Abstract
Microsomal prostaglandin E synthase-1 (MPGES1) is induced during an inflammatory reaction from low basal levels by pro-inflammatory cytokines and subsequently involved in the production of the important mediator of inflammation, prostaglandin E2. Nonsteroidal anti-inflammatory drugs prevent prostaglandin E2 production by inhibiting the upstream enzymes cyclooxygenases 1 and 2. In contrast to these conventional drugs, a new generation of NSAIDs targets the terminal enzyme MPGES1. Some of these compounds potently inhibit human MPGES1 but do not have an effect on the rat orthologue. We investigated this interspecies difference in a rat/human chimeric form of the enzyme as well as in several mutants and identified key residues Thr-131, Leu-135, and Ala-138 in human MPGES1, which play a crucial role as gate keepers for the active site of MPGES1. These residues are situated in transmembrane helix 4, lining the entrance to the cleft between two subunits in the protein trimer, and regulate access of the inhibitor in the rat enzyme. Exchange toward the human residues in rat MPGES1 was accompanied with a gain of inhibitor activity, whereas exchange in human MPGES1 toward the residues found in rat abrogated inhibitor activity. Our data give evidence for the location of the active site at the interface between subunits in the homotrimeric enzyme and suggest a model of how the natural substrate PGH2, or competitive inhibitors of MPGES1, enter the active site via the phospholipid bilayer of the membrane.
Keywords: Enzyme Mechanisms, Enzyme Mutation, Inflammation, Membrane Proteins, Prostaglandins, Chimeric Protein, Inhibitor-binding Site, MPGES1 Inhibitor, Nonsteroidal Anti-inflammatory Drugs (NSAIDs), Prostaglandin E2 (PGE2)
Introduction
Nonsteroidal anti-inflammatory drugs (NSAIDs)3 are pharmaceuticals widely used in the treatment of pain, inflammation, and fever. They prevent the formation of important lipid mediators, the prostaglandins (PGs), via inhibition of cyclooxygenase (COX) enzymes. COX catalyzes the conversion of free arachidonic acid to the unstable endoperoxide intermediate PGH2, which in turn is a common substrate for a number of different terminal enzymes downstream of COX, leading to the formation of PGE2, PGD2, PGF2α, prostacyclin (PGI2), and thromboxane A2 (TXA2). Traditional NSAIDs are unselective inhibitors of both COX isozymes and prevent the formation of all prostaglandins, which can lead to severe adverse drug effects, mainly in the gastrointestinal tract. On the other hand, a second generation of NSAIDs, the COXIBs, was developed to specifically inhibit COX-2, the isozyme that is connected with inflammatory diseases and coregulated with microsomal prostaglandin E synthase-1 (MPGES1). This was believed to prevent inflammation and pain without impairing the physiological functions that COX-1-derived PGs have. Use of COXIBs, however, was accompanied with an increased incidence of severe cardiovascular side effects leading to myocardial infarction and stroke caused by a change in the ratio between PGI2 and TXA2. Nowadays, direct pharmacological inhibition of the downstream enzyme MPGES1 is therefore favored as a promising strategy for the treatment of inflammatory diseases.
In contrast to other enzymes that have been associated with PGE2 formation, namely MPGES2 (1) and cytosolic PGE2 synthase (2), targeted knock-out of MPGES1 in mice shows that MPGES1 plays a pivotal role in the production of PGE2 that mediates acute pain during an inflammatory response and in the pathogenesis of collagen-induced arthritis, a disease model of human rheumatoid arthritis (3). Moreover, mice deficient in MPGES1 do not induce fever after injection of bacterial lipopolysaccharide (4). In addition, more PGI2 than TXA2 is produced in these mice, and atherogenesis is retarded, suggesting a favorable cardiovascular side effect profile for the inhibition of MPGES1 compared with COX-2 (5).
MPGES1 inhibitors are currently developed by several pharmaceutical companies. Some of these compounds potently inhibit the human MPGES1 enzyme but do not have any effect on rat or mouse MPGES1 (6), which complicates animal testing in preclinical studies. To get insights into the molecular basis for this species difference and to define the inhibitor-binding site of MPGES1, we investigated the effect of MPGES1 inhibitors on a rat/human chimeric form of the enzyme and located crucial residues for inhibitor binding to transmembrane helix (TM) 4. Site-directed mutagenesis in combination with the recently solved protein structure (7) revealed that residues Thr-131, Leu-135, and Ala-138 in human MPGES1 are lining the entrance to the putative active site of MPGES1, a crevice between TM1 and TM4 of two adjacent subunits of the MPGES1 trimer, and that the respective residues Val-131, Phe-135, and Phe-138 prevent inhibitor binding in the rat enzyme.
EXPERIMENTAL PROCEDURES
Materials
Restriction endonuclease enzymes BtgI and NcoI, T4 DNA ligase, GeneTailorTM site-directed mutagenesis kit, Escherichia coli strains DH5α-T1R and BL21StarTM (DE) pLysS, LB and Terrific Broth media, as well as NuPAGE polyacrylamide gels and buffers were purchased from Invitrogen. Recombinant Pfu DNA polymerase, isopropyl β-d-thiogalactopyranoside, and PageRuler prestained protein ladder were purchased from Fermentas GmbH (St. Leon-Rot, Germany). Complete protease inhibitor was purchased from Roche Diagnostics GmbH. PVDF membrane was purchased from Pall Life Sciences (Pensacola, FL). Rabbit polyclonal antiserum raised against purified human MPGES1 was described before (8). Horseradish peroxidase-linked anti-rabbit IgG from donkey and Amersham Biosciences Hyperfilm ECL were purchased from GE Healthcare AB (Stockholm, Sweden). Supersignal West Pico ECL substrate was purchased from Thermo Fisher Scientific Inc. (Göteborg, Sweden). All other chemicals were obtained from Sigma-Aldrich or Merck. PGH2 was obtained from Lipidox (Lidingö, Sweden). MPGES1 inhibitors compound I (1-C-(2-chlorobenzene)-3-N-(naphtalen-1-yl)benzene-1,3-dicarboxamide) and compound II ((4E)-4-[2-(3-bromophenyl)hydrazin-1-ylidene]-1-ethanethioyl-3-methyl-4,5-dihydro-1H-pyrazol-5-one) were kindly provided by NovaSAID AB (Solna, Sweden).
Construction of Chimeric MPGES1 Enzyme
Chimeric protein was constructed from existing expression vectors pSP19T7LT harboring the coding sequence for human (9) and rat MPGES1 by endonuclease digestion using BtgI and NcoI. DNA fragments were isolated by agarose gel electrophoresis and ligated into the digested vector backbone containing the coding sequence of the respective other species. The correct sequence of the construct was verified by DNA sequencing (Seqlab Sequence Laboratories Göttingen GmbH, Göttingen, Germany).
Site-directed Mutagenesis
Mutations in MPGES1 were created with the help of the GeneTailorTM site-directed mutagenesis system (Invitrogen). The expression vectors pSP19T7LT harboring the coding sequence for human (9), rat MPGES1, or respective chimeric protein were methylated, and the methylated plasmid DNA was amplified in the mutagenesis polymerase chain reaction using two overlapping primers, one of which contained the target mutation. The resulting unmethylated product DNA containing the mutation was transformed into E. coli DH5α-T1R-competent cells. These cells circularize the DNA and digest residual methylated template DNA, leaving only unmethylated, mutated product, which can be isolated and used for further processing. All of the mutations were verified by DNA sequencing.
Protein Expression and Subcellular Fractionation
The expression constructs containing the correct coding sequence were transformed into E. coli BL21StarTM (DE) pLysS expression hosts. 6 ml of LB medium containing ampicillin (100 μg/ml) and chloramphenicol (20 μg/ml) were inoculated with a single colony of freshly transformed bacteria and incubated overnight at 37 °C with 300 rpm shaking. The cultures were diluted 1:50 into 200 ml of Terrific Broth medium containing ampicillin (100 μg/ml) and chloramphenicol (20 μg/ml) in a 500-ml culture flask. The cultures were grown at 37 °C with 300 rpm shaking, until the A600 reached 0.45–0.6. At this point expression was induced by the addition of 1 mm isopropyl β-d-thiogalactopyranoside, and the temperature was lowered to 30 °C. The cultures were allowed to grow for another 7 h, after which the cells were harvested at 4600 rpm for 15 min at 4 °C.
The cell pellet from 200 ml of culture was resuspended in 1 ml of TSEGP buffer (15 mm Tris/HCl, pH 8.0, 0.25 m sucrose, 1 mm EDTA, 1 mm GSH, 1× complete protease inhibitor). Lysozyme was added to a final concentration of 10 mg/ml, and the mixture was incubated on ice for 20 min. 7 ml of TEGP buffer (TSEGP buffer without sucrose) was added, and the cells were lysed by six 30-s sonication pulses on ice (Bandelin Sonoplus HD2070) at 60% of the maximum power. Cell debris was removed by centrifugation at 10,000 × g for 15 min at 4 °C. The supernatant was further centrifuged at 200,000 × g for 1 h at 4 °C. The membrane pellet was washed once and then resuspended in 1 ml of resuspension buffer (0.1 m potassium phosphate buffer, pH 7.5, 10% glycerol, 2.5 mm GSH, 1× complete protease inhibitor). Total protein concentration was determined using a Bradford protein assay according to the manufacturer's instructions (Bio-Rad), and aliquots were stored at −20 °C.
Immunoblot Analysis
Samples were diluted to appropriate concentrations with 1× NuPAGE SDS sample buffer and heated to 70 °C for 10 min. The proteins were then separated on a 4–12% NuPAGE polyacrylamide gels and electroblotted onto a PVDF membrane. The transfer efficiency was visualized by staining the membrane with Ponceau red. Additional protein-binding sites on the membrane were blocked overnight at 4 °C or for 1 h at room temperature, respectively, with 5% (w/v) nonfat dried milk protein in 0.1% TTBS. The membranes were washed three times for 10 min each with 0.1% TTBS and incubated for 1 h at room temperature with rabbit polyclonal antiserum directed against purified human MPGES1 at 1:2,500 dilution in 0.05% TTBS. After additional washing the membranes were incubated for 1 h at room temperature with horseradish peroxidase-linked anti-rabbit IgG at 1:100,000 dilution in 0.05% TTBS. The membranes were finally washed, and chemiluminescence detection was performed according to the manufacturer's instructions.
MDA-TBA Enzyme Activity Assay
PGH2 converting activity of the heterologously expressed enzyme was assayed based on a previously described method (10). In short, appropriate dilutions of total protein were made in activity assay buffer (0.1 m potassium phosphate buffer, pH 7.4, 2.5 mm GSH) and incubated on ice with inhibitors as indicated. PGH2 was added to a final concentration of 20 μm and subsequently incubated for 75 s at room temperature. The reaction was stopped by the addition of stop solution (25 mm FeCl2, 50 mm citric acid) to break down the remaining PGH2 to 12-(S)-hydroxy-8,10-trans-5-cis-heptadecatrienoic acid and malondialdehyde (MDA) (11). TBA solution (128.4 mm TBA, 148.15 mm citric acid) was added, and the samples were incubated for 30 min at 80 °C to convert MDA into a stable, pink-colored MDA-TBA complex. Fluorescence of the MDA-TBA complex was measured using a Victor3V 1420 Multilabel counter (PerkinElmer Life Sciences) with an excitation wavelength of 485 nm and an emission wavelength of 545 nm. The data were analyzed using GraphPad Prism version 5.00.
Molecular Modeling
The rat MPGES1 homology model is based on the structure of human MPGES1 (Protein Data Bank code 3DWW) as the template and was built using Modeler 9v2 with default settings and scripts. All images of the MPGES1 protein structure were created with PyMol version 0.98.
RESULTS
Differences in Inhibitor Sensitivity between Human and Rat MPGES1
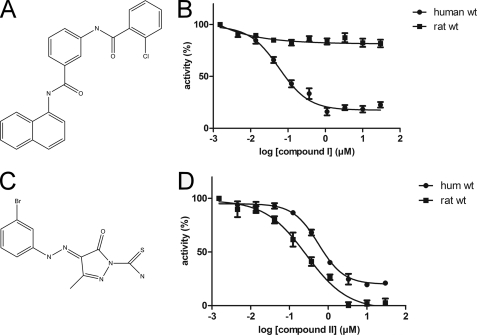
Compound I (Fig. 1A) potently inhibits human MPGES1 with an IC50 value of 58 nm. It has, however, no effect on the activity of rat MPGES1 (Fig. 1B and Table 1). On the other hand, inhibitors like compound II (Fig. 1C) inhibit both the human and the rat form of MPGES1 but display only moderate potency, with a factor 10 difference in IC50 for compound II as compared with compound I (Fig. 1D).
FIGURE 1.
MPGES1 inhibitor compound I is selective for human MPGES1. A, molecular structure of compound I. B, compound I fails to inhibit rat MPGES1, whereas it is fully active on the human enzyme. C, molecular structure of compound II. D, the enzymatic activity of both human and rat MPGES1 is inhibited by compound II.
TABLE 1.
Specific activity and IC50 values for different forms of MPGES1
| Protein | Specific activitya | IC50 compound I | 95% confidence interval |
|---|---|---|---|
| nmol min−1mg−1 | nm | nm | |
| Human WT MPGES1 | 214 | 58 | 45.0–74.7 |
| Rat WT MPGES1 | 59 | NIb | |
| rat115hum140rat | 64 | 300 | 194.8–455.2 |
| hum115rat140hum | 57 | NI | |
| Rat V131T,F135L,F138A | 86 | 590 | 435.4–796.6 |
| Human T131V,L135F,A138F | 54 | NI | |
| Rat V131T,F135L | 87 | NI | |
| Rat V131T,F138A | 50 | NI | |
| Rat F135L,F138A | 86 | NI | |
| rat115hum140rat-A29V | 60 | 450 | 268.1–738.9 |
| rat115hum140rat-V32I | 63 | 310 | 177.4–530.7 |
| rat115hum140rat-A29V,V32I | 63 | 270 | 131.9–551.3 |
a Related to total protein in E. coli membrane preparations (n ≥ 8).
b NI, no inhibition.
The two orthologue MPGES1 enzymes from rat and human share 77% identical amino acids in their sequence. Differences between the enzymes of these two species are mainly located at the N terminus as well as in TM3 and TM4 (Fig. 2). Almost all amino acids that differ between human and rat MPGES1 are located on the membrane face of the enzyme and point outwards, toward the phospholipid bilayer, as can be seen in the crystal structure of human MPGES1 (supplemental Fig. S1). There are no substitutions in TM2, which forms the core of the functionally active trimeric form of the enzyme and harbors many of the interactions with the cofactor GSH. This is in agreement with phylogenetic conservation analyses of other membrane proteins that show residues with a low degree of sequence variation to be located at strategic positions and lipid-facing positions for variable residues (12).
FIGURE 2.
The human and rat forms of MPGES1 differ mainly in TM3 and TM4. Amino acid sequences of human and rat MPGES1 were aligned using ClustalW. Every 10th residue is indicated by underlined letters. Based on the known structure of MPGES1, the positions of the four transmembrane helices were assigned, indicated with strings of H and the respective numbers of the helices. The consensus sequence denotes identical residues (*), strong conservation (:), weak conservation (.), and unconserved replacements (blank). Species differences are also highlighted in yellow. Differences are located mainly within TM3 and TM4.
Mapping of the Inhibitor-binding Site to TM4
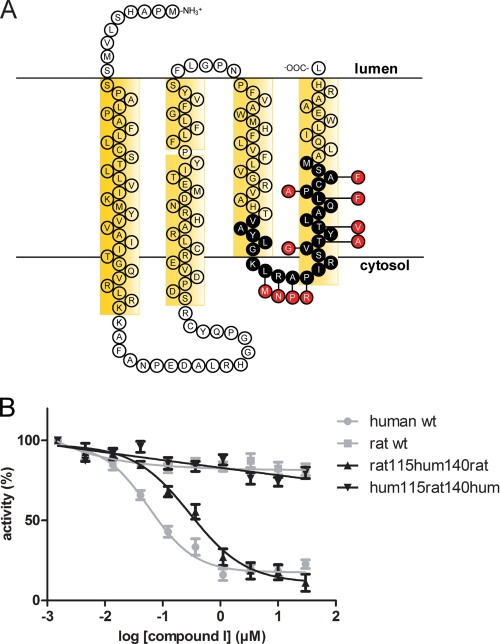
To investigate the species discrimination of compound I toward MPGES1, we first concentrated on the differences within TM4. Evidence from the crystal structure of MPGES1 shows that these residues are in close proximity to GSH and the predicted active site (7). Because compound I is competitive toward PGH2,4 we expected it to bind in this area. We therefore created a chimeric enzyme by replacing residues 115–140 from human MPGES1 with the respective residues from rat (Fig. 3A). Expression of this and all of the following chimeric and mutated proteins was verified by immunoblot analysis and their PGH2 converting activity. Furthermore, their general ability to be inhibited was confirmed with the species-indiscriminative inhibitor compound II (data not shown). The chimeric enzyme hum115rat140hum completely lost its ability to be inhibited by compound I (Fig. 3B). On the other hand, rat MPGES1 containing residues 115–140 from the human enzyme, chimera rat115hum140rat, gained inhibitory potential and was affected by compound I with an IC50 value of 300 nm (Fig. 3B and Table 1).
FIGURE 3.
Residues 115–140 are important for binding of compound I to MPGES1. A, parts of TM3 and TM4 were exchanged to the respective sequence of the other species, as indicated in the topology map of human MPGES1. Black circles represent exchanged residues; red circles represent the respective residues in rat MPGES1. B, the chimeric enzyme rat115hum140rat, containing the human residues 115–140, is inhibited by compound I in a similar way as the human WT MPGES1 (shown in gray). Exchange of the same residues in human MPGES1 toward the rat sequence, however, abolishes inhibitor action on chimeric enzyme hum115rat140hum.
Identification of Residues in TM4 Involved in Inhibitor Binding
The part of MPGES1 between residues 115 and 140, which was exchanged between human and rat MPGES1 in the chimeric proteins, contains 10 amino acids that differ between the two species: four residues situated in the loop connecting TM3 and TM4 and six residues within TM4 (Fig. 3A). Because exchange of this part confers inhibitor sensitivity to rat MPGES1, one of these 10 residues or a combination of several residues must play a crucial role in inhibitor binding. We went on to locate individual residues that are important for inhibitor binding by exchanging single amino acid residues in rat MPGES1 toward the corresponding residue in the human enzyme and tested their ability to be inhibited by compound I.
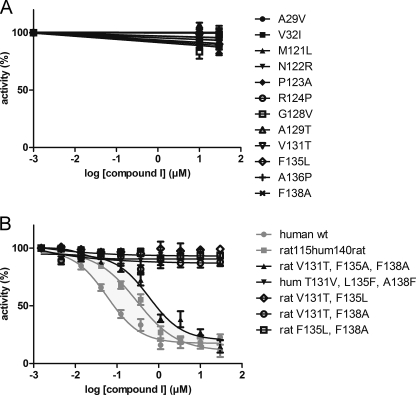
At the highest inhibitor concentration tested (50 μm), all single amino acid mutants retained their full activity (Fig. 4A). A combination of three amino acid substitutions in the triple mutant rat V131T,F135L,F138A, however, showed an effect on the inhibition profile. Like human WT MPGES1 or the chimeric protein rat115hum140rat, this mutant was inhibited by compound I, albeit with a higher IC50 value of 590 nm (Fig. 4B and Table 1). When human MPGES1 was mutated at these three positions toward the corresponding rat residues (triple mutant hum T131V,L135F,A138F), the enzyme became unresponsive to the inhibitor (Fig. 4B). Rat enzyme substituted at only two of these positions in different combinations, giving rise to the respective double mutants rat V131T,F135L, rat V131T,F138A, and rat F135L,F138A, resulted also in unresponsive enzyme (Fig. 4B).
FIGURE 4.
Only the triple mutant rat V131T,F135L,F138A is sensitive to inhibition by compound I. A, mutation of single amino acid residues within TM1, TM4, or the second cytosolic loop of rat MPGES1 toward the respective human residues has no effect on the binding of compound I. B, the triple mutant rat V131T,F135L,F138A is, however, inhibited by compound I in a similar way as the human WT MPGES1 or the chimeric protein rat 115hum140rat (shown in gray). If these residues are mutated in human MPGES1, on the other hand (triple mutant hum T131V,L135F,A138F), the enzyme becomes unresponsive to the inhibitor. Rat MPGES1 substituted at only two of these positions for the respective human residues (double mutants rat V131T,F135L, rat V131T,F138A, or rat F135L,F138A) remains unresponsive to compound I.
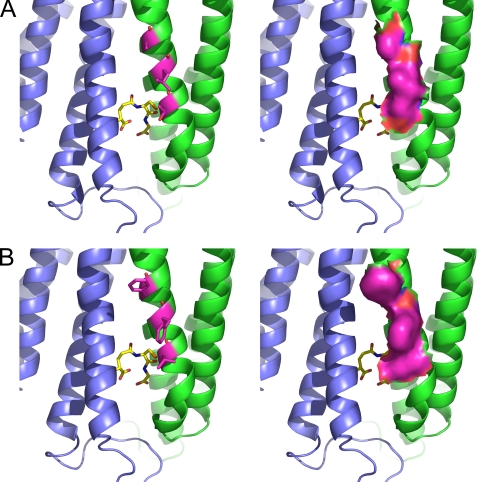
The protein structure reveals that these three residues are located at the same side of TM4, all being one turn of the helix apart from each other and pointing toward TM1 of the adjacent subunit (Fig. 5). They are thus lining the entrance of the crevice between TM4 and TM1 of the neighboring subunit, which is believed to be the putative substrate-binding site. Although these residues are relatively small in human MPGES1, they are mostly substituted for bulky aromatic residues in the rat enzyme. Apparently, the inhibitor is sterically hindered to access the active site of rat MPGES1 and thus fails to inhibit the enzyme.
FIGURE 5.
Residues 131, 135, and 138 occlude the entrance to the active site in rat MPGES1. Residues 131, 135, and 138 (represented with carbon atoms colored in magenta in the stick model on the left side and in space-filling model on the right side) are located in TM4, each one turn of the helix apart from the other, thus lining the crevice between TM4 and TM1 of an adjacent subunit. This crevice is believed to be the putative substrate-binding site. Although these residues are relatively small in human MPGES1, they are substituted for bulky aromatic residues in the rat enzyme. A, human MPGES1. B, rat MPGES1. Shown are two neighboring subunits in blue and green and one molecule of GSH as a stick model with carbon atoms colored in yellow. The top and bottom are the luminal side and the cytosolic side, respectively, of the ER membrane.
Interspecies Differences in TM1
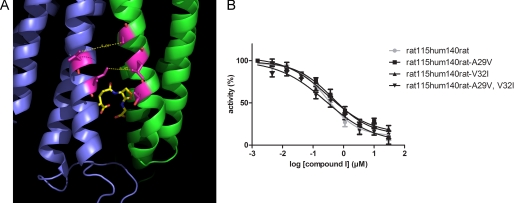
Both the chimeric enzyme hum115rat140hum and the rat-to-human triple mutant rat V131T,F135L,F138A were affected by compound I, however, with IC50 values that were 5 and 10 times higher compared with human WT MPGES1 (Table 1). We therefore investigated whether interspecies differences in TM1 have an additional effect on binding of compound I. Val-29 and Ile-32 are the only two residues in TM1 substituted between human and rat that are in the same region as the putative active site (the corresponding residues in rat MPGES1 are Ala-29 and Val-32). In the protein structure of MPGES1, they are lining the crevice between subunits in a similar way as residues Thr-131, Leu-135, and Ala-138 in TM4, with an estimated distance of 6.5 Å between Val-32 and Leu-135 and 8.6 Å between Ile-32 and Ala-138 (Fig. 6A).
FIGURE 6.
Residues Val-29 and Ile-32 in TM1 do not contribute to inhibitor binding. A, Val-29 and Ile-32 are lining TM1 at the entrance to the inhibitor binding cleft in the structure model of human MPGES1. Val-29 is directly opposing Leu-135, whereas Ile-32 is situated opposite to Ala-138. B, when these two residues were mutated in the chimeric protein rat115hum140rat, this did, however, not change the inhibitor binding characteristic of the enzyme.
Like the single amino acid residue mutations in TM4, mutation of rat WT MPGES1 at these positions toward the human residues did not convey inhibitor sensitivity to the enzyme (Fig. 4A). We therefore mutated the chimeric protein rat115hum140rat at these two positions, one residue at a time and both in combination. However, the resulting mutants displayed similar IC50 values as the nonmutated chimeric protein (Fig. 6B and Table 1). Hence, the amino acid differences between rat and human in TM1 do not play a crucial role for inhibitor binding of MPGES1.
DISCUSSION
Although inhibition of COX-2 is an effective way to shut down the production of induced pro-inflammatory PGE2 and thus prevent symptoms of inflammation like pain and fever, the use of COXIBs has been limited because of their adverse side effects on cardiovascular systems. By selectively targeting COX-2, which is expressed in vascular endothelial cells along with PGI2 synthase, COXIBs suppress production of anti-thrombotic PGI2 (as measured by its stable urinary metabolite 2, 3-dinor 6-keto PGF1α) to a similar degree as traditional nonselective NSAIDs. In contrast to these isoform unspecific NSAIDs, however, COXIBs do not affect levels of COX-1-derived thromboxane A2 and fail to inhibit platelet aggregation ex vivo. Thus, use of COXIBs removes a protective constraint on thrombogenesis, hypertension, and atherogenesis in vivo (13–15).
MPGES1 on the other hand is a terminal prostaglandin synthase and is not coupled to any downstream enzymes in the enzymatic cascade. Furthermore, constitutive levels of MPGES1 are normally low, and MPGES1 is highly up-regulated by pro-inflammatory stimuli. Therefore it is generally regarded as a target for pharmaceutical intervention of inflammatory conditions with less severe side effects (16).
In this study we used the MPGES1 inhibitor compound I to probe a rat/human chimeric enzyme as well as different mutants to gain insight in the active site of the enzyme. We particularly made use of two distinctive properties of the inhibitor that helped us to draw general conclusions. First, compound I discriminates between the human WT form of MPGES1 and orthologous rodent forms, like the rat WT enzyme. Changes in the amino acid sequence that occurred during evolution, as humans and rodents developed apart, must be regarded as the reason for this species discrimination of compound I. This makes it possible to identify specific residues that differ between the species and are crucial for inhibitor binding. Second, compound I acts as a competitive inhibitor toward the natural substrate PGH2 and thus binds to the corresponding location of the active site of MPGES1. Interspecies differences that account for inhibitor binding are therefore likely to be located within or in close proximity to the actual active site.
We found that exchange of residues 115–140 in the inhibitor-insensitive rat WT MPGES1 results in a chimeric enzyme that is inhibited by compound I with an IC50 similar to the one of human WT MPGES1. Exchange of the same residues 115–140 in human WT MPGES1 abrogated inhibitor binding. Furthermore, we identified three specific residues in TM4 that have a particularly important role. Substitution of rat WT MPGES1 at these three positions for the respective human residues was sufficient to gain inhibitory activity, albeit at a higher IC50 value, whereas mutation of these residues in human WT MPGES1 rendered the resulting triple mutant unresponsive toward the inhibitor.
Whereas the human enzyme contains small and aliphatic residues in all three positions, the corresponding residues in rat MPGES1 are aromatic and bulky in two of three positions. By their strategic positioning at the edge of TM4, pointing toward TM1 of the adjacent subunit in the catalytically active trimer, these aromatic residues occlude the active site and thus function as gate keepers that regulate access for the inhibitor tested in this study.
The sensitive rat enzyme mutants were clearly and almost completely inhibited by compound I. However, the IC50 values differed by factors of 2 (chimeric enzyme rat115hum140rat) and 10 (triple mutant rat V131T,F135L,F138A), respectively, from the IC50 value of human WT MPGES1. Other substitutions between rat and human MPGES1 remote from the active site thus also contribute to the differences in inhibitor potency, albeit less dramatically. It was suggested that MPGES1 alternates between an open and a closed conformation by dynamic flexibility of TM1, regulating access for PGH2, or a competitive inhibitor, respectively, to the active site (7). Clearly such a dynamic opening of the active site in the rodent enzyme does not allow inhibitor entrance because it is occluded by larger residues (residues 131, 135, and 138). The smaller residues in the human enzyme could allow more rapid dynamics, perhaps accounting for the higher activity observed.
The substitutions at positions 131, 135, and 138 occur interestingly only in rat and mouse MPGES1, but not in other rodent orthologues of known sequence (supplemental Fig. S2A). As a consequence of this, rat and mouse are unsuitable rodent models of inflammation when it comes to testing the effects of newly developed MPGES1 inhibitors. However, other rodents most likely represent alternatives because their MPGES1 protein is more closely related to the human enzyme (supplemental Fig. S2A), allowing MPGES1 inhibitors with species selectivity to bind. The phenanthrene imidazole MF63 possesses similar properties as our inhibitor compound I in that it inhibits human MPGES1 in the low nanomolar range but not the mouse or rat enzyme, indicating that this species discrimination is a general problem for MPGES1 inhibitors with high potency for the human orthologue. MF63 was shown to inhibit guinea pig MPGES1 with a similar IC50 value as the human enzyme and could be further used in guinea pig models of hyperalgesia, pyresis, and osteoarthritic pain (6). Structural analogues of MF63 showed an improved pharmacokinetic profile and superior in vitro and in vivo activities in a guinea pig hyperalgesia model when compared with MF63 (17). This demonstrates that guinea pigs might be a useful animal model in preclinical tests of MPGES1 inhibitors. In addition, guinea pigs have been used to study inflammatory diseases in several models of arthritis, including antigen-induced arthritis (18) and adjuvant-induced arthritis (19, 20). Although guinea pigs and humans share the same amino acid residues in positions 131, 135, and 138 of MPGES1, allowing for species discriminating MPGES1 inhibitors to bind, the overall identity of the two enzymes is only 79%. Thus, it is similar to the overall identity between the human enzyme and the enzyme of rat and mouse (77% and 78%, respectively). Because of the moderate overall identity between these species and man, other compounds might fail to inhibit the enzyme in a rodent model, depending on the class of molecule. Therefore, a knock-in mouse expressing human MPGES1 might be a preferable rodent animal model to test the anti-inflammatory properties of newly developed MPGES1 inhibitors (6).
Four members of the protein superfamily of membrane-associated proteins in eicosanoid and glutathione metabolism (MAPEG) have been structurally characterized and show the same overall fold and organization as homotrimers. In addition to the structures of MPGES1 (Protein Data Bank code 3DWW) (7) and its closest homologue microsomal glutathione transferase 1 (MGST1, Protein Data Bank code 2H8A) (21), which were both solved by electron crystallography, the x-ray structures of Leukotriene C4 Synthase (LTC4S, Protein Data Bank codes 2UUH, 2UUI, and 2PNO) (22, 23), and 5-Lipoxygenase-Activating Protein (FLAP, Protein Data Bank code 2Q7M) (24) are available.
The observation of a detergent molecule, bound between TM1 and TM4 of neighboring subunits in LTC4S, led to the suggestion that it mimics LTA4, the substrate of this enzyme, and thus indicates the active site region of LTC4S (22). This location between subunits of the homotrimer is in agreement with the location of the residues in MPGES1, which we identified in this study as being important for inhibitor binding and for occluding the active site in the rat enzyme. In fact, residue Trp-116 in LTC4S, which is proposed to play a key role in positioning the aliphatic chain of the substrate LTA4 by forming a lid over the ω-end (22), corresponds to residue Ala-138 in MPGES1. Furthermore, in a structure of LTC4S that was independently solved by a second group, residues Tyr-109 and Trp-116 (corresponding to Thr-131 and Ala-138, respectively, in MPGES1) but not Arg-113 (corresponding to Leu-135 in MPGES1) have been identified to compose part of the hydrophobic interior of the crevice to which the aliphatic chain of a detergent molecule was bound (23).
Additional supporting evidence in agreement with our findings can be obtained from the FLAP crystal structure. FLAP does not possess known catalytic activity and thus differs from all other MAPEG members in that it has no active site in an enzymological sense. However, it is essential for the activity of the enzyme 5-lipoxygenase, leading to generation of lipid mediators of the leukotriene family. It is suggested that FLAP provides arachidonic acid in a way that 5-lipoxygenase can use it as substrate. This activity can be blocked by FLAP inhibitors. Despite this difference from other MAPEG members, FLAP shares the structural characteristics of the MAPEG family. The structure of FLAP was solved in complex with the leukotriene biosynthesis inhibitors MK-591 and an iodinated analogue of MK-591 and is to date the only member of the protein family on which structural information of an inhibitor-binding site is available. Among the residues that were found to make contact with MK-591 are Ile-113 in the second cytosolic loop and Leu-120 in TM4 (24), which correspond to residues Thr-131 and Ala-138, respectively, in TM4 of MPGES1 (supplemental Fig. S3). Further residues in close proximity to the inhibitor include Lys-116, Ile-119, and Phe-123 of TM4 (corresponding to Glu-134, Cys-137, and Ala-141 in MPGES1) and Val-20, Val-21, Asp-23, Glu-24, Phe-25, and Ala-27 of TM1 (corresponding to Val-24, Ile-25, Met-27, Tyr-28, Val-29, and Ala-31, respectively, in MPGES1). The location of the active site and/or the inhibitor-binding site therefore seems to be highly conserved throughout the MAPEG superfamily.
Although evidence for the substrate and/or inhibitor-binding site in the protein structures of other MAPEG members fit very well with the findings in this work, there is no direct structural data on inhibitor binding in MPGES1 to date. However, two homology models of MPGES1 (25, 26) tried to predict this interaction. Because both models are based on the structure of rat MGST1, they employ the elongated conformation of GSH that was found for this enzyme, instead of the U-shaped conformation as determined for human MPGES1. Xing et al. (26) located the endoperoxide moiety of PGH2 close to the glycine carboxylate of GSH. Hamza et al. (25), on the other hand, made the assumption that the highly conserved residue Arg-110 participates in the reaction and hence located the substrate/inhibitor-binding site close to this residue. Thus, both homology models deviate to a certain degree from the experimentally determined structure of MPGES1, which has a direct impact on the prediction of the substrate/inhibitor-binding site. Therefore, the residues we identified in this work to be important for inhibitor binding were not suggested in either of these two homology models.
In conclusion, we have identified crucial residues in TM4 of MPGES1 that function as gate keepers and regulate access to the active site of the enzyme for the inhibitor compound I, coming from the lipid bilayer of the membrane. Differences in these positions in rat and mouse MPGES1 but not in other rodent orthologues to the human enzyme explain the selective inhibition pattern of compound I. Furthermore, the corresponding residues in FLAP and LTC4S have been determined to bind to an inhibitor that was cocrystallized, or to a substrate surrogate, respectively, and hence support a high degree of conservation of the substrate/inhibitor-binding site within the MAPEG superfamily.
Supplementary Material
This work was supported by funds from the Swedish Research Council, Karolinska Institutet, The Swedish County Council, The Swedish Rheumatism Association, the King Gustaf V 80-Year Fund, the Marianne and Marcus Wallenberg Foundation, and the Börje Dahlin Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
L. Spahiu, unpublished results.
- NSAID
- nonsteroidal anti-inflammatory drug
- COXIB
- COX-2 specific inhibitor
- MPGES1
- microsomal prostaglandin E synthase-1
- PG
- prostaglandin
- COX
- cyclooxygenase
- TM
- transmembrane helix
- MDA
- malondialdehyde
- MAPEG
- membrane-associated proteins in eicosanoid and glutathione metabolism
- LTC4S
- leukotriene C4 synthase
- FLAP
- 5-lipoxygenase-activating protein
- TBA
- 2-thiobarbituric acid.
REFERENCES
- 1.Jania L. A., Chandrasekharan S., Backlund M. G., Foley N. A., Snouwaert J., Wang I. M., Clark P., Audoly L. P., Koller B. H. (2009) Prostaglandins Other Lipid Mediat. 88, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lovgren A. K., Kovarova M., Koller B. H. (2007) Mol. Cell Biol. 27, 4416–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trebino C. E., Stock J. L., Gibbons C. P., Naiman B. M., Wachtmann T. S., Umland J. P., Pandher K., Lapointe J. M., Saha S., Roach M. L., Carter D., Thomas N. A., Durtschi B. A., McNeish J. D., Hambor J. E., Jakobsson P. J., Carty T. J., Perez J. R., Audoly L. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9044–9049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engblom D., Saha S., Engström L., Westman M., Audoly L. P., Jakobsson P. J., Blomqvist A. (2003) Nat. Neurosci. 6, 1137–1138 [DOI] [PubMed] [Google Scholar]
- 5.Wang M., Zukas A. M., Hui Y., Ricciotti E., Puré E., FitzGerald G. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14507–14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu D., Rowland S. E., Clark P., Giroux A., Côté B., Guiral S., Salem M., Ducharme Y., Friesen R. W., Méthot N., Mancini J., Audoly L., Riendeau D. (2008) J. Pharmacol. Exp. Ther. 326, 754–763 [DOI] [PubMed] [Google Scholar]
- 7.Jegerschöld C., Pawelzik S. C., Purhonen P., Bhakat P., Gheorghe K. R., Gyobu N., Mitsuoka K., Morgenstern R., Jakobsson P. J., Hebert H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11110–11115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westman M., Korotkova M., af Klint E., Stark A., Audoly L. P., Klareskog L., Ulfgren A. K., Jakobsson P. J. (2004) Arthritis Rheum. 50, 1774–1780 [DOI] [PubMed] [Google Scholar]
- 9.Jakobsson P. J., Thorén S., Morgenstern R., Samuelsson B. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7220–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basevich V. V., Mevkh A. T., Varfolomeev S. D. (1983) Bioorg. Khim. 9, 658–665 [PubMed] [Google Scholar]
- 11.Hamberg M., Samuelsson B. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 3400–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleishman S. J., Unger V. M., Ben-Tal N. (2006) Trends Biochem. Sci. 31, 106–113 [DOI] [PubMed] [Google Scholar]
- 13.Grosser T., Fries S., FitzGerald G. A. (2006) J. Clin. Invest. 116, 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAdam B. F., Catella-Lawson F., Mardini I. A., Kapoor S., Lawson J. A., FitzGerald G. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 272–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patrignani P., Sciulli M. G., Manarini S., Santini G., Cerletti C., Evangelista V. (1999) J. Physiol. Pharmacol. 50, 661–667 [PubMed] [Google Scholar]
- 16.Samuelsson B., Morgenstern R., Jakobsson P. J. (2007) Pharmacol. Rev. 59, 207–224 [DOI] [PubMed] [Google Scholar]
- 17.Giroux A., Boulet L., Brideau C., Chau A., Claveau D., Côté B., Ethier D., Frenette R., Gagnon M., Guay J., Guiral S., Mancini J., Martins E., Massé F., Méthot N., Riendeau D., Rubin J., Xu D., Yu H., Ducharme Y., Friesen R. W. (2009) Bioorg. Med. Chem. Lett. 19, 5837–5841 [DOI] [PubMed] [Google Scholar]
- 18.Kuwabara K., Jyoyama H., Fleisch J. H., Hori Y. (2002) Inflamm. Res. 51, 541–550 [DOI] [PubMed] [Google Scholar]
- 19.Clark P., Rowland S. E., Denis D., Mathieu M. C., Stocco R., Poirier H., Burch J., Han Y., Audoly L., Therien A. G., Xu D. (2008) J. Pharmacol. Exp. Ther. 325, 425–434 [DOI] [PubMed] [Google Scholar]
- 20.Vermeirsch H., Biermans R., Salmon P. L., Meert T. F. (2007) Pharmacol. Biochem. Behav. 87, 349–359 [DOI] [PubMed] [Google Scholar]
- 21.Holm P. J., Bhakat P., Jegerschöld C., Gyobu N., Mitsuoka K., Fujiyoshi Y., Morgenstern R., Hebert H. (2006) J. Mol. Biol. 360, 934–945 [DOI] [PubMed] [Google Scholar]
- 22.Martinez Molina D., Wetterholm A., Kohl A., McCarthy A. A., Niegowski D., Ohlson E., Hammarberg T., Eshaghi S., Haeggström J. Z., Nordlund P. (2007) Nature 448, 613–616 [DOI] [PubMed] [Google Scholar]
- 23.Ago H., Kanaoka Y., Irikura D., Lam B. K., Shimamura T., Austen K. F., Miyano M. (2007) Nature 448, 609–612 [DOI] [PubMed] [Google Scholar]
- 24.Ferguson A. D., McKeever B. M., Xu S., Wisniewski D., Miller D. K., Yamin T. T., Spencer R. H., Chu L., Ujjainwalla F., Cunningham B. R., Evans J. F., Becker J. W. (2007) Science 317, 510–512 [DOI] [PubMed] [Google Scholar]
- 25.Hamza A., Abdulhameed M. D., Zhan C. G. (2008) J. Phys. Chem. B 112, 7320–7329 [DOI] [PubMed] [Google Scholar]
- 26.Xing L., Kurumbail R. G., Frazier R. B., Davies M. S., Fujiwara H., Weinberg R. A., Gierse J. K., Caspers N., Carter J. S., McDonald J. J., Moore W. M., Vazquez M. L. (2009) J. Comput. Aided Mol. Des. 23, 13–24 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.