Abstract
Protein kinases are potential targets for the prevention and control of UV-induced skin cancer. T-cell-originated protein kinase (TOPK) is highly expressed in skin cancer cells, but its specific function is still unknown. We investigated the role of TOPK in UVB-induced apoptosis in RPMI7951 human melanoma cells. Liquid chromatography-tandem mass spectrometry analysis was used to identify proteins that bind with TOPK. Immunofluorescence, Western blot, and flow cytometry were used to assess the effect of UVB on TOPK, peroxiredoxin 1 (Prx1), and apoptosis in RPMI7951 cells. TOPK binds with Prx1 and its phosphorylation of Prx1 at Ser-32 is important for regulation of H2O2-mediated signal transduction. Analysis of the CD spectra of Prx1 and mutant Prx1 (S32A) proteins showed that the secondary structure of Prx1 was significantly altered by phosphorylation of Prx1 at Ser-32. UVB irradiation induced phosphorylation of TOPK in RPMI7951 human melanoma cells and phosphorylated TOPK co-localized with Prx1 in the nucleus. UVB induced the peroxidase activity of Prx1 in vitro and ex vivo. Following treatment with UVB, H2O2 levels and apoptosis were increased in RPMI7951 cells stably expressing TOPK siRNA or stably mutant Prx1 (S32A). Phosphorylation of Prx1 (Ser-32) by TOPK prevents UVB-induced apoptosis in RPMI7951 melanoma cells through regulation of Prx1 peroxidase activity and blockade of intracellular H2O2 accumulation.
Keywords: Apoptosis, Cell Sorting, E2F Transcription Factor, MAP Kinases (MAPKs), Polymerase Chain Reaction (PCR), Prx1, TOPK, Peroxidase Activity
Introduction
PDZ-binding kinase/T-LAK cell-originated protein kinase (PBK/TOPK)3 is a novel MEK3/6-related MAPKK family member that phosphorylates p38 and is involved in H-Ras signaling (1–3). During mitosis, TOPK is phosphorylated at Thr-9 by Cdk1/cyclin B (4, 5), and phosphorylation is required for its kinase activity (6). TOPK expression is regulated by cell cycle-specific transcription factors E2F and CREB/ATF (7). TOPK is found in activated T-LAK cells, lymphoid tumor cells, and normal testicular tissue (1). It is highly expressed in hematologic tumors such as leukemia (8), lymphoma (9), and myeloma (10); and its expression corresponds with malignant potential of these tumors. TOPK is also overexpressed in breast cancer (11) and human colorectal cancer and colorectal cancer cell lines (12). Recent studies suggest a role for TOPK in DNA damage sensing and repair through phosphorylation of histone H2AX (2, 13, 14). Earlier we showed that TOPK is involved in preventing apoptosis in melanoma cells (13) and is a positive regulator of c-Jun-NH2-kinase 1 (JNK1) signaling and H-Ras-induced cell transformation (15). We reported that a positive feedback loop between TOPK and ERK2 increases the carcinogenic properties of HCT116 colorectal cancer cells, and therefore TOPK-regulated signaling might be a potential therapeutic target in colorectal cancer (12).
The 25-kDa protein peroxiredoxin 1 (Prx1) is an antioxidant enzyme that participates in the regulation of hydrogen peroxide-mediated signal transduction and is also implicated in the immune response, cell proliferation, differentiation, and apoptosis (16–20). Prx1 plays a negative role in apoptosis signal-regulating kinase 1 (ASK1)-induced apoptosis (21), and its overexpression in cancer cells could enhance survival. Prx1 is a dual function protein because its phosphorylation at Thr-90 by Cdc2 either decreases its peroxidase activity or its function as a molecular chaperone (22, 23). However, the role of Prx1 in cancer development is not clear.
We report that Prx1 is newly discovered direct target of TOPK. Our results demonstrate that TOPK phosphorylation of Prx1 at Ser-32 inhibits UVB-induced apoptosis in RPMI7951 melanoma cells by increasing Prx1 peroxidase activity and decreasing the intracellular accumulation of H2O2.
MATERIALS AND METHODS
Reagents, Antibodies, and Cell Culture
SK-MEL-5, SK-Mel-28, and RPMI7951 human malignant melanoma epithelial-like cell lines were from the American Type Culture Collection (Manassas, VA). RPMI7951 cells stably expressing control siRNA or TOPK siRNA were prepared as described (13). Cells were cultured in MEM supplemented with 10% FBS, 2 mm l-glutamine, 25 μg/ml gentamicin, 0.1 mm non-essential amino acids, 1 mm sodium pyruvate, and 200 μg/ml G418. Active PBK/TOPK kinase and antibodies that detect cleaved caspase-3, PBK/TOPK and phospho-PBK/TOPK (Thr-9) were from Cell Signaling Technology, Inc. (Beverly, MA). Active TOPK was from Signal Chem (Richmond, BC, Canada). β-Actin was from Sigma/Aldrich and antibodies to detect Prx1, Prx2, and Prx3 were from Upstate (Lake Placid, NY). Anti-peroxiredoxin-SO3 was from Abcam Inc. (Cambridge, MA).
Identification of Proteins Binding with TOPK (24)
Liquid chromatography-tandem mass spectrometry analysis to determine TOPK-binding partners was performed as described (13). Product ion spectra were correlated with theoretical peptides and proteins using Protein Pilot v.2.0.1 (ABI) and the Paragon algorithm (25).
UVB and Western Blotting
Cells (7 × 105) were treated with UVB (4 kJ/m2) as described (13, 26). Protein concentration was determined by the Bradford method (27). Antibody-bound proteins were detected by chemiluminescence (ECF, Amersham Biosciences, Piscataway, NJ) and analyzed using the Storm 840 Scanner (Molecular Dynamics, Sunnyvale, CA). Untreated cells were used as negative controls.
Construction of Expression Vectors and Stable Cells
RPMI7951 cells stably expressing control siRNA or TOPK siRNA were prepared and cultured as described (13). The sense siRNA template sequence for TOPK was 5′-GATCCGAGGTTTGTCTCATTCTCCTTCAAGAGAGGAGAATGAGACAAACCTCTTTTTTGGAAA-3′ and the antisense siRNA template sequence was 5′-AGCTTTTCCAAAAAAGAGGTTTGTCTCATTCTCCTCTCTTGAAGGAGAATGAGACAAACCTCG-3′. The sense and antisense oligonucleotides were annealed and cloned into the pSilencer 3.1-H1 neo vector (Amnion, TX) at the BamHI and HindIII sites following instructions provided by the manufacturer. The resulting pSilencer 3.1-H1-siRNA plasmids were transfected into RPMI7951 cells, and stable cells were obtained by G418 screening. For purification of His-Prx1 or His-Prx3 fusion proteins, the pCMV6-XL5 plasmid (OriGene Technologies, Inc.; Rockville, MD) was amplified by PCR using 5′-GACGACGACAAGATGTCTTCAGGAAATGCTAAAATTGGGC-3′ as the sense primer and 5′-GAGGAGAAGCCCGGTTCACTTCTGCTTGGAGAAATATTCTTTG-3′ as the antisense primer for Prx1 and 5′-GACGACGACAAGATGGCGGCTGCTGTAGGACGGTTGCTCC-3′ as the sense primer and 5′-GAG GAGAAGCCCGGTCTACTGATTTACCTTCTGAAAGTACTCT-3′ as the antisense primer for Prx3 and cloned into the pET-46Ek/LIC vector (Novagen, Madison, WI).
Ser-32 and Ser-126 in Prx1 were mutated to alanine (S32A or S126A) using the QuikChange II Site-directed Mutagenesis kit (Stratagene, La Jolla, CA). Prx1-Wt or mutant Prx1-S32A was inserted into the pcDNA4/His Max (Invitrogen, Carlsbad, CA) vector using jetPEITM (Qbiogen, Inc., Montreal, Quebec, Canada) for transfection into RPMI7951 cells following the suggested protocol to generate stable cells. Transfected cells were selected in normal medium for RPMI7951 cells with 200 μg/ml of zeocin (Invitrogen) for 10–14 days. Mutant Prx1-S32A and Prx1-S126A fusion proteins were also inserted into the pET-46Ek/LIC vector (Novagen) to generate His-Prx1-S32A and His-Prx1-S126A fusion proteins, respectively.
Deletion mutants of Prx-1 were constructed by a PCR-based method. Sense and antisense primers were used to amplify DNA and then clone each construct into the pET-46Ek/Lic vector as follows. For the N-terminal Prx1 deletion mutant (D1–56, Ndel), sense: 5′-GACGACGACAAGatgtcttcaggaaatgctaaaattg-3′ and antisense: 5′-GAGGAGAAGCCCGGTgatctccgtggggcacacaaaggtg-3′; for the C-terminal Prx1 deletion mutant (D128–199, Cdel), sense: 5′-GACGACGACAAGatgattgctttcagtgatagggcagaag-3′ and antisense: 5′-GAGGAGAAGCCCGGTgaacgagatgccttcatcagcctttaag-3′; for Δ-Prx-1 deletion mutant (D 57–127, Δdel), sense: 5′-GACGACGACAAGatgaggggcctttttatcattgatgata-3′ and antisense: 5′-GAGGAGAAGCCCGGTcttctgcttggagaaatattctttgctc-3′. The capitalized bases are sequences required for pET-46 LIC cloning.
Bacterial Expression and Purification of His-Prx1 or Mutant Prx1 or Prx3 or Prx1 Deletion Fusion Proteins
His-Prx1-Wt (Wt), His-Prx1-S32A (S32A), His Prx1-S126A (S126A), His-Prx3 fusion proteins or His-Prx1-deletion mutant proteins (Ndel, Cdel or Δdel) were expressed in BL21 (DE3) bacteria (Novagen). Bacteria were grown at 37 °C to an absorbance of 0.9–1.0 at 600 nm and induced with 0.5 mm IPTG overnight at 15 °C and harvested by centrifugation. Cell pellets were suspended in 50 mm Tris-HCl (pH 8.0) lysis buffer containing 200 mm NaCl, 10 mm imidazole, and 10 mm mercaptoethanol. After sonication and centrifugation, the supernatant fraction was incubated with Ni-NTA-agarose beads (Qiagen, Valencia, CA) overnight at 4 °C. Beads were washed with lysis buffer and PBS and His-proteins eluted with 100 mm imidazole. His-Prx1-Wt, His-Prx1-S32A or His-Prx1-S126A was purified by FPLC on a HiLoad 16/60 system using Superdex-75 gel filtration chromatography (GE Healthcare, Piscataway, NJ) in buffer (20 mm Tris-HCl, 100 mm NaCl). Proteins were concentrated using centrifugal filter tubes with Biomax 5K NMWL membranes (Millipore Corp., Billerica, MA).
Pulldown Assay
The supernatant fraction prepared from RPMI7951 melanoma cells (500 μg of protein) was incubated with TOPK-Ni-NTA-agarose beads (50 μl), prepared as described (13). After incubation with gentle rocking overnight at 4 °C, beads were washed with 50 ml of 1% Tween in PBS (once) and then with PBS (3 times) followed by centrifugation at 13,000 rpm for 2–3 min, and proteins bound to the beads were analyzed by 15% SDS-PAGE and Western blotting with anti-Prx1.
Construction of the GST-TOPK Plasmid and Preparation of the GST-TOPK Protein Beads
A cDNA encoding TOPK was generated by PCR and subcloned into the BamHI/XhoI sites of the pGEX-5X-1 vector (Amersham Biosciences) to produce the glutathione S-transferase (GST)-TOPK fusion protein. Escherichia coli BL21 (DE3) (Novagen) freshly transformed with the GST-TOPK plasmid were grown in 300 ml LB with 100 μg/ml ampicillin at 37 °C to OD600 of 0.8 and induced by IPTG at a final concentration of 0.5 mm. After induction, the cells were cultured at 26 °C for another 4 h. The bacterial cells were harvested by centrifugation, resuspended in 1× PBS buffer, and sonicated. After centrifugation for 30 min at 15,000 rpm at 4 °C, the supernatant fraction was added to GST-agarose beads (Pierce) and binding was allowed to occur for 1 h at 4 °C. Resin-bound GST-TOPK fusion proteins were washed five times with PBS. GST-TOPK beads were used for an in vitro protein binding assay.
In Vitro Protein Binding Assay
TOPK-phosphorylated or nonphosphorylated His-Prx1-Wt or His-Prx1S32A mutant proteins (2 μg each) or His-Prx1 deletion mutants (Ndel, Cdel or Δdel) were allowed to bind with 20 μl of GST-TOPK-beads in 500 μl of PBS overnight at 4 °C. Beads were washed with PBS 3–5 times. PBS (40 μl) and 6× SDS loading buffer (10 μl) were directly added to the beads. After heating at 95 °C for 5 min and centrifugation at 14,000 rpm for 10 min, supernatant fractions were separated by 15% SDS-PAGE. The His-probe (H3) mouse antibody (Santa Cruz Biotechnology, Inc.) was used for detection of Prx1.
Immunoprecipitation (IP)
RPMI7951 cells were treated or not treated with UVB (4 kJ/m2) and then disrupted in CHAPS lysis buffer (30 mm Tris-Cl, pH 7.5, 150 mm NaCl, 1% CHAPS, 1× protease inhibitors). Cell lysates (1 mg of total protein/800 μl of lysis buffer) were incubated with 4 μg of TOPK (BD Biosciences, San Jose, CA) or with 2 μg of anti-Prx1 (Santa Cruz Biotechnology, Inc.) at 4 °C overnight and 50 μl of protein A/G-Sepharose beads (Santa Cruz Biotechnology, Inc.) for an additional 5 h. After centrifugation at 3,000 rpm for 3 min, beads were washed (1 ml each) once with of a high concentration of salt (500 mm NaCl), once with a low concentration of salt (50 mm, NaCl), and 3× with PBS. Proteins bound to the beads were analyzed by 15% SDS-PAGE and Western blotting with anti-TOPK or anti-Prx1.
In Vitro Kinase Assay
To detect γ-32P incorporation, 2–3 μg of His-Prx1, mutant His-Prx1-S32A or His-Prx1-S126A, or His-Prx3 proteins were mixed with active PBK/TOPK (0.1 μg/20 μl reaction) in 10× kinase buffer containing 10 μmol/liter unlabeled ATP and 10 μCi of [γ-32P]ATP, and incubated at 30 °C for 30 min. The reaction was stopped by adding 5× SDS sample buffer. Samples were separated by 15% SDS-PAGE, and proteins visualized by autoradiography or Western blot with anti-TOPK or anti-Prx1 as loading controls.
Peroxidase Activity
To determine peroxidase activity, we used the Amplex Red Hydrogen Peroxide/Peroxidase assay kit (Molecular Probes, Inc., Eugene, OR). To determine peroxidase activity in vitro, active TOPK (1.0 μg) was mixed with 20 μl of 5× kinase buffer containing 10 μmol/liter unlabeled ATP and 10 μg of Prx1-Wt or Prx1-S32A in 20 μl of FPLC buffer and incubated at 30 °C for 1 h. Peroxidase activity was assessed using a microplate reader and an absorbance of 570 nm according to the protocol provided by the manufacturer. To convert the absorbance (OD570) to peroxidase activity (mU/ml), we used a peroxidase standard curve, which was prepared according to the protocol provided in the Amplex Red Hydrogen Peroxide/Peroxidase assay kit. For determination of peroxidase activity ex vivo, we used cells (2 × 104) stably expressing control siRNA or TOPK siRNA in 100 μl of Krebs-Ringer phosphate buffer (KRPB: 145 mm NaCl, 5.7 mm sodium phosphate, 4.86 mm KCl, 0.54 mm CaCl2, 1.22 mm MgSO4, 5.5 mm glucose, pH 7.35). Cells were harvested after UVB at 0 (control), 0.25, 0.5, and 24 h at 37 °C as described above.
Measurement of H2O2
RPMI7951 cells (2 × 104 in 100 μl KRPB) stably expressing mock, Prx1-Wt or Prx1-S32A were incubated at 37 °C for various times after UVB. Measurement of H2O2 released from cells was determined as above for peroxidase activity except to convert the absorbance (A570) to H2O2 level (μm), we used an H2O2 standard curve, prepared according to the protocol provided with the Amplex Red Hydrogen Peroxide/Peroxidase Assay kit. Oxidation of Prx1 was detected by Western blotting using anti-peroxiredoxin-SO3.
Confocal Laser-scanning Fluorescence Microscopy
To determine whether phosphorylated TOPK and Prx1 co-localized in the nucleus, RPMI951 melanoma cells (2 × 105) were treated with UVB (4 kJ/m2) and incubated for 30 min at 37 °C. Cells were fixed in methanol (−20 °C) and blocked for 15 min in 1% donkey serum in 0.3% Tween in PBS. Cells were incubated 1 h with anti-p-TOPK (Thr-9) rabbit (1:25 dilution) and anti-Prx1 goat (1:100 dilution). Then, after washing with PBS, cells were incubated 1 h with anti-cyanine Cy2 donkey anti-goat IgG (for Prx1) or anti-cyanine Cy3 donkey anti-rabbit IgG (for p-TOPK). Co-localization of proteins was observed by laser scanning confocal microscopy (NIKON C1si Confocal Spectral Imaging System, NIKON Instruments Co., Melville, NY) using a CFI Plan Fluor 40× oil objective. The channels were collected individually to avoid bleed-through between the 2 channels, and to “balance” the fluorescence intensity in each channel. Confocal Z-sections of 0.6 μm thickness were imaged.
Tissue Array
Two human skin tissue arrays (U.S. Biomax, Inc.) were prepared and analyzed according to the protocol provided. The samples were blocked with 5% goat serum albumin in 800 μl 1× PBS/0.03% Triton X-100, (pH 6.0) in a humidified chamber for 1 h at room temperature, and then incubated with the TOPK rabbit or the Prx1 goat antibody (1:100 dilutions in 800 μl 1× PBS/0.03% Triton X-100, pH 6.0) at 4 °C in a humidified chamber overnight. The slides were washed and hybridized 2 h at room temperature in the dark with the secondary antibody (anti-rabbit, donkey antibody) conjugated with Cy3 (1:200 dilution) for TOPK and (anti-goat, donkey antibody) conjugated with Cy2 (1:200 dilution) for Prx1. Slides were washed with PBS (2×, 5 min) and by high salt/PBS (2×, 2 min).
Circular Dichroism (CD) Spectra
Prx1-Wt and Prx1-S32A proteins (50 μg each) were mixed with active PBK/TOPK (3 μg/50 μl reaction) in 10× kinase buffer containing 50 μmol/liter of unlabeled ATP and the reaction was incubated at 30 °C for 60 min. Phosphorylated and nonphosphorylated Prx1-Wt and Prx1-S32A were used for far UV CD spectrometry. CD spectra were recorded (in triplicate) at room temperature with a JASCO 500A spectropolarimeter (Jasco Co., Tokyo, Japan). The wavelength range measured was from 250 to 190 nm at a scan rate of 20 nm/min. The CD spectra are expressed in terms of ellipticity, [θ] (degrees × cm2 × decimol−1), which is related to the mean residue molecular weight of 110 Da. The following formula was used to generate as [θ] = [θ]observed × S × 110/10 × C × 1, where [θ]observed is expressed in degrees; S = sensitivity of spectropolarimeter; C = concentration of protein, mg/ml; and 1 = path length, mm. Calibration of the spectrometer was prepared with 0.06% ammonium salt of d-camphor-10-sulfonic acid (Katayama Chemical, Japan). The CDPro software package was used for calculation of elements of secondary structure (28, 29).
Flow Cytometry Analysis
UVB-induced apoptosis was determined using the Annexin V-FITC Apoptosis Detection kit (MBL International Corp., Watertown, MA). Cells were harvested with 0.025% trypsin in PBS/5 mm EDTA. After washing with MEM containing serum, cells were incubated 5 min at room temperature with Annexin V-FITC plus propidium iodide according to the protocol provided by the manufacturer. Apoptosis was analyzed on a Becton Dickinson FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
Statistical Analysis
Comparisons were analyzed by one-way ANOVA, and a p value of < 0.05 was considered statistically significant.
RESULTS
Prx1 Binds TOPK
We previously identified 26 proteins that could bind with TOPK (13). Human peroxiredoxin was identified by tandem mass spectrometry as potential binding partner for TOPK. The protein was identified from 15 distinct peptide assignments (54% coverage by amino acid sequence), of which 8 (33% coverage) showed > 95% confidence in peptide identification, according to the Paragon scoring algorithm. All tandem mass spectra were validated by visual inspection. A fragment composed of 10 amino acids, LVQAFQFTDK, was identified with a Pro ID confidence of 99% as a peptide from the typical 2-Cys peroxiredoxin family (17, 19, 20) (Fig. 1A). The gene sequence of this peptide was 100% homologous to the sequence of Prx1, which indicated that TOPK could bind with Prx1. Thus, we focused on elucidating the function and physiologic significance of the interaction of TOPK and Prx1.
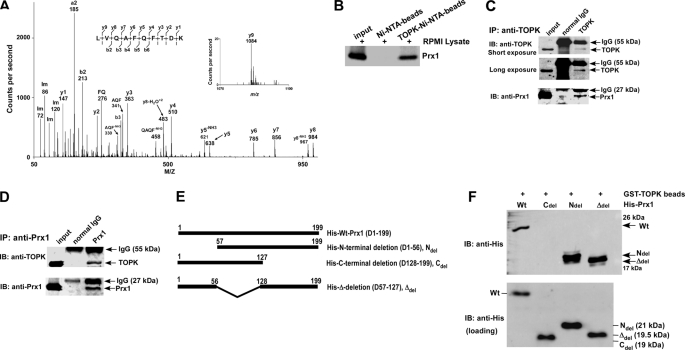
FIGURE 1.
TOPK interacts with Prx1 in vitro and ex vivo. A, tandem mass spectrum for the doubly charged ion from the peptide sequence LVQAFQFTDK (theoretical, monoisotopic neutral MW of 1195.62 Da), labeled with select experimental product ions, corresponding m/z values, and select ammonium ions (Im). The precursor mass error is 40 ppm. The experimentally determined diagnostic b- and y-type product ions with S/N >3 are shown on the peptide sequence above the spectrum (S/n = signal to noise). B, TOPK binding with Prx1 ex vivo. Lane 1: input, whole-cell lysate from RPMI7951 human melanoma cells; lane 2: negative control, whole-cell lysate from RPMI7951 cells precipitated with Ni-NTA-agarose beads; lane 3: whole-cell lysate from RPMI7951 cells precipitated with TOPK-Ni-NTA-agarose beads. TOPK and Prx1 binding was confirmed by immunoblotting with anti-Prx1. C and D, the endogenous TOPK-Prx1 complex was immunoprecipitated from RPMI7951 human melanoma cells with a TOPK (C) or a Prx1 (D) antibody, and TOPK or Prx1 were detected by Western blotting with a TOPK or Prx1 antibody. Precipitation with normal IgG served as a negative control. E, schematic diagrams for construction of Prx1 deletion mutants. Ndel (D1–56), Cdel (D128–199), or Δdel (D57–127) were each introduced into the pET46 Ek/LIC His fusion vector. F, Prx1 (Wt) and deletion mutants (Ndel, Cdel, or Δdel) purified from BL21 (bottom) were used for binding with GST-TOPK beads (top). Protein abundance were confirmed by Western blotting with an antibody against the His tag.
TOPK Interacts and Co-localizes with Prx1
To confirm the interaction of TOPK and Prx1, RPMI7951 melanoma cell lysates were incubated with Ni-NTA-agarose beads (negative control) or with His-TOPK-Ni-NTA-agarose beads at 4 °C overnight. The binding interaction was analyzed by Western blot, and results confirmed that TOPK binds with Prx1 (Fig. 1B). To determine whether the interaction of TOPK with Prx1 occurs endogenously, we used RPMI7951 cell lysates to perform immunoprecipitation experiments with anti-TOPK or anti-Prx1. The results shown that TOPK binds with Prx1 ex vivo (Fig. 1, C and D).
To determine the specific binding region of Prx1 that binds with TOPK, we first generated three Prx1 deletion constructs: N-terminal deletion (D1–56) as Ndel; C-terminal deletion (D128–199) as Cdel; and Δ-deletion (D57–127) as Δdel (Fig. 1E). Prx1-Wt and all deletion mutant proteins were used for assessment of binding with GST-TOPK beads. Results indicated that the Ndel and Δdel mutant proteins interacted efficiently with TOPK (Fig. 1F). The results indicated that, deletion of the C-terminal amino acids (D128–199) of Prx1 abrogated the binding of TOPK and Prx1. The crystal structure of Prx1 (30) shows that the C-terminal region contains a potential TOPK-binding site that is located close to Ser-32, which may be important for Prx1 phosphorylation.
Immunocytofluorescence analysis was used to examine the localization and interaction of TOPK and Prx1. Results indicated that following UVB treatment, most phosphorylated TOPK (red) and Prx1 (green) co-localized in the nucleus (supplemental Fig. S1; Merge, bottom right). Overall, the data show that TOPK interacts with Prx1.
TOPK Phosphorylates Prx1 in Vitro
The gene sequence of the LVQAFQFTDK peptide is also 60% homologous to the sequence of Prx3. Prx1 is located in the cytosol, whereas Prx3 is only found in the mitochondria (31, 32). To test the specificity of TOPK to phosphorylate Prx1, we determined whether TOPK can phosphorylate Prx3. The Prx proteins were purified from BL21 bacteria using Ni-NTA-agarose beads, and purity was confirmed by Coomassie Blue R-250 staining (Fig. 2A). These proteins were used as a substrate for active TOPK. Prx phosphorylation was visualized by autoradiography in the presence of [γ-32P]ATP. Results strongly indicated that TOPK phosphorylates Prx1 and not Prx3 (Fig. 2B).
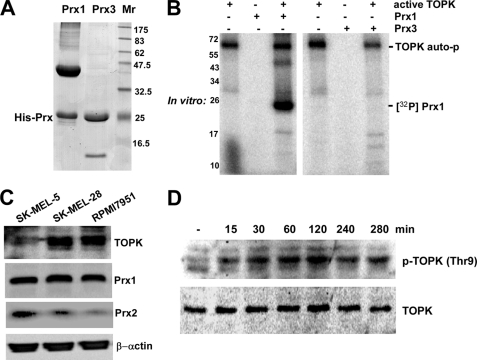
FIGURE 2.
TOPK phosphorylates Prx1 in vitro and UVB induces phosphorylation of TOPK in RPMI7951 melanoma cells. A, His-Prx1 and His-Prx3 fusion proteins were purified from BL21 bacteria using Ni-NTA-agarose beads and purity was confirmed by Coomassie Blue R-250 staining. B, in vitro kinase assay to determine whether TOPK phosphorylates Prx1 or Prx3. Results are visualized by autoradiography with [γ-32P]ATP. C, different human melanoma cell lines were screened by Western blot to determine total protein abundance of endogenous TOPK, Prx1, or Prx2. β-Actin verified equal protein loading. D, time-dependent UVB-induced phosphorylation of TOPK (Thr-9). RPMI7951 cells were harvested at various times after UVB (4 kJ/m2) treatment. Phosphorylation of TOPK (Thr-9) or nonphosphorylated TOPK was detected. Data for C and D are representative of three independent experiments.
TOPK and Prx1 Are Expressed in Cancer Tissues and Human Melanoma Cells
Our earlier study showed that TOPK is highly expressed in human colorectal cancer tissues and cell lines and plays an important role in colorectal cancer (12). Furthermore, melanoma cells expressing high levels of TOPK were more resistant to arsenite-induced apoptosis (13). These results suggested that the TOPK protein level might be higher in malignant tissues compared with normal tissues. Results of a human cancer tissue array analysis indicated that the abundance of TOPK and Prx1 proteins was greater in malignant melanoma tissues (supplemental Fig. S2). We also examined the levels of TOPK and Prx1 in different melanoma cell lines. Results indicated that the expression of TOPK was highest in SK-MEL-28 or RPMI7951 cell lines compared with SK-MEL-5 cells (Fig. 2C). The abundance of Prx1 was similar in the three cell lines. Human Prx1 and Prx2 share >90% homology in their amino acid sequences (19), but our results suggest that Prx2 abundance is very low in RPMI7951 cells (Fig. 2C). Based on these results, we chose the RPMI7951 melanoma cell line to study the function and relationship of TOPK and Prx1.
UVB Induces Phosphorylation of TOPK in RPMI7951 Melanoma Cells
UV is an important etiological factor in human skin cancer (33). The effect of UVB irradiation on phosphorylation of TOPK (Thr-9) in RPMI7951 melanoma cells was examined by Western blot. Results indicated that UVB (4 kJ/m2) induced a strong time-dependent phosphorylation of TOPK at Thr-9 (Fig. 2D).
TOPK Phosphorylates Prx1 at Ser-32
Because TOPK is a serine/threonine kinase, we examined potential phosphorylation of serine and threonine residues in Prx1 using NetPhos 2.0 (34) (Table 1). We compared sequences (Fig. 3A) of Prx1 and Prx3 (negative control) because TOPK phosphorylated Prx1 but not Prx3. We found that only Ser-32 and Ser-126 are potential sites for phosphorylation by TOPK. To confirm the possibility that TOPK phosphorylates these serine residues, we constructed and expressed His-Prx1-Wt or His-Prx1-S32A or His-Prx1-S126A mutant fusion proteins in BL21 bacteria. Proteins were purified by FPLC and purification of proteins was confirmed by Coomassie Blue R-250 staining (supplemental Fig. S3A). Purified protein fraction 3 for each protein was used for an in vitro kinase assay (Fig. 3B). The results showed that mutation of Ser-32 to Ala (S32A) abrogated the phosphorylation of Prx1, whereas mutation of Ser-126 to Ala (S126A) had no effect on phosphorylation of Prx1. This result indicated that TOPK phosphorylated Prx1 at Ser-32 only.
TABLE 1.
Identification of phosphorylation sites on Prx1
| Amino acid | Sequence | NetPhos 2.0 Score |
|---|---|---|
| Ser-2 | MSSGNA | 0.014 |
| Ser-3 | MSSGNAK | 0.002 |
| Ser-30 | FKDISLSDY | 0.970 |
| Ser-32 | DISLSDYKG | 0.959 |
| Ser-60 | IIAFSDRAE | 0.930 |
| Ser-77 | VIGASVDSH | 0.152 |
| Ser-80 | ASVDSHFCH | 0.575 |
| Ser-106 | IPLVSDPKR | 0.006 |
| Ser-126 | DEGISFRGL | 0.736 |
| Ser-152 | PVGRSVDET | 0.942 |
| Ser-181 | WKPGSDTIK | 0.056 |
| Ser-191 | DVQKSKEYF | 0.954 |
| Ser-196 | KEYFSKQK | 0.290 |
| Thr-18 | NFKATAVMP | 0.133 |
| Thr-49 | PLDFTFVCP | 0.026 |
| Thr-54 | FVCPTEIIA | 0.013 |
| Thr-90 | AWVNTPKKQ | 0.966 |
| Thr-111 | DPKRTIAQD | 0.287 |
| Thr-143 | LRQITVNDL | 0.404 |
| Thr-156 | SVDETLRLV | 0.790 |
| Thr-166 | AFQFTDKHG | 0.123 |
| Thr-183 | PGSDTIKPD | 0.077 |
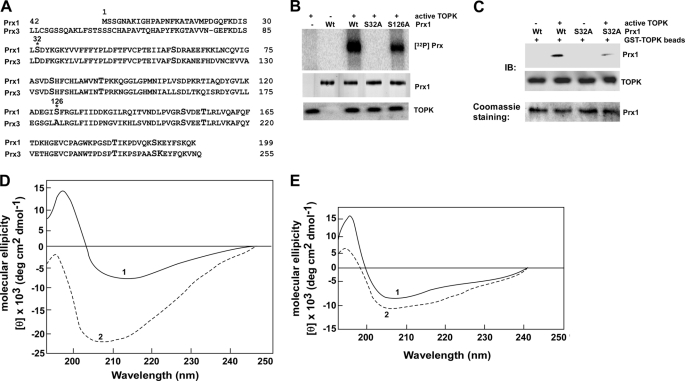
FIGURE 3.
TOPK phosphorylates Prx1 (Ser-32). A, comparison of amino acid sequences of Prx1 and Prx3. Amino acids Ser-32 and Ser-126 in Prx1 correspond with Asp-32 and Ala-126 in Prx3, respectively. B, in vitro kinase assay using [γ-32P]ATP to determine whether TOPK phosphorylates Wt or mutant (S126A or S32A) Prx1. Proteins were visualized by autoradiography. C, TOPK binds with phosphorylated Prx1. TOPK-phosphorylated or nonphosphorylated Prx1 (Wt) or Prx1 mutant (S32A) protein (in vitro kinase assay) were used for binding with GST-TOPK beads. TOPK and Prx1 binding was confirmed by immunoblotting with anti-Prx1. D and E, CD spectra in the far UV range were prepared to determine changes in the secondary structure of Prx1 induced by phosphorylation. D, Prx1-Wt (solid line, 1) or phosphorylated Prx1-Wt (dashed line, 2). E, Prx1-S32A (solid line) or phosphorylated Prx1-S32A (dashed line).
Phosphorylation Prx1 at Ser-32 by TOPK Mediates Changes in the Secondary Structure of Prx1
TOPK-phosphorylated or nonphosphorylated wild-type or mutant (S32A) Prx1 was used to assess binding with GST-TOPK beads to determine whether phosphorylation of Prx1 is necessary for the interaction. The data indicated after mutation, the binding decreased dramatically (Fig. 3C). Next we analyzed the secondary structure of TOPK-phosphorylated or nonphosphorylated Prx1 using far UV CD spectra from 190 to 250 nm. Prx1-Wt and Prx1-S32A proteins were purified by FPLC (supplemental Fig. S3A) and Fraction 3 was further purified by FPLC (supplemental Fig. S3B) and used for the far UV CD spectra analysis. The CD spectra of nonphosphorylated Prx1-Wt showed a negative minima at 210 nm and a maximum at 195 nm (Fig. 3D, line 1). This CD spectrum was altered by phosphorylation of Prx1-Wt by TOPK (Fig. 3D, line 2). The ellipticity of the negative area increased more than 4-fold compared with nonphosphorylated Prx1 and showed two negative minima at 205 and 212 nm but the positive maximum was absent. The CD spectra of mutant Prx1-S32A showed negative minima at 205 nm and maximum at 195 nm (Fig. 3E, line 1). TOPK had little effect on the mutant Prx-S32A CD spectrum (Fig. 3E, line 2). The Prx1 protein comprises a mixture of α/β or α+β secondary structure. In the secondary structure of phosphorylated Prx1, only the β-strand structure decreased by 12% (supplemental Table S1). The CD spectrum results indicated that TOPK binds with Prx1 and phosphorylation of Prx1 by TOPK leads to significant changes in the secondary structure of Prx1 that affects the CD spectrum in the far UV CD (Fig. 3D, line 2). This result was confirmed by the calculation of the elements of secondary structure (supplemental Table S1).
Phosphorylation of Prx1 (Ser-32) Increases Peroxidase Activity
To investigate the effect of phosphorylation at Ser-32 on the peroxidase activity of Prx1 in vitro, we first performed a kinase assay and following phosphorylation, Prx1 peroxidase activity was assessed. The peroxidase activity of Prx1-Wt after phosphorylation by TOPK was increased more than 2-fold compared with the nonphosphorylated Prx1-Wt (Fig. 4A). In contrast, TOPK had no effect on peroxidase activity of the mutant Prx1-S32A protein. To investigate the effect of phosphorylation of Prx1 by TOPK on Prx1 peroxidase activity ex vivo, we compared RPMI7951 cells stably expressing TOPK siRNA or control siRNA. Cells were treated with UVB and peroxidase activity was determined at 37 °C at various times after UVB. At 24 h, cells expressing TOPK siRNA exhibited about a 2-fold decrease in peroxidase activity compared with cells expressing control siRNA (Fig. 4B). Overall, these results show that phosphorylation of Prx1 (Ser-32) by TOPK increases the peroxidase activity of Prx1 and suggests that this phosphorylation is important for optimal function of Prx1.
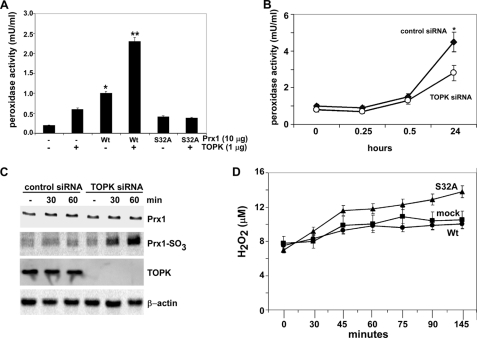
FIGURE 4.
TOPK increases the peroxidase activity of Prx1 and suppresses H2O2 accumulation. A, in vitro peroxidase activity was determined as described under “Materials and Methods” using active TOPK and FPLC purified His-Prx1-Wt (Wt) or His-Prx1-S32A (S32A) fusion proteins. Peroxidase activities of nonphosphorylated Wt and S32A were used as controls. To convert the spectrophotometric data (A570) to peroxidase activity (mU/ml), a peroxidase standard curve was prepared according to the manufacturer's instructions. B, peroxidase activity was determined as for (A), but using RPMI7951 cells (2 × 104) transfected with control siRNA or TOPK siRNA. Samples were harvested at different times following UVB (4 kJ/m2) treatment. Data are shown as means ± S.D. of three samples from two independent experiments. The asterisk (*) indicates a significantly increased (p < 0.001) activity induced by UVB in control siRNA cells compared with untreated or UVB-treated TOPK siRNA cells. C, oxidation of Prx1 to Prx1-SO3 was assessed by Western blotting using anti-Prx1-SO3 (recognizes oxidized Cys-52 in Prx1) after UVB (4 kJ/m2) in RPMI7951 cells transfected with control siRNA or TOPK siRNA. Total TOPK, Prx1, or β-actin protein levels were used as internal controls to confirm TOPK deficiency and equal protein loading. D, effect of TOPK on H2O2 accumulation was measured as for (A) in mock, Prx1 Wt or Prx1 mutant (S32A) stably transfected cells. To convert the spectrophotometric data (A570) to reflect the levels of H2O2 (μm), an H2O2 standard curve was prepared according to the manufacturer's instructions.
TOPK Regulates the Level of H2O2 through Its Phosphorylation of Prx1
Prx1 can be inactivated by oxidation of the active Cys-52 site to sulfinic acid (Cys-SO2) or sulfonic acid (Cys-SO3). The significance of this inactivation as a means to regulate intracellular H2O2 levels has generated interest in peroxiredoxin as a mediator of H2O2-regulated signaling pathways. The sulfinic or sulfonic forms of Prx1 lose their anti-peroxidase activity, which allows cells to accumulate H2O2 for signaling or pathogenesis in inflammation or cancer (35–37). The anti-Prx1-SO3 antibody recognizes both the sulfinic or sulfonuc forms of Prx1 and detects oxidized Prx with high sensitivity and specificity (38–40). We used RPMI7951 cells expressing control siRNA or TOPK siRNA for determining the oxidation status of Prx1 (Fig. 4C) using Western blotting and the Prx1-SO3 antibody. The level of Prx1-SO3 in TOPK siRNA cells was dramatically increased after UVB, indicating that the peroxidase activity of Prx1 decreased and H2O2 level was accumulating. After UVB, the level of H2O2 in Prx1-S32A cells was increased compared with mock or Prx1-Wt cells (Fig. 4D), indicating that TOPK suppresses H2O2 accumulation and the phosphorylation of Prx1 (Ser-32) is important for this function.
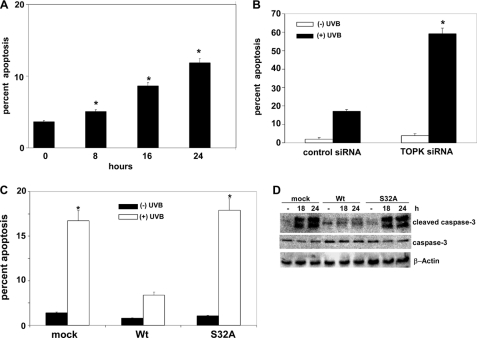
TOPK Inhibits UVB-induced Apoptosis in RPMI7951 Melanoma Cells
We previously reported that high levels of TOPK in different melanoma cells were associated with resistance to arsenite-induced apoptosis (13). RPMI7951 melanoma cells treated with UVB show an approximate 12% increase in early apoptosis (Fig. 5A). At 24 h after UVB, a significant (p < 0.0001) increase in early apoptosis was observed in TOPK siRNA cells compared with control siRNA-transfected cells (Fig. 5B). This suggested that the absence of TOPK sensitizes cells to UVB-induced apoptosis. Cells overexpressing Prx1 (Wt) exhibit a significant (p < 0.001) decrease in UVB-induced early apoptosis compared with cells stably expressing mock or mutant Prx1-S32A (S32A) (Fig. 5C). This result was further confirmed with the observation of markedly decreased levels of cleaved caspase-3 in Prx1-Wt cells compared with mock or mutant Prx1-S32A cells (Fig. 5D). Taken together, these results indicated that TOPK plays an important role in suppressing UVB-induced apoptosis that is mediated through its phosphorylation of Prx1 at Ser-32.
FIGURE 5.
TOPK inhibits UVB-induced apoptosis in RPMI7951 melanoma cells, mediated through phosphorylation of Prx1 at Ser-32. A, analysis of apoptosis by flow cytometry in RPMI7951 melanoma cells; B, RPMI7951 melanoma cells transfected with control siRNA or TOPK siRNA. C, RPMI7951 melanoma cells stably expressing mock, Prx1 wild type (Wt), or Prx1 mutant (S32A) constructs. For A–C, cells were incubated with annexin V-conjugated FITC at 24 h after UVB (4 kJ/m2); data are shown as means ± S.D. from three independent experiments. The asterisk (*) indicates a significant increase (p < 0.0001) in UVB-induced apoptosis. D, RPMI7951 melanoma cells stably expressing mock, Prx1 Wt or Prx1 mutant (S32A) constructs were treated or not treated with UVB and harvested after 18 or 24 h. The level of cleaved caspase-3 was assessed by Western blot, and β-actin was used as an internal control to monitor equal protein loading.
DISCUSSION
UV irradiation activates various signaling pathways that are oncogenic or protective or both (33). Progress in understanding the mechanisms of UV-induced signal transduction could lead to the use of protein kinases as specific targets for the prevention and control of skin cancer. The mitogen-activated protein (MAP) kinase pathway is activated after UVB treatment. Previous studies indicated that TOPK is heavily involved in MAP kinase signaling. Yeast two-hybrid screening analysis suggested a possible association between Raf and TOPK (41); and p38 MAP kinase and c-Myc are potential substrates of TOPK (1, 8). TOPK appears to play an important role in melanoma. We found that TOPK was more abundant in malignant melanoma tissues compared with squamous and basal carcinoma tissues and that UVB increased the phosphorylation of TOPK in RPMI7951 melanoma cells. We previously identified 26 individual proteins that could bind with TOPK (13), including peroxiredoxin 1. Prx1 is a member of the redox-regulating peroxiredoxin protein family and has been shown to be elevated in several cancers. Its ability to enhance the survival and progression of cancer cells has been suggested (42–45). Prx1 plays a role in regulating hydrogen peroxide levels, which might be important in melanoma. The RPMI7951 cells express high levels of Prx1, but not Prx2, which makes these cells a good model to study Prx1 function.
Our results clearly indicate that TOPK phosphorylates Prx1 at Ser-32 and increases its peroxidase activity. Phosphorylation of Prx1 at Thr-90 was reported to decrease Prx1 activity and the resulting intracellular accumulation of H2O2 was suggested to be important for progression of the cell cycle in HeLa cells (22). A mutant Prx1-T90D (i.e. a mimic form of phosphorylated-Prx1) showed higher chaperone activity (23, 46). In the present study, we showed that UVB induced phosphorylation of TOPK at Thr-9. However, TOPK did not phosphorylate Prx1 at Thr-90, but instead phosphorylated Prx1 at Ser-32, resulting in increased peroxidase activity of Prx1 and reduced accumulation of H2O2 in vitro and ex vivo. No change was observed in chaperone activity (data not shown). Prx1 is reportedly involved in ASK1-mediated apoptosis induced by H2O2 and negatively regulates ASK1-induced apoptosis in human embryonic kidney 293 and HeLa cells (21). Our results suggest that phosphorylation of Prx1 (Ser-32) by TOPK is associated with an inhibition of UVB-induced apoptosis in RPMI7951 melanoma cells that is mediated by increased Prx1 peroxidase activity associated with a decreased level of H2O2. Based on our results, increased TOPK and Prx1 in melanoma might play a critical role in providing resistance against UVB-induced apoptosis. Further studies on the mechanisms of the regulation of TOPK activity and the development of TOPK- or Prx1-targeting drugs might contribute to treatment of melanoma.
Supplementary Material
Acknowledgments
We thank Dr. J. Abe (Department of Pathology, Division of Molecular Pathology, Ehime University School of Medicine, Toh-on, Ehime, Japan) for pcDNA3-HA-TOPK. The mass spectrometric analysis was performed at the Center for Mass Spectrometry and Proteomics in the Department of Biochemistry, Molecular Biology, and Biophysics at the University of Minnesota. The authors have no conflicts of interest. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA 077646 (ARPA), R37 CA 081646, CA 120388, and ES 016548 and The Hormel Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- PBK/TOPK
- PDZ-binding kinase/T-LAK cell-originated protein kinase
- Prx
- peroxiredoxin
- ASK
- apoptosis signal-regulating kinase.
REFERENCES
- 1.Abe Y., Matsumoto S., Kito K., Ueda N. (2000) J. Biol. Chem. 275, 21525–21531 [DOI] [PubMed] [Google Scholar]
- 2.Ayllón V., O'Connor R. (2007) Oncogene 26, 3451–3461 [DOI] [PubMed] [Google Scholar]
- 3.Dougherty J. D., Garcia A. D., Nakano I., Livingstone M., Norris B., Polakiewicz R., Wexler E. M., Sofroniew M. V., Kornblum H. I., Geschwind D. H. (2005) J Neurosci 25, 10773–10785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abe Y., Takeuchi T., Kagawa-Miki L., Ueda N., Shigemoto K., Yasukawa M., Kito K. (2007) J. Mol. Biol. 370, 231–245 [DOI] [PubMed] [Google Scholar]
- 5.Matsumoto S., Abe Y., Fujibuchi T., Takeuchi T., Kito K., Ueda N., Shigemoto K., Gyo K. (2004) Biochem. Biophys. Res. Commun. 325, 997–1004 [DOI] [PubMed] [Google Scholar]
- 6.Gaudet S., Branton D., Lue R. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5167–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nandi A. K., Rapoport A. P. (2006) Leuk Res 30, 437–447 [DOI] [PubMed] [Google Scholar]
- 8.Nandi A., Tidwell M., Karp J., Rapoport A. P. (2004) Blood Cells Mol. Dis. 32, 240–245 [DOI] [PubMed] [Google Scholar]
- 9.Simons-Evelyn M., Bailey-Dell K., Toretsky J. A., Ross D. D., Fenton R., Kalvakolanu D., Rapoport A. P. (2001) Blood Cells Mol Dis 27, 825–829 [DOI] [PubMed] [Google Scholar]
- 10.Côté S., Simard C., Lemieux R. (2002) Cytokine 20, 113–120 [DOI] [PubMed] [Google Scholar]
- 11.Park J. H., Lin M. L., Nishidate T., Nakamura Y., Katagiri T. (2006) Cancer Res. 66, 9186–9195 [DOI] [PubMed] [Google Scholar]
- 12.Zhu F., Zykova T. A., Kang B. S., Wang Z., Ebeling M. C., Abe Y., Ma W. Y., Bode A. M., Dong Z. (2007) Gastroenterology 133, 219–231 [DOI] [PubMed] [Google Scholar]
- 13.Zykova T. A., Zhu F., Lu C., Higgins L., Tatsumi Y., Abe Y., Bode A. M., Dong Z. (2006) Clin Cancer Res. 12, 6884–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nandi A. K., Ford T., Fleksher D., Neuman B., Rapoport A. P. (2007) Biochem. Biophys. Res. Commun. 358, 181–188 [DOI] [PubMed] [Google Scholar]
- 15.Oh S. M., Zhu F., Cho Y. Y., Lee K. W., Kang B. S., Kim H. G., Zykova T., Bode A. M., Dong Z. (2007) Cancer Res. 67, 5186–5194 [DOI] [PubMed] [Google Scholar]
- 16.Aran M., Ferrero D. S., Pagano E., Wolosiuk R. A. (2009) Febs. J. 276, 2478–2493 [DOI] [PubMed] [Google Scholar]
- 17.Butterfield L. H., Merino A., Golub S. H., Shau H. (1999) Antioxid. Redox. Signal 1, 385–402 [DOI] [PubMed] [Google Scholar]
- 18.Kim J. H., Bogner P. N., Baek S. H., Ramnath N., Liang P., Kim H. R., Andrews C., Park Y. M. (2008) Clin. Cancer Res. 14, 2326–2333 [DOI] [PubMed] [Google Scholar]
- 19.Rhee S. G., Kang S. W., Chang T. S., Jeong W., Kim K. (2001) IUBMB Life 52, 35–41 [DOI] [PubMed] [Google Scholar]
- 20.Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 21.Kim S. Y., Kim T. J., Lee K. Y. (2008) FEBS Lett. 582, 1913–1918 [DOI] [PubMed] [Google Scholar]
- 22.Chang T. S., Jeong W., Choi S. Y., Yu S., Kang S. W., Rhee S. G. (2002) J. Biol. Chem. 277, 25370–25376 [DOI] [PubMed] [Google Scholar]
- 23.Jang H. H., Kim S. Y., Park S. K., Jeon H. S., Lee Y. M., Jung J. H., Lee S. Y., Chae H. B., Jung Y. J., Lee K. O., Lim C. O., Chung W. S., Bahk J. D., Yun D. J., Cho M. J., Lee S. Y. (2006) FEBS Lett. 580, 351–355 [DOI] [PubMed] [Google Scholar]
- 24.Biemann K. (1988) Biomed. Environ. Mass Spectrom. 16, 99–111 [DOI] [PubMed] [Google Scholar]
- 25.Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) Mol. Cell Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 26.Zykova T. A., Zhang Y., Zhu F., Bode A. M., Dong Z. (2005) Carcinogenesis 26, 331–342 [DOI] [PubMed] [Google Scholar]
- 27.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 28.Sreerama N., Venyaminov S. Y., Woody R. W. (2000) Anal. Biochem. 287, 243–251 [DOI] [PubMed] [Google Scholar]
- 29.Sreerama N., Woody R. W. (2000) Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 30.Hirotsu S., Abe Y., Okada K., Nagahara N., Hori H., Nishino T., Hakoshima T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12333–12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knoops B., Loumaye E., Van Der Eecken V. (2007) Subcell Biochem. 44, 27–40 [DOI] [PubMed] [Google Scholar]
- 32.Hofmann B., Hecht H. J., Flohé L. (2002) Biol. Chem. 383, 347–364 [DOI] [PubMed] [Google Scholar]
- 33.Bode A. M., Dong Z. (2003) Sci. STKE 2003, RE2. [DOI] [PubMed] [Google Scholar]
- 34.Diella F., Cameron S., Gemünd C., Linding R., Via A., Kuster B., Sicheritz-Pontén T., Blom N., Gibson T. J. (2004) BMC Bioinformatics 5, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker L. M., Poole L. B. (2003) J. Biol. Chem. 278, 9203–9211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole L. B., Karplus P. A., Claiborne A. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 325–347 [DOI] [PubMed] [Google Scholar]
- 37.Yang K. S., Kang S. W., Woo H. A., Hwang S. C., Chae H. Z., Kim K., Rhee S. G. (2002) J. Biol. Chem. 277, 38029–38036 [DOI] [PubMed] [Google Scholar]
- 38.Cordray P., Doyle K., Edes K., Moos P. J., Fitzpatrick F. A. (2007) J. Biol. Chem. 282, 32623–32629 [DOI] [PubMed] [Google Scholar]
- 39.Woo H. A., Chae H. Z., Hwang S. C., Yang K. S., Kang S. W., Kim K., Rhee S. G. (2003) Science 300, 653–656 [DOI] [PubMed] [Google Scholar]
- 40.Woo H. A., Kang S. W., Kim H. K., Yang K. S., Chae H. Z., Rhee S. G. (2003) J. Biol. Chem. 278, 47361–47364 [DOI] [PubMed] [Google Scholar]
- 41.Yuryev A., Wennogle L. P. (2003) Genomics 81, 112–125 [DOI] [PubMed] [Google Scholar]
- 42.Chang J. W., Jeon H. B., Lee J. H., Yoo J. S., Chun J. S., Kim J. H., Yoo Y. J. (2001) Biochem. Biophys. Res. Commun. 289, 507–512 [DOI] [PubMed] [Google Scholar]
- 43.Chen M. F., Keng P. C., Shau H., Wu C. T., Hu Y. C., Liao S. K., Chen W. C. (2006) Int. J. Radiat. Oncol. Biol. Phys. 64, 581–591 [DOI] [PubMed] [Google Scholar]
- 44.Yanagawa T., Ishikawa T., Ishii T., Tabuchi K., Iwasa S., Bannai S., Omura K., Suzuki H., Yoshida H. (1999) Cancer Lett 145, 127–132 [DOI] [PubMed] [Google Scholar]
- 45.Yanagawa T., Iwasa S., Ishii T., Tabuchi K., Yusa H., Onizawa K., Omura K., Harada H., Suzuki H., Yoshida H. (2000) Cancer Lett 156, 27–35 [DOI] [PubMed] [Google Scholar]
- 46.Barford D. (2004) Curr. Opin. Struct. Biol. 14, 679–686 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.