Abstract
The amount of RNase R, an important degradative exoribonuclease, increases 3–10-fold under a variety of stress conditions. This elevation is due to posttranslational regulation in which the highly unstable RNase R protein becomes stabilized during stress. Here we identify two components of the trans-translation machinery, transfer-messenger RNA (tmRNA) and SmpB, that are responsible for the short half-life of RNase R in exponential phase cells. The absence of either lengthens the half-life of RNase R in vivo >6-fold. SmpB directly interacts with RNase R in vitro and is stimulated by tmRNA. The C-terminal region of RNase R, encompassing its basic region and adjacent S1 domain are required for the interaction; their removal eliminates binding and stabilizes RNase R in vivo. However, the binding of SmpB and tmRNA does not alter RNase R activity. These data define a previously unknown regulatory process in which the stability of an RNase is determined by its interaction with an RNA and an RNA-associated protein.
Keywords: Protein Stability, Protein Turnover, Protein-Protein Interactions, Ribonuclease, RNA-Protein Interaction
Introduction
RNase R is a processive 3′- to 5′-exoribonuclease (1) that in Escherichia coli plays an important role in multiple aspects of RNA metabolism including turnover of mRNA (2, 3) and quality control of stable RNAs (4, 5). The catalytic properties of RNase R are unusual because it is able to digest structured RNAs in the absence of an added RNA helicase (1, 6, 7). As a consequence, RNase R, together with polynucleotide phosphorylase and its associated RNA helicase, are the primary RNases responsible for degradation of structured RNAs. In fact, cells lacking both RNase R and polynucleotide phosphorylase are inviable, and fragments of rRNA and structured mRNAs accumulate in their absence (2, 5, 8).
The amount of RNase R in E. coli increases 3–10-fold during cold shock (9, 10), stationary phase (10, 11), and other stress conditions (10). The molecular mechanisms that underlie these changes in RNase R are of considerable interest inasmuch as little is known about regulation of RNase levels in cells and how this may impact RNA metabolism. In recent studies from our laboratory, it was shown that the elevation of RNase R is largely due to posttranslational regulation (12). RNase R was found to be a highly unstable protein in exponential phase with a half-life of ∼10 min, whereas it is stabilized under stress conditions, leading to its relative elevation (12). However, the factors or processes responsible for the instability of RNase R are not understood.
Here, we show that two components of trans-translation, tmRNA2 and its associated protein, SmpB, are required for the instability of RNase R. In the absence of either, RNase R levels in exponential phase cells increase markedly, and the half-life of RNase R increases from ∼10 min to more than 60 min. We also show that SmpB and tmRNA interact with RNase R both in vitro and in vivo and that the C-terminal region of RNase R is required for this interaction. The half-life of truncated RNase R lacking its C-terminal basic region and S1 domain increases dramatically when compared with that of full-length RNase R. On the other hand, binding of SmpB and tmRNA does not alter RNase R activity. These data indicate that RNase R is subject to a previously unknown mode of regulation requiring components of the trans-translation machinery.
EXPERIMENTAL PROCEDURES
Materials
Antibody against RNase R was prepared from purified protein (1) by Sigma-Genosys and purified using standard procedures (13). Anti-FLAG M2 mAbs and cross-linker dimethyl pimelimidate dihydrochloride (DMP) were from Sigma. His-probe (H3) monoclonal antibody, anti-rabbit, and anti-mouse IgG HRP conjugate were obtained from Santa Cruz Biotechnology. [γ-32P]ATP was purchased from PerkinElmer Life Sciences. RNeasy mini kit was from Qiagen. Plasmid pET15b and Ni-NTA His-bind resin were from Novagen. Protein A-agarose beads and protease inhibitor mixture were purchased from Calbiochem. Purified full-length RNase R and RNase RΔS1 were described previously (7). Purified RNase RΔBasic was a gift from Dr. Arun Malhotra (University of Miami).
Bacterial Strains and Growth Conditions
E. coli K12 strain MG1655(Seq)rph+ and its derivative lacking SmpB were from Dr. Kenneth Rudd, University of Miami (14). The ssrA insertion mutant deficient in tmRNA was a kind gift from Dr. David Friedman, University of Michigan (15). It was transduced into strain MG1655(Seq)rph+ by the phage P1vir. His6 and 2×FLAG sequences were fused to the N termini of chromosomal RNase R and SmpB, respectively, by recombineering (16), using primers H1 and H2 and F1 and F2 (supplemental Table S1). Recombinants were selected on LB-kanamycin plates and confirmed by PCR. Kanamycin resistance cassettes were removed (17), and the resulting gene fusions were confirmed by DNA sequencing. The basic region and S1 domain of RNase R were also removed by recombineering using primers B1 and B2 and R1 and B2 (supplemental Table S1), respectively.
Cells were grown in YT medium. Antibiotics were at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 34 μg/ml. Cells were grown at 37 °C and collected at ∼0.3 A550.
Cross-linking of RNase R
Cells were harvested and ruptured in 20 mm NaH2PO4, pH 8.0, 150 mm NaCl, 1 mm dithiothreitol (DTT), 1 mm phenylmethylsulfonyl fluoride (PMSF) by three passes through an Aminco French press at 20,000 p.s.i. After centrifugation at 12,000 × g for 15 min, the supernatant fraction was collected, and 2 μg of soluble proteins were incubated in the presence or absence of 10 mm DMP for 30 min at room temperature. The reactions were stopped by 50 mm Tris-HCl (pH 7.5) and analyzed by immunoblotting.
Co-immunoprecipitation of RNase R
Cells were disrupted as before in lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm DTT, 0.5% Nonidet P-40, 1 mm PMSF) containing protease inhibitor mixture. One mg of soluble protein was incubated in the presence or absence of 2 μg of purified RNase R antibody at 4 °C for 2 h. Protein A-agarose beads were added, and the mixture was incubated for 1 h. Beads were washed five times with 500 μl of lysis buffer, and bound proteins were eluted with 1× SDS-PAGE sample buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 2% 2-mercaptoethanol, 0.01% bromphenol blue).
Pulldown of RNase R from Extracts
MG1655(Seq)rph+ (FLAG-SmpB, His-RNase R) cells were disrupted as before in 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 10 mm imidazole, 0.5% Nonidet P-40, 1 mm PMSF containing protease inhibitor mixture. The lysates were centrifuged at 15,000 × g for 30 min, and the resulting extracts were loaded on a Ni-NTA-agarose column. After washing with 5 volumes of 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 20 mm imidazole, His-RNase R, and its interacting factors were eluted with 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 250 mm imidazole. Associated proteins and RNAs were analyzed by Western blotting and Northern blotting, respectively.
Overexpression and Purification of SmpB and tmRNA
The tandem smpB and ssrA genes (18) were amplified by PCR with primers S1 and S2 (supplemental Table S1). The PCR product was purified and digested with NdeI and BamHI and cloned into pET15b. His-SmpB and tmRNA were overexpressed and purified (19). This procedure yields His-SmpB free of tmRNA.
Pulldown of Purified Components
His-SmpB (0.2 nm) was mixed with 0.1 nm full-length or truncated RNase R protein in the presence or absence of 0.2 nm tmRNA in 500 μl of binding buffer (50 mm Tris-Cl, pH 7.6, 100 mm NaCl, 20 mm imidazole). Mixtures were gently rocked for 60 min at 4 °C, 50 μl of Ni-NTA resin were added, and the mixture was incubated for 30 min at 4 °C. The Ni-NTA resin was recovered by centrifugation, washed five times with 300 μl of binding buffer, and eluted with 50 μl of binding buffer containing 200 mm imidazole. Eluted proteins were separated by SDS-PAGE on a 12% gel and probed with RNase R antibody and His-probe monoclonal antibody.
Western Blot Analysis
Proteins were resolved on 8 or 12% gels (for detection of RNase R) or 12% SDS-PAGE (for detection of SmpB) and subjected to immunoblotting. RNase R, recombinant His-RNase R, recombinant FLAG-SmpB, and recombinant His-SmpB were detected by purified RNase R antibody (1:10,000 dilution), His-probe monoclonal antibody (H3) (1:1000 dilution), anti-FLAG M2 mAbs (1:1000 dilution), and His-probe (H3) monoclonal antibody (1:1000 dilution), respectively. Underexposed films were used for quantitation by Quantity One (Bio-Rad).
Northern Blot Analysis
RNA was extracted with an RNeasy mini kit according to the manufacturer's protocol and electrophoresed on a 6% polyacrylamide/7.5 m urea gel. Prehybridization and hybridization with a probe complementary to tmRNA was as described previously (20).
Measurement of RNase R Half-life
Cells were grown under normal conditions to an A550 of ∼0.3. A portion of cells was collected at zero time, and chloramphenicol was added to the remaining culture at 200 μg/ml. Cells were collected at the indicated times, lysed by sonication (10), and assayed by immunoblotting to determine RNase R.
RESULTS
Size Analysis of RNase R
One possibility to explain the instability of RNase R during exponential phase is that it is associated with a destabilizing protein, such as has been observed for RpoS (21). As an initial, simple test of this idea, the size of RNase R was determined by gel filtration. Interestingly, RNase R migrated as an ∼150-kDa protein in extracts (data not shown), although based on its amino acid sequence, the calculated molecular mass of RNase R should be ∼95 kDa (1). These results suggested either that RNase R is an elongated protein with a larger Stokes radius than that expected for its molecular mass or that it is associated with other components in a cell extract.
Cross-linking of RNase R
To ascertain whether RNase R might be associated with another protein, extracts were incubated with the cross-linking agent, DMP, and the total samples were analyzed by SDS-PAGE and immunoblotting (Fig. 1A). As can be seen, when compared with the control sample, a new, prominent band of ∼115 kDa appeared in extracts cross-linked with DMP. These data suggest that RNase R is associated with a protein of ∼20 kDa. Moreover, based on the amount of cross-linking, which generally is not efficient, a considerable portion of RNase R appears to be in complexed form.
FIGURE 1.
Association of RNase R with SmpB. A, chemical cross-linking of soluble proteins in extracts from strain MG1655(Seq)rph+. Samples were treated without (−) or with (+) DMP and analyzed as described under “Experimental Procedures.” The position of RNase R is indicated, and * indicates a band of about 115 kDa. The migration positions of the indicated molecular mass standards are shown on the left. This gel was overexposed to observe the cross-linked bands. B, co-immunoprecipitation of extracts prepared from MG1655(Seq)rph+ (FLAG-SmpB, His-RNase R) cells. Extracts were treated without (−) or with (+) RNase R antibody, and the resulting immunoprecipitates were analyzed by SDS-PAGE by probing with His-probe monoclonal antibody or anti-FLAG monoclonal antibody. C, pulldown of FLAG-SmpB and tmRNA with His-RNase R. Extracts from MG1655(Seq)rph+ (FLAG-SmpB, His-RNase R) and smpB derivative cells were treated with Ni-NTA, and the presence of His-RNase R, FLAG-SmpB, and tmRNA in the eluant was detected by His-probe monoclonal antibody, anti-FLAG monoclonal antibody, or tmRNA-specific probe, respectively. D, pulldown of RNase R with His-SmpB. Purified RNase R, His-SmpB, and tmRNA were prepared and used for a pulldown assay as described under “Experimental Procedures.” The proteins and tmRNA present in each assay are indicated above the lanes. The presence of RNase R protein and recombinant His-SmpB protein was detected as above.
RNase R Interacts with SmpB and tmRNA
It has been reported that a small amount of RNase R is present in a large complex with tmRNA and its associated protein, SmpB (18). Inasmuch as SmpB has a molecular mass close to 20 kDa (18, 22), it was a possible candidate for the protein that was cross-linked to RNase R. To test this possibility, His6 and 2×FLAG sequences were fused to the N termini of chromosomal RNase R and SmpB, respectively, by recombineering (16). Extracts were prepared from this strain followed by immunoprecipitation with antibody to RNase R. The resulting immunoprecipitate was analyzed by SDS-PAGE and immunoblotting using His-probe monoclonal antibody to detect RNase R and anti-FLAG monoclonal antibody to detect SmpB. As shown in Fig. 1B, both RNase R and SmpB were observed under conditions in which both proteins were present in vivo at their normal levels. No bands were observed when RNase R antibody was omitted during the immunoprecipitation.
To determine whether tmRNA is also associated with RNase R, the extract was treated with Ni-NTA resin to pull down His-RNase R. Analysis of the materials isolated in this manner using Western blot and Northern blot analysis revealed that both SmpB and tmRNA are present (Fig. 1C). These data show that tmRNA is also a part of the complex between RNase R and SmpB. However, when the same pulldown experiment was carried out with an extract prepared from an smpB mutant strain, no tmRNA was observed (Fig. 1C), indicating that the association with tmRNA requires the presence of SmpB. Note the higher amount of RNase R in the SmpB mutant (see below).
To further explore the interaction of RNase R with SmpB, His-tagged SmpB was overexpressed and purified, and its association with purified RNase R was determined using the pulldown assay with Ni-NTA resin. The data in Fig. 1D show that purified RNase R was pulled down in the presence, but not in the absence, of His-SmpB (compare lanes 1 and 3), confirming a direct association between the two purified proteins. Moreover, in the presence of tmRNA, the amount of RNase R associated with SmpB increased ∼7-fold (compare lane 4 with lane 3). These data show that SmpB can bind to RNase R and that the interaction is strongly stimulated by tmRNA.
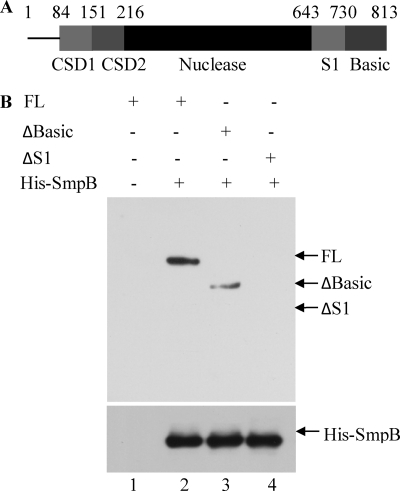
Interacting Regions of RNase R
In earlier studies, the roles of several individual domains of RNase R were identified (7, 23). However, the function of the low complexity, highly basic C-terminal domain (residues 731–813) remained unclear (Fig. 2A). In fact, its removal actually increased RNase R activity on a variety of substrates (7). To test whether this region of RNase R might play a role in the interaction with SmpB, we repeated the pulldown experiment with purified, truncated protein lacking the basic region. As shown in Fig. 2B, removal of the basic region resulted in weaker binding of SmpB (lane 3) when compared with full-length RNase R (lane 2). Additional removal of the adjacent S1 domain (residues 644–730) completely abolished the association of RNase R and SmpB (lane 4). Thus, interaction with SmpB is completely dependent on the C-terminal region of RNase R.
FIGURE 2.
Interaction between SmpB and various forms of RNase R. Full-length RNase R (FL), RNase RΔBasic (ΔBasic), and RNase RΔS1 (ΔS1) proteins (A) were purified and used in a pulldown assay (B) using Ni-NTA resin as described under “Experimental Procedures.” The proteins present are indicated above each lane. The presence of RNase R proteins and recombinant His-SmpB protein was detected as in Fig. 1.
SmpB and tmRNA Negatively Regulate RNase R Levels in Vivo
Recent studies from our laboratory revealed that RNase R is an extremely unstable protein in exponential phase and that this leads to a lower steady state level of RNase R in this phase of growth (12). To examine whether SmpB or tmRNA might affect RNase R levels, we generated smpB and ssrA (tmRNA) mutant strains and measured the amount of RNase R present in these strains using immunoblotting. In each mutant strain, the amount of RNase R was elevated about 3-fold (supplemental Fig. S1), indicating that the binding of SmpB and tmRNA does affect RNase R in vivo. These findings strongly suggest that SmpB and tmRNA play a role in the instability of RNase R observed in exponential phase.
SmpB and tmRNA Affect the Stability of RNase R
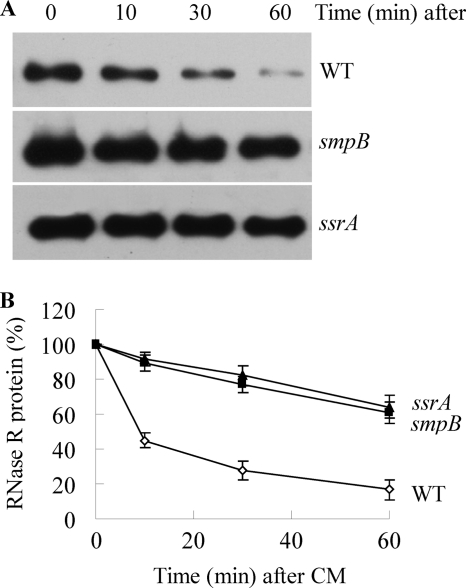
To determine whether SmpB and tmRNA affect the level of RNase R by altering its stability, we directly measured RNase R half-life in smpB and ssrA mutant cells (Fig. 3). In agreement with our previous findings (12), the half-life of RNase R in exponential phase wild type cells was ∼10 min. In contrast, the absence of either SmpB or tmRNA led to a dramatic stabilization of RNase R such that its half-life was increased to more than 60 min. These results demonstrate that SmpB and tmRNA regulate the stability of RNase R and that the absence of either component results in RNase R stabilization. Moreover, these data explain why RNase R is present at higher levels in smpB and ssrA mutant cells (supplemental Fig. S1).
FIGURE 3.
Stability of RNase R in wild type (WT) and smpB and ssrA mutant strains. The indicated cells were grown to exponential phase, treated with chloramphenicol, and assayed for RNase R by immunoblotting as described under “Experimental Procedures.” A, representative Western blot of RNase R from samples at the indicated times after the addition of chloramphenicol. B, quantitation of three independent experiments carried out as in A. RNase R levels at zero time of chloramphenicol (CM) addition were set at 100% for each strain. Error bars indicate S.D.
Stability of C-terminal Truncated RNase R
The data presented have shown that SmpB and tmRNA bind to RNase R and affect its stability. The simplest explanation for these findings is that the binding directly destabilizes RNase R. If this explanation is correct, it predicts that preventing SmpB and tmRNA binding would lead to stabilization of RNase R even when SmpB and tmRNA are present. To test this idea, we determined the half-life of C-terminal truncated RNase R molecules lacking the basic region or the basic region and the S1 domain. As shown above (Fig. 2B), these RNase R variants bind SmpB more weakly or not at all. The data in Fig. 4 show that chromosomally encoded, truncated RNase R molecules were, in fact, much more stable in vivo. Thus, when compared with the 10-min half-life of full-length RNase R, the protein lacking the basic region had a half-life of close to 40 min, and that lacking the basic region and S1 domain had a half-life of ∼60 min. Based on these data and those presented above, we conclude that binding of SmpB and tmRNA to RNase R results in its destabilization.
FIGURE 4.
Stability of full-length and truncated RNase R proteins. Experiments were carried out as in Fig. 3. A, representative Western blot of full-length (FL) and truncated RNase R proteins from exponential phase cells. CM, chloramphenicol. B, quantitation of three independent experiments carried out as shown in Panel A. RNase R levels at zero time of chloramphenicol addition were set at 100% for each RNase R. Error bars indicate S.D.
Activity of Complexed RNase R
The association of SmpB and tmRNA with RNase R raised the question of whether this interaction affected the activity of the RNase. To address this point, RNase R activity was measured in the presence and absence of SmpB and tmRNA. The data in supplemental Fig. S2 show that neither SmpB by itself (panel A) nor the SmpB-tmRNA complex (panel B) alters RNase R activity even when present at 10-fold the level of RNase R. SmpB was shown to bind to RNase R under these conditions (data not shown). These results indicate that binding of SmpB and tmRNA to the C-terminal region does not affect the activity of RNase R despite their strong influence on its stability.
DISCUSSION
The data presented in this work describe a completely novel mechanism for regulation of a cellular RNase, and they identify several factors responsible for this regulation. Thus, we have shown that RNase R associates in extracts both with the protein SmpB and with tmRNA, that purified RNase R and SmpB stably interact and that this latter association is greatly stimulated by tmRNA, and that C-terminal domains of RNase R, including the basic region and the adjacent S1 domain, are essential for the association with SmpB. Most importantly, we have found that the association of RNase R with these components has dramatic consequences, namely the rapid degradation of RNase R in exponential phase cells.
Interestingly, earlier work had shown that RNase R was a relatively minor component of a >450-kDa complex involved in “ribosome rescue” (18) and that it participates in mRNA degradation during this process (3). This large complex contained SmpB and tmRNA as well as other possible components of the trans-translation apparatus. However, our results differ in that the complex we detected by gel filtration is considerably smaller, only ∼150 kDa. We suspect that a likely reason for these differences is that Karzai and Sauer (18) prepared their extracts from cells overexpressing His6-SmpB, whereas our data were generated with normal extracts. The overexpression of SmpB may have led to binding of components that are not bound when SmpB is present at its usual intracellular concentration. Further work will be necessary to resolve this issue. Nevertheless, the data presented here clearly demonstrate a direct association of SmpB, tmRNA, and RNase R.
The consequences of this interaction in vivo are profound. Our laboratory has recently shown (12) that RNase R is subject to posttranslational regulation in which it is extremely unstable in exponential phase but is stabilized in stationary phase and other stress conditions, resulting in a dramatic elevation of the protein under stress conditions (9–11). The data presented here indicate that the instability of RNase R in exponential phase cells is critically dependent on association with SmpB and tmRNA. Elimination of either of these factors or removal of the C-terminal binding region of RNase R substantially elevates the amount of RNase R in exponential phase cells and greatly lengthens its half-life.
These data raise a number of interesting questions that remain to be answered. For example, how does binding of SmpB and tmRNA to RNase R lead to its destabilization? Our data indicate that SmpB binds directly to RNase R in its C-terminal region, and this binding is stimulated by tmRNA. However, the structural changes that occur in RNase R as a consequence of this binding that lead to its increased degradation are unknown. Detailed structural analysis of free and complexed RNase R will be needed to answer this question. Secondly, RNase R participates in mRNA degradation during trans-translation (3). How its role in this process might relate to its instability is unclear. Finally, there is the question of why exponential phase RNase R is sensitive to degradation, whereas the stationary phase enzyme is stable (12). Perhaps subtle structural differences between the two forms of RNase R, such as a covalent modification, are responsible. Studies are currently in progress to examine this possibility.
Supplementary Material
Acknowledgments
We thank Dr. Kenneth Rudd for providing strains and Dr. Richard Myers for assistance with the recombineering experiment. We also thank Drs. Georgeta Basturea, Tanmay Dutta, Arun Malhotra, and Chaitanya Jain for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM16317 (to M. P. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- tmRNA
- transfer-messenger RNA
- DMP
- dimethyl pimelimidate dihydrochloride
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Cheng Z. F., Deutscher M. P. (2002) J. Biol. Chem. 277, 21624–21629 [DOI] [PubMed] [Google Scholar]
- 2.Cheng Z. F., Deutscher M. P. (2005) Mol. Cell 17, 313–318 [DOI] [PubMed] [Google Scholar]
- 3.Richards J., Mehta P., Karzai A. W. (2006) Mol. Microbiol. 62, 1700–1712 [DOI] [PubMed] [Google Scholar]
- 4.Li Z., Reimers S., Pandit S., Deutscher M. P. (2002) EMBO J. 21, 1132–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng Z. F., Deutscher M. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6388–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vincent H. A., Deutscher M. P. (2006) J. Biol. Chem. 281, 29769–29775 [DOI] [PubMed] [Google Scholar]
- 7.Vincent H. A., Deutscher M. P. (2009) J. Biol. Chem. 284, 486–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Z. F., Zuo Y., Li Z., Rudd K. E., Deutscher M. P. (1998) J. Biol. Chem. 273, 14077–14080 [DOI] [PubMed] [Google Scholar]
- 9.Cairrão F., Cruz A., Mori H., Arraiano C. M. (2003) Mol. Microbiol. 50, 1349–1360 [DOI] [PubMed] [Google Scholar]
- 10.Chen C., Deutscher M. P. (2005) J. Biol. Chem. 280, 34393–34396 [DOI] [PubMed] [Google Scholar]
- 11.Andrade J. M., Cairrão F., Arraiano C. M. (2006) Mol. Microbiol. 60, 219–228 [DOI] [PubMed] [Google Scholar]
- 12.Chen C., Deutscher M. P. (2010) RNA 16, 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin C., Ahmad M., Cashmore A. R. (1996) Plant J. 10, 893–902 [DOI] [PubMed] [Google Scholar]
- 14.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Withey J., Friedman D. (1999) J. Bacteriol. 181, 2148–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherepanov P. P., Wackernagel W. (1995) Gene 158, 9–14 [DOI] [PubMed] [Google Scholar]
- 18.Karzai A. W., Sauer R. T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3040–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundermeier T., Ge Z., Richards J., Dulebohn D., Karzai A. W. (2008) Methods Enzymol. 447, 329–358 [DOI] [PubMed] [Google Scholar]
- 20.Liang W., Li C., Liu F., Jiang H., Li S., Sun J., Wu X., Li C. (2009) Cell Res. 19, 307–316 [DOI] [PubMed] [Google Scholar]
- 21.Gottesman S. (2003) Ann. Rev. Cell Dev. Biol. 19, 565–587 [DOI] [PubMed] [Google Scholar]
- 22.Karzai A. W., Susskind M. M., Sauer R. T. (1999) EMBO J. 18, 3793–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awano N., Rajagopal V., Arbing M., Patel S., Hunt J., Inouye M., Phadtare S. (2010) J. Bacteriol. 192, 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.