Abstract
Oxygen-evolving photosystem II (PSII) isolated from a marine centric diatom, Chaetoceros gracilis, contains a novel extrinsic protein (Psb31) in addition to four red algal type extrinsic proteins of PsbO, PsbQ′, PsbV, and PsbU. In this study, the five extrinsic proteins were purified from alkaline Tris extracts of the diatom PSII by anion and cation exchange chromatographic columns at different pH values. Reconstitution experiments in various combinations with the purified extrinsic proteins showed that PsbO, PsbQ′, and Psb31 rebound directly to PSII in the absence of other extrinsic proteins, indicating that these extrinsic proteins have their own binding sites in PSII intrinsic proteins. On the other hand, PsbV and PsbU scarcely rebound to PSII alone, and their effective bindings required the presence of all of the other extrinsic proteins. Interestingly, PSII reconstituted with Psb31 alone considerably restored the oxygen evolving activity in the absence of PsbO, indicating that Psb31 serves as a substitute in part for PsbO in supporting oxygen evolution. A significant difference found between PSIIs reconstituted with Psb31 and with PsbO is that the oxygen evolving activity of the former is scarcely stimulated by Cl− and Ca2+ ions but that of the latter is largely stimulated by these ions, although rebinding of PsbV and PsbU activated oxygen evolution in the absence of Cl− and Ca2+ ions in both the former and latter PSIIs. Based on these results, we proposed a model for the association of the five extrinsic proteins with intrinsic proteins in diatom PSII and compared it with those in PSIIs from the other organisms.
Keywords: Membrane Energetics, Photosynthesis, Plant, Protein Purification, Protein Structure, Chaetoceros gracilis, PSII, Psb31, Diatom, Extrinsic Protein
Introduction
Oxygen-evolving photosystem II (PSII)2 is a thylakoid membrane-located, multisubunit pigment complex catalyzing light-induced electron transfer from water to plastoquinone, with the concomitant production of molecular oxygen. The PSII complex consists of a number of intrinsic proteins and several extrinsic proteins associated with the lumenal side. So far, PSII membrane fragments and core complexes that are highly active in oxygen evolution and that retain all of the extrinsic proteins have been isolated from cyanobacteria (1–3), red alga (4, 5), Euglena (6), green alga (7), and higher plants (8–10). Among these PSII complexes from a wide variety of organisms, the major intrinsic core proteins are largely conserved, whereas the extrinsic proteins are significantly different among different plant species.
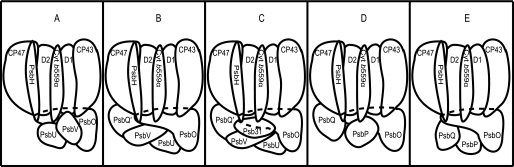
The differences in the composition of the extrinsic proteins and their binding patterns in PSII from various organisms are illustrated in the schematic models shown in Fig. 1. The direct and indirect associations of various extrinsic proteins at the lumenal side are based on various reconstitution and cross-reconstitution studies reported so far (for a recent review see Ref. 11). Purified cyanobacterial PSII (Fig. 1A) contains three extrinsic proteins of PsbO, PsbV, and PsbU, in which PsbO and PsbV can directly bind to PSII intrinsic proteins essentially independent of the presence of other proteins, whereas the binding of PsbU requires the presence of both PsbO and PsbV (1–3, 12, 13). In red algal PSII (Fig. 1B), a fourth extrinsic protein, the unique 20-kDa protein (PsbQ′), is present in addition to the three cyanobacterial extrinsic proteins (5, 14). Among the four extrinsic proteins, PsbO and PsbQ′ can directly bind to PSII intrinsic proteins essentially independent of the presence of other proteins, whereas the effective binding of PsbV and PsbU requires the presence of both PsbO and PsbQ′ (5). These results suggested that the binding property of PsbV is different between cyanobacterial and red algal PSIIs in that the former binds directly to PSII but the latter binds to PSII through its interaction with the other three extrinsic proteins. This difference of the binding property was shown to depend mainly on the structure of PSII intrinsic proteins but not that of PsbV by cross-reconstitution experiments using PsbV and PSII from the two organisms (15).
FIGURE 1.
Schematic models of PSIIs from cyanobacteria (A), red algae (B), diatom (C), green alga and Euglena (D), and higher plants (E) showing the species of various extrinsic proteins and their direct and indirect associations with PSII at the lumenal side based on various reconstitution and cross-reconstitution studies. See text for details.
In contrast to cyanobacterial and red algal PSII, PSIIs of Euglena, green algae, and higher plants contain PsbP and PsbQ proteins instead of PsbV and PsbU (6–10). In green algal and Euglena PSIIs (Fig. 1D), PsbP and PsbQ as well as PsbO can directly bind to PSII independent of the other extrinsic proteins, and PsbO functionally re-binds to PSII, which has been reconstituted with PsbP and PsbQ (6, 7), indicating that each of the three extrinsic proteins has their own binding sites in PSII that are independent of the other extrinsic proteins (7). Higher plant PSII also contains PsbO, PsbP, and PsbQ as extrinsic proteins. However, their binding properties are largely different from those of the corresponding proteins in PSII of Euglena and green algae. In higher plant PSII, only PsbO can directly bind to PSII, whereas PsbP cannot bind to PSII directly and associates with PSII only through its interaction with PsbO, and PsbQ functionally associates with PSII only through its interaction with both PsbO and PsbP (16). Detailed analysis on the differences of the binding properties between green algal and higher plant PSIIs has been performed by cross-reconstitution experiments using extrinsic proteins and PSIIs from the two organisms (17). The results showed that the green algal PsbP and PsbQ proteins can directly bind to green algal PSII but not to higher plant PSII in the absence of PsbO, whereas the higher plant PsbP and PsbQ proteins cannot bind to both higher plant and green algal PSIIs in the absence of PsbO and functionally bind to higher plant PSII only in the presence of higher plant PsbO but not in the presence of green algal PsbO (17).
Among the variously different extrinsic proteins, PsbO is present in all of the oxygenic photosynthetic organisms and plays an important role in maintaining the stability and activity of the Mn4Ca cluster (11, 18, 19). On the other hand, the PsbV and PsbU proteins in cyanobacterial and red algal PSIIs or the PsbP and PsbQ proteins in green algal and higher plant PSIIs function to optimize the availability of Ca2+ and Cl− cofactors for water oxidation (1, 4–7, 12, 20–22). The PsbQ′ protein in red algal PSII is not involved directly in oxygen evolution but is required for effective binding of the PsbV and PsbU proteins (5). These facts imply that PsbV and PsbU were replaced by PsbP and PsbQ during evolution from prokaryotic cyanobacteria and the primitive eukaryotic red algae to the green lineage Euglena, green algae, and higher plants. The distribution of these extrinsic proteins in various organisms was examined using a variety of antibodies, which showed that they have been diverged into cyanobacterial type (PsbO, PsbV, and PsbU), red algal type (PsbO, PsbQ′, PsbV, and PsbU), and green algal type (PsbO, PsbP, and PsbQ) during early phases of evolution after a primary endosymbiosis (23).
It should be pointed out that PsbP- and PsbQ-like proteins have been found in cyanobacterial thylakoid membranes and purified PSII (3). The binding characteristics and functions of these proteins are different from those of the extrinsic PsbP, PsbQ, and PsbQ′ proteins (11), and they have been suggested to be lipoproteins and to play regulatory roles in maintaining the PSII activity in nutrient-limiting media depleted of Cl−, Ca2+, or iron in the prokaryotic cyanobacteria (24, 25). The major function of PsbQ-like protein was shown to stabilize PsbV, thereby contributing to the protection of the catalytic Mn4Ca-Cl cluster of the water oxidation machinery (26). The specific binding of PsbQ-like protein to cyanobacterial PSII was also suggested based on the co-purification of PSII with a His-tagged PsbQ-like protein (27). The binding properties and functions of some other hydrophilic, peripheral PSII subunits, including PsbP- and PsbQ-like proteins, Psb27 and PsbR, have been reviewed by Roose et al. (28). PsbQ′ in red algal PSII may be an intermediate between the PsbQ-like proteins in cyanobacteria and the mature PsbQ protein in higher plants.
Recently, we succeeded for the first time in preparation of PSII retaining a high oxygen evolving activity from a marine centric diatom, Chaetoceros gracilis (29, 30). The diatom PSII was found to contain a novel fifth extrinsic protein (referred to as Psb31 following the nomenclature for PSII subunits (3)) in addition to the four red algal type extrinsic proteins PsbO, PsbQ′, PsbV, and PsbU (Fig. 1C) (29, 30). The gene encoding the novel Psb31 protein was cloned and sequenced (31), which showed that the deduced protein contained three characteristic leader sequences targeted for chloroplast endoplasmic reticulum membrane, chloroplast envelope membrane, and thylakoid membrane, indicating that Psb31 is encoded in the nuclear genome and constitutes one of the extrinsic proteins located on the lumenal side (Fig. 1C) (31). The function of the Psb31 protein, however, remains to be clarified. For understanding of the oxygen-evolving complex in diatom PSII, it is important to elucidate the binding and functional properties of the novel Psb31 protein. In this study, the five extrinsic proteins were purified from the diatom PSII, and reconstitution experiments using the purified extrinsic proteins in various combinations with PSII were performed to examine the binding and functional properties of the five extrinsic proteins, especially the Psb31 protein.
EXPERIMENTAL PROCEDURES
Preparation of Oxygen-evolving PSII from a Marine Centric Diatom, C. gracilis
A marine centric diatom, C. gracilis, was grown in artificial seawater as described previously (29). Thylakoid membranes and oxygen-evolving PSII were prepared according to Nagao et al. (29, 30). The cells suspended in a medium containing 1 m betaine, 50 mm MES-NaOH (pH 6.5), and 5 mm MgCl2 were disrupted by freeze-thawing and then incubated in the presence of DNase I and 1 mm PMSF for 30 min at 0 °C in the dark. The supernatants after centrifugation at 3,000 × g for 3 min were centrifuged at 40,000 × g for 10 min, and its precipitates (thylakoid membranes) were suspended in a medium containing 1 m betaine, 50 mm MES-NaOH (pH 6.5), and 1 mm EDTA (buffer A). The thylakoid membranes were treated with 1% Triton X-100 in buffer A at 1 mg of chlorophyll (Chl)/ml for 5 min at 0 °C in the dark and then fractionated by differential centrifugations according to Ref. 29. The resulting oxygen-evolving PSII particles (crude PSII) contained a large amount of fucoxanthin chlorophyll a/c-binding protein and were remarkably unstable as a significant inactivation of oxygen evolution, Chl bleaching and degradation of PSII subunits were observed during incubation at 25 °C in the dark (30). The crude PSII (1 mg of Chl/ml) suspended in buffer A was then immediately solubilized with 1% Triton X-100 for 20 min at 0 °C in the dark and applied to a DEAE-Toyopearl 650 M column equilibrated with the buffer A containing 0.03% Triton X-100. After the column was washed with 2–3 bed volumes of the equilibrating buffer, a green fraction (purified PSII lacking major components of fucoxanthin chlorophyll a/c-binding protein) was eluted at 180 mm NaCl according to Ref. 30. The purified PSII was concentrated by centrifugation at 40,000 × g for 20 min after addition of 10% PEG 6000, suspended in a medium containing 0.4 m sucrose and 40 mm MES-NaOH (pH 6.5) (buffer B), and stored at −196 °C. The purified PSII lacking major components of fucoxanthin chlorophyll a/c-binding protein was more stable as compared with the crude PSII, because the inactivation of oxygen evolution, Chl bleaching, and degradation of PSII subunits observed in the crude PSII were largely suppressed in the purified PSII (30). Thus, the purified PSII was used for release-reconstitution experiments in this study. Chl concentrations were determined in 90% acetone using the equation of Jeffrey and Humphrey (32).
Purification Procedures of Five Extrinsic Proteins
The crude PSII suspended in 10 mm MES-NaOH (pH 6.5) (buffer C) was treated with 1 m Tris-HCl (pH 8.5) at 0.5 mg of Chl/ml for 1 h on ice in the dark to release all of the extrinsic proteins of PsbO, PsbQ′, PsbV, Psb31, and PsbU (29, 30, 33). The treated samples were centrifuged at 227,000 × g for 30 min after addition of 14% PEG 1450. The supernatants were dialyzed using Spectra/Por dialysis membrane (molecular weight cutoff, 8000) (Spectrum Laboratories) against buffer C after addition of 1 mm PMSF, 1 mm EDTA, and 10 μl of benzamidine-Sepharose 6B (GE Healthcare) to prevent cleavages of the extrinsic proteins by endogenous proteases. The dialyzed samples were applied to a DEAE-Toyopearl 650 M column (pH 6.5) equilibrated with buffer C, and then the fraction containing PsbO and PsbV was eluted at 0.2 m NaCl. The fraction passed through the DEAE column was applied to a CM-Toyopearl 650 M column (pH 6.5) equilibrated with buffer C, and Psb31 was then eluted at 0.2 m NaCl. The fraction passed through the CM column (pH 6.5) was dialyzed against 10 mm citric acid/NaOH (pH 5.5) and applied to another CM-Toyopearl 650 M column (pH 5.5) equilibrated with the same buffer. PsbQ′ was eluted at 0.1 m NaCl. The fraction passed through the CM column was dialyzed against 10 mm citric acid/NaOH (pH 4) and applied to a CM-Toyopearl 650 M column (pH 4) equilibrated with the same buffer. PsbU was eluted at 1 m NaCl. The fraction containing PsbO and PsbV eluted at 0.2 m NaCl from the DEAE-Toyopearl 650 M column (pH 6.5) was dialyzed against 10 mm citric acid/NaOH (pH 5.5) and applied to a CM-Toyopearl 650 M column (pH 5.5) equilibrated with the same buffer. PsbV was passed through the column, and PsbO was eluted at 1 m NaCl. A flow chart for the purification procedure of the five extrinsic proteins from the crude PSII is shown in supplemental Fig. S1.
The purified extrinsic proteins were concentrated by a small size of each column and/or ultrafiltration and then dialyzed against 10 mm MES-NaOH (pH 6.5). The concentrations of purified extrinsic proteins were determined by BCA protein assay kit (Pierce) (34).
Dissociation of Extrinsic Proteins
The purified PSII was treated either with 1 m NaCl, 1 m MgCl2, 1 m Tris-HCl (pH 8.5), 2.6 m urea plus 0.2 m NaCl or 4 m urea plus 0.2 m NaCl at 0.25 mg of Chl/ml for 20 min on ice in the dark. The treated samples were centrifuged at 40,000 × g for 10 min after addition of 10% PEG 6000. The polypeptides of the resulting precipitates and supernatants were analyzed by SDS-PAGE.
SDS-PAGE
Samples were solubilized with 5% lithium lauryl sulfate and 75 mm dithiothreitol for 30 min at room temperature. The solubilized samples (1 μg of Chl) were applied to a gradient gel containing 16–22% acrylamide and 7.5 m urea according to Ikeuchi and Inoue (35). After electrophoresis at a constant current of 8 mA for 15 h, gels were stained with Coomassie Brilliant Blue R-250 and photographed.
Reconstitution Experiments
For reconstitution, the purified PSII was treated with 4 m urea plus 0.2 m NaCl to remove all of the extrinsic proteins. The urea/NaCl-treated PSII was incubated with the extrinsic proteins in various combinations for 20 min on ice at 0.05 mg of Chl/ml. The extrinsic protein/PSII ratio used was 3:1 during reconstitution, based on the estimation that each PSII reaction center contains 42 molecules of Chl a (30). The reconstitution experiments were performed in buffer B containing 10 μm 2,6-dichloroindophenol, 2 mm MnCl2, 10 mm MgCl2, and 10 mm CaCl2 on ice at room light (7–8 μmol photons/m2/s) (Mn-photoactivation condition (36)). After reconstitution, the mixtures were centrifuged at 40,000 × g for 5 min after addition of 10% PEG 6000, and the resulting precipitates were suspended in buffer B and used for electrophoretic analysis and oxygen evolution measurements.
Immunological Assays
The relative amounts of the PsbV and PsbU proteins rebound by reconstitution experiments were estimated by immunological assays with antibodies raised against PsbV from a red alga, Cyanidium caldarium (23), and against PsbU from the diatom, C. gracilis. For immunological assays, proteins on the SDS-polyacrylamide gel were transferred onto a polyvinylidene fluoride (PVDF) membrane and reacted with respective antibodies, and then horseradish peroxidase (HRP)-conjugated anti-IgG was used as a secondary antibody. Chemiluminescent detection of HRP was carried out using ECL Western blotting detection reagents (GE Healthcare) and an ECL camera system using FP300B film (Fuji film). The peak area corresponding to each band was calculated with the NIH image software (ImageJ, rsb.info.nih.gov).
Assay of Oxygen Evolving Activity
Oxygen evolution was measured with a Clark-type electrode at 25 °C in buffer B with 0.4 mm phenyl-p-benzoquinone as the electron acceptor in the absence or presence of 10 mm NaCl or 5 mm CaCl2.
RESULTS
Purification of PsbO, PsbQ′, PsbV, Psb31, and PsbU
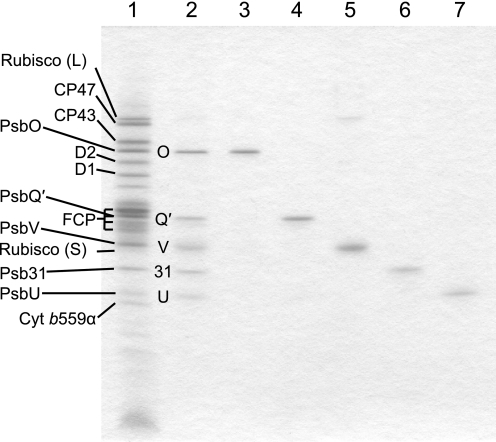
As shown in Fig. 2, the five extrinsic proteins (lane 2) were released from the crude PSII (lane 1) by alkaline Tris treatment and then purified from the Tris extracts by anion and cation exchange chromatographic columns at different pH values (see supplemental Fig. S1). The procedures we used yielded highly purified PsbO (Fig. 2, lane 3), PsbQ′ (lane 4), PsbV (lane 5), Psb31 (lane 6), and PsbU (lane 7), which were identified by immunoblotting and N-terminal sequencing analysis according to the methods described in our previous papers (29, 30). However, a small amount of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) was contaminated in the fraction of PsbV (Fig. 2, lane 5), which will not interfere with our reconstitution experiments.
FIGURE 2.
Purification of the five extrinsic proteins from crude PSII of a marine centric diatom, C. gracilis. Lane 1, crude PSII; lane 2, five extrinsic proteins extracted from the crude PSII by alkaline Tris treatment; lane 3, purified PsbO; lane 4, purified PsbQ′; lane 5, purified PsbV; lane 6, purified Psb31; and lane 7, purified PsbU. The gel was stained with Coomassie Brilliant Blue R-250, and each polypeptide was identified by immunological and/or N-terminal sequence analyses according to the methods described previously (29, 30). L, large subunit; S, small subunit; Cyt, cytochrome.
Dissociation of Extrinsic Proteins by Various Treatments
The extrinsic proteins of PSII have been reported to be released by various treatments. In spinach PSII, the PsbP and PsbQ proteins are selectively released with 1 m NaCl-wash (37), and all of the three extrinsic proteins of PsbO, PsbP, and PsbQ can be released with 1 m CaCl2 (38), 2.6 m urea plus 0.2 m NaCl (39), or 1 m Tris (pH 8.5) treatment (33). In red algal PSII, no extrinsic proteins are released by 1 m NaCl wash, whereas treatments with 1 m CaCl2, 2.6 m urea plus 0.2 m NaCl, or 1 m Tris (pH 8.5) released all of the extrinsic proteins (4, 5). In green algal PSII, the PsbP and PsbQ proteins are only partially released by 1 m NaCl-wash, and most of the three extrinsic proteins are released by treatments with 2.6 m urea plus 0.2 m NaCl, 1 m Tris (pH 8.5), or 1 m CaCl2, although a small amount of the PsbO protein remains bound after these treatments (7). These indicate that the binding properties of the extrinsic proteins are different among different organisms.
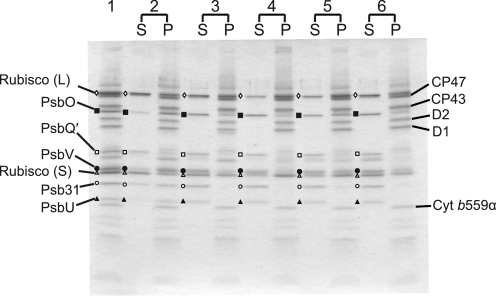
Thus, we first examined the release of the five extrinsic proteins in diatom PSII by various treatments. For release-reconstitution experiments, we used the purified PSII because the purified PSII was more stable compared with the crude PSII as described above. Fig. 3 shows dissociation of the extrinsic proteins from the purified PSII by various treatments. The PsbO, PsbQ′, PsbV, and Psb31 proteins were only partially released together with the large and small subunits of Rubisco by 1 m NaCl-wash (Fig. 3, lane 2). Treatments with 1 m MgCl2 (Fig. 3, lane 3), 1 m Tris (pH 8.5) (lane 4), or 2.6 m urea plus 0.2 m NaCl (lane 5) completely released the PsbQ′, Psb31, and PsbU proteins, but a considerable amount of the PsbO and PsbV proteins remained bound to PSII after these treatments. When the purified PSII was treated with 4 m urea plus 0.2 m NaCl, most of the PsbO and PsbV proteins were released together with a complete release of the other extrinsic proteins, although a trace amount of PsbO remained bound (Fig. 3, lane 6). We attempted to remove the PsbO protein completely by various treatments, and we found that the protein could be completely released by two treatments with 4 m urea plus 0.2 m NaCl. However, the PSII treated twice with 4 m urea plus 0.2 m NaCl showed a very low oxygen evolving activity even after complete rebinding of all of the extrinsic proteins, suggesting that the twice treatments may have damaged the Mn4Ca cluster itself. Thus, the purified PSII treated once with 4 m urea plus 0.2 m NaCl was used as the extrinsic protein-deleted PSII for reconstitution experiments in this study.
FIGURE 3.
Dissociation of extrinsic proteins from purified PSII (lane 1) of a marine centric diatom, C. gracilis, by treatment with 1 m NaCl (lane 2), 1 m MgCl2 (lane 3), 1 m Tris-HCl (pH 8. 5) (lane 4), 2.6 m urea plus 0.2 m NaCl (lane 5), or 4 m urea plus 0.2 m NaCl (lane 6). S, supernatant; P, pellet; Cyt, cytochrome; Rubisco (L), large subunit; Rubisco (S), small subunit.
Restoration of Oxygen Evolution by Reconstitution With All of the Extrinsic Proteins under Normal or Mn-photoactivation Conditions
Table 1 shows restoration of oxygen evolution by reconstitution with all of the extrinsic proteins of PsbO, PsbQ′, PsbV, Psb31, and PsbU to the extrinsic protein-deleted PSII prepared by 4 m urea plus 0.2 m NaCl treatment. When the reconstitution experiments were performed in a medium containing 0.4 m sucrose and 40 mm MES-NaOH (pH 6.5) (buffer B) on ice in the dark (normal condition), the restoration of oxygen evolution was 20–23% of the original activity in the absence and presence of Cl− and Ca2+ ions. This level of restoration is considerably lower than those observed with PSIIs from other organisms. For example, upon reconstitution of all of the extrinsic proteins under normal conditions, restoration of the oxygen evolution has been reported to be 42% in spinach PSII (12), 42% in green algal PSII (7), 40% in Euglena PSII (6), 59–74% in red algal PSII (5, 12), and 79–87% in PSII from a thermophilic cyanobacterium (1, 12). This may be due in part to the harsh treatment of 4 m urea plus 0.2 m NaCl required to release all of the extrinsic proteins from the diatom PSII, which may have damaged the Mn4Ca cluster to some extent. To obtain a higher level of restoration of the oxygen evolution, we screened various conditions for reconstitution, and we found that the oxygen evolution restored to 46–50% of the original activity when reconstitution experiments were performed in buffer B containing 10 μm 2,6-dichloroindophenol, 2 mm MnCl2, 10 mm MgCl2, and 10 mm CaCl2 on ice at room light (7–8 μmol photons/m2/s) (Table 1), which is comparable with the restoration level observed in spinach, green algal, Euglena, and red algal PSIIs. This reconstitution condition is the same as that used for Mn-photoactivation (36). Thus, in this study, we performed reconstitution experiments under the Mn-photoactivation condition to achieve the maximum restoration of oxygen evolution.
TABLE 1.
Oxygen evolution of the urea/NaCl-treated PSII reconstituted with all of the five extrinsic proteins under normal conditions and the Mn-photoactivation condition
| Oxygen evolution (μmol of O2/mg of Chl/h)a |
|||
|---|---|---|---|
| − ions | n+10 mm NaCl | +5 mm CaCl2 | |
| Untreated PSII | 2143 ± 123 (100) | 2240 ± 117 (100) | 2240 ± 140 (100) |
| Urea/NaCl-treated PSII | 0 (0) | 90 ± 9 (4) | 134 ± 19 (6) |
| + all of the extrinsic proteins (normal conditions)b | 450 ± 43 (21) | 448 ± 43 (20) | 515 ± 40 (23) |
| + all of the extrinsic proteins (Mn-photoactivation conditions)c | 988 ± 74 (46) | 1113 ± 66 (50) | 1130 ± 90 (50) |
a Values are shown as mean ± S.D., with n = 3 (three independent experiments).
b Reconstitution experiments were carried out in a medium containing 0.4 m sucrose and 40 mm MES-NaOH (pH 6.5) (buffer B) on ice in the dark.
c Reconstitution experiments were carried out in buffer B containing 10 μm 2,6-dichloroindophenol, 2 mm MnCl2, 10 mm MgCl2, and 10 mm CaCl2 on ice at room light (7–8 μmol of photons/m2/s).
Binding Properties of the Five Extrinsic Proteins
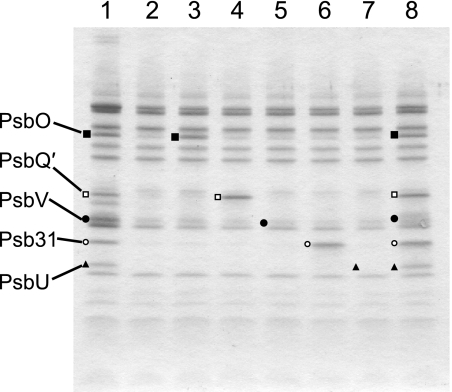
To examine which proteins in the five extrinsic proteins of diatom PSII directly interact with PSII intrinsic proteins, each of the extrinsic proteins was separately reconstituted with the extrinsic protein-deleted PSII. The results obtained are shown in Fig. 4. As described above, treatment of the purified PSII (Fig. 4, lane 1) with 4 m urea plus 0.2 m NaCl released most of the five extrinsic proteins (lane 2). Reconstitution with PsbO (Fig. 4, lane 3), PsbQ′ (lane 4), or Psb31 (lane 6) alone resulted in the binding of the respective proteins to the extrinsic proteins-depleted PSII to a level equal to those in the purified PSII, whereas reconstitution with PsbV (Fig. 4, lane 5) or PsbU (lane 7) alone resulted in almost no re-binding of these two proteins. When all of the five extrinsic proteins were reconstituted together, the PsbV and PsbU proteins rebound, together with the other three proteins, to a level equal to those in the purified PSII (Fig. 4, lane 8). It is noted that the band of PsbV became somewhat broad after reconstitution, although the reason for this is unknown at present. The broad band was confirmed to be PsbV but not the small subunit of Rubisco by immunoblotting analysis, as shown in supplemental Fig. S2. These results indicate that the PsbO, PsbQ′, and Psb31 proteins can directly bind to PSII intrinsic proteins independent of the other extrinsic proteins, whereas the effective binding of the PsbV and PsbU proteins requires the presence of other extrinsic proteins. To examine the extrinsic proteins that are required for the binding of PsbV and PsbU to PSII, we carried out reconstitution experiments with each of the other four proteins in combination with PsbV or PsbU. First, we carried out the reconstitution experiments with various extrinsic proteins in combination with PsbV.
FIGURE 4.
Reconstitution of urea/NaCl-treated PSII with each or all of the five extrinsic proteins. Lane 1, purified PSII from a marine centric diatom, C. gracilis; lane 2, 4 m urea plus 0.2 m NaCl-treated PSII; lanes 3–8, the urea/NaCl-treated PSII reconstituted with PsbO (lane 3), PsbQ′ (lane 4), PsbV (lane 5), Psb31 (lane 6), PsbU (lane 7), and all of the five extrinsic proteins (lane 8).
Because the band of the small subunit of Rubisco appeared close to the band of PsbV (Fig. 3) and the band of PsbV became broad after reconstitution, the relative amounts of PsbV rebound were determined by immunoblotting assay with an antibody against PsbV (supplemental Fig. S2A), and the results obtained are summarized in Table 2. The amount of the PsbV protein rebound in the presence of the PsbO, PsbQ′, or Psb31 protein increased to 42, 56, or 54% from 14% in the absence of the other extrinsic proteins, respectively. When reconstitution was performed in combination with PsbO and PsbQ′, PsbQ′ and Psb31, or PsbO and Psb31, the amount of the PsbV protein rebound increased to 65, 72, or 61%, respectively, which further increased to 84% when reconstitution was performed in combination with the three extrinsic proteins of PsbO, PsbQ′, and Psb31. When all of the five extrinsic proteins were reconstituted together, the amount of the PsbV protein rebound reached to 100%. These results indicate that the PsbV protein partially interacts with each of the PsbO, PsbQ′, Psb31, and PsbU proteins, and its complete binding requires the presence of all of the other four extrinsic proteins. Alternatively, interactions among the other four proteins may be necessary to create an optimum site for the binding of PsbV.
TABLE 2.
Relative binding of PsbV reconstituted to urea/NaCl-treated PSII in various combinations with the other extrinsic proteins
| Amount of PsbV rebounda | |
|---|---|
| % | |
| PsbV only | 14 ± 2 |
| PsbV + PsbO | 42 ± 8 |
| PsbV + PsbQ′ | 56 ± 11 |
| PsbV + Psb31 | 54 ± 10 |
| PsbV + PsbO + PsbQ′ | 65 ± 10 |
| PsbV + PsbQ′ + Psb31 | 72 ± 11 |
| PsbV + PsbO + Psb31 | 61 ± 12 |
| PsbV + PsbO + PsbQ′ + Psb31 | 84 ± 15 |
| PsbV + PsbO + PsbQ′ + Psb31 + PsbU | 101 ± 3 |
a Values are shown as mean ± S.D., with n = 6 (two times of Western analysis for each of three independent samples).
Second, we carried out the reconstitution experiments with each of the other four proteins in combination with the PsbU protein. Because the band of the α subunit of cytochrome b559 appeared close to the band of PsbU (Fig. 3), the relative amounts of PsbU rebound were determined by immunoblotting assay with an antibody against PsbU (supplemental Fig. S2B), and the results obtained are summarized in Table 3. The amount of the PsbU protein rebound in the presence of the PsbO, PsbQ′, or Psb31 proteins increased to 34, 30, or 34% from 5% in the absence of the other extrinsic proteins, respectively. In the presence of PsbO and PsbQ′, PsbQ′, and Psb31, or PsbO and Psb31, the amount of the PsbU protein rebound increased to 41, 54, or 66%, respectively, and in the presence of the three extrinsic proteins of PsbO, PsbQ′, and Psb31, it increased to 78%. When all of the five extrinsic proteins were reconstituted together, the PsbU protein completely rebound. These results indicate that the PsbU protein also partially interacts with each of the PsbO, PsbQ′, Psb31, and PsbV proteins, and its complete binding requires the presence of all of the other four extrinsic proteins. Here again, the possibility exists that interactions among the other four proteins may be necessary to create an optimum site for the binding of PsbU.
TABLE 3.
Relative binding of PsbU reconstituted to urea/NaCl-treated PSII in various combinations with the other extrinsic proteins
| Amount of PsbU rebounda | |
|---|---|
| % | |
| PsbU only | 5 ± 3 |
| PsbU + PsbO | 34 ± 8 |
| PsbU + PsbQ′ | 30 ± 8 |
| PsbU + Psb31 | 34 ± 8 |
| PsbU + PsbO + PsbQ′ | 41 ± 9 |
| PsbU + PsbQ′ + Psb31 | 54 ± 10 |
| PsbU + PsbO + Psb31 | 66 ± 8 |
| PsbU + PsbO + PsbQ′ + Psb31 | 78 ± 11 |
| PsbU + PsbO + PsbQ′ + Psb31 + PsbV | 103 ± 4 |
a Values are shown as mean ± S.D., with n = 6 (two times of Western analysis for each of three independent samples).
Third, we carried out the reconstitution experiments with each of the other three proteins in combination with both the PsbV and PsbU proteins to examine the possible interaction between PsbV and PsbU. The result of immunoblotting assay obtained is shown in supplemental Fig. S2C and is summarized in Table 4. The amount of PsbV rebound significantly increased from 14% in the absence of PsbU (Table 2) to 56% in the presence of PsbU (Table 4), and the amount of PsbU also increased from 5% in the absence of PsbV (Table 3) to 23% in the presence of PsbV (Table 4). This suggests that PsbV and PsbU only partially interact with PSII intrinsic proteins by themselves, and an interaction between them exists, which enhanced their direct binding to PSII significantly. When reconstitution was carried out together with PsbO, PsbQ′, or Psb31, the amount of PsbV rebound increased from 42, 56, or 54% in the absence of PsbU (Table 2) to 68, 76, or 72% in the presence of PsbU (Table 4), respectively, and that of PsbU increased from 34, 30, or 34% in the absence of PsbV (Table 3) to 55, 52, or 61% in the presence of PsbV (Table 4), respectively. These results are in agreement with the existence of an interaction between PsbV and PsbU that facilitates binding of the respective proteins to PSII. All of the extrinsic proteins were, however, required for the complete binding of the PsbV and PsbU proteins (Tables 2–4). These suggest that the PsbV and PsbU proteins partially interact with all of the other extrinsic proteins, and the interaction between PsbV and PsbU is indispensable for their complete binding to PSII.
TABLE 4.
Relative binding of PsbV and PsbU reconstituted to urea/NaCl-treated PSII in various combinations with the other extrinsic proteins
| Amount of PsbV rebounda | Amount of PsbU rebounda | |
|---|---|---|
| % | % | |
| PsbV + PsbU | 56 ± 11 | 23 ± 6 |
| PsbV + PsbU + PsbO | 68 ± 13 | 55 ± 8 |
| PsbV + PsbU + PsbQ′ | 76 ± 14 | 52 ± 8 |
| PsbV + PsbU + Psb31 | 72 ± 15 | 61 ± 8 |
| PsbV + PsbU + PsbO + PsbQ′ | 80 ± 15 | 73 ± 9 |
| PsbV + PsbU + PsbQ′ + Psb31 | 87 ± 15 | 77 ± 8 |
| PsbV + PsbU + PsbO + Psb31 | 82 ± 16 | 84 ± 12 |
| PsbV + PsbU + PsbO + PsbQ′ + Psb31 | 100 ± 3 | 102 ± 3 |
a Values are shown as mean ± S.D., with n = 6 (two times of Western analysis for each of three independent samples).
Functions of the Five Extrinsic Proteins
Table 5 shows restoration of the oxygen evolving activity upon reconstitution with the extrinsic proteins in various combinations. The extrinsic proteins-depleted PSII obtained by treatment with 4 m urea plus 0.2 m NaCl showed no oxygen evolution in the absence of Cl− and Ca2+ ions. When the extrinsic proteins-depleted PSII was reconstituted with PsbO alone, the oxygen evolving activity was restored to 8% of the original activity in the absence of Cl− and Ca2+ ions, to 24% in the presence of Cl− ion, and to 44% in the presence of both Cl− and Ca2+ ions. The requirements for Cl− and Ca2+ ions in the PSII reconstituted with PsbO in the absence of the other extrinsic proteins were similar to those in PSIIs of spinach (37, 38), a green alga (7), a red alga (5, 12), and a cyanobacterium (1, 2, 12). In the PSII reconstituted with PsbQ′ alone, no restoration of the oxygen evolution was detected, indicating that the PsbQ′ protein is not involved directly in oxygen evolution but is required for effective binding of the PsbV and PsbU proteins (Tables 2–4), which are similar to those of red algal PSII (5). Interestingly, PSII reconstituted with Psb31 alone showed an oxygen evolving activity of 128 μmol of O2/mg of Chl/h in the absence of Cl− and Ca2+ ions (6% of the original activity), but the activity was scarcely stimulated in the presence of Cl− and Ca2+ ions, which will be described in detail later.
TABLE 5.
Oxygen evolution of the urea/NaCl-treated PSII reconstituted with the extrinsic proteins in various combinations under the Mn-photoactivation condition
| Oxygen evolution (μmol of O2/mg of Chl/h)a |
|||
|---|---|---|---|
| − ions | +10 mm NaCl | +5 mm CaCl2 | |
| Untreated PSII | 2143 ± 123 (100) | 2240 ± 117 (100) | 2240 ± 140 (100) |
| Urea/NaCl-treated PSII | 0 (0) | 90 ± 9 (4) | 134 ± 19 (6) |
| + PsbO | 173 ± 17 (8) | 537 ± 35 (24) | 985 ± 62 (44) |
| + PsbQ′ | 0 (0) | 112 ± 13 (5) | 134 ± 18 (6) |
| + Psb31 | 128 ± 12 (6) | 150 ± 15 (7) | 156 ± 19 (7) |
| + PsbO + PsbQ′ | 186 ± 13 (9) | 542 ± 38 (24) | 1044 ± 78 (47) |
| + PsbO + Psb31 | 169 ± 17 (8) | 588 ± 41 (26) | 979 ± 77 (44) |
| + PsbO + PsbQ′ + Psb31 | 173 ± 24 (8) | 603 ± 40 (27) | 1070 ± 61 (48) |
| + PsbO + PsbQ′ + Psb31 + PsbV | 283 ± 27 (13) | 808 ± 42 (36) | 1012 ± 72 (45) |
| + PsbO + PsbQ′ + PsbV + Psb31 + PsbU | 988 ± 74 (46) | 1113 ± 66 (50) | 1130 ± 90 (50) |
a Values are shown as mean ± S.D., with n = 3 (three independent experiments).
The PSII reconstituted with PsbO in combination with PsbQ′, Psb31, or PsbQ′ plus Psb31 showed similar activity to that of PSII reconstituted with PsbO alone, suggesting that there is no cooperative activation of oxygen evolution between PsbO and Psb31. On the other hand, the activity of PSII reconstituted with PsbO in combination with the three extrinsic proteins of PsbQ′, Psb31, and PsbV was restored to 13% of the original activity in the absence of Cl− and Ca2+ ions, to 36% in the presence of Cl− ion, and to 45% in the presence of Cl− and Ca2+ ions. This indicates that rebinding of PsbV largely decreased the requirement for Ca2+ ion on oxygen evolution, suggesting that PsbV mainly functions to optimize the availability of Ca2+ cofactor. In PSII reconstituted with all of the extrinsic proteins, the activity was restored to 46% of the original activity even in the absence of Cl− and Ca2+ ions, and scarcely stimulated in the presence of Cl− and Ca2+ ions. These indicate that the PsbV and PsbU proteins function to optimize the availability of Cl− and Ca2+ cofactors for water oxidation, with PsbV mainly functioning to optimize the availability of Ca2+ and PsbU mainly functioning to optimize the availability of Cl−.
The observation that PSII reconstituted with Psb31 alone showed an oxygen evolving activity of 128–156 μmol of O2/mg of Chl/h even in the absence of PsbO (Table 5) is significant. To further examine the function of Psb31, we compared the restoration of oxygen evolving activity in PSII reconstituted with Psb31 in combination with the other extrinsic proteins in the absence of PsbO with that in PSII reconstituted with PsbO in combination with other proteins. The results obtained are shown in Table 6. PSII reconstituted with Psb31 together with PsbQ′ showed similar activity to that of PSII reconstituted with Psb31 alone. On the other hand, when PSII was reconstituted with Psb31 together with both of PsbQ′ and PsbV, the activity increased to 292 μmol of O2/mg of Chl/h (13% recovery) in the presence of Cl− ion and 320 μmol of O2/mg of Chl/h (14% recovery) in the presence of both Cl− and Ca2+ ions, whereas the activity in the absence of Cl− and Ca2+ ions was scarcely increased. This indicates that when the PsbV protein was rebound to PSII reconstituted with Psb31, it functions to activate the oxygen evolution in the presence of Cl− ion but in the absence of Ca2+ ion. Furthermore, the activity of PSII reconstituted with Psb31 together with the three extrinsic proteins of PsbQ′, PsbV, and PsbU increased to 343 μmol of O2/mg of Chl/h (16% recovery) in the absence of Cl− and Ca2+ ions, to 452 μmol of O2/mg of Chl/h (20% recovery) in the presence of Cl− ion, and to 470 μmol of O2/mg of Chl/h (21% recovery) in the presence of Cl− and Ca2+ ions. These results indicate that when the PsbV and PsbU proteins were rebound to PSII reconstituted with Psb31, they were able to activate the oxygen evolution in the absence of Cl− and Ca2+ ions.
TABLE 6.
Oxygen evolution of the urea/NaCl-treated PSII reconstituted with the other extrinsic proteins in various combinations with the Psb31 or PsbO protein under the Mn-photoactivation condition
| Oxygen evolution (μmol of O2/mg of Chl/h)a |
|||
|---|---|---|---|
| − ions | +10 mm NaCl | +5 mm CaCl2 | |
| Untreated PSII | 2143 ± 123 (100) | 2240 ± 117 (100) | 2240 ± 140 (100) |
| Urea/NaCl-treated PSII | 0 (0) | 90 ± 9 (4) | 134 ± 19 (6) |
| + Psb31 | 128 ± 12 (6) | 150 ± 15 (7) | 156 ± 19 (7) |
| + Psb31 + PsbQ′ | 151 ± 19 (7) | 186 ± 16 (8) | 183 ± 21 (8) |
| + Psb31 + PsbQ′ + PsbV | 159 ± 14 (7) | 292 ± 23 (13) | 320 ± 26 (14) |
| + Psb31+ PsbQ′ + PsbV + PsbU | 343 ± 22 (16) | 452 ± 31 (20) | 470 ± 31 (21) |
| + PsbO | 173 ± 17 (8) | 537 ± 35 (24) | 985 ± 62 (44) |
| + PsbO + PsbQ′ | 186 ± 13 (9) | 542 ± 38 (24) | 1044 ± 78 (47) |
| + PsbO + PsbQ′ + PsbV | 257 ± 20 (12) | 672 ± 36 (30) | 1075 ± 80 (48) |
| + PsbO + PsbQ′ + PsbV + PsbU | 711 ± 56 (33) | 784 ± 62 (35) | 1081 ± 86 (48) |
a Values are shown as mean ± S.D., with n = 3 (three independent experiments).
On the other hand, PSII reconstituted with PsbO together with both PsbQ′ and PsbV showed the oxygen evolving activity of 257 μmol of O2/mg of Chl/h (12% recovery) in the absence of Cl− and Ca2+ ions, 672 μmol of O2/mg of Chl/h (30% recovery) in the presence of Cl− ion, and 1075 μmol of O2/mg of Chl/h (48% recovery) in the presence of Cl− and Ca2+ ions (Table 6). Upon reconstitution with PsbO together with the three extrinsic proteins of PsbQ′, PsbV, and PsbU, the activity reached to 711 μmol of O2/mg of Chl/h (33% recovery) in the absence of Cl− and Ca2+ ions, to 784 μmol of O2/mg of Chl/h (35% recovery) in the presence of Cl− ion, and to 1081 μmol of O2/mg of Chl/h (48% recovery) in the presence of Cl− and Ca2+ ions (Table 6). Furthermore, PSII reconstituted with all of the extrinsic proteins showed 988 μmol of O2/mg of Chl/h (46% recovery) in the absence of Cl− and Ca2+ ions, 1113 μmol of O2/mg of Chl/h (50% recovery) in the presence of Cl− ion, and 1130 μmol of O2/mg of Chl/h (50% recovery) in the presence of Cl− and Ca2+ ions, as shown in Table 5.
These data indicate that the function of PsbO on oxygen evolution can partially be replaced by the novel Psb31 protein in diatom PSII, and all of the extrinsic proteins including PsbO and Psb31 are required for the maximal oxygen evolving activity.
DISCUSSION
Binding Features of the Five Extrinsic Proteins in Diatom PSII
This study demonstrated that, among the five extrinsic proteins of PSII from a marine centric diatom, C. gracilis, the PsbO, PsbQ′, and Psb31 proteins can separately bind to PSII independent of the other extrinsic proteins (Fig. 4). This indicates that these three extrinsic proteins have their own binding sites on PSII intrinsic proteins. On the other hand, the other two proteins, PsbV and PsbU, scarcely bound to PSII unless reconstituted in combinations with the other extrinsic proteins (Fig. 4). The complete binding of PsbV and PsbU to PSII required the presence of all of the other extrinsic proteins (Tables 2–4). On the basis of these results, we propose a simplified model for the association of the five extrinsic proteins with PSII intrinsic proteins (Fig. 1C). In this model, it is shown that the PsbO, PsbQ′, and Psb31 proteins have direct contacts with PSII intrinsic proteins, whereas the PsbV and PsbU proteins mainly associate with PSII through their contacts with the PsbO, PsbQ′, and Psb31 proteins. Recently, we showed a close association of the Psb31 protein with the PsbH protein and/or cytochrome b559α in the diatom PSII core complex by cross-linking experiments using a water-soluble carbodiimide, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (31). Based on these facts and the crystal structure of cyanobacterial PSII currently available (11, 40–42), PsbO was placed on the lumenal surface with a broad contact with CP47, CP43, D1, and D2 subunits in Fig. 1C. Psb31 is also situated in close to the D1/D2 proteins through its direct association with PsbH and/or cytochrome b559α. PsbQ′ is tentatively located in the CP47 side, because there is no space in the CP43 side because of overlapping of the structure of PsbO with CP43 (11, 40–42). PsbV and PsbU have contacts with all of the other extrinsic proteins and partially with some of the PSII intrinsic proteins. Although the detailed organization of these extrinsic proteins in diatom PSII has to wait for a high resolution crystal structure to be determined, before such a structure is available, the model proposed in this study will provide useful information for the arrangement of the relevant subunits in diatom PSII.
Function of the Five Extrinsic Proteins in Diatom PSII
One significant feature of the diatom PSII revealed by this study is that diatom PSII reconstituted with the novel Psb31 protein can evolve oxygen even in the absence of the PsbO protein (Tables 5 and 6). Among the extrinsic proteins in PSIIs from a wide variety of organisms, the PsbO protein plays an important role in maintaining the stability and activity of the Mn4Ca cluster, and it has been considered to be absolutely essential for oxygen evolution (1–17, 29, 30, 37–39). This study is the first report demonstrating that PSII lacking PsbO can evolve oxygen in the absence of Cl− and Ca2+ ions. As shown in Tables 5 and 6, PSII reconstituted with Psb31 alone showed an oxygen evolving activity of 128 μmol of O2/mg of Chl/h in the absence of Cl− and Ca2+ ions, compared with 173 μmol of O2/mg of Chl/h in PSII reconstituted with PsbO alone. Furthermore, PSII reconstituted with Psb31 in combination with PsbQ′, PsbV, and PsbU showed the activity of 343–470 μmol of O2/mg of Chl/h in the absence of PsbO (Table 6). These indicate that the novel Psb31 extrinsic protein partially serves as a substitute for the PsbO protein in supporting oxygen evolution. The significant difference between PSIIs reconstituted with Psb31 alone and with PsbO alone is that the oxygen evolving activity of the former is scarcely stimulated by Cl− and Ca2+ ions but that of the latter is largely stimulated by these ions (Tables 5 and 6). This may suggest that the binding of Psb31 has shielded the access of Cl− and Ca2+ to their functional sites close to the Mn4Ca cluster, whereas binding of PsbO did not. In any event, it is an interesting subject to be elucidated as to why the activity of PSII reconstituted with PsbO was largely stimulated by Cl− and Ca2+ ions but that of PSII reconstituted with Psb31 was not. In PSII reconstituted with PsbO, rebinding of PsbV or PsbV and PsbU largely activated the oxygen evolution in the absence of Ca2+ ion, or both Cl− and Ca2+ ions, respectively (Table 6), which is similar to that reported in the cyanobacterial and red algal PSIIs (1, 2, 5, 12). These have been interpreted to indicate that PsbV and PsbU function to optimize the availability of Cl− and Ca2+ cofactors for water oxidation, with PsbV mainly functioning to optimize the availability of Ca2+ and PsbU mainly functioning to optimize Cl−, respectively. The oxygen evolution in the absence of Ca2+ ion or in the absence of both Cl− and Ca2+ ions was also activated in PSII reconstituted with Psb31 by rebinding of PsbV, or PsbV and PsbU, respectively (Table 6). These indicate that the function of the PsbV and PsbU proteins in activating oxygen evolution in the absence of Cl− and Ca2+ ions is retained in PSII reconstituted with Psb31, similar to those of PSII reconstituted with PsbO.
The restoration of oxygen evolution in PSII reconstituted with all of the five extrinsic proteins was only 20–23% of the original activity when release-reconstitution experiments were carried out under normal conditions, whereas the restoration increased to 46–50% when the experiments were performed under Mn-photoactivation condition (Table 1). This suggests that Mn ions are partially liberated by the 4 m urea plus 0.2 m NaCl treatment or during incubation following the treatment. So far, it has been reported that two of the four Mn atoms per PSII are gradually liberated after 24–48 h when the extrinsic protein-depleted PSII was incubated at 0 °C in the dark, and the Mn liberation is suppressed by rebinding of PsbO (43, 44). Our preliminary experiments on inactivation of oxygen evolution during incubation on ice in the dark, however, showed that all of the extrinsic proteins in addition to PsbO were required for stabilization of the oxygen evolving activity of the diatom PSII, suggesting that the Psb31 protein may contribute to stabilize the Mn4Ca cluster together with PsbO.
Supplementary Material
This work was supported in part by Grants-in-aid for Scientific Research from the Ministry of Education of Japan 18570049 (to I. E.) and 21570038 and 22370017 (to T. T.), and Research Fellow (to R. N.) from the Japan Society for the Promotion of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PSII
- photosystem II
- Chl
- chlorophyll
- Rubisco
- ribulose-1,5-bisphosphate carboxylase/oxygenases.
REFERENCES
- 1.Shen J. R., Inoue Y. (1993) Biochemistry 32, 1825–1832 [DOI] [PubMed] [Google Scholar]
- 2.Shen J. R., Burnap R. L., Inoue Y. (1995) Biochemistry 34, 12661–12668 [DOI] [PubMed] [Google Scholar]
- 3.Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H. B. (2002) Biochemistry 41, 8004–8012 [DOI] [PubMed] [Google Scholar]
- 4.Enami I., Murayama H., Ohta H., Kamo M., Nakazato K., Shen J. R. (1995) Biochim. Biophys. Acta 1232, 208–216 [DOI] [PubMed] [Google Scholar]
- 5.Enami I., Kikuchi S., Fukuda T., Ohta H., Shen J. R. (1998) Biochemistry 37, 2787–2793 [DOI] [PubMed] [Google Scholar]
- 6.Suzuki T., Tada O., Makimura M., Tohri A., Ohta H., Yamamoto Y., Enami I. (2004) Plant Cell Physiol. 45, 1168–1175 [DOI] [PubMed] [Google Scholar]
- 7.Suzuki T., Minagawa J., Tomo T., Sonoike K., Ohta H., Enami I. (2003) Plant Cell Physiol. 44, 76–84 [DOI] [PubMed] [Google Scholar]
- 8.Berthold D. A., Babcock G. T., Yocum C. F. (1981) FEBS Lett. 134, 231–234 [Google Scholar]
- 9.Kuwabara T., Murata N. (1982) Plant Cell Physiol. 23, 533–539 [Google Scholar]
- 10.Enami I., Kamino K., Shen J. R., Satoh K., Katoh S. (1989) Biochim. Biophys. Acta 977, 33–39 [Google Scholar]
- 11.Enami I., Okumura A., Nagao R., Suzuki T., Iwai M., Shen J. R. (2008) Photosynth. Res. 98, 349–363 [DOI] [PubMed] [Google Scholar]
- 12.Enami I., Yoshihara S., Tohri A., Okumura A., Ohta H., Shen J. R. (2000) Plant Cell Physiol. 41, 1354–1364 [DOI] [PubMed] [Google Scholar]
- 13.Shen J. R., Qian M., Inoue Y., Burnap R. L. (1998) Biochemistry 37, 1551–1558 [DOI] [PubMed] [Google Scholar]
- 14.Ohta H., Suzuki T., Ueno M., Okumura A., Yoshihara S., Shen J. R., Enami I. (2003) Eur. J. Biochem. 270, 4156–4163 [DOI] [PubMed] [Google Scholar]
- 15.Enami I., Iwai M., Akiyama A., Suzuki T., Okumura A., Katoh T., Tada O., Ohta H., Shen J. R. (2003) Plant Cell Physiol. 44, 820–827 [DOI] [PubMed] [Google Scholar]
- 16.Miyao M., Murata N. (1989) Biochim. Biophys. Acta 977, 315–321 [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T., Ohta H., Enami I. (2005) Photosynth. Res. 84, 239–244 [DOI] [PubMed] [Google Scholar]
- 18.Seidler A. (1996) Biochim. Biophys. Acta 1277, 35–60 [DOI] [PubMed] [Google Scholar]
- 19.Bricker T. M., Burnap R. L. (2007) in Photosystem II: The Light-driven Water: Plastoquinone Oxidoreductase (Wydrzynski T. J., Satoh K. eds) pp. 95–120, Springer, Dordrecht, The Netherlands [Google Scholar]
- 20.Miyao M., Murata N. (1984) FEBS Lett. 168, 118–120 [Google Scholar]
- 21.Ghanotakis D. F., Babcock G. T., Yocum C. F. (1984) FEBS Lett. 167, 127–130 [Google Scholar]
- 22.Ghanotakis D. F., Babcock G. T., Yocum C. F. (1985) FEBS Lett. 192, 1–33915890 [Google Scholar]
- 23.Enami I., Suzuki T., Tada O., Nakada Y., Nakamura K., Tohri A., Ohta H., Inoue I., Shen J. R. (2005) FEBS J. 272, 5020–5030 [DOI] [PubMed] [Google Scholar]
- 24.Thornton L. E., Ohkawa H., Roose J. L., Kashino Y., Keren N., Pakrasi H. B. (2004) Plant Cell 16, 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summerfield T. C., Shand J. A., Bentley F. K., Eaton-Rye J. J. (2005) Biochemistry 44, 805–815 [DOI] [PubMed] [Google Scholar]
- 26.Kashino Y., Inoue-Kashino N., Roose J. L., Pakrasi H. B. (2006) J. Biol. Chem. 281, 20834–20841 [DOI] [PubMed] [Google Scholar]
- 27.Roose J. L., Kashino Y., Pakrasi H. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2548–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roose J. L., Wegener K. M., Pakrasi H. B. (2007) Photosynth. Res. 92, 369–387 [DOI] [PubMed] [Google Scholar]
- 29.Nagao R., Ishii A., Tada O., Suzuki T., Dohmae N., Okumura A., Iwai M., Takahashi T., Kashino Y., Enami I. (2007) Biochim. Biophys. Acta 1767, 1353–1362 [DOI] [PubMed] [Google Scholar]
- 30.Nagao R., Tomo T., Noguchi E., Nakajima S., Suzuki T., Okumura A., Kashino Y., Mimuro M., Ikeuchi M., Enami I. (2010) Biochim. Biophys. Acta 1797, 160–166 [DOI] [PubMed] [Google Scholar]
- 31.Okumura A., Nagao R., Suzuki T., Yamagoe S., Iwai M., Nakazato K., Enami I. (2008) Biochim. Biophys. Acta 1777, 1545–1551 [DOI] [PubMed] [Google Scholar]
- 32.Jeffrey S. W., Humphrey G. F. (1975) Biochem. Physiol. Pflanzen 167, 191–194 [Google Scholar]
- 33.Yamamoto Y., Doi M., Tamura N., Nishimura M. (1981) FEBS Lett. 133, 265–268 [Google Scholar]
- 34.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 35.Ikeuchi M., Inoue Y. (1988) FEBS Lett. 241, 99–104 [DOI] [PubMed] [Google Scholar]
- 36.Ono T. A., Mino H. (1999) Biochemistry 38, 8778–8785 [DOI] [PubMed] [Google Scholar]
- 37.Miyao M., Murata N. (1983) Biochim. Biophys. Acta 725, 87–93 [Google Scholar]
- 38.Ono T., Inoue Y. (1983) FEBS Lett. 164, 255–260 [Google Scholar]
- 39.Miyao M., Murata N. (1984) FEBS Lett. 170, 350–354 [Google Scholar]
- 40.Kamiya N., Shen J. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 42.Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009) Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 43.Ono T., Inoue Y. (1984) FEBS Lett. 168, 281–286 [Google Scholar]
- 44.Enami I., Tomo T., Kitamura M., Katoh S. (1994) Biochim. Biophys. Acta 1185, 75–80 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.