Abstract
Aquaporins facilitate efficient diffusion of water across cellular membranes, and water homeostasis is critically important in conditions such as cerebral edema. Changes in aquaporin 1 and 4 expression in the brain are associated with cerebral edema, and the lack of water channel modulators is often highlighted. Here we present evidence of an endogenous modulator of aquaporin 1 and 4. We identify miR-320a as a potential modulator of aquaporin 1 and 4 and explore the possibility of using miR-320a to alter the expression of aquaporin 1 and 4 in normal and ischemic conditions. We show that precursor miR-320a can function as an inhibitor, whereas anti-miR-320a can act as an activator of aquaporin 1 and 4 expressions. We have also shown that anti-miR-320a could bring about a reduction of infarct volume in cerebral ischemia with a concomitant increase in aquaporins 1 and 4 mRNA and protein expression.
Keywords: Brain, MicroRNA, RNA Silencing, RNA Interference (RNAi), Water Channel
Introduction
MicroRNAs (miRNAs)2 regulate mRNA expression by binding to the 3′-UTR (1). As simple as it sounds, identifying targets for miRNAs remains a challenging task mainly because each miRNA has hundreds of mRNA targets (2). Increasing this complexity are emerging reports on miRNAs binding at 5′-UTR as well as promoter regions (3–5). Recent studies also reveal the possibility of miRNAs functioning as transcriptional or splicing regulators within the nucleus (6) and being involved in genetic exchange via exosomes with adjacent cells (7). Because of this complexity in regulation, of the hundreds of miRNAs identified in the human species, only a handful have been assigned their specific targets.
Comparison of miRNA profiles between normal and diseased samples allows the identification of crucial miRNAs that are altered during disease conditions and enables one to search for possibilities of altering these expression profiles in an attempt to normalize or reduce the patho-physiological conditions of the disease. Changes in the expression of miRNAs have been reported in several diseases (8–10) including cerebral ischemia (11–13). Cerebral ischemia is a highly debilitating condition, and ischemia-induced edema further increases complications and morbidity (14). The movement of water into and out of an ischemic brain is thought to be mainly modulated by aquaporins (AQPs), especially aquaporin 1 (AQP1) and aquaporin 4 (AQP4). AQPs are a family of transmembrane proteins involved in transport of water, glycerol, ions, and even CO2 (15, 16). The 13-member family is ubiquitously expressed in almost all parts of the human body. Often more than one homolog is found to be present in any tissue or organ. In the brain AQP1, 3, 4, 5, 8, and 9 have been reported, with AQP1, 4, and 9 being expressed abundantly (17). The importance of AQP1 and AQP4 regulation in edema has been highlighted in several studies (18, 19). AQP4 knockouts in mice showed increased protection against cytotoxic edema caused by water intoxication and permanent focal cerebral ischemia, whereas in conditions leading to vasogenic brain edema, it had a deleterious effect (18, 19). siRNA-based studies have paralleled these findings in which AQP4 knockdown was shown to reduce the amount of water influx but yet delay its clearance in astrocytes during hypoxic and subsequent reoxygenation events (20). Expression of AQP1 and AQP4 were induced in astrocyte cells surrounding the edematous region (21). These findings serve to relay the importance of modulating both AQP1 and AQP4 during cerebral edema, yet with the exception of mercury, gold, lithium, and ammonium quaternary compounds, no other natural modulators of AQPs are available for in vivo use. In this paper we explore the possibility of using endogenous miRNAs as riboregulators to modulate the expression of AQP1 and AQP4.
EXPERIMENTAL PROCEDURES
Transient Focal Cerebral Ischemia and Quantitation of Infarct Volume
Transient focal cerebral ischemia was induced in male Sprague-Dawley rats through middle cerebral artery occlusion (MCAo) as described by Armugam et al. (22). The animals were handled according to the guidelines of the Council for International Organization of Medical Sciences on Animal Experimentation (World Health Organization, Geneva, Switzerland) and the National University of Singapore. The animal protocols were approved (approval code 081/09) by the National University of Singapore Institutional Animal Care and Use Committee. Transient ischemic stroke was created in rat models via MCAo for a period of 1 h. After 1 h of occlusion, the suture was removed to allow for reperfusion. Intracerebroventricular administration of anti- or pre-miR-320a was carried out immediately after the removal of suture. For infarct volume quantitation, whole rat brain slices were stained in 2,3,5-triphenyltetrazolium chloride (Sigma) and fixed in 10% buffered formalin. Stained brain slices were scanned, and the images were analyzed using Scion Image analysis software (22).
Transfection of miRNAs in Astrocytoma Cells
Transfection procedures of miRNAs were adapted from Cheng et al. (23). Human anti- or pre-miR320a (anti-miR-320a; 5′-UUUUCGACCCAACUCUCCCGCU-3′ and pre-miR-320a 5′-GCUUCGCUCCCCUCCGCCUUCUCUUCCCGGUUCUUCCCGGAGUCGGGAAAAGCUGGGUUGAGAGGGCGAAAAAGGAUGAGGU-3′) at 30 nm final concentration (in 50 μl of Opti-MEM) was complexed with 1 μl of NeoFx in 50 μl of Opti-MEM (Ambion, Inc.). The cells were transfected with these complexes and maintained for 48 h prior to subsequent work.
Oxygen and Glucose Deprivation
Human astrocytoma cells (CRL-1718TM, ATCC) were cultured in RPMI 1640 medium (Hyclone Laboratories) supplemented with 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin-streptomycin (Invitrogen) and maintained in a 37 °C incubator with 5% CO2. The cells were seeded at a density of 1.0 × 105 (24-well plates) and subjected to oxygen and glucose deprivation where the cells were maintained in an incubation chamber for 6 h at 37 °C, in the absence of oxygen, glucose, and serum. Oxygen in the chamber was displaced with a nitrogen gas flow rate of 1.3 liters/min. Corresponding control cell cultures were maintained at 37 °C in a 5% CO2 incubator. After 6 h, the cells were left to recover at 37 °C in a 5% CO2 incubator for 16–18 h prior to further work.
Extraction of Total RNA and Protein
Total RNA (including miRNA) and protein were extracted from cells by a single-step method using TRIzol (Invitrogen) according to the manufacturer's protocol. The concentration and integrity of RNA were determined using Nanodrop ND-1000 spectrophotometry (Nanodrop Tech, Rockland, DE) and RNA gel electrophoresis.
Reverse Transcription and Real Time Quantitative PCR
Reverse transcription followed by real time quantitative PCR were carried out according to Jeyaseelan et al. (12). Quantitation of AQP1 and AQP4 mRNAs was performed using SYBR green assay. Specific primer sequences were generated using PrimerExpress (Applied Biosystems). (AQP1 forward primer, 5′-GACACCTCCTGGCTATTGACTACA-3′; AQP1 reverse primer, 5′-CCGCGGAGCCAAAGG-3′; AQP4 forward primer, 5′-AGCCTGGGATCCACCATC-3′; and AQP4 reverse primer, 5′-TGCAATGCTGAGTCCAAAGC-3′). For miRNA detection, reverse transcription followed by stem-loop real time quantitative PCRs were performed according to the manufacturer's protocols using miR-320a-specific primers (Applied Biosystems). All of the reactions were conducted on an Applied Biosystems 7900 sequence detection system.
mRNA and μParafloTM MicroRNA Microarray Assay and Analysis
The oligonucleotide (DNA) microarray was performed according to the manufacturer's (Illumina, San Diego, CA) protocol using 500 ng of total RNA. The μParafloTM microRNA microarray was performed as described by Jeyaseelan et al. (12). The raw data were subtracted from control samples and further filtered for signal log ratio (>1 and <−1) determination at a detection probability value of <0.01.
SDS-PAGE and Western Blot Analysis
40 μg of total protein was resolved using 12% Tris-Tricine SDS-PAGE and Western blot was carried out as described by Satoh et al. (24). The membranes were probed with primary antibodies (rabbit anti-AQP1 and AQP4; Santa Cruz Biotechnology) at a concentration of 1 μg/ml in 0.5% blocking solution. β-Actin was used as a loading control (Bio-Rad). Secondary antibodies (horseradish peroxidase-conjugated goat anti-rabbit; Bio-Rad) were used at a dilution of 1:10,000 in 0.5% blocking solution. The membranes were visualized via enhanced chemiluminescence (SuperSignal West; Thermo Scientific) with variable exposures (Kodak-MS film). Films of Western blots were scanned (Acer SWZ3300U), and the labeling intensities of the bands were quantitated using ImageJ software (National Institutes of Health).
Immunocytochemistry
Immunocytochemistry was performed on astrocyte cell cultures treated with anti- or pre-miR-320a. Briefly, astrocytes co-transfected with either AQP1 or AQP4 plasmids and anti- or pre-miR-320a were fixed with 4% formaldehyde in phosphate-buffered saline for 20 min, permeabilized with 0.1% Triton X-100 in PBS for 30 min, and blocked with 5% FBS in PBST for 30 min. AQP1 and AQP4 (Santa Cruz Biotechnology) were probed according to the procedure adapted from Satoh et al. (24). FITC-coupled secondary antibody was used at dilutions of 1:200. DAPI was used as a nuclear stain. The images were viewed and analyzed using LSM510 confocal imaging software (Carl Zeiss MicroImaging Inc.)
Luciferase Assays
Gene-specific primers were used to amplify the miR-320a binding sites predicted on AQP1 and AQP4 3′-UTR (AQP1 forward primer, 5′-ATTAACTAGTCATTCCCTAGCA-3′; AQP1 reverse primer, 5′-TATGAAGCTTCAGGCAGGGGGT-3′; AQP4 forward primer, 5′-ATTAACTAGTTTTCCTAAAGTG-3′; and AQP4 reverse primer, 5′-TATGAAGCTTTCACAGGCTAT-3′). The PCR products were cloned into the Firefly luciferase expressing vector (pMIR-REPORTTM; Ambion) at the SpeI and HindIII sites. Plasmid transfection procedure was adapted from Cheng et al. (23). HeLa cells were transfected with 50 nm anti- or pre-miR-320a for 3 h followed by 100 ng/well pMIR-REPORTTM vector for 3 h. The cells were lysed 48 h later for measurement of luciferase activity. Dual luciferase assay (Promega) was used to quantitate the effects of anti- or pre-miR-320a interaction with the 3′-UTR of AQP1 and AQP4. The assay was performed according to the manufacturer's protocol. In all experiments, transfection efficiencies were normalized to those of cells co-transfected with the Renilla luciferase expressing vector (pRL-CMV; Promega) at 10 ng/well.
RESULTS
Identification of miRNAs Binding to AQP1 and AQP4 mRNAs
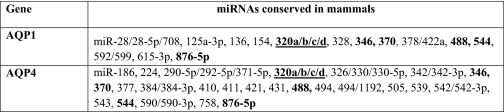
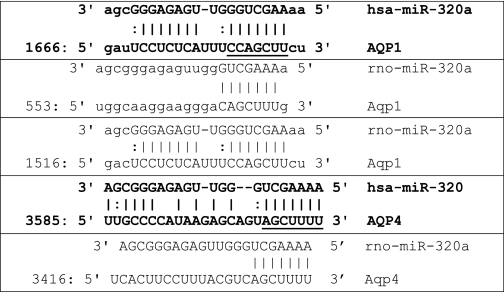
To assess the miRNAs that could target AQP1 and AQP4, a bioinformatic search was performed in the Targetscan and microRNA.org databases (25–28). Only miRNAs conserved in mammals were selected for this study. The search yielded 14 miRNAs against AQP1 and 25 miRNAs against AQP4 (Table 1). Among these miRNAs only miR-320 was reported to be differentially expressed in miRNA profiles of both human stroke patients and rat ischemic models (12, 13). miRNA profiling of human astrocytoma cells also revealed the presence of miR-320 among the 148 miRNAs detected (supplemental Table S1). More recently four different transcripts have been reported in the database for miR-320 (miR-320a, b, c, and d) (25–28). The sequence of miR-320 previously reported in the human and rat ischemic profiles (12, 13) matched that of miR-320a. Variations between the sequences of miR-320a, b, c, and d were only observed at the 5′-UTR. The seed region considered crucial for target binding remains the same for miR-320a, b, c, and d, and all four isoforms have been predicted to target the same region on the AQP1 and AQP4 transcripts. Thus in this study, we focused on miR-320a. The predicted interaction regions of miR-320a with the 3′-UTR of AQP1 and AQP4 in humans and rats are shown in Table 2.
TABLE 1.
TABLE 2.
Predicted binding sites of miR-320a in 3′UTR of AQP1 and AQP4
The binding sites for human and rat are indicated. The underlined nucleotides were subsequently mutated (see Fig. 2 legend) for the 3′-UTR-miRNA binding studies.
miR-320a Modulates AQP1 and AQP4 mRNA Expression
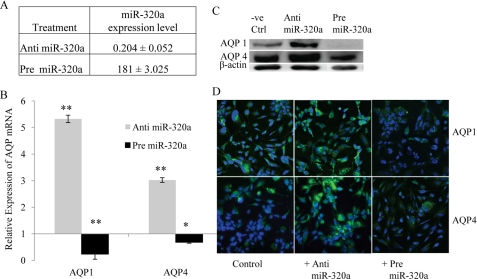
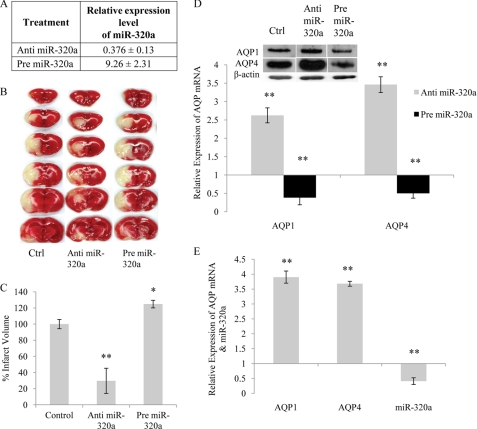
Based on the assumption that miR-320a binds to the 3′-UTR of AQP1 and AQP4, changes in miR-320a levels should be reflected in AQP1 and AQP4 gene expression. Hence in an attempt to change the cellular levels of miR-320a, human astrocytoma cells were transfected with miR-320a inhibitor (anti-miR-320a) and precursor (pre-miR-320a) independently. Changes in mRNA and miRNA levels were determined. In anti-miR-320a transfected cells, the relative expression miR-320a levels was ∼0.204 ± 0.052. This signifies ∼80% reduction in expression compared with normal cells (Fig. 1A). Simultaneously an increase in the AQP1 and AQP4 mRNA expression was observed (Fig. 1B). Likewise, introduction of pre-miR-320a increased miR-320a levels by ∼180-fold and resulted in a reduction in AQP1 and AQP4 mRNA expression (Fig. 1, A and B). Total RNA extracted from anti- or pre-miR-320a-treated cells were also subjected to mRNA array analysis. The AQP1 and AQP4 expression data extracted from the mRNA array (supplemental Table S2) correlated with the results obtained in this study. The AQP1 and AQP4 protein production (Fig. 1C) also correlated with the gene expression studies (Fig. 1B). Immunocytochemistry showed increased immunoreactivity for AQP1 and AQP4 in astrocytes treated with anti-miR-320a and reduced immunoreactivity in cells treated with pre-miR-320a (Fig. 1D). These findings suggest that miR-320a functions as a modulator of AQP1 and AQP4 gene expression.
FIGURE 1.
Expression of AQP1 and AQP4 in astrocytoma cell line transfected with anti- or pre-miR-320a. A, relative miR-320a expression in astrocytoma cell line transfected with anti- or pre-miR-320a at a concentration of 30 nm. B, relative mRNA expression of AQP1, AQP4 in astrocytoma cell line transfected with anti- or pre-miR-320a at a concentration of 30 nm. Statistical analyses were done using t tests. *, p < 0.05; **, p < 0.01 compared with respective negative controls. The data shown are the means ± S.D., n = 3. C, AQP1 and AQP4 protein expression in astrocytoma cell line transfected with anti- or pre-miR-320a. Changes observed in mRNA expression were reflected in protein expression as well. β-Actin was used as a loading control. D, AQP immunoreactivites in astrocytoma cells transfected with anti- or pre-miR-320a. Human astrocytes were co-transfected with AQP1 or AQP4 plasmids and anti- or pre-miR-320a. The cells were fixed and immunolabeled with either anti-AQP1 or anti-AQP4 antibodies (green) and nuclear stain DAPI (blue). Anti-miR-320a-treated cells showed increased immunoreactivity for AQP1 and AQP4, whereas pre-miR-320a-treated cells showed a reduction in immunoreactivity.
miR-320a Directly Targets AQP1 and AQP4
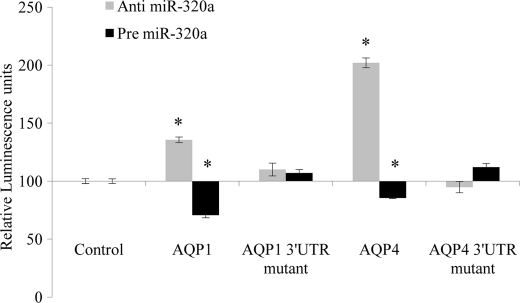
To validate direct interaction of miR-320a with AQP1 and AQP4 mRNAs, their 3′-UTR target sites were cloned into firefly luciferase reporter plasmids independently. These constructs were co-transfected with anti- or pre-miR-320a into HeLa cells. Cells transfected with anti-miR-320a exhibited an increase in the relative luciferase expression. Likewise, pre-miR-320a caused a reduction in luciferase expression (Fig. 2). Site-directed mutagenesis of the miR-320a recognition site located on the AQP1 and AQP4 3′-UTR abolished interactions between the miRNA and its AQP targets. These results indicate that miR-320a can directly target AQP1 and AQP4.
FIGURE 2.
Relative luminescence in plasmid constructs containing miR-320a target sites. miR-320a target regions in the 3′-UTR of AQP1 and AQP4 were identified using TargetScan and microRNA.org. These regions were cloned into luciferase reporter plasmids. Mutated 3′-UTR constructs were generated using site-directed mutagenesis. The miRNA recognition sites at cDNA corresponding to 3′-UTR (underlined) were mutated as follows: AQP1 (5′-CTGATTCCTCTCATTTAATTTGGCT-3′) and AQP4 (5′-TTGCCCCATAAGAGCAGTCGTCCGG-3′). The plasmid constructs together with anti- or pre-miR-320a were co-transfected into HeLa cells. Luciferase luminescence readings were obtained 48 h post-transfection. Relative luminescence was obtained by normalizing the values against control plasmids, pMIR-REPORTTM without any 3′-UTR insert. Statistical analyses were done using t tests. *, p < 0.05; **, p < 0.01 compared with control. The data shown are the means ± S.D., n = 3.
miR-320a Can Modulate AQP1 and AQP4 in Oxygen- and Glucose-deprived Conditions
The results obtained thus far indicate that miR-320a can modulate AQP1 and AQP4 expression under normal conditions. Modulation of AQP1 and AQP4 is considered crucial during edematous and ischemic conditions. Thus astrocytes were subjected to oxygen and glucose deprivation for 6 h, in the presence or absence of anti- and pre-miR-320a, after which the changes in the AQP1 and AQP4 expression levels were quantitated. AQP1 and AQP4 mRNA and protein expression were up-regulated after 6 h of oxygen and glucose deprivation (Fig. 3A). Conversely, of the 84 miRNAs that were differentially expressed because of oxygen and glucose deprivation, miR-320a was found to be down-regulated (supplemental Table S1). Decreasing miR-320a levels during oxygen and glucose deprivation with anti-miR-320a further elevated both AQP1 and AQP4 gene expression, whereas pre-miR-320a suppressed them (Fig. 3B).
FIGURE 3.
Expression of AQP1 and AQP4 in astrocytoma cells subjected to 6 h of oxygen and glucose deprivation (OGD). A, changes in AQP1 and AQP4 mRNA and protein expression in cells subjected to oxygen and glucose deprivation. Total cellular RNA and protein were used to quantify AQP1 and AQP4 levels. Up-regulation of AQP1 and AQP4 mRNA and protein (see inset) were observed. B, relative AQP expression in cells transfected with anti- or pre-miR-320a and subjected to 6 h of oxygen and glucose deprivation. The cells were transfected with anti- or pre-miR-320a 48 h prior to oxygen and glucose deprivation. All of the values were expressed relative to negative controls. Statistical analyses were done using t tests. *, p < 0.05; **, p < 0.01 compared with control. The data shown are the means ± S.D., n = 3. Ctrl, control.
Anti-miR-320a Reduces Infarct Volume
To determine the correlation between AQP1, AQP4, and miR-320a expression in vivo, ischemic animal models were used. Cerebral ischemia was induced in rats by occluding the middle cerebral artery for 60 min. 24 h after restoration of reperfusion, the animals were sacrificed. Expression levels of AQP1, AQP4, and miR-320a were determined from these MCAo brain samples. An increase in AQP1 and AQP4 mRNA expression was mirrored by a decrease in the miR-320a expression (Fig. 4, A and D). To understand the impact of modulation of AQP1 and AQP4 during ischemia, intracerebroventricular injections of anti- or pre-miR-320a were administered to ischemic rats immediately after MCAo. In anti-miR-320a-injected rats, the relative expression of miR-320a levels was ∼0.452 ± 0.23, which signifies a ∼55% reduction in expression compared with ischemic rats. Likewise in pre-miR-320a injected rats, miR-320a expression increased to 9.26 ± 2.31 (Fig. 4A). Administration of anti-miR-320a resulted in a reduction in the infarct volume, whereas pre-miR-320a caused a further increase in infarct volume (Fig. 4, B and C). Real time analyses showed that AQP1 and AQP4 expression was increased with a corresponding reduction in infarct volume, suggesting that the increased expression was facilitating edema clearance or recovery (Fig. 4D). An N-methyl-d-aspartate antagonist MK-801 is often used as a positive control for recovery from ischemia in animal models. When MK-801 was injected as described by Armugam et al. (22), the animals exhibited increased expression of AQP1 and AQP4 with a concomitant reduction in expression of miR-320a (Fig. 4E).
FIGURE 4.
Analyses of brain sections of MCA occluded rats injected with anti- or pre-miR-320a. A, expression level of miR-320a in ischemic rat injected with anti- or pre-miR-320a. Changes in miR-320a expression levels in the brain samples were quantitated using stem-loop real time quantitative PCR. B, histological analysis of brain sections. 2,3,5-Triphenyltetrazolium chloride-stained coronal brain sections (2 mm thick) of rats injected with 50 pmol of anti- or pre-miR-320a. Intracerebroventricular injections were given immediately after the removal of the suture (n = 10). Surviving cells stained red, whereas dead cells remained white. C, infarct volume of rat treated with anti- or pre-miR-320a. Infarct volumes are expressed as percentages of the control ± S.E. D, AQP1 and AQP4 mRNA levels in rats injected with anti- or pre-miR-320a. The data shown are the means ± S.D., n = 10. mRNA expression correlated with protein expression as well (see inset). E, relative AQP1, AQP4, and miR-320a expression in MK-801 administered ischemic (MCAo) rats. The rats with MCA occlusion were injected with MK-801 to reduce cerebral infarct. All of the values were expressed relative to ischemic rats. Statistical analyses were done using t tests. **, p < 0.01. The data shown are the means ± S.D., n = 10. Ctrl, control.
DISCUSSION
AQP1 and AQP4 Are Direct Targets of miR-320a
This study examined the possibility of exploiting miRNAs as natural endogenous modulators of AQP1 and AQP4 expression. With the help of bioinformatics-based databases (25–28) and published reports (12, 13), the search was narrowed down to miR-320a as a possible modulator of AQP1 and AQP4. Recent studies have established miR-320a as an inhibitor of the cell cycle gene (POLR3D) and the transferrin receptor gene (TFRC) (4, 29). Our AQP1 and AQP4 gene expression profiles were similar to those of these known targets of miR-320a (supplemental Table S2). This inhibitory effect was reflected in our in vitro systems (using astrocytes) under both normal and ischemic environment. The changes seen in AQP1 and AQP4 mRNA levels caused by the modulation of miR-320a were also reflected in the protein expression (immunoblotting and immunocytochemistry) in a target-specific manner. These observations together with luciferase binding assays demonstrate that AQP1 and AQP4 modulation is possible with the manipulation of miR-320a. The ability to modulate these two water channels can have a major impact on the outcome of edema-associated pathologies. AQP1 and AQP4 are considered the major contributors for maintaining cerebral fluid balance. In the brain, AQP1 is concentrated around the apical membrane of the choroid plexus epithelia and moderately present in the hippocampus and ependymal cells (30). Expression of AQP4 is extensive in the astrocytic processes adjacent to cerebral capillaries and in the pial membranes lining the subarachnoid space (31). The expression of these two water channels at strategic locations in the brain emphasizes their importance in cerebral water transport.
Up-regulation of AQP1 and AQP4 Expression in Edema: A Survival Mechanism?
Enhanced expression of AQP1 is observed in pathological states, and it is thought to augment susceptibility to pathological volume changes and promote edema formation. Increased AQP1 gene and protein expression was also observed in cerebral edema induced by traumatic brain injury (32). Even though AQP1 in the choroid plexus epithelia contributes to only 25% of the total cerebrospinal fluid production, diminishing its expression improved survival rates drastically from 25 to 87% in ischemic rats (33). Furthermore, reduction in AQP expression has been reported to enhance the resistance of cells against apoptotic stimulus (34). During apoptosis, cellular volume decrease is thought to be mediated via AQP1 (35). The initial up-regulation of AQP1 expression followed by an inactivation of the water channel is considered crucial for the proper progression of apoptosis. This suggests that AQP1 expression is biphasic and highlights the need for a modulator that could alter its expression accordingly.
Changes in AQP4 expression with respect to cerebral edema were reported in several studies. These studies report a peak in the accumulation of cerebral water content that can occur anywhere from 24 h to 3 days after MCAo (18, 36). Ribeiro et al. (36) reported a biphasic trend where the water content peaked at 1 and 4 h after MCAo. Interestingly, although a range of timings have been reported, these observations correlated with the increase in AQP4 gene expression levels. These authors proposed that the early up-regulation of AQP4 is associated with the increase in cerebral edema. Moreover, AQP4 null mice exhibited a 35% reduction in cerebral water content upon induction of ischemia (37).
In our study we show that introduction of anti-miR-320a causes a further increase in AQP1 and AQP4 expression with a consequent reduction in the infarct volume, and an introduction of pre-miR-320a results in an opposite effect of reducing AQP expression and increasing the infarct volume. Hence the increase in AQP expression, at least at the 24 h reperfusion time point, appears to be a part of the survival and recovery process. Interestingly, a recent study by Hirt et al. (38) proposed that the early up-regulation of AQP4 could indeed be a survival mechanism to reduce edema. Although thrombin preconditioning had no effect on AQP4 expression in normal rats, upon ischemia these rats exhibited increased AQP4 expression and reduced cerebral edema, suggesting that AQP4 increase might be facilitating edema clearance (38). In a study using a neutral anticoagulant secretory phospholipase A2 as a neuroprotectant, it has been shown that administration of secretory phospholipase A2 reduced infarct volume in rats subjected to focal transient cerebral ischemia. Secretory phospholipase A2 also alleviated the neuronal damage in organotypic hippocampal slices subjected to oxygen glucose deprivation (22). The authors also reported an increase in expression of AQP4 together with several anti-apoptotic and pro-survival genes.
In this study we have also observed an up-regulation in AQP1 and AQP4 expression levels when MK-801 was administered to MCAo ischemic models. MK-801 is an NMDA antagonist widely used in animal models to rescue or reduce cerebral infarct and mimic recovery (39). Ischemic rats injected with MK-801 exhibited increased AQP1 and AQP4 profiles with reduced miR-320a expression, paralleling expression patterns of those injected with anti-miR-320a. These findings suggest that up-regulation of AQP1 and AQP4 could possibly assist in edema clearance. A recent report on ischemic human patients with good recovery and good clinical outcome (modified Rankin scale, ≤2) also showed a reduction in miR-320 expression when compared with patients with poor clinical outcome (modified Rankin scale, >2) (13).
Other Targets of miR-320a and Their Possible Implications
Administration of anti-miR-320a was found to be beneficial in ischemic rats with a reduction in infarct volume. Although we show that miR-320a can be used to modulate the expression of AQP1 and AQP4, one has to accept that introduction of a miRNA would have a widespread impact on several other genes. Using miRNApath to gauge the impact of miR-320a modulation revealed ∼145 pathways being affected with as many as 1500 target genes (40, 41). To narrow down the list, 22 pathways (in relation to ischemia) were selected (41). Within these selected pathways 172 genes were predicted to be targets of miR-320a (40).
Expression profiles of these 172 predicted targets were compared with our anti- and pre-miR-320a array. 77 genes (including AQP1 and AQP4) were found to be up-regulated in response to anti-miR-320a, whereas they were down-regulated in response to pre-miR-320a treatments, suggesting that these genes may be affected by miR-320a via the RNAi mechanism. From the results shown in supplemental Table S2, we could hypothesize several possible mechanisms that could have aided in the reduction of infarct volume caused by anti-miR-320a administration.
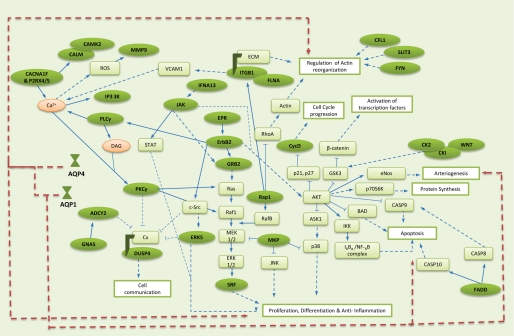
Genes such as protein kinase Cγ and MAP kinase 7 (ERK5) and tubulin (DUSP4) were up-regulated in response to anti-miR-320a (Fig. 5). Protein kinase Cγ and ERK5 have been reported to target gap junction protein connexin and affect its gating status (see review in Ref. 42). Dephosphorylation of connexins opens their configuration and allows the transfer of molecules as well as signaling information between adjacent cells, whereas phosphorylation of connexins closes their conformation, marking them for degradation and internalization. Dephosphorylation of connexins during ischemia has been reported, and this open gating state is thought to propagate ischemic injury. Studies have shown that the uncoupling of connexins reduces ischemia-related injuries (43). Up-regulation of protein kinase Cγ and ERK5 by miR-320a might play a role in uncoupling connexins, thus resulting in protection during ischemia. Cytoskeletal protein DUSP4 interacts with connexin channels and is considered important for translocation of the gap junction protein. The increase in DUSP4 could be due to the need for the association and internalization of inactive connexin proteins (42). Although co-localization of connexin with AQP4 has been reported, the extent of interaction between these two proteins is not clear. Mixed reports about their interaction exist (44, 45).
FIGURE 5.
Possible pathways/genes affected by miR-320a modulation. Compilation of possible genes affected by miR-320a modulation was adapted from the KEGG Pathway Database. Green ovals, targets of miR-320a; green rectangles, other genes in pathway; pink ovals, secondary messenger; blue right arrows, direct interaction; dashed blue right arrows, indirect interaction; broken red right arrows, AQP interaction.
Expression of genes such as Janus kinase (JAK) and receptor tyrosine kinase (ERBB2) were also increased in response to anti-miR-320a. These genes are involved in signaling pathways and have an indirect effect on serine/threonine protein kinase (AKT) (Fig. 5). AKT, also known as protein kinase B, is implicated in cell survival, induction of protein production, and proliferation. ERBB2 interacts with growth factor receptor bound protein 2 (GRB2) to stimulate cellular proliferation. Both ERBB2 and GRB2 are predicted targets of miR-320a. Both ERBB2 and GRB2 expression are considered crucial for activation of AKT-mediated survival pathways (46). AKT pathway activation can inhibit pro-apoptotic proteins such as Bcl-2-associated death promoter and caspase 9 and thus promote survival. Influx of Ca2+ ions during ischemia directly or indirectly triggers a variety of cell responses, one of which is the activation of the inflammatory pathway. Activation of the AKT pathway results in the inhibition of glycogen synthase kinase 3, which can enhance the operation of various pro-inflammatory signaling molecules (47). Thus increasing ERBB2 and GRB2 expression using miR-320a not only enhances proliferation of ischemic cells but also suppresses the inflammatory pathway possibly via AKT activation. Furthermore ERBB2 activation is often associated with cell migration, a process where AQP1 and AQP4 are also implicated (44, 48). Increased expression of AQP1 and AQP4 are crucial for the formation of cellular processes, subsequent cell migration, and proliferation (44, 48).
Several cytoskeletal proteins that aid cell migration or processes such as actin remodeling and axon guidance were also up-regulated because of anti-miR-320a (Fig. 5). Some of these proteins with predicted target regions for miR-320a are cofilin, slit homolog 3 protein (SLIT3), and filamin A. SLIT3 is a repulsive axon guidance molecule often expressed and secreted by both endothelial cells and vascular smooth muscle cells. They are considered crucial for promoting angiogenesis, for they have been shown to stimulate blood vessel growth in vivo. Apart from angiogenesis, SLIT3 also stimulates endothelial cell proliferation and promotes cell motility and chemotaxis (49). Extensive AQP1 expression has also been reported in vascular endothelia. AQP1 null mice transplanted with cerebral tumors exhibited reduced vascularity and widespread necrosis, suggesting that AQP1 expression is crucial for proper angiogenesis. The fact that anti-miR-320a up-regulates both AQP1 and SLIT3 genes suggests that the ischemic recovery process via angiogenesis could have been triggered. Angiogenesis requires the controlled remodeling of the cytoskeleton, and this is evident from the up-regulation of cytoskeletal proteins such as DUSP4, filamin A, and cofilin. Filamin A regulates reorganization of the actin cytoskeleton by interacting with integrins, which were also up-regulated because of anti-miR-320a introduction. Up-regulation of these genes with the removal of miR-320a in ischemic samples at 24 h of reperfusion suggests that survival mechanisms are triggered and recovery is underway.
Conclusion
In this study, we have established that miR-320a can directly modulate AQP1 and AQP4 gene expression in both in vitro and in vivo conditions. We have also observed that administration of anti-miR-320a could bring about a reduction in infarct volume as well as an increase in the expression of aquaporins 1 and 4 in animal models of cerebral ischemia. miR-320a is generally implicated with anti-angiogenesis. However, because of the large number of possible targets, its mode of action is not well established. Nevertheless, we have highlighted several other genes that could possibly be targets of miR-320a in cerebral ischemia.
Supplementary Material
This work was supported by National Research Foundation Grant R-184-002-165-281, National Medical Research Council Grant EDG: R-183-000-230-275, and National Kidney Foundation Grant NKFRC/2008/10.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- miRNA
- microRNA
- miR-320a
- microRNA-320a
- AQP
- aquaporin
- MCAo
- middle cerebral artery occlusion
- ERBB2
- receptor tyrosine kinase
- AKT
- serine/threonine protein kinase.
REFERENCES
- 1.Lee R. C., Feinbaum R. L., Ambros V. (1993) Cell 75, 843–854 [DOI] [PubMed] [Google Scholar]
- 2.Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 3.Li L. C., Okino S. T., Zhao H., Pookot D., Place R. F., Urakami S., Enokida H., Dahiya R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim D. H., Saetrom P., Snøve O., Jr., Rossi J. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Place R. F., Li L. C., Pookot D., Noonan E. J., Dahiya R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang H. W., Wentzel E. A., Mendell J. T. (2007) Science 315, 97–100 [DOI] [PubMed] [Google Scholar]
- 7.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 8.Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. (2005) Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 9.van Rooij E., Sutherland L. B., Liu N., Williams A. H., McAnally J., Gerard R. D., Richardson J. A., Olson E. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatsuguchi M., Seok H. Y., Callis T. E., Thomson J. M., Chen J. F., Newman M., Rojas M., Hammond S. M., Wang D. Z. (2007) J. Mol. Cell Cardiol. 42, 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dharap A., Bowen K., Place R., Li L. C., Vemuganti R. (2009) J. Cereb. Blood Flow Metab. 29, 675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeyaseelan K., Lim K. Y., Armugam A. (2008) Stroke 39, 959–966 [DOI] [PubMed] [Google Scholar]
- 13.Tan K. S., Armugam A., Sepramaniam S., Lim K. Y., Setyowati K. D., Wang C. W., Jeyaseelan K. (November2, 2009) PLoS ONE 10.1371/journal.pone.0007689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marmarou A. (September27, 2007) Neurosurgery, Focus 10.3171/foc.2007.22.5.2 [Google Scholar]
- 15.Agre P., Kozono D. (2003) FEBS Lett. 555, 72–78 [DOI] [PubMed] [Google Scholar]
- 16.Carbrey J. M., Agre P. (2009) Handb. Exp. Pharmacol. 190, 3–28 [DOI] [PubMed] [Google Scholar]
- 17.Yang M., Gao F., Liu H., Yu W. H., Sun S. Q. (2009) Brain Res. 1290, 121–132 [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi M., Yamashita T., Kumura E., Tamatani M., Kobayashi A., Yokawa T., Maruno M., Kato A., Ohnishi T., Kohmura E., Tohyama M., Yoshimine T. (2000) Mol. Brain Res. 78, 131–137 [DOI] [PubMed] [Google Scholar]
- 19.Papadopoulos M. C., Manley G. T., Krishna S., Verkman A. S. (2004) FASEB J. 18, 1291–1293 [DOI] [PubMed] [Google Scholar]
- 20.Fu X., Li Q., Feng Z., Mu D. (2007) Glia 55, 935–941 [DOI] [PubMed] [Google Scholar]
- 21.Suzuki R., Okuda M., Asai J., Nagashima G., Itokawa H., Matsunaga A., Fujimoto T., Suzuki T. (2006) Acta Neurochir. Suppl. 96, 398–401 [DOI] [PubMed] [Google Scholar]
- 22.Armugam A., Cher C., Lim K., Koh D., Howells D., Jeyaseelan K. (September23, 2009) BMC Neurosci. 10.1186/1471-2202-10-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng A. M., Byrom M. W., Shelton J., Ford L. P. (2005) Nucleic Acids Res. 33, 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh J., Tabunoki H., Yamamura T., Arima K., Konno H. (2007) Neuropathology 27, 245–256 [DOI] [PubMed] [Google Scholar]
- 25.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 26.Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 27.Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008) Nucleic Acids Res. 36, D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaar D. G., Medina D. J., Moore D. F., Strair R. K., Ting Y. (2009) Exp. Hemat. 37, 245–255 [DOI] [PubMed] [Google Scholar]
- 30.Mobasheri A., Marples D. (2004) Am. J. Physiol. Cell Physiol. 286, C529–C537 [DOI] [PubMed] [Google Scholar]
- 31.Aoki K., Uchihara T., Tsuchiya K., Nakamura A., Ikeda K., Wakayama Y. (2003) Acta Neuropathol. 106, 121–124 [DOI] [PubMed] [Google Scholar]
- 32.Tran N. D., Kim S., Vincent H. K., Rodriguez A., Hinton D. R., Bullock M. R., Young H. F. (2010) J. Neurosurg. 112, 1095–1104 [DOI] [PubMed] [Google Scholar]
- 33.Oshio K., Watanabe H., Song Y., Verkman A. S., Manley G. T. (2005) FASEB J. 19, 76–78 [DOI] [PubMed] [Google Scholar]
- 34.Jablonski E. M., Mattocks M. A., Sokolov E., Koniaris L. G., Hughes F. M., Jr., Fausto N., Pierce R. H., McKillop I. H. (2007) Cancer Lett. 250, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jablonski E. M., Webb A. N., McConnell N. A., Riley M. C., Hughes F. M., Jr. (2004) Am. J. Physiol. Cell. Physiol. 286, C975–C985 [DOI] [PubMed] [Google Scholar]
- 36.Ribeiro M. C., Hirt L., Bogousslavsky J., Regli L., Badaut J. (2006) J. Neurosci. Res. 83, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 37.Manley G. T., Fujimura M., Ma T., Noshita N., Filiz F., Bollen A. W., Chan P., Verkman A. S. (2000) Nat. Med. 6, 159–163 [DOI] [PubMed] [Google Scholar]
- 38.Hirt L., Ternon B., Price M., Mastour N., Brunet J. F., Badaut J. (2009) J. Cereb. Blood Flow Metab. 29, 423–433 [DOI] [PubMed] [Google Scholar]
- 39.Bertorelli R., Adami M., Di Santo E., Ghezzi P. (1998) Neurosci. Lett. 246, 41–44 [DOI] [PubMed] [Google Scholar]
- 40.Chiromatzo A. O., Oliveira T. Y., Pereira G., Costa A. Y., Montesco C. A., Gras D. E., Yosetake F., Vilar J. B., Cervato M., Prado P. R., Cardenas R. G., Cerri R., Borges R. L., Lemos R. N., Alvarenga S. M., Perallis V. R., Pinheiro D. G., Silva I. T., Brandão R. M., Cunha M. A., Giuliatti S., Silva W. A., Jr. (2007) Genet. Mol. Res. 6, 859–865 [PubMed] [Google Scholar]
- 41.Kanehisa M., Goto S. (2000) Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dbouk H. A., Mroue R. M., El-Sabban M. E., Talhouk R. S. (March12, 2009) Cell Commun. Signal 10.1186/1478-811X-7-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frantseva M. V., Kokarovtseva L., Perez Velazquez J. L. (2002) J. Cereb. Blood Flow Metab. 22, 453–462 [DOI] [PubMed] [Google Scholar]
- 44.Kong H., Fan Y., Xie J., Ding J., Sha L., Shi X., Sun X., Hu G. (2008) J. Cell Sci. 121, 4029–4036 [DOI] [PubMed] [Google Scholar]
- 45.Nicchia G. P., Srinivas M., Li W., Brosnan C. F., Frigeri A., Spray D. C. (2005) FASEB J. 19, 1674–1676 [DOI] [PubMed] [Google Scholar]
- 46.Lim S. J., Lopez-Berestein G., Hung M. C., Lupu R., Tari A. M. (2000) Oncogene 19, 6271–6276 [DOI] [PubMed] [Google Scholar]
- 47.Yin W., Signore A. P., Iwai M., Cao G., Gao Y., Johnnides M. J., Hickey R. W., Chen J. (2007) Stroke 38, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 48.Saadoun S., Papadopoulos M. C., Hara-Chikuma M., Verkman A. S. (2005) Nature 434, 786–792 [DOI] [PubMed] [Google Scholar]
- 49.Zhang B., Dietrich U. M., Geng J. G., Bicknell R., Esko J. D., Wang L. (2009) Blood 114, 4300–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.