Abstract
The p53 regulatory network is critically involved in preventing the initiation of cancer. In unstressed cells, p53 is maintained at low levels and is largely inactive, mainly through the action of its two essential negative regulators, HDM2 and HDMX. p53 abundance and activity are up-regulated in response to various stresses, including DNA damage and oncogene activation. Active p53 initiates transcriptional and transcription-independent programs that result in cell cycle arrest, cellular senescence, or apoptosis. p53 also activates transcription of HDM2, which initially leads to the degradation of HDMX, creating a positive feedback loop to obtain maximal activation of p53. Subsequently, when stress-induced post-translational modifications start to decline, HDM2 becomes effective in targeting p53 for degradation, thus attenuating the p53 response. To date, no clear function for HDMX in this critical attenuation phase has been demonstrated experimentally. Like HDM2, the HDMX gene contains a promoter (P2) in its first intron that is potentially inducible by p53. We show that p53 activation in response to a plethora of p53-activating agents induces the transcription of a novel HDMX mRNA transcript from the HDMX-P2 promoter. This mRNA is more efficiently translated than that expressed from the constitutive HDMX-P1 promoter, and it encodes a long form of HDMX protein, HDMX-L. Importantly, we demonstrate that HDMX-L cooperates with HDM2 to promote the ubiquitination of p53 and that p53-induced HDMX transcription from the P2 promoter can play a key role in the attenuation phase of the p53 response, to effectively diminish p53 abundance as cells recover from stress.
Keywords: Apoptosis, DNA Damage, p53, Transcription Factors, Tumor Suppressor, HDM2, HDMX, MDM2, MDM4, p53-responsive
Introduction
The tumor suppressor protein p53 functions primarily as a stress-inducible transcriptional activator of genes that promote cell cycle arrest and apoptosis (1). Stress-induced p53 activation can form a rate-limiting barrier to tumorigenesis (2, 3), and the manipulation of p53 function is key to the mechanism of action of many cancer chemotherapeutic strategies (4, 5). In unstressed cells, p53 is maintained at low levels and inactive, largely through the action of several p53-inducible negative feedback pathways, the most extensively studied of which involves the oncoproteins HDM2 and HDMX (also called MDM4) (MDM2 and MDMX/MDM4 in mice) (6, 7). Considerable research effort has been applied to understanding the mechanisms whereby these two proteins regulate p53 function. HDM2 and HDMX both contain an N-terminal pocket that binds to the primary transactivation domain of p53; they can, therefore, function independently of each other to repress p53-dependent transcription (8–10). HDM2 also forms both HDM2-HDM2 homodimers and HDM2-HDMX heterodimers. These function as E3 ubiquitin ligases for p53; monoubiquitination of p53 by HDM2 inhibits p53 activity by both inhibiting acetylation and promoting nuclear export, whereas polyubiquitination promotes proteasome-mediated p53 degradation and is largely responsible for the rapid turnover of p53 protein that occurs in proliferating cells (11). HDMX itself lacks E3 ligase activity and does not readily homodimerize; however, because HDMX-HDM2 heterodimerize with higher affinity than do HDM2-HDM2 homodimers, HDMX can effectively function to promote cellular HDM2 E3 ubiquitin ligase activity when cellular HDM2 concentrations are limiting (12–14). Conversely, at higher HDMX concentration, monomeric HDMX can potentially inhibit p53 ubiquitination by competing with the dimeric proteins for p53 binding (15). Thus, both the absolute and relative abundance of HDM2 and HDMX in cells are critical determinants of p53-dependent transcriptional activity and hence cellular proliferation and survival.
Germ line genetic changes that cause relatively modest increases or decreases in HDM2/mdm2 expression promote (16) and protect (17) from tumorigenesis, respectively. Furthermore, many separate studies have identified both HDM2 and HDMX as being overexpressed in diverse tumors, through a variety of mechanisms, including but not limited to gene amplification (18). The mechanisms regulating expression of HDM2/mdm2 have now been quite extensively studied. The HDM2/mdm2 gene is transcribed from two promoters, one (P1) “constitutive” and the second (P2), located 5′ to exon 2 which is inducible by both p53 and mitogens (19–21). The transcripts from these two promoters are translated into full-length (p90) HDM2/MDM2 and N-terminally truncated, p53 binding-incompetent, HDM2/MDM2 proteins. The mRNA transcript from the P2 promoter is ∼8-fold more efficiently translated into full-length (p90) HDM2/MDM2 than that from P1 (22–24). Following genotoxic stress, such as ionizing radiation, the abundance of both HDM2 and HDMX proteins initially decreases, due to an ATM- and HDM2 E3 ligase-dependent increase in their degradation, thus promoting activation of p53 (25–28). HDM2 levels subsequently increase rapidly, due to p53-dependent transcription from the HDM2-P2 promoter, facilitating the attenuation of the p53 response. Stress-induced reduction in HDMX protein abundance is more sustained, and HDMX transcription is not reported to be induced by p53. Indeed, although the overall gene structure of HDMX/mdmx is very similar to that of HDM2/Mdm2, HDMX/mdmx, an equivalent of the p53-inducible P2 promoter 5′ to a non-coding exon 2, has not been reported in the HDMX/mdmx genes (6).
HDMX abundance can affect the level of the p53-dependent cellular response to ionizing radiation and ribosomal stress as well as to a chemical inhibitor of the p53-HDM2 interaction (nutlin-3) that is under development as a promising novel cancer therapeutic (7, 29, 30). There is, therefore, a clear necessity for an understanding of the pathways that regulate HDMX protein levels and how they may regulate the cellular response to both established and experimental cancer therapies.
Specific forms of genotoxic stress, such as UV radiation, doxorubicin, and cisplatin can induce aberrant splicing of HDMX mRNA as well as promoting the degradation of the full-length HDMX mRNA, together resulting in the loss of expression of the full-length protein (31, 32). These studies as well our original report first describing mdmx (9) have shown that total HDMX/mdmx mRNA abundance does not generally increase in response to DNA damage-induced p53 activation. This as well as the increased rates of HDM2-dependent degradation of HDMX protein that follows p53 activation, means that HDMX protein abundance does not increase in response to genotoxic p53-activating signals and that HDMX had not been identified as a p53-inducible gene. However, it noteworthy that, when the up-regulation of p53-responsive genes is studied in, for example, mouse tissues in response to ionizing radiation, total mdm2 mRNA levels increase by a maximum of 2-fold, even in tissues such as spleen and thymus, where up-regulation of another p53-responsive gene, p21WAF1 is ∼10- and 50-fold, respectively (33). This is because in these tissues, basal levels of the mdm2-P1 transcripts are up to 10-fold higher than those derived from the P2 promoter, and the -fold increases in mdm2-P2 transcript levels in response to radiation are only sufficient to cause modest changes in total mdm2 mRNA abundance (34). MDM2 protein synthesis can increase substantially in response to radiation, due to the increased translation potential of the mdm2-P2 transcript, and thus clearly the lack of substantial changes in total mRNA abundance in this situation is potentially deceptive. Despite the overall similarity in the structure of the HDMX/mdmx and HDM2/mdm2 genes, this possibility of the existence of alternate transcripts with quantitatively different translational potential within the total pool of HDMX mRNA in cells has not, to date, been investigated.
A study that aimed to identify novel p53-responsive genes by global genomic profiling of chromatin fragments bound by p53 identified a p53-binding region within the first intron of HDMX (35), and very recently synthetic reporter constructs containing this region have been shown to drive expression of the reporter gene in a p53-dependent manner (36), suggesting that HDMX is indeed a p53-regulated gene. In this paper, we show that, like HDM2, the HDMX gene contains a p53-responsive promoter in its first intron that drives the expression of mRNA transcripts with quantitatively and qualitatively different translation potential, which participate in an autoregulatory feedback loop to control the abundance and activity of p53 in cancer cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
MCF-7, SAOS-2, SAOS-2/p53 Tet-On (37), NARF, and 174-2 cells (p53/mdm2 double knock-out murine embryo fibroblasts (MEFs)3) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum. Early passage p53+/+ and p53−/− MEFs were maintained in DMEM, supplemented with 15% fetal calf serum and 0.5 mm 2-mercaptoethanol, and grown at 3% oxygen. H1299, the breast carcinoma cell lines MPE600 and ZR75-30, the uveal melanoma cell lines MEL285 and 92.1 (38), N-TERA-2, 833KE, and mouse melanoma B16F10 cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum. The generation and culture conditions of MCF-10A (M1) and MCF-10AT (M2) cells have been described (39).
To generate stable p53 knockdown and control knockdown cell lines, cells were infected with lentiviral vectors expressing shRNA targeting human p53 or mouse mdmx and conferring puromycin resistance. The latter does not target the HDMX mRNA. After puromycin selection, polyclonal cell lines were established. Nutlin-3 (Alexis Biochemicals) was dissolved in ethanol at 5 mm, and MG-132 (Sigma) was dissolved in DMSO at 10 mm, before being adding to the medium where stated. 5-Fluorouracil (Sigma) was in aqueous solution. Etoposide (Sigma) was dissolved in DMSO at 10 mm, leptomycin B (BIOMOL) was dissolved in ethanol at 10 μm, and actinomycin D (Calbiochem) was dissolved in ethanol at 1 mg/ml. Neocarzinostatin was obtained from Sigma.
Protein Analysis
Cells were washed with phosphate-buffered saline, pelleted by centrifugation at 1000 × g, snap-frozen, and stored at −80 °C. Immunoblotting was performed as described previously (40), and membranes were probed for HDMX (A300-287A, Bethyl Laboratories), HDM2 (monoclonal antibody 2A9 or 4B2 (41)), p53 (DO-1, Serotec), GFP (Cancer Research UK), PUMA (Cell Signaling Technology), p21WAF-1 (EA10 (Calbiochem) or CP74 (Millipore)), KAP1 and phospho-KAP1/Ser6824 (A300-274A and A300767A), poly(ADP-ribose) polymerase (Cell Signaling Technology), and HAUSP (A300-033A, Bethyl Laboratories). Anti-phospho-H2AX was obtained from Millipore. Mouse MDM2, MDMX, p53, and HAUSP were detected with mouse monoclonal 4B2, (41), MX-82 (Sigma), 1C12 (Cell Signaling), and mouse monoclonal 1G7 (42), respectively. Equal protein loading was confirmed on all immunoblots using rabbit anti β-actin or anti-tubulin antibodies (Sigma-Aldrich). Bands were visualized by chemiluminescence (Supersignal, Pierce) using a Fluor-S MAX system (Bio-Rad) or by exposure to x-ray films (Fuji). In the IP/Western analysis of HDMX-p53 interactions, the IPs were performed with either anti-HA rabbit polyclonal (Abcam) or anti-FLAG rabbit polyclonal (Sigma), after which the blots were incubated with either anti-FLAG monoclonal antibody M2 (Sigma) or anti-HA monoclonal antibody HA.11 (Covance). Detection of p14ARF by immunofluorescence was performed with anti-p14ARF monoclonal antibody 4C6 (gift of Gordon Peters).
RNA Analysis
For RT-PCR analysis of transcripts, 0.5–2 μg of RNA was reverse transcribed in a 20–25-μl volume using Superscript II RNase H− reverse transcriptase (Invitrogen) and oligo(dT) primer. 2 μl of cDNA product were used as a target in 50 μl of PCRs using GoTaq DNA polymerase (Promega). RT-PCR analysis of HDM2-P2 and β-actin transcripts was as described previously (21). Primer sets used in the various RT-PCR experiments are presented in supplemental Table S1.
Plasmids
Genomic HDMX-P2 sequence was amplified from normal human liver DNA and ligated into pGL3-Basic using the MluI/XhoI sites (Promega) to generate reporter construct HDMXP2luc01. The sequence of the inserted 1332-bp region (−1535 to −202, relative to the start of exon 2) was identical to RefSeq NT_004487. Additional constructs containing deletions of the HDMX-P2 promoter (luc02-08) were generated using additional primers. 3-bp substitutions in the putative p53 binding site were introduced into HDMXP2luc01 to give HDMXP2luc01Δp53-RE using the QuikChange mutagenesis kit (Stratagene) and verified by sequencing. Expression vectors containing cDNA (including 5′-UTR, coding sequence, and a C-terminal mychis tag) for both HDMX-P1 and HDMX-P2 were created by ligation of NheI/XhoI-digested pcDNA3.1(−)mychisB with HpaI/XhoI-digested PCR product 1 (amplified from pT7.7MDMX using primer pair 5′-GCTAGCTGTTTTCGTTGTTGGGCCTTGA-3′/5′-CTCGAGTGCTATAAAAACCTTAATAACCAGCTGA-3′) and NheI/HpaI-digested PCR product 2 or 3 (amplified from MCF-7 cDNA using the following primer pairs: PCR 2, HDMXP1, 5′-GGGAGGCCGGAAGTTGCG-3′/5′-CAGTGATATCAGACGTGGAGAGAGAATGGGTTAAC-3′; PCR 3, HDMXP2, 5′-GCTAGCAGTTGGAGGTTGGAGCGTGC-3′/5′-CAGTGATATCAGACGTGGAGAGAGAATGGGTTAAC-3′) to give pP1-HDMXmh and pP2-HDMXmh, respectively. pP1-HDMX and pP2-HDMX were created using site-directed mutagenesis to introduce a stop codon immediately 5′ of the mychis tag. p21-luc reporter vector and pC53SN3 expressing human p53 were from Bert Vogelstein. pCMVDDp53 was from Moshe Oren. pHDM2 (pCMVMDM2) containing cDNA for human MDM2 was from A. J. Levine. pHis6Ub was made available by S. Mittnacht. HDM2luc01 reporter vector was described previously (21), as was the FLAG-p53 expression vector (43).
RNAi, Transfections, and Reporter Gene Assays
RNAi-mediated knockdown of HDMX-P2 was performed using the siRNA 5′-GCUUGGACGAUUCUUACUCdTdT-3′/3′-dTdTCGAACCUGCUAAGAAUGAG-5′ obtained from Qiagen. Appropriate control siRNAs, as described previously (44) were as follows: HDMX-P2ctrl1 containing a 4-nucleotide mismatch in the seed region (5′-GCUUGGACGAUUAGCAAUCdTdT-3′/3′-dTdTCGAACCUGCUAAUCGUUAG-5′) and HDMX-P2ctrl2 containing a 4-nucleotide mismatch in the central region (5′-GCUACGGUGAUUCUUACUCdTdT-3′/3′-CGAUGCCACUAAGAAUGAG-5′; 75 nm). siRNA to the HDMX coding region was from Ambion (MDM4; catalog no. 121374). p53 siRNA was obtained from Qiagen (Hs_TP53_9 HP validated; 25 nm). Negative control siRNA 1 (Ambion) was used at the appropriate concentration for experimental controls, and total siRNA concentration was equalized in all samples using negative control siRNA. siRNA was transfected for 4 h using INTERFERin reagent (Polyplus Transfection). The construction of lentiviral vectors expressing specific shRNAs and the production of lentivirus particles have been described recently (45). The target sequence for HDMX-P2 mRNA was the same as the siRNA mentioned above. The sequences targeting human and mouse p53 have been published (46, 47). For transfection of plasmid DNA, Lipofectamine 2000 (Invitrogen) was used. Unless stated otherwise, reporter assays were performed in triplicate and assayed 48 h after transfection using a Dual-GloTM luciferase assay (Promega) on cells transfected in 96-well plates, with normalization to Renilla luciferase expressed from pRLSV40 (Promega). Data pooled from at least two independent experiments are shown as mean ± S.E.
In Vivo Ubiquitination Assay
24 h post-transfection, H1299 cells were exposed to 25 μm MG132 (Sigma) for 4 h before protein was extracted by denaturing urea buffer and quantified as described above. 20 μg of total extracted proteins were analyzed by direct Western blotting, and 120 μg of proteins were used to extract His6-ubiquitinated conjugates as described (48).
In Vitro Transcription and Translation
RNA was transcribed from 3.3 μg of linearized HDMX expression vectors using T7 RNA polymerase (Promega). Template DNA was removed by digestion with RQ1 RNase-free DNase (Promega) before RNA purification using RNAbee reagent (Biogenesis Inc.). The indicated amounts of RNA were used as templates in in vitro translation reactions using nuclease-treated rabbit reticulocyte lysate (Promega). 10% of reactions were separated by SDS-PAGE before HDMX expression levels were determined by Western blotting.
Chromatin Immunoprecipitation
The protocol was adapted from Ref. 49. Cells were cross-linked in 1% formaldehyde for 30 min at room temperature, after which cross-linking was stopped by adding glycine to an end concentration of 125 mm. Cells were put on ice, rinsed twice with ice-cold PBS, and scraped in HEPES lysis buffer (10 mm HEPES, pH 7.6, 1% Nonidet P-40, 1 mm EDTA, 400 mm NaCl, 10% glycerol supplemented with protease and phosphatase inhibitors). Lysates were centrifuged at 11,000 rpm for 10 min at 4 °C. Pellets were resuspended in 500 μl of HEPES lysis buffer and spun again for 5 min at 11,000 rpm and 4 °C. Pellets were resuspended and left on ice for about 30 min and subsequently sonicated in a Bioruptor sonicator (30 min on, 30 min off; 2 × 10 min; high power). Insoluble material was removed by centrifugation at 13,000 rpm for 10 min at 4 °C. Supernatant was transferred to a new tube and diluted 1:1 with HEPES dilution buffer (10 mm HEPES, pH 7.6, 1 mm EDTA, 10% glycerol supplemented with protease and phosphatase inhibitors). Aliquots were taken and stored at 4 °C to represent input material. 300 μl of chromatin solution was used for immunoprecipitation, with a combination of DO-1 and PAb1801 (Santa Cruz Biotechnology, Inc. Santa Cruz, CA) anti-p53 antibodies (4 μg of antibody/IP; bound to 10 μl of protein G beads) for human cells and FL-393 rabbit polyclonal antibody (Santa Cruz Biotechnology, Inc.; 2 μg/IP; bound to 10 μl of protein A beads) for mouse cells. IPs were performed overnight at 4 °C, in a total volume of 400 μl, in the presence of 0.1 μg/μl BSA. Beads were then washed (three times) in wash buffer (10 mm HEPES, pH 7.6, 0.5% Nonidet P-40, 1 mm EDTA, 200 mm NaCl, 10% glycerol, supplemented with protease and phosphatase inhibitors). Beads were eluted for 20 min at room temperature (rotating) in elution buffer (1% SDS, 0.1 m NaHCO3), after which beads were spun down, and supernatant was transferred into a new tube. 16 μl of 5 m NaCl was added, and cross-linking of samples (including input chromatin; 50 μl + 350 μl elution buffer) was reversed for 4–5 h 65 °C. Chromatin was purified by phenol/chloroform/isoamyl alcohol (25:24:1) extraction, followed by chloroform/isoamyl alcohol (24:1) extraction and subsequent ethanol precipitation in the presence of 2 μg/μl glycogen. Pellets were dissolved in milliQ water, and these chromatin samples were used for analysis by quantitative PCR. Primers used to amplify the specific genomic regions are given in supplemental Table S1.
RESULTS
The HDMX Gene Contains a Functional p53-responsive Promoter in Intron 1
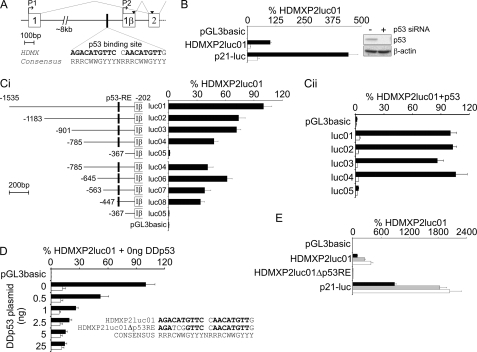
As an initial step in the analysis of the regulation of HDMX gene expression, we used BLAST to search human EST databases for mRNAs that contain the first coding exon of HDMX, exon 2. In addition to ESTs that matched the published HDMX cDNA sequence (50), two sequences included at their 5′-end a novel exon spliced into the start of exon 2. Both ESTs had been identified from a thymus library using a method that aims to find the extreme 5′-ends of cDNAs; of the two, DB137351 extended furthest in the 5′ direction. This exon is located in intron 1, and we have termed it exon 1β (Fig. 1A). Global genomic profiling previously identified a p53-binding region in intron 1 (35). We identified a good match to the p53-binding site consensus sequence (51) 151 bp 5′ to the likely 5′ limit of exon 1β (Fig. 1A). Thus, this bioinformatics analysis suggested that HDMX might contain a second, p53-responsive promoter in intron 1, analogous to the P2 promoter in the HDM2 gene (Fig. 1A). To determine whether this putative HDMX-P2 promoter is functional, we cloned 1334 bp of genomic promoter sequence into a luciferase reporter vector and tested the activity of this construct (HDMXP2luc01) in the MCF-7 cell line (Fig. 1B). These cells express endogenous wild-type p53 that becomes activated in response to DNA transfection. The promoter showed robust activity, which was ∼25% of that of the highly p53-responsive p21WAF1 promoter. HDMX-P2 promoter activity was reduced by >85% when p53 protein expression was inhibited using siRNA. HDMX-P2 activity in MCF-7 cells is strictly dependent upon an 80-bp region that includes the predicted p53 response element (p53-RE) (compare HDMXP2luc08 with HDMXP2luc05 in Fig. 1C, i). Similar findings were obtained when a subset of these vectors were tested in the p53 null H1299 cell line, in the absence (open bars) or presence (solid bars) of co-transfected p53 expression vector (Fig. 1C, ii). A targeted 3-bp substitution in the predicted p53-RE reduced promoter activity in MCF-7 cells as effectively as was achieved by inactivation of endogenous p53 using a dominant negative p53 fragment (Fig. 1D). Activation by exogenous p53 transfected into H1299 cells was also completely abrogated by this mutation (supplemental Fig. S1A). Therefore, the predicted p53-RE is indeed essential for p53-dependent HDMX-P2 promoter activity.
FIGURE 1.
A novel promoter in intron 1 of HDMX contains a functional p53 binding site. A, map of the 5′-end of the HDMX gene, showing the position of the novel exon 1β as defined by EST DB137351. A potential p53-binding site in intron 1 is compared with the consensus p53-binding sequence. Inverted triangles show the known translation start site in exon 2 and an in-frame ATG in exon 1β, initiation of translation from which would incorporate 18 additional amino acids at the N terminus of HDMX (MQNLSKVLPTDCSFFTTK). B, MCF-7 cells were transfected with 25 nm control (solid bars) or p53 siRNA (open bars), followed 24 h later by transfection with pGL3basic, HDMXP2luc01, or p21-luc reporter plasmids. Reporter activity was assayed after a further 48 h (n = 6). Western blotting demonstrates efficacy of the siRNA. C, i, MCF-7 cells were transfected with the HDMXP2luc deletion constructs shown. Numbering is relative to the start of exon 2 (n ≥ 9). ii, H1299 cells were transfected with the HDMXP2luc deletion constructs plus 25 ng of pc53SN3 (black bars) or empty vector control (white bars) (n = 5). D, MCF-7 cells were transfected with HDMXP2luc01 (solid bars) or HDMXP2luc01Δp53RE (open bars) along with increasing amounts of dominant negative p53 fragment (DDp53) (n = 6). E, MCF-7 cells were transfected with the stated reporter plasmid. Following removal of transfection mix after 4 h, cells were exposed to media only (black bars), 200 μm 5-fluorouracil (gray bars), or 5 μm nutlin-3 (white bars) for 24 h before luciferase activity was determined (n = 6). Error bars, S.E.
p53-dependent transcriptional activity can be activated in response to a wide range of cellular stresses and pharmacological agents. We therefore examined the effects of two different p53-activating agents on HDMX-P2 promoter activity in MCF-7 cells (Fig. 1E). 5-Fluorouracil (gray bars) and nutlin-3 (open bars) both increased its activity in MCF-7, through a p53-RE-dependent mechanism. Finally, we noted that the p53-RE in the HDMX-P2 promoter was as good a match to the p53 consensus sequence as that found in the highly p53-responsive p21WAF1 promoter and better than weaker response elements found in, for example, the BAX promoter. We therefore examined the relative p53 responsiveness of the HDMX-P2 promoter compared with other p53-responsive promoters (supplemental Fig. S1B). The HDM2-P2 promoter contains two p53-REs and is highly responsive to low levels of transfected p53. HDMX-P2 and p21WAF1 promoters showed comparable -fold induction by p53, this being ∼2-fold greater than the activation of the BAX promoter.
Genotoxic, Oncogenic, and Pharmacological p53-activating Signals Induce Transcription from the Endogenous p53-responsive HDMX-P2 Promoter
The results reported above clearly demonstrate that synthetic reporter constructs containing the novel HDMX-P2 promoter region do exhibit p53-dependent transcription of the reporter gene when transfected into cells. This is consistent with similar findings reported by Li et al. (36) in different experimental systems. It has also been shown that p53 can bind to chromatin in this region of the endogenous HDMX gene (35, 36). However, in order to demonstrate that this is indeed a functional promoter in the context of endogenous chromatin, it was necessary to establish whether HDMX mRNA is transcribed from the HDMX-P2 promoter in response to p53-activating signals.
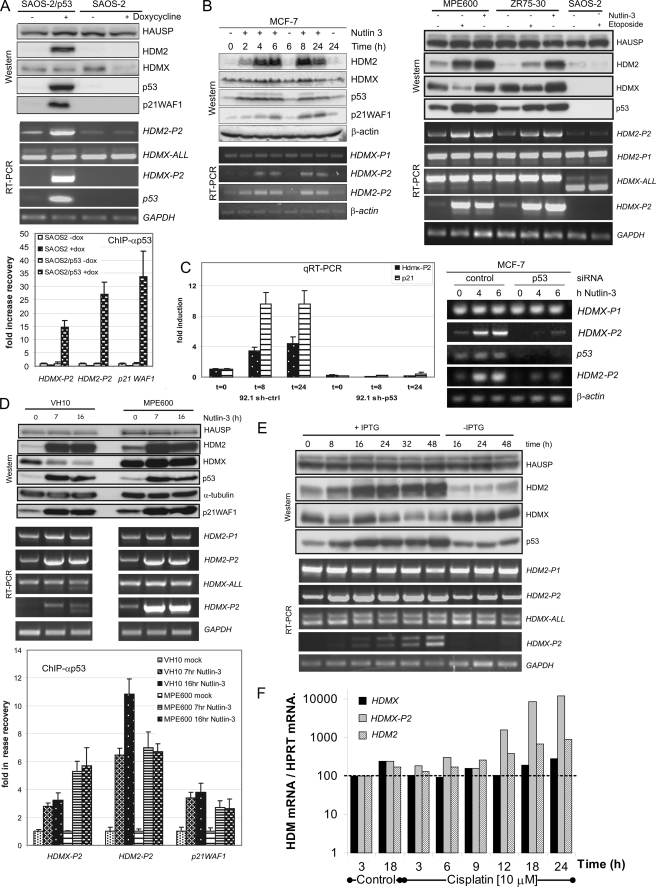
Transcript-specific PCR for mRNAs containing both exon 1β and HDMX coding sequence-containing exons can be used to identify transcripts derived from the HDMX-P2 promoter. This approach selectively identifies these transcripts because mRNA transcribed from the constitutive P1 promoter of HDMX contains exon 1 spliced directly to exon 2, exon 1β being skipped (50). Furthermore, in our EST analysis, no transcripts were detected that contained exon 1β spliced 3′ to another exon. To determine whether p53 can induce activity of the endogenous HDMX-P2 promoter, we made use of the p53 null SAOS-2 cell line containing p53 under the control of a doxycyline-inducible promoter (37) (Fig. 2A). Induction of p53 synthesis in these cells resulted in a robust increase in the expression of p21WAF1 and HDM2, the products of known p53-responsive genes. At the mRNA level, induction of p53 had no detectable effect on the abundance of total HDMX mRNA, although there was a very clear increase in the abundance of the mRNA product of the HDMX-P2 promoter, which was not detectable in the absence of p53. Thus, as we have discussed, induced HDMX-P2 transcript levels are likely to be relatively low compared with the abundance of the constitutive P1 promoter-derived transcript. Incidentally, two bands are detected by this HDMX-P2 mRNA PCR in SAOS-2 because the PCR spans exon 6, which can be alternatively spliced to produce the HDMX-S variant (52). SAOS-2 cells mainly express HDMX-S, whereas, for example, ZR75-30 and MPE600 cells predominantly express full-length HDMX mRNA (Fig. 2B). Chromatin immunoprecipitation analysis confirmed that p53 protein was recruited to the endogenous HDMX-P2 promoter in these SAOS-2/p53 cells, comparable to its recruitment to the HDM2-P2 and p21WAF1 promoters (Fig. 2A, bottom).
FIGURE 2.
The endogenous HDMX-P2 promoter is induced by p53. A, SAOS-2 cells containing a doxycyline-inducible p53 construct and control SAOS-2 cells were treated with doxycyline for 24 h, after which cells were harvested for protein analysis, mRNA analysis, and chromatin immunoprecipitation (ChIP). RT-PCR and Western blotting were used to determine expression of mRNAs and proteins. Changes in recruitment of p53 to the p53REs in HDMX-P2, HDM2-P2, and p21WAF1 promoters are indicated as -fold increase in recovery of that specific chromatin fragment. B, MCF-7 cells (left) were treated with nutlin-3 (5 μm) for the indicated times, whereas MPE600, ZR75-30, and SAOS-2 cells (right) were treated either with nutlin-3 (10 μm; 6 h) or etoposide (20 μm; 6 h) prior to harvest and analysis by Western blotting and RT-PCR. C, stable derivatives of 92.1 cells expressing either control shRNA or p53 shRNA were treated with nutlin-3 (10 μm; 24 h), after which RNA was extracted, and expression of HDMX-P2 and p21WAF1 were determined by real-time PCR. MCF-7 cells were transfected with control or p53 siRNA. 48 h later, the cells were exposed to 5 μm nutlin-3 prior to analysis of mRNA expression by RT-PCR. D, VH10hTERT and MPE600 cells were treated with nutlin-3 (10 μm) for the indicated periods, after which cells were harvested and processed for analysis of mRNA expression and protein expression by RT-PCR and Western blotting (top). In addition, ChIP was used to determine the recruitment of p53 to the p53REs in the HDMX-P2, HDM2P2, and p21WAF1 promoters (bottom). E, NARF cells (U2OS cells containing an isopropyl 1-thio-β-d-galactopyranoside-inducible p14ARF construct) were treated with isopropyl 1-thio-β-d-galactopyranoside or mock-treated for the indicated time periods. Cells were harvested, and RT-PCR and Western blotting were used to determine expression of the indicated mRNAs and proteins. p14ARF expression was investigated by immunofluorescence (supplemental Fig. S2C). F, OAW-42 ovarian cancer cells that express wild-type p53 were exposed to 10 μm cisplatin for the indicated times before being prepared for analysis by quantitative RT-PCR. HDMX-P2 induction by cisplatin in other cell lines with varying p53 status is shown in supplemental Fig. S2D. Error bars, S.E.
We next examined whether HDMX-P2 promoter-derived transcripts are synthesized in cells in response to activation of endogenous p53 in cells, by exposing a panel of wild-type p53-expressing breast cancer cell lines to either nutlin-3 or etoposide (Fig. 2B). In normally proliferating MCF-7 cells, HDMX-P2 transcripts were virtually undetectable by our RT-PCR assays. Nutlin-3 induced a robust increase in the abundance of this mRNA with kinetics consistent with it following the increase in p53 protein abundance. Peak induction of HDMX-P2 transcripts was observed after 8 h of treatment. In two other wild-type p53-expressing breast cancer lines, ZR75–30 and MPE600, detectable amounts of the HDMX-P2 transcripts were present in normally proliferating cells; nevertheless, both lines showed a similarly robust induction of HDMX-P2 transcripts in response to either nutlin-3 or etoposide (Fig. 2B). There was no such induction in p53-null SAOS-2 cells (Fig. 2B). The HDMX-P2 transcripts were also detectable in proliferating testicular germ cell tumor (TGCT) lines, in which they were strongly induced by nutlin-3 (supplemental Fig. S2A). Of note, when the data in this figure are compared with those of Li et al. (36), whose analysis was completely based on TGCT cells, both studies show that nutlin-3 causes a modest increase in total HDMX transcripts in 833 KE cells but not N-TERA-2. However, our transcript-specific analysis shows that HDMX-P2 promoter-derived transcripts are, in fact, robustly induced in both cell lines. In all of the breast cancer and TGCT lines, nutlin-3-mediated activation of p53 also caused a modest increase in HDMX protein abundance. This increase is not seen upon etoposide treatment, most likely because DNA damage triggers the HDM2-mediated degradation of HDMX proteins (53, 54).
Subsequent experiments using siRNA to p53 confirmed that both the basal and inducible expression of HDMX-P2 transcripts in these breast cancer lines and other wild-type p53-expressing cells, such as 92.1 uveal melanoma cells, is dependent upon p53 (Fig. 2C and supplemental Fig. S2, B and F) (data not shown). Chromatin immunoprecipitation experiments in VH10 (primary foreskin fibroblasts) and MPE600 showed a cell line dependent increase in the association of p53 with HDMX-P2 promoter regions in response to nutlin-3 (Fig. 2D). We next performed a number of experiments in order to determine the generality of HDMX-P2 promoter activation in response to alternative p53-activating stresses and in different cell lines. Oncogenic stress is a key activating signal, which can occur through increased expression of the HDM2 inhibitor, p14ARF. Using the U2OS-derived NARF cells (55) (a kind gift from Gordon Peters), in which p14ARF expression is inducible by isopropyl 1-thio-β-d-galactopyranoside, HDMX-P2 transcripts are clearly induced with kinetics that follow the stabilization of p53 protein (Fig. 2E). Other p53 activators, such as cisplatin (Fig. 2F) as well as neocarzinostatin, leptomycin B, and RITA (supplemental Fig. S2, E and F), also clearly induce transcription from the HDMX-P2 promoter in multiple cell lines that express wild-type p53. In general, from these and other (e.g. see supplemental Fig. S5C) experiments, we find that, compared with tumor cell lines, untransformed cells show a relatively modest increase of HDMX-P2 mRNA in response to p53 activators, despite other p53-response mRNAs, such as HDM2-P2, being relatively highly induced. It is also noteworthy that, in many examples where HDMX-P2 transcripts are robustly induced, this p53-induced transcription of HDMX is undetectable when total HDMX transcripts are analyzed. Furthermore, even when induced alternative splicing results in a decrease in full-length HDMX transcripts (e.g. in response to leptomycin B), HDMX-P2 mRNA transcripts are robustly induced, albeit in the alternatively spliced form.
Transcriptional Regulation of HDMX by p53 Is Evolutionarily Conserved
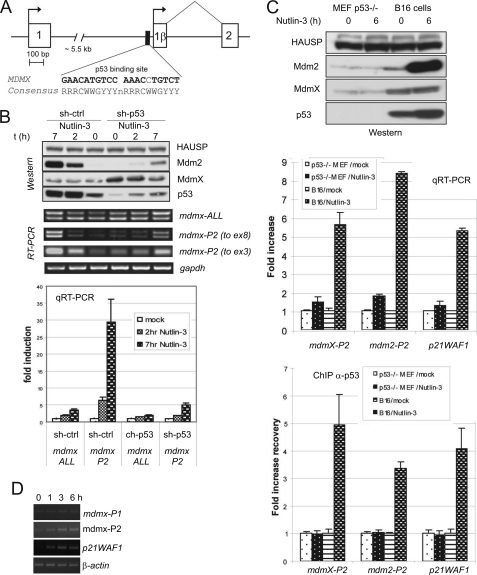
Our previous analysis of ESTs containing murine mdmx also identified transcripts containing an exon 1β (6). This exon shows limited homology to the human exon 1β and does not contain an in-frame ATG. Nevertheless, the genomic region 5′ to the murine exon 1β does contain a potential p53-response element (Fig. 3A). In order to determine whether exon 1β mdmx transcripts are inducible by p53, we infected MEFs with lentiviruses, expressing control or p53-specific shRNA, prior to exposing them to nutlin-3 (Fig. 3B). Nutlin-3 caused the expected increase in p53 protein abundance, which was reduced by the p53 shRNA. RT-PCR to detect total mdmx mRNA again detected two bands, due to alternate splicing of exon 6 (52). There was small but detectable effect of p53 activation on the abundance of the full-length mdmx mRNA transcripts, the p53 dependence of this increase being confirmed in a separate experimental system (supplemental Fig. 3B). In contrast, mdmx-P2 promoter-derived transcripts containing exon 1β were very clearly induced by nutlin-3, again in a p53-dependent manner. quantitative RT-PCR showed that the abundance of mdmx-P2 promoter-derived transcripts increased by ∼30-fold after 7 h of nutlin-3 treatment in these cells (Fig. 3B). Ionizing radiation (supplemental Fig. S3A) and etoposide (supplemental Fig. S3B) also cause p53-dependent induction of this transcript. In a separate experimental system (Fig. 3C), we showed that mdmx-P2-derived transcripts failed to be induced by nutlin-3 in p53−/− MEFs, whereas they were induced in wild-type p53-expressing B16F10 mouse melanoma cells. Chromatin immunoprecipitation experiments clearly demonstrate recruitment of p53 to the predicted mdmx-P2 promoter region in nutlin-3-treated B16F10 cells. Finally, we investigated whether p53-activating stress induces the expression of mdmx-P2-derived mRNA in normal tissues in vivo. Fig. 3D shows that mdmx-P2 mRNA transcripts are detectable in the bone marrow of C57/BL6 mice and are clearly induced in response to 4 Gy of ionizing radiation. Together, these experiments provide strong evidence that the murine mdmx gene also contains a functional p53-responsive P2 promoter in intron 1, and mdmx-P2 transcripts are clearly induced in response to diverse p53-activating signals.
FIGURE 3.
The p53-responsive promoter is conserved in the mouse Mdmx gene. A, schematic representation of the 5′-end of the Mdmx gene showing the location of the Mdmx exon 1β and the p53RE in relation to exon 1 and exon 2. B, mouse embryo fibroblasts were transduced with lentiviruses expressing control shRNA or p53 shRNA. Three days later, cells were seeded, and the next day, they were treated with nutlin-3 (10 μm) for 2 and 7 h or mock-treated. Cells were harvested, and expression of indicated proteins and mRNAs was determined by Western blotting and RT-PCR. C, B16F10 mouse melanoma cells expressing wild-type p53 or p53-null MEFs were treated with nutlin-3 (10 μm, 6 h). Subsequently, cells were harvested and processed for analysis by RT-PCR, Western blotting, and ChIP. D, cDNAs made from RNAs extracted from the bone marrow of C57/BL6 mice at the indicated time points after exposure to 4 Gy of ionizing radiation were provided by Dr. Philip Coates (University of Dundee, UK) and were analyzed by RT-PCR. Error bars, S.E.
mRNA Transcribed from the Human HDMX-P2 Promoter Is Translated into HDMX-L, a Long, Functionally Distinct Form of HDMX
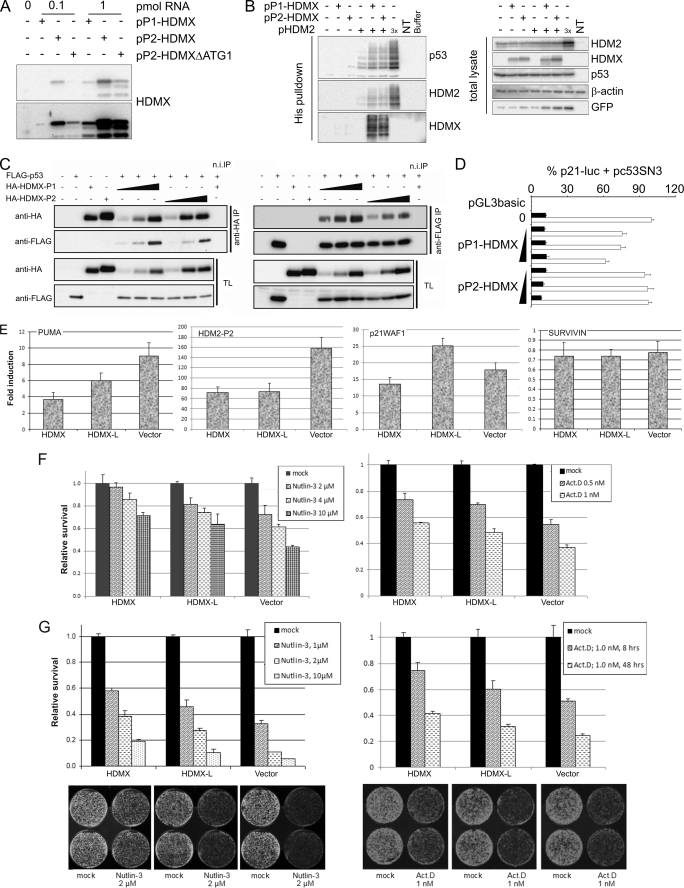
The mRNAs transcribed from the P1 and P2 promoters of human HDMX differ in the inclusion at the 5′-ends of either exon 1 or exon 1β, respectively. Exon 1 contains two potential upstream ORFs, which could potentially suppress translation of HDMX protein. Indeed, as we have already discussed, for the comparable HDM2 transcripts, HDM2-P2-derived mRNA (which lacks any upstream ORFs) can be translated up to 8-fold more efficiently than the HDM2-P1 mRNA (22–24). Exon 1β of HDMX also lacks any out-of-frame upstream ORFs but does contain an in-frame ATG that, if utilized as a translation start site, would result in the synthesis of a long form of HDMX protein with an additional 18 amino acids at its N terminus (Fig. 1A). We therefore performed a quantitative and qualitative analysis of the translation of these two HDMX transcripts. Constructs containing either exon 1 or exon 1β 5′ to exons 2–11 (pP1-HDMX and pP2-HDMX, respectively) were generated and transcribed in vitro, and equal amounts of mRNA were added to in vitro translation reactions. We also examined mRNA from pP2-HDMXΔATG1, in which the normal translation initiation site in exon 2 was mutated (Fig. 4A). Approximately 7-fold more protein was translated from the pP2-HDMX mRNA compared with pP1-HDMX mRNA. Furthermore, the protein product of pP2-HDMX-derived mRNA had a slightly reduced mobility on SDS-PAGE compared with HDMX translated from pP1-HDMX, and furthermore, the product is still present upon deletion of the AUG in exon 2. Essentially the same differences between the P1 and P2 promoter synthetic transcripts were obtained when expression vectors were transfected into human cancer cell lines (supplemental Fig. S4A; note in this experiment that the proteins had a C-terminal tag to distinguish them from endogenous cellular HDMX proteins). Thus, when expressed, the mRNA transcribed from the P2 promoter is efficiently translated from the ATG in exon 1β to generate a long form of HDMX, which has 18 additional amino acids at its N terminus, compared with HDMX. We have termed this novel protein HDMX-L.
FIGURE 4.
mRNA transcribed from the HDMX P2 promoter is efficiently translated into a long form of HDMX protein. A, RNAs transcribed from the indicated plasmids were translated in vitro using the rabbit reticulocyte lysate system. HDMX expression was determined by Western blotting. Top, short exposure; bottom, long exposure. B, H1299 cells in 6-well plates were transfected with 85 ng of pEGFP-N1, 0.33 μg of pc53SN3, and 0.67 μg of pHis6Ub. 1.33 μg of HDMX plasmid and 0.67 or 2 μg of (3×) pHDM2 were also added where stated. 24 h post-transfection cells were lysed, and His-tagged proteins were purified using Ni2+-NTA-agarose beads. HDM2, HDMX, p53, and GFP expression were determined by Western blotting. C, MCF-7 cells were transfected with the indicated constructs (HDMX-P1 and HDMX-P2, 100, 200, and 400 ng; FLAG-p53, 200 ng). The next day, cells were harvested, and protein extracts were used for immunoprecipitation with anti-HA or anti-FLAG antibodies and a nonspecific control. Immunoprecipitated proteins and total cell extracts were analyzed by Western blotting. D, 174-2 p53/mdm2 double knock-out mouse embryonic fibroblasts were transfected with p21-luc along with 300 pg of pc53SN3 (open bars) or empty vector control (solid bars) and 50, 100, or 200 ng of the indicated HDMX expression plasmid 48 h before luciferase activity was determined. Data are pooled from three independent experiments (n = 9). Expression of the ectopically expressed proteins is shown in supplemental Fig. S4E. E, MCF-7 cells stably transfected with either HDMX or HDMX-L expression vector or empty vector were treated with nutlin-3 (10 μm) for 6 h. Cells were harvested, RNA was extracted, and expression of the indicated mRNAs was determined by real-time RT-PCR. F, left, MCF-7 cells stably transfected with either HDMX- or HDMX-L expression vector or empty vector were seeded into 96-well plates (1000 cells/well), each cell line in 12 wells (left) or 9 wells (right). The next day, cells were incubated in triplicate with the indicated concentrations of nutlin-3 or actinomycin D. Relative survival of treated cells compared with mock treatment was determined after 72 h of incubation by a WST-1 assay. The experiment was repeated at least twice with similar results. Shown is a representative experiment. G, MCF-7 cells stably transfected with either HDMX or HDMX-L expression vector or empty vector were seeded into 6-well plates (10,000 cells/well). The next day, the cells were treated with the indicated concentrations of nutlin-3 for 48 h or with actinomycin D (1.0 nm) for 8 or 48 h. All conditions were in duplicate. After the treatments, medium was replaced with fresh growth medium, and cells were allowed to grow. All cells were fixed 10 days after seeding and stained with Giemsa. Plates were scanned on an Odyssey Imaging system (LI-COR Biosciences) (examples shown in the lower panels), the relative number of cells was quantified, and relative survival compared with mock-treated controls is shown in the top panels. The experiment was repeated at least twice with similar results. Error bars, S.E.
We set out to determine whether the presence of these additional amino acids has any consequence for HDMX-L regulation or function. A key point at which HDMX function is regulated is through its subcellular localization; in many proliferating cells HDMX is primarily cytoplasmic; genotoxic stress results in its ATM and 14-3-3 protein-dependent relocalization to the nucleus, where it can function to inhibit p53 (27, 56). Supplemental Fig. S4B shows that both HDMX and HDMX-L have the same subcellular distribution in both the absence and presence of etoposide-induced DNA damage. A second key point at which HDMX is regulated is through its rate of degradation via HDM2-dependent ubiquitination. HDMX and HDMX-L showed no differences in their HDM2-dependent destruction pathway, either in the absence or presence of genotoxic stress (supplemental Fig. S4, C and D).
HDMX exerts its functions through two key proteins, HDM2 and p53. HDMX-HDM2 heterodimers function as E3 ubiquitin ligases for p53, and thus HDMX can promote HDM2-dependent ubiquitination of p53 when HDM2 protein concentrations are limiting. This effect of HDMX can be seen in Fig. 4B (left, lane 5). Expression of HDMX-L from the pP2-HDMX vector had the same effect (lane 6); multiple repeats of this experiment demonstrated both HDMX-L and HDMX function comparably in this assay. Consistent with the formation of heterodimers, both HDMX and HDMX-L also promote HDM2 autoubiquitination and are themselves ubiquitinated in the presence of HDM2 (Fig. 4B). The interaction between HDMX and HDMX-L with p53 was then determined by immunoprecipitation analysis. N-terminally HA-tagged HDMX and HDMX-L were precipitated with anti-HA antibody, and the amount of FLAG-tagged p53 that was co-precipitated was determined. HDMX-L consistently pulled down less p53 protein than did HDMX (Fig. 4C, left). In the reciprocal analysis (Fig, 4C, right), immunoprecipitation of p53 clearly pulled down less HDMX-L than HDMX. These results imply that the 18-amino acid N-terminal extension of HDMX-L interferes with efficient interaction between p53 and HDMX in cells. Through its direct interaction with p53, HDMX inhibits the p53-dependent transcription from p53-responsive promoters. We therefore examined whether the reduced p53-binding efficacy of HDMX-L affects its p53-inhibitory activity (Fig. 4D). HDMX, expressed from pP1-HDMX, caused a dose-dependent reduction in the p53-dependent transcription from the p21WAF1 promoter (open bars); complete inhibition of p53 activity was not observed in this assay because HDMX requires either HDM2 binding or stress-induced 14-3-3 binding for its optimal nuclear localization that is required for its inhibition of p53. In contrast, HDMX-L expressed from the pP2-HDMX construct failed to have any effect on p53-dependent transcription in this assay. Together, these data demonstrate that, compared with HDMX translated from the constitutive P1 promoter, HDMX-L translated from the p53-inducible P2 promoter retains the ability to cooperate with HDM2 in the ubiquitination of p53 but is compromised in its ability to inhibit p53-dependent transcription through direct interaction with the transactivation domain of p53.
We subsequently established MCF-7 cell line clones stably overexpressing HDMX or HDMX-L (supplemental Fig. S4F) and vector-only cell lines as controls, distinct clones expressing equivalent amounts of the two proteins being selected for further analysis. Primarily, we have used the clones HX/C3 and HX-L/C11, which exhibit moderate expression of exogenous HDMX/HDMX-L, but HX/C6 and HX-L/C10 have also been compared with similar results. Initially, we investigated the p53 response upon treatment with nutlin-3 for 6 h by determining the induction of p53 target genes. As shown in Fig. 4E, activation of PUMA, HDM2-P2, and p21WAF1 is compromised in HDMX-expressing MCF-7 cells compared with vector-transfected controls. In HDMX-L expressing cells, HDM2-P2 induction is similarly compromised, whereas there is an intermediate inhibitory effect on PUMA induction, and p21WAF1 induction was slightly elevated compared with controls. We also investigated the SURVIVIN gene, the abundance of which was repressed by nutlin-3 to a similar extent in all three lines, and found a slight decrease, which was comparable in the different cell lines. This effect was quite modest, presumably because the 6 h time point used is too short for any transcriptional repression of the SURVIVIN gene to result in clear effects on the abundance of its mRNA.
Together, all of these results indicate that, compared with HDMX, HDMX-L is compromised in its ability to suppress the p53 response. To determine whether this effect could be recapitulated in a biological response, we determined the effect of nutlin-3 and actinomycin D on cell proliferation and survival, using both short term (72-h) cell proliferation assays (Fig. 4F) and long term colony survival assays (Fig. 4G). In both assays, HDMX overexpression conferred protection to these p53-activating compounds compared with vector-transfected controls, whereas MCF-7 cells expressing HDMX-L were also protected but to a consistently lesser extent than the HDMX-expressing cells. These results indicate that, as in the luciferase assays, the HDMX-L protein has reduced capacity to inhibit p53 activity and p53-induced antiproliferative responses.
The Role of p53-dependent Transcription of HDMX in the Feedback Control of p53
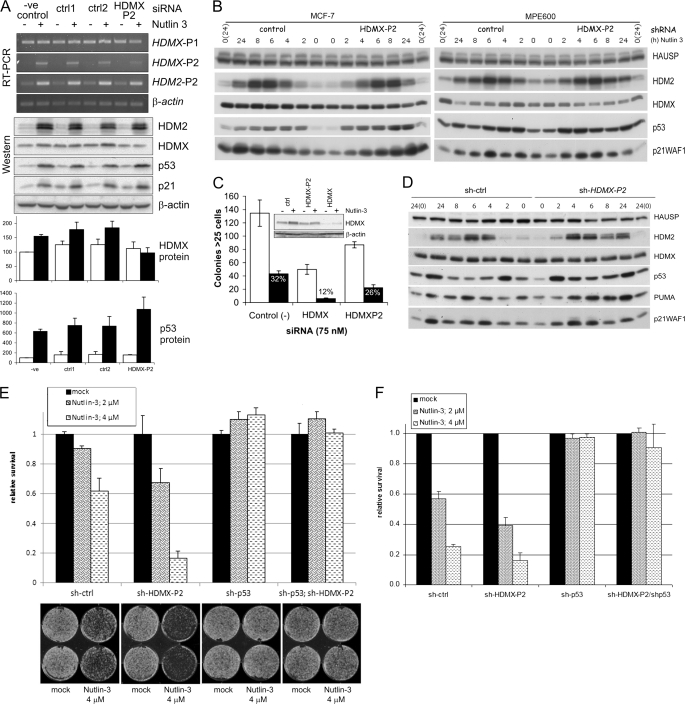
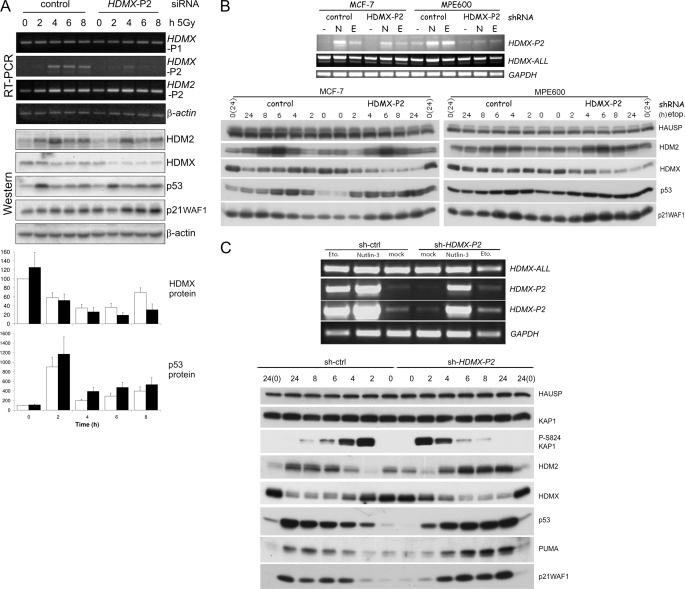
From the above data, it is clear that transcription from the HDMX-P2 promoter is inducible by a wide range of p53-activating stress in diverse human and murine cell types. We therefore wished to establish the contribution of this transcript to the abundance of HDMX proteins and the regulation of the p53 pathway in normally proliferating and stressed cells. To do this, we developed RNA interference reagents that would specifically target the HDMX-P2 transcript by recognizing sequences within the 130 bp unique to exon 1β. siRNA oligonucleotides were screened in MCF-7 cells in the absence or presence of p53-activating signals. One of the tested siRNAs most effectively reduced the abundance of p53-induced HDMX-P2 mRNA. Two further control siRNAs were synthesized based on this HDMX-P2 siRNA, which had 4 base pair mismatches in the seed and central regions, respectively. Supplemental Fig. S5A shows that although none of the control siRNAs affect either HDMX-P2 transcript levels or p53 protein abundance, the HDMX-P2 siRNA substantially reduces radiation-induced HDMX-P2 transcripts (nutlin-3 experiments are shown in Fig. 6). Exposure of MCF-7 cells to 5 Gy of ionizing radiation causes a substantial decrease in the abundance of HDMX protein (Fig. 5A and supplemental Fig. S5A), due to the activation of its ATM and HDM2-dependent degradation (note that we have used the term HDMX to refer to the endogenous ∼75-kDa HDMX proteins, which may consist of both HDMX and HDMX-L). This decrease is more pronounced in the HDMX-P2 siRNA-transfected cells than in those transfected with control siRNA (Fig. 5A and supplemental Fig. S5A); time course analysis of multiple repeated experiments (Fig. 5A, Western blot and quantification) clearly demonstrates that, in the first 2–4 h after irradiation, HDMX protein abundance drops rapidly, before leveling out at 6 h and beginning to increase again at 8 h. HDMX-P2 transcripts are up-regulated during this time frame, and the siRNA experiments clearly demonstrate that they are responsible for this early recovery of HDMX protein abundance in these cells. MCF-7 and MPE600 cells infected with a lentivirus expressing an shRNA targeting the same sequence also showed a more pronounced reduction in HDMX in response to etoposide than did control shRNA-expressing cells (Fig. 5B). Interestingly, the normal fibroblast line MRC5-hTERTneo did not detectably induce HDMX-P2 transcripts in response to 5 Gy of irradiation, and HDMX protein levels remained low for at least 24 h after radiation exposure (supplemental Fig. S5C). Therefore, in cells in which the HDMX-P2 transcript is induced in response to genotoxic stress, it makes a clear contribution to the abundance of HDMX protein and, in particular, the rate at which it recovers after its initial stress-induced degradation. This has a clear consequence for the abundance of p53 in response to DNA-damaging stress. In both of the breast cancer cell lines, the magnitude of the initial stabilization of p53 is not substantially affected by HDMX-P2 siRNA (Fig. 5, A and B); nor is the p53-dependent G1 arrest response increased (supplemental Fig. S5D). However, p53 protein stabilization is prolonged, levels remaining elevated for 24 h following etoposide treatment in HDMX-P2 knockdown cells, whereas they begin to drop toward base-line levels by 8 h in control cells (Fig. 5B). The degradation of p53 during the period following its initial stabilization in response to ionizing radiation is also delayed in HDMX-P2 siRNA-transfected MCF-7 cells (Fig. 5A). Together, these findings are consistent with a role for HDMX and HDMX-L in promoting the HDM2-dependent degradation of p53 during the attenuation phase of the stress response.
FIGURE 6.
Role of p53-dependent transcription of HDMX-P2 in the cellular response to nutlin-3. A, MCF-7 cells were transfected with siRNA to HDMX exon 1β (HDMXP2); ctr1 and ctrl2 siRNAs, which differ from HDMXP2 siRNA by 4 bases in the seed and central regions, respectively; and control siRNA. 48 h later, cells were exposed to 0 or 5 μm nutlin-3 for 6 h. RT-PCR and Western blots show results from a representative of three independent experiments. Quantifications show mean ± S.E. (error bars) changes in protein abundance for the three experiments. HDM2 and p21WAF1 data are shown in supplemental Fig. S6A. Open bars, 0 μm nutlin-3; solid bars, 5 μm nutlin-3. B, MCF-7 and MPE600 infected with lentivirus encoding either control or HDMX-P2 shRNA were treated with nutlin-3 as described in the legend to Fig. 5B. The blots show the changes in protein expression upon nutlin-3 treatment. Times shown in protein analyses are from the addition of drug. PCR analysis of mRNA transcripts following exposure of these cells to nutlin-3 is shown in Fig. 5B. C, MCF-7 cells were transfected with the indicated siRNAs; 48 h later, cells were reseeded into 6-well plates (100 cells/plate). After 24 h, cells were exposed to solvent control (open bars) or 5 μm nutlin-3 (solid bars) for 24 h. Colonies were counted after a further 11 days (n = 3). The effect of nutlin-3 on the percentage of colonies in the presence of each siRNA is shown. D, N-TERA-2 cells transduced with lentivirus encoding either control or HDMX-P2 shRNA were exposed to nutlin-3 (10 μm) as described in Fig. 5C. HDMX mRNA expression was determined 8 h after the addition of the drug and after mock treatment and is shown in Fig. 5C. Western analysis shows the changes in protein expression upon nutlin-3 treatment. Times shown in protein analyses are from the addition of drug, which was removed after 2 h. E, N-TERA-2 cells expressing the indicated shRNAs were seeded into 6-well plates (10,000 cells/well). The next day, cells were mock-treated or treated with nutlin-3 (2 or 4 μm) for 24 h, all in duplicate. Medium was replaced by fresh growth medium lacking nutlin-3, and cells were cultured for an additional 6 days. Cells were fixed, and relative survival was determined as mentioned in the legend to Fig. 4G. F, N-TERA-2 cells expressing the indicated shRNAs were seeded into 96-well plates (1000 cells/well; each cell line in 9 wells total). The next day, nutlin-3 was added (0, 2, or 4 μm), and cells were cultured for an additional 72 h. Relative survival of treated cells was determined with the use of the WST-1 assay. Data shown are the averages of three independent experiments.
FIGURE 5.
The role of p53-dependent transcription of HDMX-P2 in the cellular response to DNA damage. A, 48 h after transfection with the indicated siRNAs, MCF-7 cells were exposed to 5 Gy of ionizing radiation. Cell pellets for analysis were prepared at the indicated time points postirradiation. Quantification shows the abundance of the indicated proteins (mean ± S.E. (error bars) of seven independent experiments). HDM2 and p21WAF1 are shown in supplemental Fig. S5B. Open bars, control siRNA; solid bars, HDMX-P2 siRNA. B, MCF-7 and MPE600 infected with lentivirus encoding either control or HDMX-P2 shRNA were exposed to 20 μm etoposide (E) or 10 μm nutlin-3 (N) for 2 h or mock-treated. Drugs were then washed away, and the cells were cultured in fresh medium until lysed for analysis. Expression of HDMX mRNAs was determined 8 h after the addition of the drugs or mock treatment. The blots show the changes in protein expression upon etoposide treatment. Times shown in protein analyses are from the addition of drug. C, N-TERA-2 cells transduced with lentivirus encoding either control or HDMX-P2 shRNA were exposed to 10 μm nutlin-3 (N) or etoposide (20 μm) for 2 h, after which the drug was washed away, and the cells were cultured in fresh medium until harvested for analysis. HDMX mRNA expression was determined 8 h after the addition of the drugs and after mock-treatment. Western analysis shows the changes in protein expression upon etoposide treatment.
The above breast cancer cell lines undergo a primarily cell cycle arrest response to p53 activation (e.g. see supplemental Fig. S5D). In order to examine the role of HDMX-P2 transcripts in p53-dependent proapoptotic responses, we examined the testicular germ cell tumor line, N-TERA-2, which is highly sensitive to apoptosis induced by p53-activating DNA-damaging agents (57) or nutlin-3 (58). Furthermore, HDMX is known to be important in regulating p53 in these cells because siRNA that targets all HDMX mRNA transcripts results in the stabilization of p53 and the up-regulation of p53-responsive proteins in the absence of any other p53-activating signal (36). We therefore transduced N-TERA-2 with lentiviral constructs expressing shRNA targeting HDMX-P2 mRNA, p53, or both or a control shRNA and selected for puromycin resistance. Transduced cells were treated with etoposide for 2 h, after which medium was replaced. Cells were harvested at several time points to analyze protein and RNA expression. As shown in Fig. 5C, etoposide does increase HDMX-P2 levels in these cells, and the shRNA reduces this induction. Etoposide causes HDMX protein levels to decrease, both in the control and HDMX-P2 knockdown cells. This reduction is slightly greater in the HDMX-P2 knockdown cells, demonstrating that induction of HDMX-P2 transcripts does diminish the degree of reduction of HDMX protein abundance that occurs in these treated cells. p53 is stabilized by etoposide in control shRNA transduced cells, and p53 levels remained high for at least 24 h (Fig. 5C). In comparison, p53 abundance is more strongly increased by etoposide in the HDMX-P2 shRNA-transduced cells. In these N-TERA-2 cells, p53-induced HDMX-P2 expression is also clearly important in regulating the degree of up-regulation of p53-responsive proteins, PUMA and p21WAF1 being more strongly up-regulated in the HDMX-P2-depleted cells. Thus, as was the case in the breast cancer cells, these data demonstrate that the up-regulation of HDMX-P2 transcription is also important in attenuating the p53 response to DNA damage in this TGCT cell line.
DNA-damaging agents can have p53-independent effects on cell proliferation and survival (e.g. see Ref. 57). In order to clearly understand the role of p53-inducible HDMX-P2 promoter activity on the cellular response to p53 activation, we examined the effects of RNAi targetting HDMX-P2 mRNA in cells treated with nutlin-3, because the effects of this compound are largely p53-dependent. Treatment of MCF-7 cells caused a modest, up to 2-fold, increase in the abundance of HDMX protein (Figs. 2B and 6A). This increase is blocked by the HDMX-P2 siRNA (Fig. 6A, Western blot and quantification). HDMX-P2 siRNA also caused a reproducible enhancement of the increase in p53 protein abundance in response to nutlin-3 (Fig. 6A, Western blot and quantification). MCF-7 cells infected with a lentivirus expressing an shRNA targeting the same sequence also failed to demonstrate an increase in HDMX in response to nutlin-3, and the nutlin-3-induced increase in p53 protein abundance was enhanced (Fig. 6B). Similar effects of the shRNA were seen in nutlin-3-treated MPE600; cells expressing HDMX-P2 shRNA showed no difference in basal HDMX or p53 protein abundance, but the nutlin-3-induced reduction of HDMX and stabilization of p53 was enhanced (Fig. 6B). Together, these experiments clearly demonstrate that, in the breast cancer cells in which the HDMX-P2 transcript is induced in response to p53 activation by nutlin-3, it makes a demonstrable contribution to the abundance of HDMX protein. In contrast, when we examined MRC5-hTERTneo cells, nutlin-3 failed to detectably induce HDMX-P2 mRNA transcripts. In these cells, nutlin-3 caused a reduction, rather than increase, in HDMX protein levels (supplemental Fig. S6B), as has been reported by earlier publications using similar non-transformed fibroblast cell lines (29, 59).
We then considered the effect of HDMX-P2 RNAi on the cellular response to nutlin-3 in the breast cancer cells. In contrast to the effects on p53 protein abundance, we did not reliably detect any consistent effects of HDMX-P2 knockdown on the abundance of HDM2, p21WAF1, or PUMA (Fig. 6, A and B, and supplemental Fig. S6A) (data not shown). When subconfluent monolayers of MCF-7 cells were exposed to nutlin-3, HDMX-P2 siRNA did not enhance nutlin-3-induced cell cycle arrest or apoptosis (supplemental Fig. S6C), or long term survival (data not shown). However, when MCF-7 cells were stressed by plating at low density, their ability to form viable colonies was reduced by prior transfection with HDMX-P2 siRNA, colony formation being further reduced by the combination of the siRNA with nutlin-3 (Fig. 6C).
As mentioned above, the N-TERA-2 cells are prone to enter apoptosis upon activation of p53, whereas MCF-7 and MPE600 cells are more likely to enter a cell cycle arrest. Therefore, we tested whether N-TERA-2 would also show altered p53 activation upon nutlin-3 treatment in HDMX-P2 knockdown cells compared with controls. Cells transduced with lentiviral vectors expressing shRNA as in Fig. 5C were treated with 10 μm nutlin-3 continuously for 20 h, and RNA and protein lysates were analyzed. HDMX-P2 transcripts were strongly induced in the control cells, and HDMX-P2-specific shRNA reduces the abundance of these transcripts (supplemental Fig. S6D). This figure also shows clearly that HDMX-P2 expression is dependent on p53 in these cells. Both basal and nutlin-3-induced HDMX protein levels were marginally reduced by the HDMX-P2 shRNA; however, no differences in increase of p53 or targets were observed except when p53 shRNA was also expressed (supplemental Fig. S6D) (similar results were obtained with 6- or 8-h nutlin-3 exposure; data not shown). Nevertheless, compared with the control cells, the HDMX-P2 shRNA-expressing cells did exhibit slightly higher p53-dependent apoptosis, as determined by a poly(ADP-ribose) polymerase cleavage assay, when treated with nutlin-3. It is possible that the extended treatment with this concentration of nutlin-3 results in a nearly maximal activation of p53 that is rather insensitive to changes in HDMX abundance. Therefore, similarly to the experiment shown in Fig. 5B with the breast cancer cells, we exposed N-TERA-2 cells to nutlin-3 for only 2 h before washing it off and assaying molecular markers of the p53 response at subsequent time points (Fig. 5C). As before, HDMX-P2 transcripts are induced by nutlin-3, and this induction is reduced by HDMX-P2 shRNA. Strikingly, in control shRNA-transduced cells, p53 levels are initially induced only very transiently and decrease rapidly toward base-line levels once the drug is removed (although levels do rise again somewhat at 24 h) (Fig. 6D). In the HDMX-P2 knockdown cells, the induced levels of p53 at the 2 h time point are clearly higher than in control cells and, although p53 protein abundance does decrease upon removal of the nutlin-3, it remains elevated above base-line levels for 8 h. Furthermore, the p53 targets p21WAF1 and PUMA are more strongly induced in the HDMX-P2 knockdown cells compared with control cells, although HDM2 levels are more comparable. These effects of the HDMX-P2 shRNA on the p53 response to nutlin-3 in N-TERA-2 cells occur despite its having only a very modest effect on total HDMX protein abundance, there only being a small increase in HDMX at 2 h in the control cells that is absent in the HDMX-P2 knockdown. One possibility that is suggested by our data is that changes in the HDMX/HDMX-L ratio in these cells would occur, and these could contribute to the observed altered p53 response.
To investigate whether these effects of manipulating HDMX-P2 transcript expression on molecular aspects of the p53 response translate to an altered phenotypic response, the shRNA-expressing N-TERA-2 cells were also seeded for long term (colony assays) and short term growth assays to determine their sensitivity to nutlin-3 treatment. Based on the previously shown experiments, we reduced the concentrations of nutlin-3 used to 2 and 4 μm and treated the cells for the colony assays for only 24 h before removing nutlin-3 and replacing the medium. The results clearly show that inhibiting HDMX-P2 transcript expression sensitizes N-TERA-2 cells for nutlin-3-induced inhibition of long term cell viability (Fig. 6E). Similarly, multiple repeats of a short term growth assay show that the HDMX-P2 knockdown cells are more sensitive for nutlin-3-induced cell death (Fig. 6F). In both cases, this effect of the HDMX-P2 shRNA is entirely p53-dependent. Together, these results clearly demonstrate that the induction of HDMX-L expression in response to p53 activation suppresses the p53 response upon nutlin-3 treatment, with associated effects on cell proliferation and survival.
DISCUSSION
The activation of a p53-dependent transcriptional program is a key component of the cellular response to a diverse range of cellular stress signals. Key p53-responsive genes, such as p21WAF1 and PUMA, initiate the cell cycle arrest and proapoptotic responses; these and a wide range of other transcriptional targets of p53 implicate the p53 stress response pathway in tumorigenesis as well as other key aspects of human physiology and pathology (e.g. see Refs. 60 and 61). In proliferating cells, p53 protein is synthesized and has the potential to be active as a transcription factor (62). Cell proliferation is dependent on its abundance and activity being maintained at low levels via a dynamic equilibrium with its negative regulatory proteins HDM2 and its paralog and heterodimeric protein partner, HDMX (6, 7). A general, if not obligate, process whereby p53 is activated in response to stress involves the relief of the negative regulation of p53 by HDM2 and HDMX (7, 63). The precise mechanisms whereby this occurs depend on the nature of the stress (e.g. DNA single strand breaks trigger the ATM-dependent phosphorylation of p53, HDM2, and HDMX, promoting both p53 activation and the HDM2-dependent destruction of HDM2 and HDMX) (7, 28), whereas stresses that suppress transcription (e.g. experimentally using low dose actinomycin D) result in the binding and inhibition of HDM2 by ribosomal proteins as well as the HDM2-dependent degradation of HDMX (64). In these and other studies, HDMX, and more specifically the precise stoichiometry between p53, HDM2, and HDMX within cells (65), is revealed as a critical regulator of the response, potentially through either the ability of HDMX to bind and inhibit p53 directly or through its dimerization with HDM2 and regulation of HDM2-dependent ubiquitination of p53.
Factors influencing the abundance and activity of HDM2 and HDMX, therefore, potentially influence both the maximal intensity and duration of the p53-dependent transcriptional response to a particular stress. Regulation of the intensity of the response can be critical because p53-responsive genes differ in their sensitivity to activation by p53 (e.g. due to variations in the sequences of the p53-response elements in the promoters of the CDK inhibitor P21WAF1 versus proapoptotic genes, such as PUMA) (51). A low intensity response may induce transient cell cycle arrest, whereas a higher intensity response could induce apoptosis (66). Where transient cell cycle arrest is induced in response to acute stress, the p53 response is essentially a protect and repair signal (1) and is attenuated once the stress is relieved (63). Because prolonged p53 activation may potentially lead to apoptosis or permanent senescence, the effective attenuation of the p53 response can also be an important determinant of cellular outcome. HDM2 is known to be critical in this attenuation phase (63); the role of HDMX has not previously been determined.
HDM2 is an E3 ligase for itself as well as p53 and has a short half-life in cells; thus, changes in its rate of synthesis have an immediate and substantial effect on its cellular abundance. Its P2 promoter contains two p53-resposive elements as well as other transcription factor-binding sites, which cooperate with p53 to drive a strong transcriptional response upon p53 activation (19, 21). Furthermore, the mRNA product of the HDM2-P2 promoter can be translated into HDM2 more efficiently than that of the constitutive P1 promoter (22). With the use of conditional temperature-sensitive mutants of p53 in murine cells, a p53-induced increase of MDM2 protein abundance was readily detectable and quickly led to the identification of the p53-responsive promoter (23, 67). In contrast, HDMX is a relatively more stable protein in cells (54); it does not in itself possess significant auto-E3 ubiquitin ligase activity. Instead, its rate of turnover is dependent on its HDM2-dependent ubiquitination. Thus, upon activation of p53 in cells, the increase in HDM2 protein abundance results in increased rates of HDMX degradation. This effect has been demonstrated in experiments using nutlin-3 (29, 59) as the p53-activating agent, although the degradation of HDMX upon nutlin-3 exposure shows clear cell type specificity. In addition to this, p53-independent signaling pathways induced by DNA damage (i.e. ATM-dependent phosphorylation of HDMX) further promote its degradation by inhibiting its interaction with the deubiquitinating enzyme, HAUSP (27, 68). Thus, in many cells, stresses, such as ionizing radiation, result in a rapid decrease in HDMX protein levels that, due to relatively low rates of HDMX protein synthesis, remain low for an extended time period. HDMX protein abundance does not, therefore, substantially increase in response to stress, and HDMX had not been identified as a p53-inducible gene.
We have shown here that HDMX does indeed contain a p53-responsive promoter and that HDMX transcription can be induced in response to a wide range of p53-activating signals. The p53-RE in this HDMX-P2 promoter is a strong match to the defined optimal sequence (51). The -fold activation of the HDMX-P2 promoter by a given amount of p53 is comparable with the P21WAF1 promoter and greater than that of the BAX gene, in which the p53-RE is a weaker match to the consensus. Despite this, the absolute p53-induced activity of the HDMX-P2 promoter is lower than that of P21WAF1, and in several cell lines, we found that p53 activation does not result in substantial increases in HDMX-P2 promoter-derived transcripts. Therefore, we conclude that the HDMX/mdmX-P2 are relatively weak promoters and may require the activity of factors other than p53 that are not present in some cell types, such as the MRC5 fibroblast line. However, as is the case for HDM2, the HDMX-P2-derived mRNA is substantially more efficiently translated than that derived from the P1 promoter and does contribute to the abundance of HDMX proteins when it is expressed. Using recombinant vectors encoding synthetic cDNAs corresponding to the HDMX-P1 and HDMX-P2 promoter-derived transcripts, we determined that translation of the HDMX-P2-derived transcript is initiated from an ATG in exon 1β, giving rise to the HDMX-L form of the protein that has compromised p53 binding and compromised ability to inhibit p53-mediated transcription activation compared with HDMX. The difference in mobility of the two proteins on SDS-polyacrylamide gels is very slight, and when co-expressed as endogenous proteins, it is generally not possible to reliably distinguish between them; thus, the presence of HDMX-L within the ∼75 kDa band can only be readily determined by the reduced band intensity in cells treated with siRNA targeting the HDMX-P2 transcript.
p53-activating signals, through both the p53-induced transcription of HDM2 and post-translational modifications to HDMX itself, generally promote the degradation of HDMX protein. Other specific forms of stress (e.g. the compound leptomycin B, as we have shown here) can lead to aberrant splicing of HDMX mRNA or potentially increased degradation, as has been shown previously in response to cisplatin (32) (note that we did not observe this effect of cisplatin in our analysis of ovarian cancer cell lines, although the concentrations of the drug we used were lower than in the study by Markey and Berberich (32)). Thus, the net effect of p53-inducible HDMX transcription is, depending on the p53-activating agent and the degree of aberrant splicing of the induced transcript, to reduce the extent of DNA damage and ATM-induced reduction in HDMX proteins and promote the earlier recovery of HDMX protein abundance during the attenuation phase. In the specific case of nutlin-3, it causes a modest increase in HDMX protein abundance (in cells in which HDMX-P2 transcripts are not induced, nutlin-3 treatment actually leads to a decrease in HDMX protein abundance). An important general point, therefore, is that although the post-translational regulation of HDMX protein precludes a robust increase in its abundance in response to p53 activation, in the absence of its transcriptional induction by p53, its abundance in stressed cells is reduced, and the p53 response is enhanced or prolonged, clearly demonstrating the importance of this autoregulatory feedback mechanism of p53 regulation. The cellular response to ATM-activating DNA damage has been widely studied and has lead to the development of well defined models of the interplay between the three proteins during different stages of the response, particularly by Wahl and colleagues (7, 63). Current data indicate that in proliferating cells, HDM2-HDMX heterodimers may be a major form in which HDM2 exists as an active p53 E3 ubiqutin ligase (12, 69). In response to ATM-dependent phosphorylation events, both proteins are rapidly degraded, and p53 is consequently activated. HDM2 protein synthesis increases due to p53-dependent transcription of HDM2, further reducing HDMX levels, and, once the ATM-activating signal dissipates, HDM2 protein abundance increases to high enough levels for homodimers to form, and p53 is thus ubiquitinated and targeted for destruction, attenuating the p53 response. In cells in which HDMX transcription is not induced by p53, HDMX protein levels remain low (e.g. see supplemental Fig. S5C); this may be important to allow active HDM2 dimers to effectively bind p53 and promote its ubiquitination because monomeric HDMX could otherwise compete with HDM2 for p53 binding. Contrasting with this, however, in the presence of HDMX, active HDM2 E3 ubiquitin ligase complexes potentially form at lower concentrations of HDM2. Thus, it is of particular interest that, at least in humans, the p53-inducible HDMX mRNA encodes a form of HDMX (i.e. HDMX-L), which has reduced p53-binding activity while retaining the ability to bind HDM2 and promote its activity. Thus, the formation of HDM2-HDMX-L heterocomplexes would expedite the clearance of stress-induced p53 during the attenuation phase, in the absence of competition for p53 binding by HDMX. Precisely how the additional N-terminal 18 amino acids in HDMX-L affect p53 binding remains to be determined, although parallels may exist with the so-called lid region present at the N terminus of HDM2, which inhibits p53 binding by the p53-binding pocket of this protein (70). The results from our RNAi experiments in ionizing radiation- or etoposide-treated breast cancer cells, as compared with MRC5 fibroblasts, are entirely consistent with the above model of HDMX-L function in cells. In the TGCT cell line, N-TERA-2, p53 activation induces a primarily proapoptotic response; any role of autoregulatory feedback loops in these circumstances is less clear, and the cells essentially do not recover from the p53-activating stress. On a final note, it is interesting that, although murine mdmx does contain a p53-responsive promoter, the p53-inducible transcripts encode the MDMX protein rather than a longer, p53 binding-compromised form. The expression of HDMX-L in humans may be a relatively late evolutionary development that engenders further complexity in the p53 response.
Finally, we have in this study performed an in depth analysis of the role of p53-dependent transcription from the HDMX-P2 promoter in the cellular response to nutlin-3. Nutlin-3 is one of the first developed small molecule inhibitors of the p53-HDM2 interaction that bind HDM2 in its p53-binding pocket. HDM2 inhibitors, including nutlin-3, have proven to be promising anti-cancer agents in preclinical cancer models (7, 30) and are currently in early phase clinical trials. Although nutlin-3 is able to inhibit the HDMX-p53 interaction to some extent and can inhibit the growth of HDMX-overexpressing cell lines (71), it has also been found that it does not inhibit HDMX as effectively as it does HDM2, and the presence of high levels of HDMX or the apparent failure of nutlin-3 to induce the degradation of HDMX can provide relative resistance to nutlin-3 (29, 59, 72). Here we have shown that, rather than a failure to degrade HDMX, in the cell lines we have studied, nutlin-3 does not reduce HDMX protein levels largely due to p53-dependent transcriptional activation of HDMX, and indeed HDMX protein abundance actually increases somewhat in response to nutlin-3. RNAi-mediated knockdown of the HDMX-P2 transcript in MCF-7 cells and other breast cancer cells lines in which this transcript is induced by nutlin-3 reduces the abundance of HDMX proteins in the nutlin-3-treated cells. Interestingly, the nutlin-3-induced increase in p53 protein abundance is also increased in these RNAi-treated cells. Thus, despite the blockade of HDM2-p53 binding in these cells, HDMX is still apparently able to influence p53 degradation. Potentially, at the high levels of HDM2 present in nutlin-3-treated cells, a small proportion of this is still able to bind p53 and target it for ubiquitination, or, as has been demonstrated recently, interaction between secondary docking sites in p53 and HDM2 can be sufficient to result in p53 ubiquitination, which can thus occur in the presence of nutlin-3 (73). In either of these circumstances, the presence of HDMX or HDMX-L could potentially promote the ubiquitination of p53 by HDM2 in cells through the formation of heterodimers.
Despite this, we did not detect any substantial effects of pretreatment with RNAi to HDMX-P2 on the ability of nutlin-3 to induce p53-dependent target genes in our experiments in the breast cancer cells, nor was there a marked shift from a cell cycle arrest to apoptotic response; indeed, we only detected effects of HDMX-P2 knockdown in MCF-7 cells when the cells were subjected to single cell colony-forming assays, conditions whereby a proapoptotic response can be favored due to anoikis. One explanation for this general lack of a sensitizing effect is that, in this cell type, the effect of nutlin-3 is to induce primarily a p21WAF1 response that induces a reversible cell cycle arrest in G1 phase rather than apoptosis, as has been previously reported (29). It was interesting, therefore, to study the testicular germ cell tumor cells, in which p53 activation induces predominantly a proapoptotic response and in which Li et al. (36) have also recently provided some good evidence for the existence of a p53-HDMX autoregulatory feedback loop.
In these cells, nutlin-3 did not result in a decrease in the abundance of HDMX; in fact, as in MCF-7 cells, there was a small increase. In the N-TERA-2 cell line that we studied, this increase in HDMX was dependent upon the induction of HDMX-P2 transcripts. Of note, in their analysis, which involved the quantification of total HDMX mRNA rather than individual promoter-derived transcripts, Li et al. (36) did not identify any p53-dependent regulation of HDMX in this particular TGCT cell line; this illustrates the point that, when experiments are performed to specifically identify and manipulate HDMX-P2 promoter-derived mRNA transcripts, a functional p53-HDMX autoregulatory feedback loop is demonstrably present in a much wider range of human tumor cells than could be predicted from the analysis and manipulation of total HDMX mRNA alone. In N-TERA-2 cells, despite the failure to reduce HDMX, nutlin-3 still induces a strong apoptotic response. However, it is clear that the up-regulation of HDMX expression does limit this nutlin-3 response because both the increase in p53 protein abundance and the expression of p53-responsive proteins, notably PUMA, are up-regulated, and both short term survival and long term proliferative potential are reduced when cells are pretreated with RNAi to the HDMX-P2 transcript. In conclusion, the p53-dependent transcriptional induction of a novel HDMX mRNA transcript that is efficiently translated into HDMX-L protein is clearly able to influence the cellular response to p53 activation. A marked difference exists between the apparent ability of cells of different origins to induce HDMX expression in response to stress, with likely consequences on the eventual outcome to the cell. These novel findings will help provide further clarity to our increasing understanding of this critical stress response pathway.
Supplementary Material
Acknowledgments
We are grateful to Phil Coates for providing cDNAs from irradiated mouse tissues, Gigi Lozano for making available the p53/mdm2 double knock-out MEFs, Gordon Peters for the gift of NARF cells and anti-p14ARF antibody 4C6, and Karen Vousden for the gift of the p53-inducible SAOS-2 cells. We thank Theo van Laar for the irradiation of MEFs and Ute Rolle for excellent technical assistance.
This work was supported by Association for International Cancer Research Grants 07-0437 (to J. P. B.) and 05-273 (to A. G. J.), Dutch Cancer Society Grant UL 2006–3595, and European Community FP6 funding (Contract 503576) (to A. G. J.). This work was also supported by Wilhelm-Sander-Stiftung Grant 2006.010.1, Deutsche Krebshilfe Grant 108424, Wilhelm-Roux-Programm of the University of Halle-Wittenberg Grant 12/40, and Fritz-Thyssen-Stiftung Grant Az 10.09.2.117.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
- MEF
- murine embryo fibroblast
- IP
- immunoprecipitation
- p53-RE
- p53-response element
- TGCT
- testicular germ cell tumor
- ARF
- alternate reading frame
- EST
- expressed sequence tag
- Gy
- gray(s).
REFERENCES
- 1.Vogelstein B., Lane D., Levine A. J. (2000) Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 2.Christophorou M. A., Ringshausen I., Finch A. J., Swigart L. B., Evan G. I. (2006) Nature 443, 214–217 [DOI] [PubMed] [Google Scholar]
- 3.Wynford-Thomas D., Blaydes J. (1998) Carcinogenesis 19, 29–36 [DOI] [PubMed] [Google Scholar]
- 4.Hupp T. R., Lane D. P., Ball K. L. (2000) Biochem. J. 352, 1–17 [PMC free article] [PubMed] [Google Scholar]
- 5.Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 6.Marine J. C., Jochemsen A. G. (2005) Biochem. Biophys. Res. Commun. 331, 750–760 [DOI] [PubMed] [Google Scholar]
- 7.Toledo F., Wahl G. M. (2007) Int. J. Biochem. Cell Biol. 39, 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uesugi M., Verdine G. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14801–14806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shvarts A., Steegenga W. T., Riteco N., van Laar T., Dekker P., Bazuine M., van Ham R. C., van der Houven van Oordt W., Hateboer G., van der Eb A. J., Jochemesen A. G. (1996) EMBO J. 15, 5349–5357 [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y., Zhao W., Chen Y., Zhao Y., Gu W. (2008) Cell 133, 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks C. L., Gu W. (2006) Mol. Cell 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linares L. K., Hengstermann A., Ciechanover A., Müller S., Scheffner M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12009–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamoto K., Taya Y., Nakagama H. (2009) FEBS Lett. 583, 2710–2714 [DOI] [PubMed] [Google Scholar]
- 14.Uldrijan S., Pannekoek W. J., Vousden K. H. (2007) EMBO J. 26, 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu J., Kawai H., Nie L., Kitao H., Wiederschain D., Jochemsen A. G., Parant J., Lozano G., Yuan Z. M. (2002) J. Biol. Chem. 277, 19251–19254 [DOI] [PubMed] [Google Scholar]
- 16.Bond G. L., Hu W., Bond E. E., Robins H., Lutzker S. G., Arva N. C., Bargonetti J., Bartel F., Taubert H., Wuerl P., Onel K., Yip L., Hwang S. J., Strong L. C., Lozano G., Levine A. J. (2004) Cell 119, 591–602 [DOI] [PubMed] [Google Scholar]
- 17.Alt J. R., Greiner T. C., Cleveland J. L., Eischen C. M. (2003) EMBO J. 22, 1442–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onel K., Cordon-Cardo C. (2004) Mol. Cancer Res. 2, 1–8 [PubMed] [Google Scholar]
- 19.Zauberman A., Flusberg D., Haupt Y., Barak Y., Oren M. (1995) Nucleic Acids Res. 23, 2584–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danovi D., Meulmeester E., Pasini D., Migliorini D., Capra M., Frenk R., de Graaf P., Francoz S., Gasparini P., Gobbi A., Helin K., Pelicci P. G., Jochemsen A. G., Marine J. C. (2004) Mol. Cell. Biol. 24, 5835–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelps M., Darley M., Primrose J. N., Blaydes J. P. (2003) Cancer Res. 63, 2616–2623 [PubMed] [Google Scholar]
- 22.Brown C. Y., Mize G. J., Pineda M., George D. L., Morris D. R. (1999) Oncogene 18, 5631–5637 [DOI] [PubMed] [Google Scholar]
- 23.Barak Y., Gottlieb E., Juven-Gershon T., Oren M. (1994) Genes Dev. 8, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 24.Landers J. E., Cassel S. L., George D. L. (1997) Cancer Res. 57, 3562–3568 [PubMed] [Google Scholar]
- 25.Stommel J. M., Wahl G. M. (2004) EMBO J. 23, 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L., Gilkes D. M., Pan Y., Lane W. S., Chen J. (2005) EMBO J. 24, 3411–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereg Y., Shkedy D., de Graaf P., Meulmeester E., Edelson-Averbukh M., Salek M., Biton S., Teunisse A. F., Lehmann W. D., Jochemsen A. G., Shiloh Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y. V., Leblanc M., Wade M., Jochemsen A. G., Wahl G. M. (2009) Cancer Cell 16, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wade M., Wong E. T., Tang M., Stommel J. M., Wahl G. M. (2006) J. Biol. Chem. 281, 33036–33044 [DOI] [PubMed] [Google Scholar]
- 30.Vassilev L. T. (2007) Trends Mol. Med. 13, 23–31 [DOI] [PubMed] [Google Scholar]
- 31.Chandler D. S., Singh R. K., Caldwell L. C., Bitler J. L., Lozano G. (2006) Cancer Res. 66, 9502–9508 [DOI] [PubMed] [Google Scholar]
- 32.Markey M., Berberich S. J. (2008) Oncogene 27, 6657–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouvard V., Zaitchouk T., Vacher M., Duthu A., Canivet M., Choisy-Rossi C., Nieruchalski M., May E. (2000) Oncogene 19, 649–660 [DOI] [PubMed] [Google Scholar]
- 34.Mendrysa S. M., Perry M. E. (2000) Mol. Cell. Biol. 20, 2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei C. L., Wu Q., Vega V. B., Chiu K. P., Ng P., Zhang T., Shahab A., Yong H. C., Fu Y., Weng Z., Liu J., Zhao X. D., Chew J. L., Lee Y. L., Kuznetsov V. A., Sung W. K., Miller L. D., Lim B., Liu E. T., Yu Q., Ng H. H., Ruan Y. (2006) Cell 124, 207–219 [DOI] [PubMed] [Google Scholar]
- 36.Li B., Cheng Q., Li Z., Chen J. (2010) Cell Cycle 9, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakano K., Bálint E., Ashcroft M., Vousden K. H. (2000) Oncogene 19, 4283–4289 [DOI] [PubMed] [Google Scholar]
- 38.De Waard-Siebinga I., Blom D. J., Griffioen M., Schrier P. I., Hoogendoorn E., Beverstock G., Danen E. H., Jager M. J. (1995) Int. J. Cancer 62, 155–161 [DOI] [PubMed] [Google Scholar]
- 39.Tait L., Dawson P. J., Wolman S. R., Galea K., Miller F. R. (1996) Int. J. Oncol. 9, 263–267 [DOI] [PubMed] [Google Scholar]
- 40.Blaydes J. P., Hupp T. R. (1998) Oncogene 17, 1045–1052 [DOI] [PubMed] [Google Scholar]
- 41.Chen J., Marechal V., Levine A. J. (1993) Mol. Cell. Biol. 13, 4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kessler B. M., Fortunati E., Melis M., Pals C. E., Clevers H., Maurice M. M. (2007) J. Proteome Res. 6, 4163–4172 [DOI] [PubMed] [Google Scholar]
- 43.Wiederschain D., Gu J., Yuan Z. M. (2001) J. Biol. Chem. 276, 27999–28005 [DOI] [PubMed] [Google Scholar]
- 44.Cullen B. R. (2006) Nat. Methods 3, 677–681 [DOI] [PubMed] [Google Scholar]
- 45.Lam S., Lodder K., Teunisse A. F., Rabelink M. J., Schutte M., Jochemsen A. G. (2010) Oncogene 29, 2415–2426 [DOI] [PubMed] [Google Scholar]
- 46.Brummelkamp T. R., Bernards R., Agami R. (2002) Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 47.Dirac A. M., Bernards R. (2003) J. Biol. Chem. 278, 11731–11734 [DOI] [PubMed] [Google Scholar]
- 48.Xirodimas D., Saville M. K., Edling C., Lane D. P., Laín S. (2001) Oncogene 20, 4972–4983 [DOI] [PubMed] [Google Scholar]
- 49.Ohkubo S., Tanaka T., Taya Y., Kitazato K., Prives C. (2006) J. Biol. Chem. 281, 16943–16950 [DOI] [PubMed] [Google Scholar]
- 50.Shvarts A., Bazuine M., Dekker P., Ramos Y. F., Steegenga W. T., Merckx G., van Ham R. C., van der Houven van Oordt W., van der Eb A. J., Jochemsen A. G. (1997) Genomics 43, 34–42 [DOI] [PubMed] [Google Scholar]
- 51.Inga A., Storici F., Darden T. A., Resnick M. A. (2002) Mol. Cell. Biol. 22, 8612–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rallapalli R., Strachan G., Cho B., Mercer W. E., Hall D. J. (1999) J. Biol. Chem. 274, 8299–8308 [DOI] [PubMed] [Google Scholar]
- 53.Pan Y., Chen J. (2003) Mol. Cell. Biol. 23, 5113–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawai H., Wiederschain D., Kitao H., Stuart J., Tsai K. K., Yuan Z. M. (2003) J. Biol. Chem. 278, 45946–45953 [DOI] [PubMed] [Google Scholar]
- 55.Stott F. J., Bates S., James M. C., McConnell B. B., Starborg M., Brookes S., Palmero I., Ryan K., Hara E., Vousden K. H., Peters G. (1998) EMBO J. 17, 5001–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LeBron C., Chen L., Gilkes D. M., Chen J. (2006) EMBO J. 25, 1196–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burger H., Nooter K., Boersma A. W., van Wingerden K. E., Looijenga L. H., Jochemsen A. G., Stoter G. (1999) Int. J. Cancer 81, 620–628 [DOI] [PubMed] [Google Scholar]
- 58.Bauer S., Muhlenberg T., Leahy M., Hoiczyk M., Gauler T., Schuler M., Looijenga L. (2010) Eur. Urol. 57, 679–687 [DOI] [PubMed] [Google Scholar]
- 59.Patton J. T., Mayo L. D., Singhi A. D., Gudkov A. V., Stark G. R., Jackson M. W. (2006) Cancer Res. 66, 3169–3176 [DOI] [PubMed] [Google Scholar]
- 60.Hu W., Feng Z., Teresky A. K., Levine A. J. (2007) Nature 450, 721–724 [DOI] [PubMed] [Google Scholar]
- 61.Vousden K. H., Ryan K. M. (2009) Nat. Rev. Cancer 9, 691–700 [DOI] [PubMed] [Google Scholar]
- 62.Blaydes J. P., Wynford-Thomas D. (1998) Oncogene 16, 3317–3322 [DOI] [PubMed] [Google Scholar]
- 63.Wahl G. M. (2006) Cell Death Differ. 13, 973–983 [DOI] [PubMed] [Google Scholar]
- 64.Gilkes D. M., Chen L., Chen J. (2006) EMBO J. 25, 5614–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y. V., Wade M., Wahl G. M. (2009) Cell Cycle 8, 3443–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knights C. D., Catania J., Di Giovanni S., Muratoglu S., Perez R., Swartzbeck A., Quong A. A., Zhang X., Beerman T., Pestell R. G., Avantaggiati M. L. (2006) J. Cell Biol. 173, 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu X., Bayle J. H., Olson D., Levine A. J. (1993) Genes Dev. 7, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 68.Meulmeester E., Maurice M. M., Boutell C., Teunisse A. F., Ovaa H., Abraham T. E., Dirks R. W., Jochemsen A. G. (2005) Mol. Cell 18, 565–576 [DOI] [PubMed] [Google Scholar]
- 69.Kawai H., Lopez-Pajares V., Kim M. M., Wiederschain D., Yuan Z. M. (2007) Cancer Res. 67, 6026–6030 [DOI] [PubMed] [Google Scholar]
- 70.McCoy M. A., Gesell J. J., Senior M. M., Wyss D. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1645–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laurie N. A., Donovan S. L., Shih C. S., Zhang J., Mills N., Fuller C., Teunisse A., Lam S., Ramos Y., Mohan A., Johnson D., Wilson M., Rodriguez-Galindo C., Quarto M., Francoz S., Mendrysa S. M., Guy R. K., Marine J. C., Jochemsen A. G., Dyer M. A. (2006) Nature 444, 61–66 [DOI] [PubMed] [Google Scholar]
- 72.Hu B., Gilkes D. M., Farooqi B., Sebti S. M., Chen J. (2006) J. Biol. Chem. 281, 33030–33035 [DOI] [PubMed] [Google Scholar]
- 73.Wallace M., Worrall E., Pettersson S., Hupp T. R., Ball K. L. (2006) Mol. Cell 23, 251–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.