Abstract
The Pim-1 protein kinase plays an important role in regulating both cell growth and survival and enhancing transformation by multiple oncogenes. The ability of Pim-1 to regulate cell growth is mediated, in part, by the capacity of this protein kinase to control the levels of the p27, a protein that is a critical regulator of cyclin-dependent kinases that mediate cell cycle progression. To understand how Pim-1 is capable of regulating p27 protein levels, we focused our attention on the SCFSkp2 ubiquitin ligase complex that controls the rate of degradation of this protein. We found that expression of Pim-1 increases the level of Skp2 through direct binding and phosphorylation of multiple sites on this protein. Along with known Skp2 phosphorylation sites including Ser64 and Ser72, we have identified Thr417 as a unique Pim-1 phosphorylation target. Phosphorylation of Thr417 controls the stability of Skp2 and its ability to degrade p27. Additionally, we found that Pim-1 regulates the anaphase-promoting complex or cyclosome (APC/C complex) that mediates the ubiquitination of Skp2. Pim-1 phosphorylates Cdh1 and impairs binding of this protein to another APC/C complex member, CDC27. These modifications inhibit Skp2 from degradation. Marked increases in Skp2 caused by these mechanisms lower cellular p27 levels. Consistent with these observations, we show that Pim-1 is able to cooperate with Skp2 to signal S phase entry. Our data reveal a novel Pim-1 kinase-dependent signaling pathway that plays a crucial role in cell cycle regulation.
Keywords: CDK (Cyclin-dependent Kinase), Cell Cycle, E3 Ubiquitin Ligase, Oncogene, Protein Degradation, APC/C, Pim Kinase, Skp2, p27, Ubiquitination
Introduction
The Pim family of serine/threonine kinases regulates the growth and survival of cells and plays a role in enhancing the transformed phenotype driven by oncogenes, including Myc and Akt (1–3). As the Pim kinases are elevated in human tumors, including prostate, leukemia, and pancreatic cancer, and appear to be useful in distinguishing benign from malignant tumors (4), it has been suggested that they play a role in the growth or progression of these malignancies (5, 6). In prostate cancer, decreased Pim-1 expression correlated significantly with measures of poor outcome and was found to be associated with a higher cumulative rate of prostate-specific antigen failure and a strong predictor of prostate-specific antigen recurrence (4). Based on crystal structural analysis (7–11), the Pim family of kinases appears to be constitutively active and not regulated by a kinase cascade. To explain the ability of the Pim protein kinases to regulate growth and survival, research has initially focused on the ability of these protein kinases to regulate CDC25A and CDC25C, p21Waf1, and the C-TAK12 protein kinase (12–14). Recently, Pim-1 has been shown to increase the cyclin-dependent kinase-2 activity, by decreasing the levels of p27Kip1 (p27) protein (15). Similarly, we have demonstrated that small molecule inhibitors of Pim-1 both translocate the p27 protein to the nucleus and markedly increase its levels (16, 17), suggesting that inhibiting Pim-1 activity may regulate the cell cycle by controlling p27 levels and localization.
The SCFSkp2 ubiquitin ligase (Skp1/cullin/F-box protein) targets cell cycle negative regulators p27, p21Waf1, and p130 (18) to the proteasome for degradation and controls progression through the cell cycle. A key protein in this complex Skp2 binds phosphorylated p27 and is responsible for its destruction. The fact that increased Skp2 expression is frequently found in many cancers (19, 20) and Skp2 overexpression can drive cell transformation suggests the importance of the levels of this protein in regulating cell growth (19, 21, 22). The amount of the Skp2 protein in cells is tightly regulated by multiple pathways, including phosphorylation and proteasome degradation. The anaphase-promoting complex or cyclosome (APC/C) is active from mitosis to late G1 (23, 24) and functions as the E3 ligase for this protein when activated by Cdh1 (25, 26). Phosphorylation of Skp2 by CDK2 (27) and Akt1 (28, 29) on Ser64 and Ser72 protects it from degradation by the APC/CCdh1 complex and elevates the levels of this protein. However, the role of Skp2 Ser72 phosphorylation is under debate as contradictory findings have been reported (30, 31). Further studies are required to elucidate fully the mechanisms by which cells regulate Skp2 levels.
Here, we demonstrate that Pim-1 kinase activity stabilizes and increases the levels of Skp2 protein, thus decreasing p27 levels and promoting cell cycle progression. Pim-1 both binds Skp2 and phosphorylates it on Ser64 and Ser72, but also on a novel site, Thr417. Furthermore, Pim-1 phosphorylates Cdh1, impairing its association with CDC27 and inhibiting APC/C activity, thus protecting Skp2 from degradation.
EXPERIMENTAL PROCEDURES
Antibodies, Drugs, and Reagents
Anti-Pim-1 (19F7) antibody was produced and purified in this laboratory. Anti-cyclin E (HE12), anti-Met (25H2), anti-phospho-Met (D26), Myc tag (71D10), anti-AKT, anti-phospho-AKT (S473), and anti-polo-like kinase-1 antibodies were purchased from Cell Signaling Technology. Anti-p27 (C19), anti-CDC27 (AF3.1), and anti-cyclin B1 (H-433) were from Santa Cruz Biotechnology. Anti-β-actin (AC-15), anti-FLAG M2, anti-HA (HA-7), and anti-β-tubulin (TUB 2.1) antibodies were from Sigma. Anti-Skp2 and anti-Cks1 antibodies were from Invitrogen/Zymed Laboratories Inc.. Anti-His tag antibody was from Qiagen. Anti-Cdh1(DH01) antibody was from Abcam. Anti-lamin B antibody was from Calbiochem.
Roscovitine and reagents for in vitro ubiquitination assay were from Biomol. Cycloheximide, MG132, LY294002, wortmannin, nocodazole, and thymidine were from Sigma. GSK690693 was provided by Glaxo Smith Kline.
Recombinant human HGF was from Antigenix America. Active GST-tagged Pim-1 was from SignalChem. Active His-tagged human Pim-1 was purified from Escherichia coli using a Calbiochem nickel-nitrilotriacetic acid column. GST and GST-Skp2 proteins were purified from E. coli using glutathione-Sepharose 4B resin (GE Healthcare).
Plasmids
A Pim-1 siRNA plasmid and the control plasmid were described previously (32). pGIPZ Pim-1 shRNA constructs were from Open Biosystems.
pCMV-Skp2 plasmid expressing FLAG-tagged Skp2 was kindly provided by Dr. Liang Zhu (33). Site-directed mutants were prepared using PCR based on this plasmid. HA-Cdh1 and HA-Cdc20 plasmids were described elsewhere (34). The Ubc3 and ubiquitin plasmids have been previously described (35). The HA-Pim-1 and FLAG-Pim-1 constructs were generated by subcloning murine Pim-1 cDNA into pcDNA3 vector, and the K67M (HA-tagged) mutant was constructed using PCR. An N-terminally truncated mutant (NT81) of Pim-1 was described previously (36). Lentiviral expression constructs pLEX-Pim-1 and pLEX-Skp2 was obtained by subcloning human Pim-1 and Skp2 cDNAs into pLEX vector (Open Biosystems). A human Pim-1 construct, pcDNA3-Pim-1, was described elsewhere (32).
Cell Culture, Transfections, Transductions, and Cell Synchronization
Cell lines were grown in RPMI 1640 medium (PC3) or DMEM (HeLa, HEK293T, Rat1, and mouse embryonic fibroblasts). The triple knock-out mouse of the Pim-1, -2, -3 genes used to isolate mouse embryonic fibroblasts were described previously (17). Mouse prostate epithelial cells were isolated as described (37). HEK293T cells were transfected by the calcium phosphate method, and HeLa cells were transfected with Lipofectamine 2000 reagent. Lentiviruses were produced and transduced into Rat1 cells using kits from Open Biosystems.
For synchronization experiments, HeLa cells were treated with 2 mm thymidine for 18 h, washed, and released into fresh medium for 9 h. Then, a second thymidine treatment was applied to yield cells at the G1/S transition. Mitotic HeLa cells were obtained by treating HeLa cells with 2 mm thymidine for 24 h, washing, and releasing into fresh medium for 3 h. The cells were then treated with 100 ng/ml nocodazole for 12 h.
Ubiquitination Assays
In vitro p27 ubiquitination assays were performed essentially as described (38). In brief, the SCFSkp2 complex was expressed and purified from insect cells (39) and mixed with in vitro-translated 35S-labeled p27 that had previously been incubated with cyclin E/Cdk2 along with methylated ubiquitin and ubiquitin aldehyde for 60 min at 30 °C. The reaction was stopped with 2× SDS sample buffer and run on polyacrylamide gels. In vivo ubiquitination assays were performed as described (40). HEK293T cells were transfected with the indicated plasmids for 24 h, treated with 10 μm MG132 for 6 h, and lysed in denaturing buffer (6 m guanidine-HCl, 0.1 m Na2HPO4/NaH2PO4, 10 mm imidazole). The cell extracts were then incubated with nickel beads for 3 h, washed, and subjected to immunoblot analysis.
In Vitro and in Vivo Phosphorylation Assay
FLAG-Skp2 or its mutants were immunoprecipitated with anti-FLAG antibody from HEK293T cells. Immune complexes were washed three times in radioimmune precipitation assay lysis buffer (150 mm NaCl, 10 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS), then washed twice in 1× kinase buffer (25 mm Tris-HCl, pH 7.5, 5 mm β-glycerophosphate, 2 mm dithiothreitol, 0.1 mm Na3VO4, 10 mm MgCl2, 2 μm unlabeled ATP) and incubated with 0.5 μg of recombinant active Pim-1 kinase and 2 μCi of [γ-32P]ATP in 3 μl of total reaction buffer for 30 min at 30 °C. Phosphorylation of Cdh1 or Cdc20 was detected using in vitro translated proteins produced by TNT Coupled Reticulcyte Lysate System (Promega). Reactions were stopped by washing twice in kinase buffer and boiling in 2× SDS loading buffer. Proteins were resolved by 9% SDS-PAGE, and 32P incorporation was detected by autoradiography. For in vivo labeling experiments, HeLa cells were transfected with the indicated plasmids for 24 h, and the medium was changed to phosphate-free DMEM with 0.5% dialyzed FBS containing 200 μCi ml−1 ortho-32PO4 for 4 h. Cells were lysed by radioimmune precipitation assay buffer for immunoprecipitation, and the immune complexes were subjected to 9% SDS-PAGE followed by autoradiography analysis.
Flow Cytometry
Cell cycle distribution was monitored by FACS analysis of ethanol-fixed, propidium iodide-stained cells on a Becton Dickinson FACSCalibur Analytical Flow Cytometer.
BrdU Incorporation Assay
Rat1 cells were seeded in 96-well plates (3000 cells/well) and maintained as described in the figure legends. An ELISA BrdU kit (Roche Applied Science) was used to assay the cell cycle. Absorbance at 370 nm (reference wavelength 492 nm) was measured using a Molecular Devices microplate reader.
Densitometry Analysis
Densitometry was determined with ImageJ version 1.42q software (National Institutes of Health) with normalization to the corresponding controls (β-actin or input).
Statistical Analysis
All assays were repeated at least three times. The results of quantitative studies are reported as mean ± S.D. Differences were analyzed by Student's t test. p < 0.05 was regarded as significant, and such differences are indicated in the figures.
RESULTS
Pim-1 Stabilizes Skp2 Protein
Overexpression in HeLa cells of wild type Pim-1 but not a kinase-dead mutant, K67M, leads to a decrease in the level of the p27 protein (Fig. 1A) without any change in the mRNA level of this protein (supplemental Fig. S2D). To evaluate the mechanism by which Pim-1 functions, we focused attention on the E3 ligase SCF complex that targets p27 for proteasomal degradation and in particular the Skp2 protein which is known to directly bind p27. Western blots demonstrate that transfection of the Pim-1 kinase increases the levels of Skp2 protein (Fig. 1A), while conversely siRNA or shRNA (supplemental Fig. S1A) knockdown of endogenous Pim-1 expression reduces Skp2 levels. The interplay between these two proteins is further demonstrated by the observation that transfection of murine Pim-1 into HeLa cells in which endogenous enzyme has been knocked down again elevates the level of Skp2 (Fig. 1A). Using two small molecule Pim kinase inhibitors, SMI-4a, which has demonstrated excellent selectivity (16, 17, 41), and a structurally unrelated Pim kinase inhibitor, K00135 (27), treatment of both HeLa cells (Fig. 1B) and PC3 prostate cancer cells (supplemental Fig. S1, B and C) causes a dose-dependent reduction of Skp2 protein expression and a concomitant rise in p27. We and others have shown that Pim-1 facilitates cell cycle progression as overexpression of Pim-1 promotes G1-S transition (15) whereas Pim kinase inhibitor caused cell cycle arrest at G1 (16). Because the Akt protein kinase family is thought to control the level of Skp2 (28, 29), we evaluated whether the PI3K inhibitor, wortmannin or a pan-Akt inhibitor, GSK690693, had similar affects on Skp2 levels. However, no significant changes in the levels of Skp2 were seen after treatment with these reagents until the highest concentrations tested (supplemental Fig. S1, D and E). Interestingly, LY294002, which is both a PI3K and Pim-1 inhibitor (9), reduced Skp2 expression (supplemental Fig. S1E).
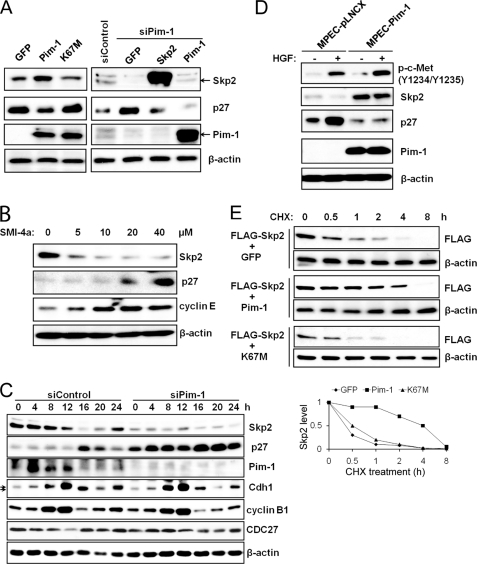
FIGURE 1.
Regulation of Skp2 protein levels by Pim-1. A, HeLa cells were transiently transfected with cDNAs encoding green fluorescent protein (GFP), Pim-1, kinase-dead Pim-1 (K67M), or a siRNA to Pim-1 together with GFP, or Skp2, or Pim-1, or a scrambled sequence. Forty-eight h after transfection, extracts of these cells were probed on Western blots with the listed antibodies. B, HeLa cells were treated with various concentrations of Pim kinase inhibitor SMI-4a for 16 h, extracts were prepared, and immunoblotting was carried out with the identified antibodies. C, HeLa cells were transfected with the indicated siRNA plasmids followed by a double-thymidine block treatment (see “Experimental Procedures”). Lysates were prepared at the indicated time points after release from the thymidine block and subjected to immunoblot analysis. Arrows indicate phosphorylated and unphosphorylated forms of Cdh1. D, mouse prostate epithelial cells (MPECs) stably transfected with a control vector (pLNCX) or a human Pim-1-expressing plasmid were treated with HGF (50 ng/ml) for 24 h followed by immunoblot analysis. E, 24 h after transfection with expression plasmids (time 0), HEK293T cells were incubated for the indicated times with cycloheximide (CHX, 100 μg/ml) followed by immunoblot analysis with FLAG or β-actin antibodies. Densitometric analysis was performed using ImageJ software to quantify the expression of Skp2. Skp2 band intensity was normalized to β-actin, then normalized to the t = 0 controls.
To test whether the effects of Pim-1 knockdown were cell cycle-specific, we transfected HeLa cells with Pim-1 siRNA, blocked them in the G1/S boundary, and then released them into the cell cycle and measured the Skp2 and p27 levels. We found that the siRNA knockdown of Pim-1 regulated these two proteins throughout the cell cycle (Fig. 1C).
To test the activity of Pim-1 in a different cellular system we examined the role of Pim-1 overexpression in mouse prostate epithelial cells. These cells respond to hepatocyte growth factor (HGF), a powerful mitogen and morphogen for epithelial and endothelial cells, through binding to its receptor the Met tyrosine kinase (42, 43). The growth inhibitory activity of HGF on cancer cells is associated with up-regulation of p27 expression (44), mediated by down-regulation of Skp2 expression (45). We found that the HGF-induced p27 up-regulation is inhibited by Pim-1 in mouse prostate epithelial cells (Fig. 1D). Similar results were also obtained in Pim-overexpressing HeLa cells when they were treated with HGF (supplemental Fig. S1F).
Finally, in HEK293T cells the coexpression of Pim-1, but not kinase-dead Pim-1, K67M, or GFP, was able to induce a longer Skp2 half-life (Fig. 1E). Taken together, these experiments suggest that Pim-1 controls the levels of Skp2 and consequently regulates the amounts of p27 protein in cells.
Pim-1 Binds Directly to Skp2 and Reduces Skp2 Ubiquitination
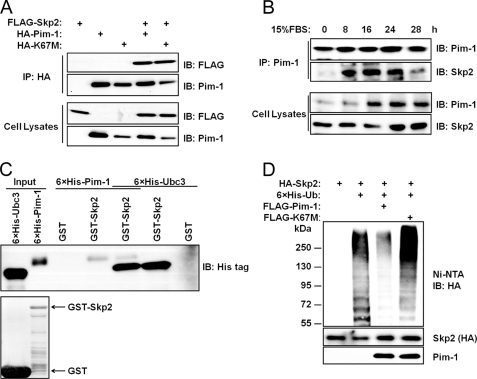
We cotransfected HEK293 cells with FLAG-Skp2 and either HA-Pim-1 or kinase-dead Pim-1 (HA-K67M) expression constructs. When cell lysates were subjected to immunoprecipitation with HA antibody, we found that Pim-1 and Skp2 are able to interact physically in cells irrespective of the Pim-1 kinase activity (Fig. 2A). In HEK293T cells that are transfected then serum-starved and finally released into 15% serum, this interaction between Pim-1 and Skp2 occurs maximally between hours 8 and 24 (Fig. 2B). We did not perform a cell cycle analysis on these cells. However, knockdown of endogenous Pim-1 in HeLa cells appears to reduce Skp2 expression throughout a full cell cycle (Fig. 1C). These observations suggest that Pim-1 may regulate other molecule(s) controlling Skp2 levels in vivo. This binding is also seen in vitro in glutathione S-transferase (GST) pulldown experiments. Recombinant His-tagged Pim-1 protein binds to Skp2; the binding of Pim-1 did not interfere with the interaction between Skp2 and Ubc3, an E2 ubiquitin enzyme that is known to interact with the Skp2 protein (Fig. 2C). Furthermore, Pim-1 did not interfere with the formation of the SCFSkp2 complex, from Skp2, Ubc3, and Rbx1 proteins (supplemental Fig. S2A).
FIGURE 2.
Pim-1 binds to Skp2. A, HEK293T cells were transfected with the indicated plasmids, protein immunoprecipitated (IP) with HA antibody and immunoblotted with FLAG or Pim-1 antibody. Lysates of cells used for this assay are probed with identical antibodies. B, HEK293T cells were serum-starved (0.2%) for 48 h prior to the addition of 15% FBS at 0 time. Cells were then harvested at the indicated time points, and coimmunprecipitation (co-IP) was performed. C, GST-Skp2 proteins were incubated overnight with His-tagged Pim-1 or Ubc3 proteins purified from E. coli at 4 °C, washed with PBS, and subjected to immunoblot analysis (upper panel). GST and GST-Skp2 were stained with Coomassie Brilliant Blue (lower panel). D, HEK293T cells were transfected with the indicated plasmids, treated with 10 μm MG132 for 6 h, and ubiquitination as measured by binding to nickel-nitrilotriacetic acid (Ni-NTA) beads (see “Experimental Procedures”) followed by an immunoblot with anti-HA antibody. Immunoblot (IB) analysis was performed on total cell lysates from these HEK293T cells (two lower panels).
Like p27, Skp2 levels are regulated by ubiquitination and proteasome degradation (25, 26), suggesting that Pim-1 could decrease the levels of Skp2 ubiquitination. Using protein extracts from HEK293T cells transfected with Pim-1 and Skp2, we found that the presence of active but not kinase-dead Pim-1 is sufficient to repress the ubiquitination of the Skp2 protein markedly (Fig. 2D). Using this same approach, consistent with the effect on Skp2, Pim-1 transfection was found to also increase p27 ubiquitination (supplemental Fig. S2C). This suggests that an increase in Skp2 levels is needed to mediate increased ubiquitination of p27 by Pim-1. Indeed, in an in vitro assay, the presence of Pim-1 did not directly influence p27 ubiquitination (supplemental Fig. S2B).
Pim-1 Phosphorylates Skp2 on Multiple Sites
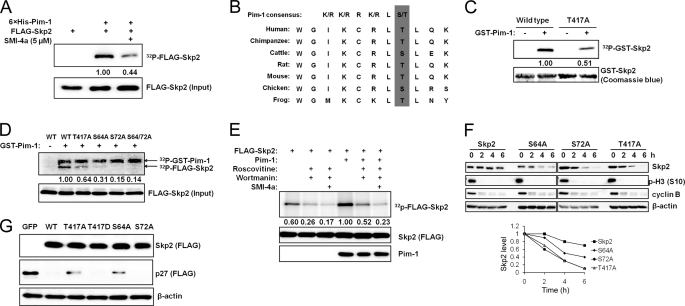
To determine whether Skp2 was a substrate for Pim-1, purified His-tagged protein kinase was incubated with immunoprecipitated FLAG-Skp2 in the presence of [γ-32P]ATP. In this assay, Skp2 was clearly phosphorylated, and this phosphorylation was decreased by the addition of a small molecule Pim-1 inhibitor, SMI-4a (Fig. 3A). Pim-1 is known to phosphorylate the sequence R-X-R-L-S/T (46). Scanning the Skp2 sequence, we identified a potential Pim-1 consensus site at the C terminus of Skp2, Thr417, which is conserved from frog to humans (Fig. 3B). Mutation of this residue from threonine to alanine (T417A) led to reduced Skp2 phosphorylation by Pim-1 in vitro, but did not completely abolish this modification (Fig. 3C). Previous studies (8–10) have demonstrated that Ser64 and Ser72 in Skp2 are CDK2 phosphorylation sites (27), and Ser72 can also be phosphorylated by Akt1 (28, 29). Using GST-Pim-1 as a kinase, mutation of either Ser64 or Ser72 to Ala markedly decreased Skp2 phosphorylation by Pim-1 with both of these changes having a somewhat greater effect than the T417A mutation (Fig. 3D), suggesting that each of these sites might also be a Pim-1 target. It appears that phosphorylation of Ser64 and/or Ser72 may be required for Thr417 phosphorylation to take place because mutation of either Ser64 or Ser72 almost completely abolished Skp2 phosphorylation in this experiment. However, a complete understanding of the relationship between these sites requires further studies.
FIGURE 3.
Pim-1 phosphorylates Skp2. A, FLAG-Skp2 was immunoprecipitated from HEK293T cells, incubated with recombinant His-tagged Pim-1 for 30 min with or without SMI-4a for an in vitro kinase assay (“Experimental Procedures”) followed by SDS-PAGE autoradiography (upper panel) and immunoblot analyses (lower panel). The phosphorylation of FLAG-Skp2 was quantified by densitometry from three independent experiments after normalization to input. B, C-terminal sequence of Skp2 contains a Pim-1 consensus site. C, GST-tagged Skp2 proteins or a T417A mutant was incubated with recombinant GST-Pim-1 and [γ-32P]ATP for 30 min, and subjected to SDS-PAGE followed by autoradiography. The phosphorylation of GST-Skp2 was quantified by densitometry from three independent experiments with normalization to Coomassie Blue staining. D, wild type (WT) FLAG-Skp2 and its mutants T417A, S64A, S72A, and S64A/S72A were immunoprecipitated from HEK293T cells, incubated with recombinant GST-tagged Pim-1 and [γ-32P]ATP for 30 min, followed by SDS-PAGE autoradiography (upper panel) and immunoblot analysis (lower panel). The phosphorylation of FLAG-Skp2 was quantified by densitometry from three independent experiments following normalization to the level of protein input. E, HeLa cells were pretreated with roscovitine (20 μm), wortmannin (1 μm), or SMI-4a (10 μm) for 1 h, transfected with human Pim-1 and Skp2, and labeled with 32Pi followed by FLAG immunoprecipitation, autoradiography (upper panel), and FLAG/Pim-1 immunoblots (two lower panels). The phosphorylation of FLAG-Skp2 was quantified by densitometry from three independent experiments along with normalization to Skp2 expression. F, HeLa cells were transfected with the indicated Skp2 constructs and synchronized in M phase by mitotic shake-off of cells obtained after release from a thymidine-nocodazole block. The cells were then replated and allowed to progress through the cell cycle in the presence of cycloheximide (100 μg/ml). Immunoblot analysis was performed at specific time points using antibodies to cyclin B and phosphohistone H3 Ser10 (p-H3 (S10)) as controls. Densitometric analysis was performed using ImageJ software to quantify the expression of Skp2. Skp2 band intensity was normalized to β-actin and then normalized to the t = 0 controls. G, HEK293T cells were transfected with a FLAG-tagged p27 Skp2 construct or a GFP control. Expression of exogenous p27 and Skp2 is measured by immunoblotting.
To test whether Pim-1 has a role in regulating Skp2 phosphorylation in vivo, HeLa cells were transfected with Pim-1 and Skp2, metabolically labeled with orthophosphate, and then treated with kinase inhibitors such as roscovitine (pan-CDK inhibitor), wortmannin (PI3K inhibitor), and SMI-4a (Pim-1 inhibitor). Treatment with roscovitine and wortmannin reduced Skp2 phosphorylation in vivo. Overexpression of Pim-1 markedly increased Skp2 phosphorylation, and this phosphorylation was inhibited by all three agents (Fig. 3E), suggesting that multiple kinases can play a role in regulating phosphorylation of this protein.
Skp2 is a degraded by the APC/CCdh1 (25, 26) which is known to have its highest activity from late mitosis to the G1 phase of the cell cycle (47). To test the impact of phosphorylation of Skp2 on protein stability, we used HeLa cells that were released from a thymidine-nocodazole block in the G1 phase of the cell cycle into media containing cycloheximide. Exit from mitosis was monitored by the loss of histone H3 phospho-Ser10 immunoreactivity and the degradation of cyclin B1 on Western blots (Fig. 3F). Using this technique, we found that all three individual Skp2 phosphorylation mutants were more efficiently degraded than the wild type Skp2 protein (Fig. 3F), suggesting that phosphorylation by protein kinases, including Pim-1, controls the rate of degradation of Skp2.
We next examined the biological activity of wild type and Skp2 phosphorylation mutants by transfecting them along with p27 into HeLa cells and then examining p27 levels by Western blotting. We found that both the Skp2 T417A and S64A mutants decreased the ability of Skp2 to stimulate the degradation of p27, but T417A retained some degrading activity (Fig. 3G). In contrast, an aspartate mutation, T417D, that mimics phosphorylation at this site was more efficient than the T417A at degrading p27. Surprisingly, S72A mutation did not cause any detectable effect on p27 degradation (Fig. 3G).
Pim-1 Impairs Cdh1 and CDC27 Interaction and Phosphorylates Cdh1
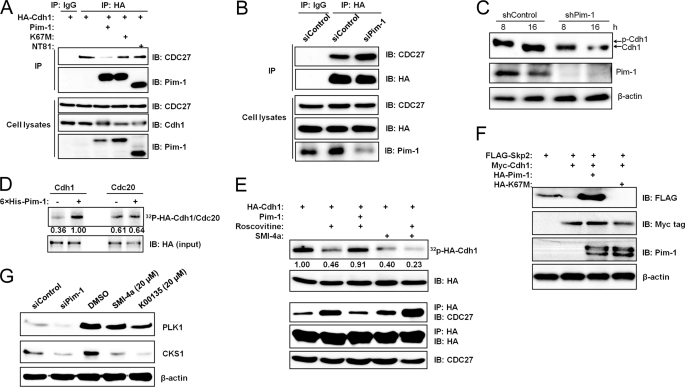
Because Pim-1 regulates Skp2 ubiquitination, we examined whether this enzyme might interact with components of the APC/C complex that are responsible for Skp2 degradation. In coimmunoprecipitation experiments done in transfected HEK293T cells, Pim-1 was found to complex with either Cdh1 or CDC20, two well known activators of APC/C (supplemental Fig. S3A). However, Pim-1 did not physically impair the interaction between Cdh1 or CDC20 and Skp2 (supplemental Fig. S3B). Using the same methodology, in contrast, we found that Pim-1 could impair the interaction between Cdh1 and CDC27, another APC/C component, in a phosphorylation-dependent manner (Fig. 4A). The two kinase-dead Pim-1 mutants, K67M and NT81 (36), are also able to form a complex with Cdh1, but only wild type Pim-1 was capable of reducing the interaction between Cdh1 and CDC27 (Fig. 4A). Additionally, incubation with the Pim-1 inhibitor, SMI-4a (supplemental Fig. S4C) or treatment with siRNA to knock down endogenous Pim-1 expression (Fig. 4B) increased the Cdh1/CDC27 interaction.
FIGURE 4.
Pim-1 kinase phosphorylates Cdh1 and impairs its binding to CDC27. A, HEK293T cells were transfected with HA-Cdh-1, Pim-1, or kinase-dead Pim-1 K67M or NT81, immunoprecipitated (IP) with HA antibody followed by Western blotting with antibodies to CDC27 and Pim-1. Lysates from these cells were immunoblotted (IB) with antibody as shown. B, HeLa cells were cotransfected with HA-Cdh1 and Pim-1 or scrambled siRNA plasmids before harvesting for coimmunoprecipitation analysis. Levels of transfected proteins in lysates were monitored by immunoblotting. C, HeLa cells were transfected with the indicated shRNA plasmids followed by a double-thymidine block. After release from the block cell, lysates were prepared at 8 and 16 h and subjected to immunoblot analysis. The arrows indicate phosphorylated and unphosphorylated Cdh-1. D, in vitro translated HA-tagged Cdh1 or Cdc20 was incubated with recombinant Pim-1 and [γ-32P]ATP in an in vitro kinase assay. Autoradiography (upper panel) and immunoblot (lower panel) analyses were performed. The phosphorylation of Cdh1 or Cdc20 was quantified by densitometry from three independent experiments after normalization to the loaded protein. E, HeLa cells were transfected with HA-Cdh1 and human Pim-1, and the kinase inhibitors roscovitine (20 μm) and SMI-4a (10 μm) were added 1 h before labeling with 32Pi. HA-Cdh-1 was immunoprecipitated, and autoradiography (top panel) and immunoblot analysis were performed (second panel). The phosphorylation of Cdh1 was quantified by densitometry from three independent experiments with normalization to Cdh1 expression (HA). A coimmunoprecipitation experiment was performed to monitor Cdh1 and CDC27 interaction under the same experimental conditions (lower three panels). F, HEK293T cells were transfected with FLAG-Skp2, myc-Cdh-1, and HA-tagged WT and kinase-dead (K67M) Pim kinase, and the extracts were immunoblotted with the specified antibodies. G, HeLa cells were transiently transfected with a siRNA to Pim-1 or a scrambled sequence. Forty-eight h after transfection, extracts of these cells were probed on Western blots with the listed antibodies (left two lanes). HeLa cells were treated with 20 μm Pim kinase inhibitor SMI-4a or K00135 for 16 h, extracts were prepared, and immunoblotting was carried out with the identified antibodies (right three lanes).
Our results are consistent with previous findings that demonstrate that phosphorylation of Cdh1 dissociates this protein from the APC/C complex (48, 49). Cdh1 is hyperphosphorylated in vivo during S, G2, and M phases, and this phosphorylation causes an electrophoretic mobility shift on SDS-polyacrylamide gels (Fig. 5A) (48, 50, 51). To explore the role of Pim-1 in phosphorylation of Cdh1 further, we first reprobed the same membrane used in Fig. 1C with antibodies against Cdh1 and CDC27. In cells treated with Pim-1 siRNA, Cdh1 displayed higher mobility at 0, 4, 8, and 12 h compared with those in control cells (Fig. 1C), suggesting that phosphorylation was reduced. To confirm this finding, we knocked down endogenous Pim-1 expression in HeLa cells using shRNA and then subjected these cells to a double-thymidine block. The cells were then released from the block (for cell cycle analysis, see Fig. 5A), and immunoblotting was performed to examine Cdh1 phosphorylation. Phosphorylation of endogenous Cdh1 at 8 h after release from a double-thymidine block was dramatically reduced in Pim-1 knockdown cells as judged by the protein mobility shift (Fig. 4C).
FIGURE 5.
Pim-1 is required for Skp2 to signal cell cycle S phase entry. A, HeLa cells were treated with a double-thymidine block and released into fresh medium. Cells were then harvested at the indicated time points and subjected to FACS (left panel) and immunoblot analysis (right panel). The arrow denotes the Pim-1 signal. * indicates a nonspecific signal. B, Rat1 cells were transduced with a lentivirus carrying the indicated cDNAs. Cells were maintained in low serum conditions (0.2%) for 48 h before harvested for FACS (upper panel) and immunoblot (lower panel) analyses. Percent of S phase cells was compared with vector control, except where indicated by a bracket. C, the experiment was performed as in B except that SMI-4a (5 μm) was added 3 h before a 20% FBS stimulation (16 h). S phase induction was also determined by BrdU incorporation assay. Brackets indicate comparison of with and without SMI-4a treatment.
Additionally, we found that recombinant Pim-1 was capable of phosphorylating in vitro translated Cdh1 but not CDC20 (Fig. 4D). Cdh1 is heavily phosphorylated by CDKs in vivo. To examine whether Pim-1 can also phosphorylate Cdh1 in vivo, we first treated HeLa cells with the CDK inhibitor roscovitine or Pim inhibitor SMI-4a and then labeled them with 32Pi. Roscovitine or SMI-4a treatment reduced Cdh1 phosphorylation, and overexpression of Pim-1 in the presence of roscovitine reversed the roscovitine effect, although the combination of roscovitine and SMI-4a treatment further decreased Cdh1 phosphorylation (Fig. 4E). We performed coimmunoprecipitation experiments to monitor the Cdh1/CDC27 interactions under these conditions. Consistently, roscovitine treatment increased the Cdh1/CDC27 interaction whereas overexpression of Pim-1 in the presence of roscovitine suppressed this effect. Combined treatment with roscovitine and SMI-4a further increased the Cdh1/CDC27 interaction compared with roscovitine or SMI-4a alone (Fig. 4E).
Because the APC/C activity to degrade Skp2 can be activated by Cdh1 overexpression (25, 26), we tested the ability of Pim-1 to reverse this effect. We found that coexpression of wild-type Pim-1, but not its mutant, K67M, is capable of blocking Cdh1-mediated degradation of Skp2 (Fig. 4F). However, data in Fig. 4F cannot distinguish whether Pim-1 blocks degradation of Skp2 by acting on Cdh1 or Skp2 or both.
Because Pim-1 expression impairs APC/CCdh1 activity, we examined whether other known APC/CCdh1 substrates are regulated by Pim-1 expression. Knockdown of endogenous Pim-1 expression or suppression of Pim-1 kinase activity with Pim kinase inhibitors SMI-4a or K00135 in HeLa cells led to reduced protein expression of both polo-like kinase-1 and CDK subunit 1 (Cks1), two proteins known to be regulated by APC/CCdh1 (Fig. 4G). This finding further demonstrates that Pim-1 is a negative regulator of APC/CCdh1 activity.
Pim-1 Is Required for Skp2 to Signal Cell Cycle S Phase Entry
Based on the activities of Pim-1, we have attempted to correlate the levels of this enzyme with other cell cycle regulatory components. HeLa cells were released from a double-thymidine block, cell cycle progression was monitored by FACS analysis (Fig. 5A), and the expression patterns of Skp2, p27, Cdh1, CDC27, cyclin B1, and cyclin A were measured by Western blotting (Fig. 5B). We found that Pim-1 levels were very high at S (4 h) and G2/M phases (8 h) of the cell cycle. Lower Pim-1 expression was seen at G1 and the G1/S boundary (0, 16, 20, and 24 h; Figs. 5A and 1C). Because Pim-1 is a constitutively active kinase (7–11), this expression pattern of Pim-1 should represent its activity profile during cell cycle progression. Interestingly, but not surprisingly, Pim-1 activity coincides with Skp2 expression (Fig. 5A) and inversely correlates with Cdh1 activity (47, 52) during the cell cycle (Fig. 5A). Because Skp2 is known to have the ability to induce S phase in quiescent fibroblasts (40, 53), we determined whether the Skp2/Pim-1 interaction is important for S phase progression. Investigation of this question was carried out using Rat1 cells because they were able to undergo complete cell cycle blockade at G0/G1 upon serum starvation. As judged by FACS analysis, we found that in the absence of serum addition, overexpressed Skp2 or Pim-1 each stimulates S phase entry (Fig. 5B), and coexpression of Skp2 and Pim-1 further enhances the S phase entry of these cells (Fig. 5B). Conversely, as revealed by both FACS analysis and BrdU incorporation, treatment with a small molecule Pim inhibitor, SMI-4a, reduced Skp2-induced S phase progression (Fig. 5C) and impaired serum-induced S phase entry (Fig. 5C). Another structurally unrelated small molecule Pim inhibitor, K00135 (54), displayed a similar effect (supplemental Fig. S5A). These observations suggest that Pim kinases are required along with Skp2 to allow cells to exit from quiescence.
DISCUSSION
The data presented suggest the novel observation that the Pim-1 protein kinase through a dual mechanism can regulate the levels and hence the activity of Skp2. Pim-1 is capable of binding and phosphorylating Skp2 and stabilizing protein levels, but does not affect the interaction of Skp2 with the E2 ligase Ubc3. Conversely, both siRNA and small molecule Pim-1 inhibitors decrease Skp2 levels and phosphorylation. Skp2 is phosphorylated by CDK2 at Ser64 and Ser72 (27) and by Akt1 at Ser72 to stabilize this protein (28, 29). Pim-1 appears capable of phosphorylating Skp2 at these two sites (Fig. 3), as well as a unique site in the C terminus, Thr417, that is highly conserved throughout the animal kingdom, including humans and mice. Phosphorylation of this site is required for maximal Skp2 activity and stabilization of Skp2 protein levels in vivo (Fig. 3). In the prostate cancer cell line PC3 that contains an activated Akt, a small molecule Pim inhibitor SMI-4a but not wortmannin or the Akt inhibitor GSK690693 decreased the levels of Skp2. LY294002, which inhibits both Akt and Pim, displayed an effect similar to that of SMI-4a, suggesting that in this cell line the Pims are essential for the regulation of Skp2 levels. Unlike Akt (28, 29), Pim-1 kinase did not appear to regulate Skp2 subcellular localization (supplemental Fig. S4). The Pim kinases share multiple similarities with AKT (1, 55, 56). It is possible that the relative abundance of each of these Skp2-phosphorylating kinases may decide which is essential to the control of Skp2 levels. It is quite surprising that our Skp2 S72A mutant did not lose p27 degradation activity compared with the wild type Skp2 (Fig. 3G) because two previous studies demonstrated that this very same Skp2 mutant completely lost ubiquitin ligase activity (28, 29). However, another two recent reports confirmed our finding (30, 31). The half-life of this mutant was indeed shorter than that of wild-type Skp2 (Fig. 3F), consistent with previous reports (27–29).
The degradation of Skp2 is regulated by APC/CCdh1 complex (25, 26) which preferentially associates with non-phospho-Ser64 form of Skp2 (27). Pim-1 kinase activity does not affect the binding of Cdh1 to total Skp2 (supplemental Fig. S3B), but does impair the interaction between Cdh1 and CDC27 (Fig. 4A), Interaction with CDC27/APC3 protein allows Cdh1 to activate the APC/C (57). Although Cdh1 is inhibited by both the Emi-1 protein and multiple phosphorylations initiated in part by cyclin A-CDK2 and cyclin B1-CDK1 (47, 52), it has been proposed that an additional kinase may play a role (58). Here, we demonstrate that Cdh1 is a phosphorylation target of Pim-1 (Fig. 4, D and E) and the knockdown of Pim-1 with siRNA reduces Cdh1 phosphorylation during S, G2, and M phases (Figs. 1C and 4C), demonstrating the critical involvement of Pim-1 in tightly controlled Cdh1 phosphorylation during the cell cycle. It remains unknown whether Pim-1 and CDKs share some phosphorylation sites on Cdh1. Further studies are required to determine the precise Pim-1 sites and how these two different types of kinases cooperate to control Cdh1 activity. Interestingly, the levels of Pim-1 protein are correlated with Cdh1 phosphorylation during cell cycle progression as high Pim-1 expression and high Cdh1 phosphorylation were seen during S, G2, and M phases, and the opposite occurred during the G1 phase (Fig. 5A). Given the role of Cdh1 in regulating mitosis, this may explain why Pim-1 is not only required for Skp2 to signal S phase entry (Fig. 5), but also plays a critical role in G2/M phase regulation. Consistent with this hypothesis, mouse embryo fibroblasts that are knocked out for all three Pim kinase isoforms display increased number of cells in the G2/M phase of the cell cycle (supplemental Fig. S5). These observations are in concert with previous discoveries suggesting that Pim-1 functions in mitosis (59–61). Therefore, the Pim-1 kinase regulates Skp2 levels through the Pim-1 kinase activity, reduces APC/CCdh1 E3 ligase activity, and thus protects Skp2 from degradation (Fig. 4F).
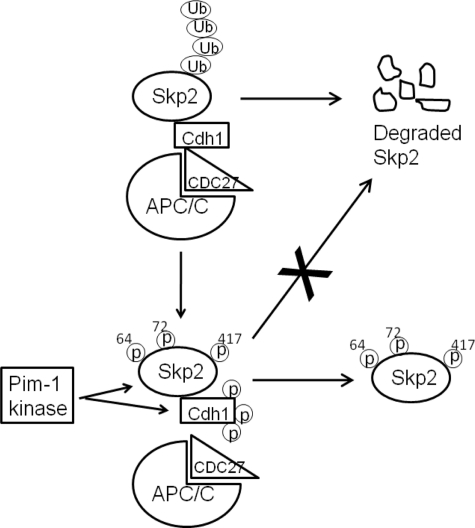
The Pim-1 protein kinase is abnormally elevated in human cancers, regulated by growth factors, and collaborates with other oncogenes to induce cell transformation (1, 2, 5, 6). The ability of this enzyme to modulate the activity of both the SCFSkp2 and APC/CCdh1 (Fig. 6) and thus control p27 levels is likely to be essential to the biological activities of this protein kinase.
FIGURE 6.
Model of Pim-1 regulation on Skp2 degradation. Nonphosphorylated Skp2 binds to E3 ligase APC/CCdh1 and gets ubiquitinated (Ub) followed by proteasome-mediated degradation. Pim-1 kinase phosphorylates Skp2 on multiple sites: Ser64, Ser72, and Thr417. Furthermore, the phosphorylation of Cdh1 by Pim-1 reduces Cdh1 and APC/C interaction. Both Pim-1 actions result in decreased Skp2 ubiquitination and consequently increased Skp2 stability.
Supplementary Material
Acknowledgments
We thank Dr. Liang Zhu (Albert Einstein College of Medicine, Yeshiva University) for providing the Skp2 expression construct and Dr. Xuedong Liu (University of Colorado-Boulder) for providing the active Skp2 complex for the in vitro ubiquitination assay.
This work was supported by Department of Defense Grants W8IXWH-08 and W81XWH-10-1-0249. The flow cytometry core received support from 1P30-CA138313.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and additional references.
- C-TAK1
- Cdc25C-associated kinase 1
- APC/C
- anaphase-promoting complex or cyclosome
- HGF
- hepatocyte growth factor
- SCF
- Skp1/cullin/F-box protein.
REFERENCES
- 1.Hammerman P. S., Fox C. J., Birnbaum M. J., Thompson C. B. (2005) Blood 105, 4477–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellwood-Yen K., Graeber T. G., Wongvipat J., Iruela-Arispe M. L., Zhang J., Matusik R., Thomas G. V., Sawyers C. L. (2003) Cancer Cell 4, 223–238 [DOI] [PubMed] [Google Scholar]
- 3.Zippo A., De Robertis A., Serafini R., Oliviero S. (2007) Nat. Cell Biol. 9, 932–944 [DOI] [PubMed] [Google Scholar]
- 4.Dhanasekaran S. M., Barrette T. R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2001) Nature 412, 822–826 [DOI] [PubMed] [Google Scholar]
- 5.Shah N., Pang B., Yeoh K. G., Thorn S., Chen C. S., Lilly M. B., Salto-Tellez M. (2008) Eur. J. Cancer 44, 2144–2151 [DOI] [PubMed] [Google Scholar]
- 6.Speers C., Tsimelzon A., Sexton K., Herrick A. M., Gutierrez C., Culhane A., Quackenbush J., Hilsenbeck S., Chang J., Brown P. (2009) Clin. Cancer Res. 15, 6327–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullock A. N., Debreczeni J., Amos A. L., Knapp S., Turk B. E. (2005) J. Biol. Chem. 280, 41675–41682 [DOI] [PubMed] [Google Scholar]
- 8.Kumar A., Mandiyan V., Suzuki Y., Zhang C., Rice J., Tsai J., Artis D. R., Ibrahim P., Bremer R. (2005) J. Mol. Biol. 348, 183–193 [DOI] [PubMed] [Google Scholar]
- 9.Jacobs M. D., Black J., Futer O., Swenson L., Hare B., Fleming M., Saxena K. (2005) J. Biol. Chem. 280, 13728–13734 [DOI] [PubMed] [Google Scholar]
- 10.Qian K. C., Wang L., Hickey E. R., Studts J., Barringer K., Peng C., Kronkaitis A., Li J., White A., Mische S., Farmer B. (2005) J. Biol. Chem. 280, 6130–6137 [DOI] [PubMed] [Google Scholar]
- 11.Bullock A. N., Russo S., Amos A., Pagano N., Bregman H., Debreczeni J. E., Lee W. H., von Delft F., Meggers E., Knapp S. (2009) PLoS One 4, e7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mochizuki T., Kitanaka C., Noguchi K., Muramatsu T., Asai A., Kuchino Y. (1999) J. Biol. Chem. 274, 18659–18666 [DOI] [PubMed] [Google Scholar]
- 13.Bachmann M., Hennemann H., Xing P. X., Hoffmann I., Möröy T. (2004) J. Biol. Chem. 279, 48319–48328 [DOI] [PubMed] [Google Scholar]
- 14.Wang Z., Bhattacharya N., Mixter P. F., Wei W., Sedivy J., Magnuson N. S. (2002) Biochim. Biophys. Acta 1593, 45–55 [DOI] [PubMed] [Google Scholar]
- 15.Morishita D., Katayama R., Sekimizu K., Tsuruo T., Fujita N. (2008) Cancer Res. 68, 5076–5085 [DOI] [PubMed] [Google Scholar]
- 16.Beharry Z., Zemskova M., Mahajan S., Zhang F., Ma J., Xia Z., Lilly M., Smith C. D., Kraft A. S. (2009) Mol. Cancer Ther. 8, 1473–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y. W., Beharry Z. M., Hill E. G., Song J. H., Wang W., Xia Z., Zhang Z., Aplan P. D., Aster J. C., Smith C. D., Kraft A. S. (2009) Blood 115, 824–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardozo T., Pagano M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 19.Gstaiger M., Jordan R., Lim M., Catzavelos C., Mestan J., Slingerland J., Krek W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5043–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Signoretti S., Di Marcotullio L., Richardson A., Ramaswamy S., Isaac B., Rue M., Monti F., Loda M., Pagano M. (2002) J. Clin. Invest. 110, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Latres E., Chiarle R., Schulman B. A., Pavletich N. P., Pellicer A., Inghirami G., Pagano M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2515–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shim E. H., Johnson L., Noh H. L., Kim Y. J., Sun H., Zeiss C., Zhang H. (2003) Cancer Res. 63, 1583–1588 [PubMed] [Google Scholar]
- 23.Reed S. I. (2003) Nat. Rev. Mol. Cell Biol. 4, 855–864 [DOI] [PubMed] [Google Scholar]
- 24.Nakayama K. I., Nakayama K. (2006) Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 25.Bashir T., Dorrello N. V., Amador V., Guardavaccaro D., Pagano M. (2004) Nature 428, 190–193 [DOI] [PubMed] [Google Scholar]
- 26.Wei W., Ayad N. G., Wan Y., Zhang G. J., Kirschner M. W., Kaelin W. G., Jr. (2004) Nature 428, 194–198 [DOI] [PubMed] [Google Scholar]
- 27.Rodier G., Coulombe P., Tanguay P. L., Boutonnet C., Meloche S. (2008) EMBO J. 27, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao D., Inuzuka H., Tseng A., Chin R. Y., Toker A., Wei W. (2009) Nat. Cell Biol. 11, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin H. K., Wang G., Chen Z., Teruya-Feldstein J., Liu Y., Chan C. H., Yang W. L., Erdjument-Bromage H., Nakayama K. I., Nimer S., Tempst P., Pandolfi P. P. (2009) Nat. Cell Biol. 11, 420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bashir T., Pagan J. K., Busino L., Pagano M. (2010) Cell Cycle 9, 971–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boutonnet C., Tanguay P. L., Julien C., Rodier G., Coulombe P., Meloche S. (2010) Cell Cycle 9, 975–979 [DOI] [PubMed] [Google Scholar]
- 32.Zemskova M., Sahakian E., Bashkirova S., Lilly M. (2008) J. Biol. Chem. 283, 20635–20644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji P., Jiang H., Rekhtman K., Bloom J., Ichetovkin M., Pagano M., Zhu L. (2004) Mol. Cell 16, 47–58 [DOI] [PubMed] [Google Scholar]
- 34.Biggs J. R., Peterson L. F., Zhang Y., Kraft A. S., Zhang D. E. (2006) Mol. Cell. Biol. 26, 7420–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cen B., Li H., Weinstein I. B. (2009) J. Biol. Chem. 284, 5265–5276 [DOI] [PubMed] [Google Scholar]
- 36.Aho T. L., Sandholm J., Peltola K. J., Mankonen H. P., Lilly M., Koskinen P. J. (2004) FEBS Lett. 571, 43–49 [DOI] [PubMed] [Google Scholar]
- 37.Barclay W. W., Cramer S. D. (2005) Prostate 63, 291–298 [DOI] [PubMed] [Google Scholar]
- 38.Carrano A. C., Eytan E., Hershko A., Pagano M. (1999) Nat. Cell Biol. 1, 193–199 [DOI] [PubMed] [Google Scholar]
- 39.Wang W., Ungermannova D., Chen L., Liu X. (2003) J. Biol. Chem. 278, 32390–32396 [DOI] [PubMed] [Google Scholar]
- 40.Kim S. Y., Herbst A., Tworkowski K. A., Salghetti S. E., Tansey W. P. (2003) Mol. Cell 11, 1177–1188 [DOI] [PubMed] [Google Scholar]
- 41.Xia Z., Knaak C., Ma J., Beharry Z. M., McInnes C., Wang W., Kraft A. S., Smith C. D. (2009) J. Med. Chem. 52, 74–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balkovetz D. F., Lipschutz J. H. (1999) Int. Rev. Cytol. 186, 225–260 [DOI] [PubMed] [Google Scholar]
- 43.Bottaro D. P., Rubin J. S., Faletto D. L., Chan A. M., Kmiecik T. E., Vande Woude G. F., Aaronson S. A. (1991) Science 251, 802–804 [DOI] [PubMed] [Google Scholar]
- 44.Nagahara H., Vocero-Akbani A. M., Snyder E. L., Ho A., Latham D. G., Lissy N. A., Becker-Hapak M., Ezhevsky S. A., Dowdy S. F. (1998) Nat. Med. 4, 1449–1452 [DOI] [PubMed] [Google Scholar]
- 45.Zhang H., Ozaki I., Mizuta T., Yoshimura T., Matsuhashi S., Hisatomi A., Tadano J., Sakai T., Yamamoto K. (2003) Hepatology 38, 305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachmann M., Möröy T. (2005) Int. J. Biochem. Cell Biol. 37, 726–730 [DOI] [PubMed] [Google Scholar]
- 47.Peters J. M. (2006) Nat. Rev. Mol. Cell Biol. 7, 644–656 [DOI] [PubMed] [Google Scholar]
- 48.Jaspersen S. L., Charles J. F., Morgan D. O. (1999) Curr. Biol. 9, 227–236 [DOI] [PubMed] [Google Scholar]
- 49.Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., Amon A. (1998) Mol. Cell 2, 709–718 [DOI] [PubMed] [Google Scholar]
- 50.Zachariae W., Schwab M., Nasmyth K., Seufert W. (1998) Science 282, 1721–1724 [DOI] [PubMed] [Google Scholar]
- 51.Kramer E. R., Scheuringer N., Podtelejnikov A. V., Mann M., Peters J. M. (2000) Mol. Biol. Cell 11, 1555–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Leuken R., Clijsters L., Wolthuis R. (2008) Biochim. Biophys. Acta 1786, 49–59 [DOI] [PubMed] [Google Scholar]
- 53.Sutterlüty H., Chatelain E., Marti A., Wirbelauer C., Senften M., Müller U., Krek W. (1999) Nat. Cell Biol. 1, 207–214 [DOI] [PubMed] [Google Scholar]
- 54.Pogacic V., Bullock A. N., Fedorov O., Filippakopoulos P., Gasser C., Biondi A., Meyer-Monard S., Knapp S., Schwaller J. (2007) Cancer Res. 67, 6916–6924 [DOI] [PubMed] [Google Scholar]
- 55.Choudhary C., Olsen J. V., Brandts C., Cox J., Reddy P. N., Böhmer F. D., Gerke V., Schmidt-Arras D. E., Berdel W. E., Müller-Tidow C., Mann M., Serve H. (2009) Mol. Cell 36, 326–339 [DOI] [PubMed] [Google Scholar]
- 56.Amaravadi R., Thompson C. B. (2005) J. Clin. Invest. 115, 2618–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kraft C., Vodermaier H. C., Maurer-Stroh S., Eisenhaber F., Peters J. M. (2005) Mol. Cell 18, 543–553 [DOI] [PubMed] [Google Scholar]
- 58.Hall M. C., Warren E. N., Borchers C. H. (2004) Cell Cycle 3, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 59.Bhattacharya N., Wang Z., Davitt C., McKenzie I. F., Xing P. X., Magnuson N. S. (2002) Chromosoma 111, 80–95 [DOI] [PubMed] [Google Scholar]
- 60.Roh M., Gary B., Song C., Said-Al-Naief N., Tousson A., Kraft A., Eltoum I. E., Abdulkadir S. A. (2003) Cancer Res. 63, 8079–8084 [PubMed] [Google Scholar]
- 61.Roh M., Song C., Kim J., Abdulkadir S. A. (2005) J. Biol. Chem. 280, 40568–40577 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.