 7.5), 5% glycerol, 0.025% Nonidet P-40, 10 mm magnesium acetate.
Protein Determinations
Protein concentrations were determined by the method of Bradford using the Coomassie Plus Bradford assay reagent (Pierce) according to the manufacturer's instructions (bovine serum albumin as a standard).
Expression and Purification of Mutant DnaX Proteins
The plasmid pETdnaX.1-M2 expressing mutant γ and τ was constructed by site-directed mutagenesis of pET-dnaX.1 to change Lys-51 to Glu (AAA to GAA). pET-dnaX.1 was derived from an earlier DnaX overproducer (pBBMD11) (6). The TTG immediately preceding the initiating ATG was changed to a CAT to avoid occasional premature initiation detected by mass spectrometry,3 and a 2250-bp XbaI-HindIII fragment was moved into plasmid pET29b. pET-dnaX.1-M2 was transferred into Escherichia coli strain BL21 (DE3). Cells were grown overnight at 37 °C in a fermentor (12.5 liters) in F medium lacking added glucose (7) but containing kanamycin at 35 mg/liter. Overproduction was performed at 37 °C in an 180-liter fermentor in F medium with kanamycin added at 10 mg/ml and glucose at 1%. Cells were induced with IPTG at A600 = 1.0 and harvested at A600 = 3.8. Cells (900 g) were lysed as described (11) with one backwash in 0.125× Fraction I volume of Buffer A with 0.2 g of ammonium sulfate added to each ml. SP-Sepharose chromatography was used to purify both γ and τ as described (6). Elution from the SP-Sepharose column gave two peaks; the first peak was γ (94 mg), and the second peak was τ (40 mg). The subunits were further individually purified by gel filtration on an FPLC Superose-12 column equilibrated in Buffer S. The proteins were eluted as single peaks and were identified by 4–20% SDS-PAGE. Only one-half of the γ peak was loaded onto the column, yielding 25 mg of the purified protein. The purification yield for the whole τ peak was 17.5 mg.
Reconstitution of DnaX Complexes Containing Mixtures of Wild-type and K51E DnaX Protomers and Purification of the Resulting Complexes
Complexes were prepared by mixing K51E τ or γ with wild-type γ or τ, respectively, at a molar ratio of τ:γ 2:1 and incubation for 1 h at 15 °C followed by the addition of δ-δ′ and χψ at the ratio of 1:1:1:1, premixed for 1 min as described (9). Proteins were used as follows: 128 nmol of τ K51E (1.8 ml, 5 mg/ml) or 128 nmol of wild-type τ (0.9 ml, 10 mg/ml), 64 nmol of γ K51E (0.27 ml, 11.3 mg/ml) or 64 nmol of wild-type γ (0.4 ml, 7.4 mg/ml), 64 nmol of δ (0.51 ml, 4.8 mg/ml), 64 nmol of δ′ (0.11 ml, 20.7 mg/ml), 64 nmol of χψ (0.1 ml, 20 mg/ml). Complexes, (1.9–3.5 ml and 16–19 mg), were diluted (up to 32 ml) to a conductivity of buffer T/20 + 20 mm NaCl and loaded onto an 8-ml FPLC Mono-S column equilibrated with buffer T/20 + 20 mm NaCl. After loading and washing the column, separation was achieved by using a two-step gradient: a 6-column volume gradient of 0.02–0.135 m NaCl followed by a 20-column volume gradient of 0.135–0.2 m NaCl. γ3 complex elutes during the first step, and the second step separates the remaining three complexes in the following order: γ2τ1, τ2γ1, and τ3. The column was run at a flow rate of 0.5 ml/min; 0.3-ml fractions were collected. Fractions containing at least one-half peak height of absorbance were pooled, and their components were confirmed by SDS-PAGE.
DnaX Complex Activity Assay
The specific activity of the DnaX complexes was determined using a holoenzyme reconstitution assay as described (8). The assay contained 40 nm pol III, 10 nm β2, 540 pmol M13Gori (as nucleotide), 0.9 μm SSB4, and 60 units of DnaG primase. The unit definition for DnaX complex is defined as the amount of complex required to catalyze the incorporation of 1 pmol of total deoxyribonucleotide per min at 30 °C.
Titration of β and DnaX
Titrations of β and DnaX were performed using the pol III HE reconstitution assay (7) with the following differences. The assay was performed in the absence of primase. Instead, M13Gori single-stranded circular DNA (425 pmol of total nucleotide/assay) primed with a 30-mer oligonucleotide (3′-TCTAATGAGAACTACTTCCAGTCGGTCGGA-5′) was used. The primer was annealed to M13Gori by mixing a 1:1 molar ratio of primer to template in the presence of 0.1 m NaCl and 10 mm HEPES (pH 7.5) and then heating to 75 °C and slowly cooling to room temperature. The assay contained 40 nm pol III, 0.9 μm SSB4, and 200 μm ATP. In β titrations, DnaX complex was set at 0.8 nm (sufficient to be at the upper end of the linear range). In DnaX complex titrations, for complexes containing two or three γs (see Fig. 2A), β2 was used at 210 nm (6 × K0.5 determined for the weakest binding complex (τγm2),4 see Table 1). For complexes containing two or three τs, β2 was set at 140 nm, (9 × K0.5 determined for the weakest binding complex (τm2γ), see Table 1).
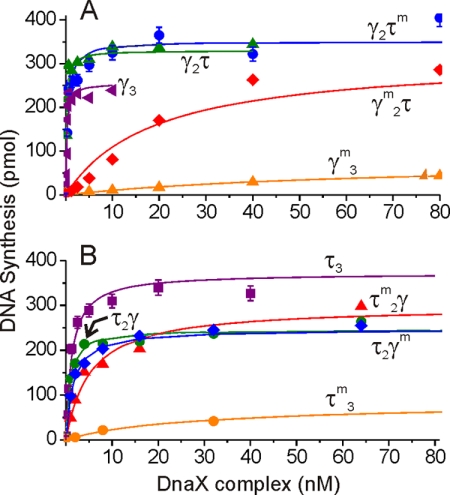
FIGURE 2.
DnaX complexes containing at least one DnaX subunit competent for ATP binding support processive DNA replication. DnaX was titrated into replication reactions containing singly primed single-stranded circular templates as described under “Experimental Procedures.” A, titration curves for a series of DnaX complexes containing two or three γ subunits. B, titration curves for a series of DnaX complexes containing two or three τ subunits. Error bars indicate S.E.
TABLE 1.
Properties of DnaX complexes with various combinations of wild-type and K51E DnaX protomers
| DnaX Complex | No. of active DnaX | β2K0.5 | Specific activity of replicase | ATPase (units/pmol of DnaXcx)a |
|---|---|---|---|---|
| nm | units/mg × 106 | |||
| τ3 | 3 | 1.5 | 4.1 | 520 |
| γ3 | 3 | 1.2 | 96 | 400 |
| τ2γ | 3 | 0.4 | 1.9 | 470 |
| τγ2 | 3 | 1.4 | 24 | 230 |
| τ2γm | 2 | 11 | 1.5 | 21 |
| τmγ2 | 2 | 6.6 | 13 | 7.9 |
| τm2γ | 1 | 15 | 1.0 | 3.3 |
| τγm2 | 1 | 37 | 1.2 | 4.4 |
| τm3 | 0 | NDb | 0.04 | −1.2 |
| γm3 | 0 | NDb | 0.03 | 0.2 |
a Unit = pmol of ATP hydrolyzed/min. Assays represent the average of three or more assays.
b ND, not detectable.
Estimation of K0.5 for the β Interaction with DnaX
To determine from our functional assay an apparent dissociation constant for the interaction between β and DnaX complexes, Prizm software was used to perform nonlinear regression analysis. Hyperbolic data obtained from titration experiments were fit with the following equation: Y = Ymax × X/(X + K0.5), where Y represents the amount of DNA synthesis (pmol of nucleotides) at a given concentration of the protein being titrated, X. The two constants that are obtained from the nonlinear regression are K0.5 and Ymax, the latter of which is the plateau amount of DNA synthesis reached at high concentrations of protein X.
ATPase Assay
This assay was based on a modification of a published procedure (12). The DNA template (50-mer/70-mer complementary synthetic oligonucleotides) was mixed with DnaX complex and β (20 μl) in ATPase reaction buffer. The reaction was initiated by adding 5 μl of ATP dissolved in ATPase assay buffer and incubating at 37 °C. The incubation time varied from 5 to 60 min as required to keep the response in a range that was proportional to the amount of DnaX complex added. After incubation, 65 μl of 27.5 mm ammonium molybdate in 2 m H2SO4 was added followed by 160 μl of water and 50 μl of 0.72 mm malachite green in 0.35% polyvinyl alcohol. Next, 290 μl of the reaction was transferred onto a transparent 96-well microplate. The plate was incubated in the dark for 5 min and read at 645 nm on a SpectraMax Plus microplate spectrophotometer. For assays where the final concentration was above 0.25 mm orthophosphate, dilutions were made with EDTA-containing buffer prior to the addition of molybdate to bring the final reading into photometric linearity. The primer template used as an effector of the ATPase activity was made by annealing a 70-mer (5′-TAA AAA AAA AAA AAA AAA AAG GAT TAC TGG ATC CGA AGG TCA GCC AGC CTA TGC GCC TTC ATC TGA ACA A-3′) with a complementary 50-mer (5′-TTG TTC AGA TGA AGG CGC ATA GGC TGG CTG ACC TTC GGA TCC AGT AAT CC-3′) (6 μm final concentration).
The DnaX complexes were titrated under conditions of saturating DNA, β2, and ATP. For β2 and DNA, saturating conditions were 1 and 0.5 μm, respectively. ATP concentrations were 1.5 and 2 mm for wild-type complexes and complexes containing mutant K51E subunits, respectively. Specific activities (pmol of ATP hydrolyzed per min/pmol of DnaX complex) were determined over ranges where the response was linearly proportional to DnaX complex added (0.4–1.6 pmol/25-μl assay).
RESULTS AND DISCUSSION
Purification of 10 DnaX Complexes Containing Various Combinations of Wild-type and K51E τ and γ Subunits
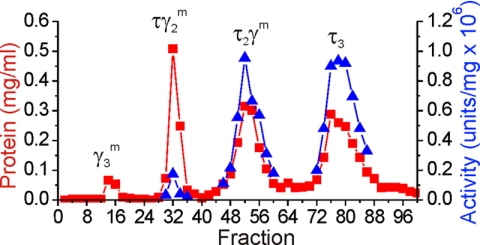
To investigate the role of ATP binding and hydrolysis in the pol III HE reaction, we constructed plasmids carrying dnaX genes that express γ and τ with the critical lysine in the Walker A box mutated to glutamate (K51E). It would be expected that mutation of a positively charged phosphate-binding residue to a negatively charged residue would lead to severe defects in ATP binding and abolish the ability of the affected subunit to hydrolyze ATP. We purified both wild-type and K51E forms of τ and γ and used them to reconstitute DnaX complexes. Because DnaX complexes contain three DnaX subunits and the complexes formed in vitro can contain any proportion of γ or τ subunits, 10 different DnaX complexes resulted (listed in Table 1). Each of the 10 complexes were purified using high resolution Mono S chromatography (Fig. 1). The separation procedure exploits the presence of the basic domain IV present in τ but not in γ, permitting purification of complexes based on their τ content. Rechromatography of the purified complexes was conducted to ensure that they were free of other contaminating DnaX complexes that might confuse analysis (supplemental Fig. 1). This procedure provided the reagents necessary to determine the minimal number of ATP binding/hydrolysis-competent subunits required to support processive DNA replication by the pol III HE.
FIGURE 1.
High resolution chromatographic separation of DnaX complex forms. Mono S chromatography was used to separate DnaX complexes based upon their τ content as described under “Experimental Procedures.” Shown is a separation of DnaX complexes reconstituted from K51E γ and wild-type τ.
DnaX Subunits Containing a Walker A K51E Mutation Do Not Bind ATP
We would expect a mutation of the lysine residue in the Walker A motif, normally involved in phosphate binding and hydrolysis, to lead to charge repulsion and loss of binding when changed to a glutamate residue. To confirm this effect, we performed nitrocellulose filter binding experiments, using [35S]ATPγS. We observed binding to wild-type γ and τ complexes with affinities (1–2 μm) consistent with published results (13). However, the corresponding complexes containing K51E DnaX subunits failed to bind ATPγS (supplemental Fig. 2).
DnaX Complexes Containing Only One DnaX Subunit Competent for ATP Binding Support Activity of Reconstituted pol III HE
We analyzed each of the 10 DnaX complexes for their ability to support processive DNA replication on singly primed single-stranded circular templates coated with SSB. Titration of DnaX complexes to saturating levels showed at least two-thirds of the maximal level of synthesis by all complexes that contained at least one wild-type DnaX subunit (Fig. 2). Thus, although ATP hydrolysis by multiple subunits might enhance the clamp loading and initiation complex assembly reactions, these results show that hydrolysis of more than one ATP is not essential for basic processive replication by pol III HE.
To calculate a specific activity of each of the purified DnaX complexes, we examined the overall level of DNA synthesis at low DnaX complex concentrations, where the observed activity was proportional to DnaX complex concentration (Table 1). Complexes containing a single K51E DnaX subunit (and two fully active DnaX subunits) exhibited 50–80% of the activity of their all wild-type counterparts. Complexes having only one active DnaX subunit show a lower specific activity than complexes containing two active DnaX proteins. The DnaX complex containing two K51E τ subunits (τm2γδδχψ) exhibits 2-fold lower activity than its wild-type counterpart (τ2γδδχψ). A complex containing two K51E γ subunits (τγm2δδ′χψ) suffers an additional 4-fold drop in specific activity (relative to τm2γδδχψ), yet at increased concentrations, it supports nearly the same level of synthesis as wild-type DnaX complexes (Fig. 2A).
Comparison of the activities of all complexes reveals that those that have two or more active γ subunits show a higher level of activity than species that possess two or more active τ subunits (Table 1). The activity of γ3 complex has been routinely observed to have much higher levels of activity in simple SS → RF replicative assays (where SS is single-stranded DNA and RF is duplex replicative form DNA), presumably because it is not part of the final initiation complex and can cycle off to catalyze the formation of additional initiation complexes. τ-Containing complexes remain bound to pol III in initiation complexes and therefore are not available to support the formation of additional initiation complexes (14). Accordingly, in this study, γ3 complex exhibits a 23-fold higher specific activity than τ3 complex. The higher activities of τγ2 complex relative to τ2γ complex and τmγ2 complex relative to τ2γm complex (13- and 9-fold, respectively) may be due to a similar cycling activity.
One ATP Binding-competent DnaX Subunit Is Sufficient to Promote a DnaX Complex State with High Affinity for β2, but Three Are Required for Maximal Affinity
ATP binding to DnaX complex markedly increases its affinity for β2 (15), so β2 was titrated against the 10 forms of DnaX complex to determine whether the absence of ATP-binding sites in some of the DnaX subunits influenced β2 binding. Using our processive replication assay as a functional readout of β2 binding, we determined an apparent Kd (termed K0.5) for binding to β2 for each of the DnaX complexes (Table 1). Complexes composed of entirely wild-type τ and/or γ subunits exhibited K0.5 values of 0.4–1.5 nm, consistent with reported values (15). Complexes containing only two wild-type subunits bound β2 less tightly (K0.5 = 7–11 nm), and complexes having only one wild-type subunit bound β2 even more weakly (K0.5 = 15–37 nm).
Together, these results are consistent with the model that ATP binding to the DnaX complex favors a conformation with high affinity for β2. Our results show that binding of a single ATP is sufficient to significantly stabilize a high affinity conformation because the observed K0.5 of ≤37 nm is significantly less than the KD of 135 nm for DnaX complex binding to β2 in the absence of ATP (15). However, all three ATP-binding sites are required for the highest β2 affinity, even at high ATP concentrations. This argues that in addition to a global conformational change, presumably exposing the β2-binding site of δ as proposed (13), there are additional, possibly local, conformational changes. These local changes might be promoted by binding of the second and third ATPs that increase β2 binding, presumably through additional interactions within the DnaX complex, as proposed (16).
A Lys → Glu change in the Walker A box could drastically decrease ATP binding but not completely ablate it. Thus, we performed ATP titrations under two conditions to determine whether high concentrations of ATP resulted in the allosteric effects of ATP binding that would normally occur at low μm concentrations. In the first approach, performed with a representative subset of DnaX complexes, the concentration of the DnaX complex was set at the level that gave 50% maximal synthesis. This would permit detection of a stimulation of DnaX replication activity as ATP concentration was increased. Varying ATP up to 1 mm did not result in any additional stimulation of activity (supplemental Fig. 3A).
In a second set of experiments, β2 was set at the K0.5 determined for each DnaX complex (Table 1), ATP was varied, and processive elongation was monitored. This approach would permit an increased affinity for β2 at high ATP concentrations to be detected. Again, varying ATP up to 1 mm did not result in any further stimulation of the pol III HE reaction (supplemental Fig. 3B).
DnaX Complexes Containing a Single K51E DnaX Subunit Are Severely Deficient in ATP Hydrolysis
In contrast to the high levels of activity observed for complexes containing a mixture of wild-type and K51E DnaX subunits, K51E-containing complexes are severely deficient in ATPase activity when assayed under similar conditions (Table 1). Inclusion of a single K51E subunit reduces hydrolytic activity 28-fold, on average, relative to the average activity of the wild-type DnaX complexes. Inclusion of two K51E DnaX subunits drops activity further, reducing it to near background (100-fold reduction relative to wild-type complexes).
Our results demonstrate that binding of a single ATP can drive β2 loading onto DNA and the formation of processive replication complexes. These findings are inconsistent with models where a wave of hydrolysis by three coupled ATPases is required for β2 loading (17). Our observations do not preclude multisite hydrolysis affording significant kinetic or other advantages, but they do demonstrate that this is not essential for initiation in vitro. Thus, ATP hydrolysis enhances β2 loading but is not an intrinsically fundamental part of the β2 loading process.
The ATP hydrolysis defects caused by inclusion of one or two K51E subunits within a DnaX complex are much more severe than defects for initiation complex formation and ensuing processive replication. This difference in effects is consistent with other observations that ATP binding is more important than ATP hydrolysis in initiation complex formation. ATPγS, which is not hydrolyzed by the DnaX complex, supports the initiation complex formation provided that the τ form of DnaX is present (4, 5). Recently, we have demonstrated that τ3 complexes chaperone the associated pol III onto a newly loaded β2 with the kinetics and affinity required to support rapid sequential Okazaki fragment synthesis (5). That study supported a model in which pol III, if bound to τ, could participate in initiation complex formation in the presence of ATPγS by attacking transiently loaded β2 and driving initiation complex formation to completion. This suggests that the pathway taken for β2 loading could be different in the presence and absence of polymerase.
This study demonstrates that only one active, ATP-binding DnaX subunit must be present within a DnaX complex to support efficient initiation complex formation on the timescale of our assay. The fact that a drastic drop in ATPase activity in K51E DnaX-containing complexes greatly exceeds the modest drop in replication activity, coupled with previous observations that the nonhydrolyzed ATP analog ATPγS supports initiation complex formation catalyzed by τ-containing DnaX complexes, suggests that the presence of a binding site for an allosteric effector (ATP) is more important than hydrolysis per se. Our findings do not diminish a potential critical role for ATP hydrolysis to accelerate the rate of β loading and initiation complex formation, DnaX complex regulation, or other undiscovered functions that may be essential for cellular replication. ATP hydrolysis could be critical for β2 loading in the absence of pol III bound and positioned for β2 binding by τ, as in the γ3 complexes typically employed in most previous studies of the β2-loading mechanism. However, our results do suggest either that ATP hydrolysis occurs in reaction steps separate from the mechanistically essential (as opposed to rate-enhancing or auxiliary) steps for β2 loading or that the hydrolysis steps can be bypassed through alternative pathways.
Our results also impact previously proposed models for ordered ATP hydrolysis during β2 loading. Based on ablation of ATPase activity in arginine finger mutants that are part of the γ ATPase sites 2 and 3, but not γ site 1 that is provided by δ′, it was proposed that ATP hydrolysis must occur first in site 3 and then site 2 before hydrolysis occurs in site 1 (17). In our DnaX complexes containing a single K51E DnaX protomer, two-thirds of the complexes should have active DnaX subunits in positions 2 and 3. The 16-fold or greater drop in ATPase activity that we observe for single mutants is not consistent with the expected 33% reduction in the single mutant. More likely, a conformation with all three sites occupied by ATP promotes hydrolysis most effectively.
Our result that initiation complex formation does not require hydrolysis by each ATPase subunit is analogous to previous results for the bacteriophage T4 gp44/62 clamp loader (18). In that work, it was argued that ATPase activity observed in simple model clamp loading reactions may not reflect the true activity required for formation of authentic, full processive replication complexes. It was proposed that in authentic T4 replication, initiation complex formation requires hydrolysis of as few as one ATP to destabilize the clamp loader/loaded clamp interaction and enable return to a ground state conformation with decreased affinity for primed DNA. In the T4 system, like our E. coli system, full occupancy of all ATP-binding sites was required for maximal affinity for the clamp protein.
Supplementary Material
Acknowledgments
We thank Diane Hager for figure preparation and Melissa Stauffer, Ph.D., of Scientific Editing Solutions, for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM060273 from the NIGMS (to C. S. M.) and by National Institutes of Health Grant F32GM084697 (postdoctoral fellowship for C. D. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
A. Wieczorek, J. A. Thompson, and C. S. McHenry, unpublished data.
In τ2γm, the superscript m designates a DnaX subunit containing the K51E mutation.
- pol III HE
- DNA polymerase III holoenzyme
- pol III
- DNA polymerase III
- SSB
- single-stranded DNA-binding protein
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1.Indiani C., O'Donnell M. (2006) Nat. Rev. Mol. Cell. Biol. 7, 751–761 [DOI] [PubMed] [Google Scholar]
- 2.McHenry C. S. (2003) Mol. Microbiol. 49, 1157–1165 [DOI] [PubMed] [Google Scholar]
- 3.Anderson S. G., Thompson J. A., Paschall C. O., O'Donnell M., Bloom L. B. (2009) Biochemistry 48, 8516–8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glover B. P., McHenry C. S. (2001) Cell 105, 925–934 [DOI] [PubMed] [Google Scholar]
- 5.Downey C. D., McHenry C. S. (2010) Mol. Cell 37, 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dallmann H. G., Thimmig R. L., McHenry C. S. (1995) J. Biol. Chem. 270, 29555–29562 [PubMed] [Google Scholar]
- 7.Song M. S., Pham P. T., Olson M., Carter J. R., Franden M. A., Schaaper R. M., McHenry C. S. (2001) J. Biol. Chem. 276, 35165–35175 [DOI] [PubMed] [Google Scholar]
- 8.Olson M. W., Dallmann H. G., McHenry C. S. (1995) J. Biol. Chem. 270, 29570–29577 [PubMed] [Google Scholar]
- 9.Pritchard A. E., Dallmann H. G., Glover B. P., McHenry C. S. (2000) EMBO J. 19, 6536–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D. R., McHenry C. S. (1996) J. Biol. Chem. 271, 20681–20689 [DOI] [PubMed] [Google Scholar]
- 11.Cull M. G., McHenry C. S. (1995) Methods Enzymol. 262, 22–35 [DOI] [PubMed] [Google Scholar]
- 12.Cogan E. B., Birrell G. B., Griffith O. H. (1999) Anal. Biochem. 271, 29–35 [DOI] [PubMed] [Google Scholar]
- 13.Hingorani M. M., O'Donnell M. E. (1998) J. Biol. Chem. 273, 24550–24563 [DOI] [PubMed] [Google Scholar]
- 14.Kim S., Dallmann H. G., McHenry C. S., Marians K. J. (1996) J. Biol. Chem. 271, 4315–4318 [DOI] [PubMed] [Google Scholar]
- 15.Thompson J. A., Paschall C. O., O'Donnell M., Bloom L. B. (2009) J. Biol. Chem. 284, 32147–32157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simonetta K. R., Kazmirski S. L., Goedken E. R., Cantor A. J., Kelch B. A., McNally R., Seyedin S. N., Makino D. L., O'Donnell M., Kuriyan J. (2009) Cell 137, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson A., O'Donnell M. E. (2003) J. Biol. Chem. 278, 14406–14413 [DOI] [PubMed] [Google Scholar]
- 18.Pietroni P., von Hippel P. H. (2008) J. Biol. Chem. 283, 28338–28353 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.