Abstract
Two-pore domain potassium (K2P) channels modulate neuronal excitability throughout the entire CNS. The stretch-activated channel TREK1 (K2P2.1) is expressed widely in brain and has been linked to depression, neuroprotection, pain perception, and epilepsy. Little, however, is known about the regulation of TREK1 expression on the transcriptional and translational level or about its trafficking to the plasma membrane. Here we have used PCR techniques to identify a splice variant of TREK1 expressed in the brain, which encodes a heavily truncated TREK1 protein retaining a single transmembrane domain. Functional expression of this splice variant TREK1ΔEx4 in tsA201 cells in the presence or absence of wild type TREK1 revealed that TREK1ΔEx4 has no channel activity itself but reduced TREK1 whole cell current amplitude. Confocal analysis of the expression of fluorescently tagged TREK1 variants revealed that TREK1ΔEx4 is translated, but it is retained in the intracellular compartment. Additionally, TREK1ΔEx4 reduced the level of TREK1 expression in the plasma membrane. Long and short forms of TREK1 derived from alternative translation initiation are differentially affected by TREK1ΔEx4, with the short form (lacking the first 41 amino acids at its N terminus) unaffected. This differential regulatory role of TREK1ΔEx4 will alter the functional profile of TREK1 current in neurons where they are expressed. These results indicate that the N-terminal domain and first transmembrane domain of TREK1 are likely to be important for channel dimerization and trafficking to the plasma membrane.
Keywords: Brain, Fluorescence, Gene Expression, Ion Channels, Potassium Channels, Protein-Protein Interactions, Trafficking, K2P Channel, KCNK Channel, Splice Variant
Introduction
Modulation of the activity of 2-pore-domain potassium (K2P) channels is an important means of regulating neuronal excitability (1, 2). The majority of the K2P channels (3) that are encoded by the KCNK genes have distinct expression patterns within the CNS (4–6). Activity of these “leak” potassium channels is highly regulated by physical parameters such as heat, stretch, or pH as well as by lipids, G-protein-coupled receptors, and anesthetics (1, 7–11). Particularly, members of the TASK and TREK channel subfamilies have been implicated in such diverse processes as anesthetic sensitivity, thermosensitivity, neuroprotection, pain perception, and peripheral chemosensitivity (12–19).
K2P2.1, or TREK1, is widely expressed in the brain (4–6), and genetic ablation has revealed involvement of this channel in depression, neuroprotection, pain perception, and ischemia-precipitated epilepsy (3, 14, 15, 20–22). In rodents, TREK1 is prominently expressed not only in the brain but also in the lung, whereas expression levels in the heart and kidney are much lower (23). Functionally, this channel is characterized by mechanosensitivity, heat sensitivity, G-protein regulation, and activation by lipids, arachidonic acid, and volatile anesthetics (11, 13, 24, 25).
Although TREK1 is probably the most studied two-pore domain potassium channel to date (21), there is still little known about the regulation of its functional expression at a transcriptional, translational, and post-translational level. A few recent studies have addressed its interaction with AKAP150 (the A kinase anchoring protein 150) or the microtubule-associated protein Mtap2 and the consequences for trafficking and targeting to specific regions of the cell membrane (26, 27). Furthermore, both TREK1 and TREK2 have recently been shown to be the subject of alternative translation initiation (ATI),3 which produces channels differing in the length of their intracellular N-terminal domain and in their functional properties (28, 29). These studies highlighted the importance of the N-terminal part of the channel for ion selectivity and single channel conductance. In contrast, two TREK1 splice variants of exon 1 isolated from rat cardiac muscle tissue, which differed by 15 amino acid residues in length of the N terminus, showed identical biophysical properties (30).
In the course of using PCR techniques to identify TREK-1 mRNA in specific brain regions, we detected a further splice variant within the first pore loop of TREK1, which leads to a predicted protein that is truncated before the second transmembrane domain. We demonstrate in tsA201 cells that this mRNA is not degraded (31) but translated into a peptide that down-regulates functional expression of TREK1 channels.
EXPERIMENTAL PROCEDURES
Molecular Biology
For mRNA isolation, adult male C57bl6 mice and 18–30-day-old Sprague-Dawley rats of either sex were sacrificed in accordance with the Animals (Scientific Procedures) Act 1986. Briefly, animals were anesthetized with halothane and decapitated, and the brain, lungs, heart, pancreas, and liver were rapidly removed and frozen on liquid nitrogen. The frozen tissue was homogenized, and mRNA was isolated using a kit (Micro Fast Track, Qiagen). First strand cDNA was synthesized for 1 h at 37 °C in reverse transcription buffer containing 5 μm random hexamer primers, 20 mm dithiothreitol, 0.5 mm (each) dNTPs, 20 units of ribonuclease inhibitor (Promega), and 100 units of Superscript II reverse transcriptase (Invitrogen). PCR amplification (for primers, see Table 1) was carried out using a hot start protocol with Taq polymerase in a PerkinElmer Life Sciences Thermal Cycler 9700 (30 cycles: 30 s at 94 °C, 1 min at 58 °C, 2 min at 72 °C). PCR samples were analyzed by agarose gel electrophoresis and visualized using ethidium bromide. The PCR products were verified by direct sequencing (Eurofins MWG Operon, Ebersberg, Germany). Exons were numbered according to the reference sequences for mice (NM_001159850.1) and humans (NM_014217.3). This numbering includes an exon 1 before the start of the coding sequence. Previous studies (e.g. Ref. 30) had exon 1 as the exon that includes the start codon. Consequently, the splice variant TREK1ΔEx4 described here would have been named TREK1ΔEx3 under the alternative nomenclature.
TABLE 1.
PCR primers used to amplify TREK1 fragments from rat and mouse cDNA
The forward primer was designed to bind within exon 3, and the reverse primer was designed to bind in exon 5′.
| Forward primer | Reverse primer | Product size | |
|---|---|---|---|
| bp | |||
| Mouse TREK1 | GTCCTCTACCTGATCATCGGAGC | CCTAGCTGATCACCAACCCC | 383 |
| Rat TREK1 | GTCCTCTACCTGATCATCGGAGC | CCACAAAGTAGATGGCGTCCAGG | 574 |
Mutagenesis
To generate a TREK1 cDNA lacking either exon 4 or its first 41 amino acids, a deletion mutagenesis strategy was utilized. A pair of complementary oligonucleotide primers, 30–40 bp long, composed of 15–20 bp of complementary sequence prior to the region being deleted and 15–20 bp of complementary sequence after the deletion region, were synthesized (Eurofins MWG Operon). PCR was then performed using a QuikChange site-directed mutagenesis kit and procedure (Stratagene, La Jolla, CA).
Wild type TREK1 was cloned into the MCS of pAcGFP1-N1 vector (Clontech-Takara Bio Europe) to create a fusion construct with the N terminus of AcGFP1. The terminating stop codon of TREK1 was removed by site-directed mutagenesis and maintained in the same reading frame with the start codon of AcGFP1.
TREK1ΔEx4 was cloned into the MCS of pDsRed2-N1 vector (Clontech-Takara Bio Europe) to create a fusion construct with the N terminus of DsRed2. Site-directed mutagenesis was used to delete all of the TREK1 sequence from and including the stop codon at position 142, leaving 105 amino acids of homologous TREK1 sequence and the 37-amino acid frame-shifted tail in frame with the DsRed2 protein.
pDsRed-Monomer-Mem vector (Clontech-Takara Bio Europe) encodes a fusion protein consisting of the DsRed monomer red fluorescent protein and the N-terminal 20 amino acids of neuromodulin (or Gap 43). This fragment of neuromodulin contains a signal for post-translational palmitoylation of cysteines 3 and 4, which targets DsRed monomer to cellular membranes and the plasma membrane in particular. Point mutations were produced using a site-directed mutagenesis strategy (Stratagene). All constructs were fully sequenced (Eurofins MWG Operon) to ensure correct sequence incorporation, deletion, and mutation.
Tissue Culture
Modified HEK293 cells (tsA201) were maintained in growth medium (minimum essential medium enriched with 10 ml/500 ml penicillin/streptomycin solution, 5 ml/500 ml MEM nonessential amino acids, 50 ml/500 ml fetal bovine serum, and 2 mm l-glutamine) in 5% CO2, 95% O2 in a humidified incubator at 37 °C. When the cells were 80% confluent, they were plated onto glass coverslips coated with poly-d-lysine (1 mg/ml). Transient transfection was carried out using the calcium phosphate method (32, 33). In brief, 0.2–0.5 μg of cDNA encoding wild type or mutant human TREK1 (GenBankTM AF171068) (34) (isoform b, the shortest N-terminal human transcript, same start codon position as rodent) or TASK3 (GenBankTM Q9NPC2-1) (35) or mouse Kir6.2 (GenBankTM D50581) (36, 37) and 0.2 μg of cDNA encoding for green fluorescent protein (GFP) were added to each well. Cells were then incubated at 3% CO2, 97% O2 for 12–18 h. Subsequently, cells were washed twice with PBS and incubated in growth medium in 5% CO2, 95% O2 in a humidified incubator at 37 °C for 24–96 h until used for electrophysiological recordings or confocal microscopy.
Confocal Microscopy
Cells were fixed with 4% paraformaldehyde and then washed with PBS before application of 5 μg/ml Hoechst 33258 nuclear dye (Invitrogen). Coverslips were rinsed with PBS and mounted onto slides using Mowiol mounting medium with 0.05% p-Phenylenediamine. Images were taken with a Leica TCS SP2 confocal microscope and processed using Image J software. Co-expression of transcripts was verified by randomly selecting >50 cells that expressed one transcript (e.g. GFP) and then evaluating what percentage of these expressed the other transcript (e.g. DsRed). This revealed co-expression in >95% of successfully transfected cells. Fluorescence intensity was read as an 8-bit value (from 0 to 255) for each pixel as obtained from the camera. Membrane colocalization was quantified via Pearson's correlation coefficient, determined using Image J WCIF software and the colocalization plug-in (colocalization test plug-in, Tony Collins and Wayne Rasband). Pearson's colocalization coefficients were calculated for five membrane sections per cell and displayed as the average of five cells ± S.E.
Electrophysiology
For electrophysiological recordings, a coverslip with transfected tsA201 cells was transferred into a recording chamber mounted under an inverted microscope (Nikon Diaphot) with epifluorescence. During the course of the experiment, cells were constantly superfused at 3–5 ml/min with extracellular solution of the following composition: 145 mm NaCl, 2.5 mm KCl, 3 mm MgCl2, 1 mm CaCl2, 10 mm HEPES (pH adjusted to 7.4 with NaOH). Only cells that were successfully transfected with GFP as evident from green fluorescence (excitation, 395–440 nm; emission, 470–600 nm) were selected for electrophysiological recordings.
Patch pipettes were pulled from thin walled borosilicate glass (GC150TF, Harvard Apparatus, Edenbridge, UK) and had resistances of 3–5 megaohms when filled with pipette solution. The pipette solution contained 150 mm KCl, 3 mm MgCl2, 5 mm EGTA, 10 mm HEPES (pH adjusted to 7.4 with KOH). Whole cell currents were recorded at a holding potential of −60 mV at 20–24 °C. Cells were hyperpolarized to −80 mV for 100 ms and then subjected to a step to −40 mV for 500 ms, followed by a step to −120 mV for 100 ms, followed by a 500-ms voltage ramp to +20 mV and a step back to −80 mV for another 100 ms before being returned to the holding potential of −60 mV. This protocol was repeated every 5 s. Currents were recorded using an Axopatch 1D patch clamp amplifier (Molecular Devices, Sunnyvale, CA), filtered at 0.3 kHz, digitized at 1 kHz, and analyzed using Clampfit software (Molecular Devices) running on a personal computer. Whole cell capacitance was determined for every recording, and current densities were calculated.
Statistical Analysis
Statistical analysis was performed using Origin7.5 (OriginLab). All mean data are expressed as mean current density ± S.E. To test for differences between groups, two-tailed unpaired t test or one-way analysis of variance followed by Bonferroni's multiple comparison test were used as appropriate.
RESULTS
Identification of TREK1ΔEx4 Splice Variant by PCR
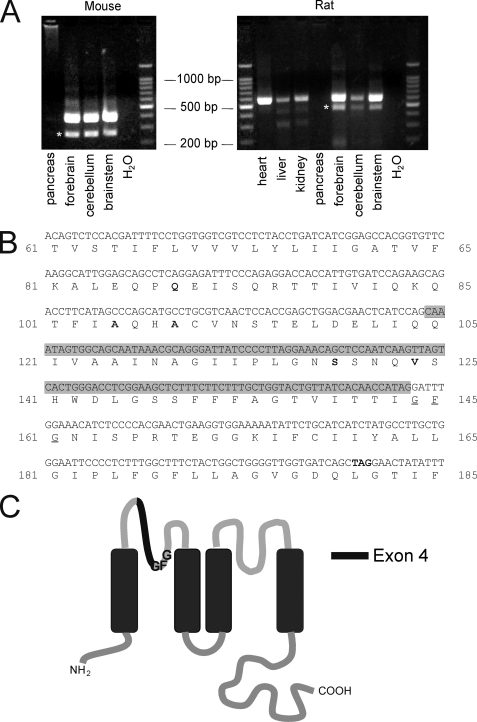
PCR analysis of TREK1 mRNA expression in rats and mice using primers that anneal in exon 3 and exon 5 revealed a prominent additional product of shorter length in brain tissue. Direct sequencing of both the long and short product revealed that the long product was the expected TREK1 PCR product, whereas the shorter product lacked exon 4 of TREK1. Interestingly, in other organs (heart, liver, and kidney), the short product was either absent or produced at a much lower level. cDNA from pancreas was negative for TREK1 (Fig. 1A). In mice, exon 4 of TREK1 encodes a portion of the extracellular/pore loop between transmembrane domains 1 and 2. It ends within the codon for the first “G” of the “GFG” signature sequence for two-pore domain potassium channels (Fig. 1, B and C). Deletion of exon 4 (gray box) will not only destroy this signature sequence but also cause a frameshift, which generates a premature stop codon after 37 amino acid residues of exon 5 (Fig. 2A). Consequently, translation of TREK1ΔEx4 mRNA is expected to yield a protein with one transmembrane domain that is identical to the first 104 amino acids of TREK1, followed by a tail of 37 amino acids with a novel sequence due to the frameshift caused by deletion of exon 4. This protein is not expected to form functional channels, but the seemingly robust expression of TREK1ΔEx4 in rodent brain suggested to us that it might have some functional role. Consequently, this splice variant was mimicked by removing the sequence corresponding to exon 4 from the cDNA encoding human TREK1, and the mutant plasmid was transfected into tsA201 cells for electrophysiological analysis.
FIGURE 1.
Expression pattern of TREK1ΔEx4. A, RT-PCR analysis of TREK1 (K2P2.1, KCNK2) expression in mice and rats. In mice, a 383 bp band corresponds to TREK1, and a 265 bp band corresponds to TREK1ΔEx4(*). In rats, a 574 bp band signifies TREK1, and a 456 bp band indicates TREK1ΔEx4(*). TREK1 was amplified from the brain, heart, liver, and kidney but not from the pancreas. TREK1ΔEx4 was more prominent in the brain than in the heart, liver, and kidney, where it was seen only as a spurious product. B, nucleotide and amino acid sequence of mouse TREK1 (GenBankTM U73488) in the vicinity of exon 4. Nucleotides contributing to exon 4 are shown in a gray box. Note that exon 4 ends with the first nucleotide of the triplet encoding for the first glutamine of the “GFG” potassium channel signature sequence. The loss of exon 4 leads to a frameshift and a premature stop codon (TAG) after a further 37 amino acids. This suggests that TREK1ΔEx4 encodes a heavily truncated protein consisting of only one TM and an extracellular tail. Amino acid residues that are different in the human sequence are indicated in boldface type. Numbering according to the human sequence is shown on the right (GenBankTM AF171068). C, membrane topology of TREK1. The position of exon 4 between TM1 and the pore sequence GFG is indicated in black.
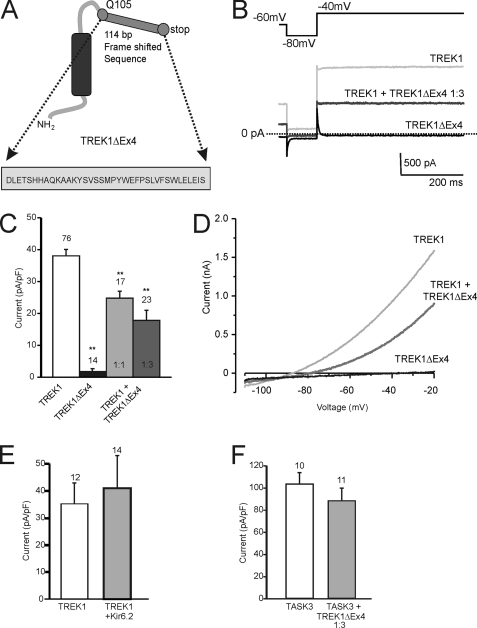
FIGURE 2.
TREK1Δ Ex4 produces no functional current and reduces TREK1 currents. A, topology of TREK1ΔEx4. This variant consists of a homologous N terminus, TM1, and small part of the M1P1 loop up to Glu105 (Q105), followed by 114 bp of frameshifted sequence, which encodes for 37 amino acid residues (see inset) and a stop codon. In total, TREK1ΔEx4 is composed of 675 bp of coding sequence. B, representative traces from TREK1- and TREK1ΔEx4-transfected tsA201 cells with the voltage protocol employed in this study. C, mean current densities for TREK1 and TREK1ΔEx4 and co-expression of TREK1 and TREK1ΔEx4 at the ratios indicated, calculated from the difference in holding current between −80 and −40 mV. TREK1 current density was significantly reduced in a concentration-dependent manner, when co-expressed with TREK1ΔEx4. D, current-voltage relationships for TREK1, TREK1 co-expressed (1:3 ratio) with TREK1ΔEx4, and TREK1ΔEx4 alone. E, co-expression of the inward rectifier Kir6.2 with TREK1 does not reduce the TREK1 current density. Note that Kir6.2 does not form functional K+ channels in the absence of its β-subunit SUR. F, mean current density of TASK3 (K2P3.2, KCNK9) was not significantly affected by co-transfection with TREK1ΔEx4 at a 1:3 ratio. Numbers (n) are given above the bars. *, p < 0.05; **, p < 0.01. Error bars, S.E.
Effects of TREK1ΔEx4 on TREK1 Currents
Whole cell recordings from tsA201 cells transfected with wild type TREK1 cDNA under normal conditions gave current densities of 38 ± 2 pA/pF (n = 76) (measured as the difference in holding current between that at −40 mV and that at −80 mV; Fig. 2, B and C). Ramp changes in holding potential between −110 and −20 mV confirmed that the whole cell current was outwardly rectifying and had a tendency to run down with time. This current had a reversal potential of −83 ± 1 mV (n = 70), close to the equilibrium potential for potassium ions under these recording conditions (Fig. 2D).
In contrast, the TREK1 splice variant TREK1ΔEx4, with the whole of exon 4 missing, was found to be non-functional when expressed alone in tsA201 cells. The average current density amounted to 2 ± 1 pA/pF (n = 14) and a reversal potential of −37 ± 3 mV in 2.5 mm potassium (Fig. 2, A–D). The current recorded from TREK1ΔEx4 alone is indistinguishable from the current recorded from untransfected tsA201 cells (data not shown).
Co-expression of wild type TREK1 and TREK1ΔEx4 somewhat surprisingly gave reduced (by 34%) whole cell currents of 25 ± 2 pA/pF (n = 17) when expressed in a 1:1 ratio. A further decrease (to 53% of wild type TREK1 expressed alone) in current (18 ± 3 pA/pF, n = 23) was seen when wild type TREK1 and TREK1ΔEx4 were expressed in a 1:3 ratio (Fig. 2, B and C). These results suggested that TREK1ΔEx4 might have some dominant-negative effect on TREK1 expression.
We also wanted to ensure that the dominant negative effect of TREK1ΔEx4 on TREK1 in tsA201 cells was not caused by the overexpression of a second non-functional potassium channel. To do this, we made use of the properties of the inward rectifier potassium channel Kir6.2. Like TREK1ΔEx4, Kir6.2 does not form functional channels when expressed alone. The protein is made but does not reach the plasma membrane in the absence of its regulatory subunit SUR1 (38). Here we demonstrate that, unlike TREK1ΔEx4, the presence of Kir6.2 does not affect the functional expression of TREK1 currents. TREK1 expressed on its own exhibited a current density of 35 ± 7 pA/pF (n = 12), and in the presence of Kir6.2, the current density for the outwardly rectifying TREK1 current amounted to 41 ± 12 pA/pF (n = 14; Fig. 2E). This difference was not significant and demonstrates that the expression of an unrelated protein that is unlikely to interact with TREK1 has no effect on the functional expression of TREK1. Second, to determine whether TREK1ΔEx4 affected K2P channels from other subfamilies, we co-expressed it with wild type TASK3 channels. Measurement of current through TASK3 channels resulted in current densities of 104 ± 11 pA/pF (n = 10) with a reversal potential of −89 ± 1 mV. Measurement of current from cells containing wild type TASK3 channels, co-expressed with TREK1ΔEx4 in a 1:1 ratio, resulted in current amplitudes of 89 ± 10 pA/pF (n = 11) with a reversal potential of −83 ± 2 mV, demonstrating no significant change in current amplitude compared with wild type TASK3 channels alone (Fig. 2F). This agrees with previous data indicating that K2P channels from other subfamilies are unable to heterodimerize across subfamilies, and consequently, we would postulate that TREK1ΔEx4 interacts specifically with TREK channels (39–43).
Analysis of Cellular Localization of TREK1 and TREK1ΔEx4
Next we explored further the nature of the dominant negative effect of TREK1ΔEx4 upon TREK1. In order to determine whether TREK1ΔEx4 is translated and whether it is trafficked to the plasma membrane, GFP was fused to the C terminus of TREK1. Similarly, DsRed2 was fused to the C terminus of TREK1ΔEx4. C-terminal insertion of GFP did not interfere with TREK1 channel function. TREK1-GFP expressed large outwardly rectifying potassium currents (data not shown).
Cellular localization of TREK1-GFP and TREK1ΔEx4-DsRed2 was examined under a confocal microscope. GFP fluorescence was observed both at the surface membrane and internally in tsA201 cells transfected with TREK1-GFP (Fig 3A, i). First, mean whole cell fluorescence intensity from TREK1-GFP was determined for 14 cells without and 10 cells with co-expression of TREK1ΔEx4. The values of 92.29 ± 10.73 and 83.44 ± 7.97, respectively, were not statistically significantly different (p = 0.545). These data suggested that the reduction in TREK1 whole cell current observed in the presence of TREK1ΔEx4 was not due to reduced translation of the protein.
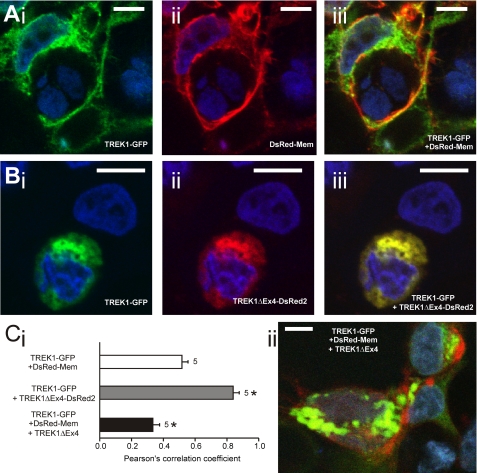
FIGURE 3.
Evaluation of cellular localization of fluorescently labeled TREK1 variants. A, photomicrographs showing cellular localization of TREK1 tagged with GFP (TREK1-GFP) (i) relative to the location of the membrane label DsRed-Mem (ii), determined by confocal microscopy. Nuclei were stained with the blue fluorescent dye Hoechst 33258. Overlay (iii) indicates strong co-localization of TREK1-GFP and DsRed-Mem in yellow. B, co-localization of TREK1-GFP and TREK1ΔEx4 tagged with DsRed2 (TREK1ΔEx4-DsRed2) in individual tsA201 cells. Cells were transfected with both constructs in a 1:1 ratio. Note the strong co-localization indicated by yellow fluorescence (iii). C, i, quantification of the co-localization observed in experiments as shown in A and B. A correlation coefficient of 1 indicates complete overlap of fluorescence, whereas a coefficient of −1 indicates no co-localization at all. C, ii, example of reduced co-localization between TREK1-GFP and DsRed-Mem in the presence of unlabeled TREK1ΔEx4 (1:1 ratio with TREK1-GFP). White scale bar, 10 μm. *, p < 0.05 as compared with TREK1-GFP + DsRed-Mem. Error bars, S.E.
Expression in the membrane was confirmed by coexpression with a fusion protein consisting of the DsRed red fluorescent protein monomer and the N-terminal 20 amino acids of neuromodulin (DsRed-Mem), which is specifically targeted to membranes (Fig. 3A, ii). Fig. 3A, iii, demonstrates visually the overlap in localization between TREK1-GFP and DsRed-Mem. Similarly, Fig. 3B shows that TREK1 and TREK1ΔEx4 are highly co-localized when co-expressed, but expression of TREK1 in the plasma membrane is greatly reduced. Co-localization was quantified using Pearson's correlation (Fig 3C, i). The highest level of co-localization was found between TREK1 and TREK1ΔEx4, consistent with the idea that these two interact. A good correlation was detected between TREK1 and DsRed-Mem, indicating that TREK1 is targeted to the plasma membrane. This correlation was significantly (p < 0.05) reduced by co-expression with TREK1ΔEx4, consistent with the hypothesis that TREK1ΔEx4 subdues targeting of TREK1 to the plasma membrane. This is illustrated in Fig 3C, ii, which shows reduced co-localization of TREK1-GFP and DsRed-Mem in the presence of unlabeled TREK1ΔEx4 (reduced levels of yellow at the plasma membrane). In contrast, when mean whole cell TREK1-GFP fluorescence was compared with mean whole cell DsRed-Mem fluorescence, the ratio was 1.00 ± 0.07 and 1.14 ± 0.09 in the absence and presence of TREK1ΔEx4, respectively, and thus remained unchanged (p = 0.222). Again, this result indicated that TREK1ΔEx4 did not interfere with translation of TREK1.
These experiments confirmed that the TREK1ΔEx4 protein is made but that its trafficking to the plasma membrane is impaired. TREK1 cannot act as a chaperone, but the reduction in TREK1 whole cell currents suggests that TREK1 is likely to be retained in the intracellular compartment by interaction with TREK1ΔEx4.
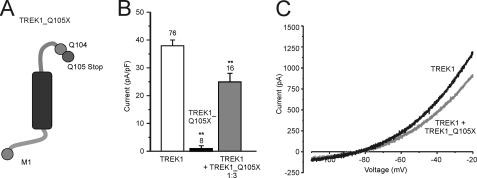
Truncated TREK1_Q105X Mimics TREK1ΔEx4
The TREK1ΔEx4 protein is identical to TREK1 up to residue Gln104, which marks the end of exon 3. After this position, TREK1ΔEx4 has a novel non-homologous sequence of 37 residues. In order to determine whether this novel tail contributes to the properties of TREK1ΔEx4, a straight truncation was produced at the end of exon 3 (TREK1_Q105X; Fig. 4A). This truncated protein was expressed in tsA201 cells either by itself or in a 3-fold excess together with TREK1. Analysis of whole cell currents from these cells revealed that TREK1_Q105X expressed by itself is non-functional (2 ± 1 pA/pF; reversal potential −37 ± 3 mV; n = 14) and that it is capable of significantly reducing TREK1 currents by 34% when co-expressed in a 3:1 ratio with TREK1 (Fig. 4, B and C). These results indicate that TREK1_Q105X interferes with TREK1 expression in a way similar to TREK1ΔEx4 and, thus, that the novel tail of TREK1ΔEx4 plays only a minor role in its function.
FIGURE 4.
Truncation of TREK1 at glutamine 105 replicates dominant negative effect of TREK1ΔEx4 on TREK1. A, topology of TREK1_Q105X, an N-terminal fragment of TREK1 with a stop codon inserted at glutamine 105 (Q105X). It differs in structure from TREK1ΔEx4 by lacking the additional 37 amino acids of frameshifted sequence following from the glutamine at position 104 (see also Fig. 2). B, current density of TREK1, TREK1_Q105X, and TREK1 co-expressed (1:3 ratio) with TREK1_Q105X. TREK1 whole cell current was significantly reduced when co-expressed with TREK1_Q105X. C, current-voltage relationships for TREK1 and TREK1 co-expressed with TREK1_Q105X. Numbers (n) are given above the bars. *, p < 0.05; **, p < 0.01. Error bars, S.E.
Effects of Alternative Translation Initiation
TREK1 channels are subject to ATI. The human splice variant used in this study has an alternative translation initiation codon 42 amino acids upstream from the first methionine (Fig. 5A). A study exploring the properties of such an N-terminal truncated TREK1 variant found evidence for reduced potassium selectivity (28). We used this property to determine whether the N-terminal truncated ATI variant is made to a significant extent in our heterologous expression system.
FIGURE 5.
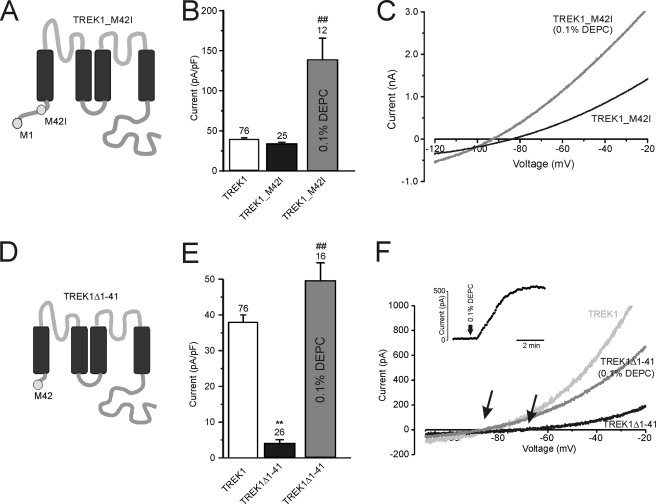
Effects of alternative translation initiation on TREK1 currents and their modification by DEPC. A, topology of TREK1_M42I. This mutation removes the alternative start codon at position 41, ensuring that only a complete full-length channel is transcribed. B, mean current densities of TREK1, TREK1_M42I, and TREK1_M42I after exposure to 0.1% DEPC. C, current-voltage relationships for TREK1_M42I before and after exposure to 0.1% DEPC. D, topology of TREK1Δ1–41, where the first 41 amino acids have been deleted, resulting in an N-terminally truncated TREK1 channel, with Met42 becoming the only translation start point of the channel. E, current density of TREK1, TREK1Δ1–41, and TREK1Δ1–41 after DEPC modification. F, current-voltage relationships for TREK1, TREK1Δ1–41, and TREK1Δ1–41 after DEPC exposure. Arrows indicate the shift in reversal potential observed for TREK1Δ1–41 currents after DEPC toward that observed for TREK1 currents. The inset shows the time course of DEPC-induced increase in TREK1Δ1–41 current. Numbers (n) are given above the bars. **, p < 0.01 compared with TREK1; ##, p < 0.01 compared with absence of DEPC. Error bars, S.E.
A point mutation was introduced into TREK1 that changed the ATI methionine into an isoleucine (TREK1_M42I), thus rendering the ATI non-functional. When expressed in tsA-201 cells, TREK_M42I produced whole cell current densities of 34 ± 2 pA/pF with a reversal potential of −84 ± 1 mV (n = 18), which is not significantly different from TREK1 (Fig. 5, B and C). In contrast, the N-terminal truncation TREK1Δ1–41 expressed alone showed a current density of 4 ± 1 pA/pF and a reversal potential of −70 ± 3 mV (n = 18; Fig. 5, D–F). This current density was too low to meaningfully test the effects of TREK1ΔEx4 on this mutant.
In order to enhance these currents, we made use of the histidine-modifying agent DEPC. DEPC has been shown to prevent inhibition of TREK1 by protons (44), thus leading to an increased current density at physiological extracellular pH. The irreversible modification by DEPC followed a relatively slow time course that required application for several minutes for the maximal effect (see inset in Fig. 5F). The DEPC-enhanced current was outwardly rectifying, reversed at −100 ± 4 mV (n = 4), and was not observed in untransfected tsA201 cells (n = 5). Treatment with 0.1% DEPC led to a severalfold increase in current density for both TREK1_M42I and TREK1Δ1–41 (Fig. 5). In fact, after DEPC treatment, TREK1Δ1–41 exhibited a mean current density of 50 ± 5 pA/pF and a reversal potential of −87 ± 2 mV (n = 15), thus allowing analysis of the effects of TREK1ΔEx4 on TREK1Δ1–41 currents.
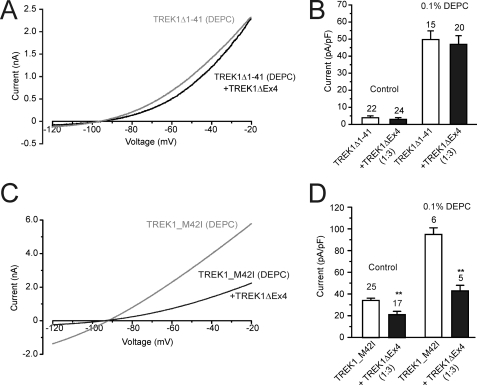
Fig. 6, A and B, demonstrates that co-expression of TREK1ΔEx4 in a 3-fold excess failed to significantly reduce TREK1Δ1–41 currents both in the absence and presence of DEPC enhancement. In contrast, equivalent to TREK1, TREK1_M42I currents were significantly inhibited by co-expression with TREK1ΔEx4 at a 1:3 ratio (Fig. 6, C and D). Current density under these conditions was 32% smaller at 23 ± 3 pA/pF (n = 13) with a reversal potential of −84 ± 2 mV. This effect persisted after DEPC enhancement (Fig. 6, C and D). These results suggest that TREK1ΔEx4 fails to interact with TREK1Δ1–41 channels.
FIGURE 6.
TREK1ΔEx4 does not affect TREK1Δ1–41 currents. A, current-voltage relationships for TREK1Δ1–41 alone and co-expressed with TREK1ΔEx4, following preincubation with 0.1% DEPC. B, mean current density (pA/pF) for TREK1Δ1–41 was not significantly affected by co-expression with TREK1ΔEx4 at a 1:3 ratio with or without preincubation in DEPC. C, TREK1ΔEx4 significantly reduced TREK1_M42I currents after preincubation with 0.1% DEPC. D, mean current density (pA/pF) for TREK1_M42I alone and co-expressed with TREK1ΔEx4 at a 1:3 ratio with or without preincubation in DEPC. Numbers (n) are given above the bars. *, p < 0.05; **, p < 0.01. Error bars, S.E.
DISCUSSION
We have identified in rodent brain a splice variant of TREK1 that lacks exon 4. This splice variant removes part of the extracellular loop following TM1 and truncates the potassium channel signature sequence GFG of the first pore loop. It also causes a frameshift resulting in a premature stop codon after an additional 37 residues. The resulting truncated protein has only one TM. In accordance with this, we observed no functional TREK-like currents when this splice variant was expressed in modified HEK cells. Surprisingly, however, co-expression of this splice variant with wild type TREK1 led to a reduction in TREK1 current density. This finding is indicative of a potential regulatory role of this splice variant upon stretch-activated or temperature-sensitive K+ currents.
This splice variant has been isolated from human hippocampus cDNA before and has been posted in GenBankTM (AK123409) as “moderately similar to Homo sapiens TREK1 potassium channel (KCNK2) mRNA.” No functional analysis had been described. The fact that, despite minor sequence differences, this splice variant is consistently amplified from native brain tissue of different species (mice, rats, and humans) strongly supports the view that this is indeed a splice variant rather than some sort of PCR artifact. Because both products will be amplified in one reaction sharing the same primers, it seems to be a valid assumption that the relative amount of PCR product obtained for the two cDNAs gives an indication of the relative abundance of the two mRNA species in the tissue they were isolated from. Based on this assumption, it would appear that the splice variant constitutes a sizeable fraction of the TREK1 mRNA in the brain, but not in the heart, liver, or kidney. However, because our study points at a direct interaction between the two splice variants of TREK1, accurate assessment of the potential impact of the splice variant in the brain would require a quantitative comparison of TREK1 and TREK1ΔEx4 protein expression at the level of the individual neuron.
To date, one other report has described splice variants for TREK1; in the heart, two variants of exon 1 are used, which leads to a differential proximal N terminus differing in length by 15 amino acid residues (30). However, there might well be more variation expected by analogy with TREK2 (45).
Functional Effects of the Splice Variant TREK1ΔEx4 on Potassium Currents
TREK1ΔEx4 expression in tsA201 cells had a dominant negative effect on TREK1, reducing whole cell currents. Taken together with the lack of effect of Kir6.2 expression on TREK1 currents and the finding that TREK1ΔEx4 had no effect on TASK3 currents, this result suggests that this is not due to some nonspecific effect on transcription or translation of TREK1.
A similar observation has been made for TASK1 channels. Hsu et al. (46) noticed a high sequence similarity between part of the HIV-1 accessory protein Vpu and the N-terminal portion of TASK1. They noticed that Vpu suppressed TASK1 currents and discovered that a truncated version of TASK1, consisting only of the N terminus and the first TM, also strongly suppressed TASK1 currents heterologously expressed in HEK293 cells. Our discovery of the TREK1 splice variant suggests that this suppression of current by a truncated channel protein comprising only the first TM is indeed utilized by neurons in vivo to limit/regulate current density for K2P channels.
Given that TREK1ΔEx4 is not a functional potassium channel itself, it could feasibly reduce TREK1 currents either by reducing the number of TREK1 channels that reach the plasma membrane (i.e. interfere with trafficking), or it could bind to the functional TREK1 channel like a β-subunit to alter its biophysical properties (e.g. reduce the open probability of the channel).
From our confocal microscopy experiments with fluorescently tagged TREK1, we know that TREK1ΔEx4 is translated but that the protein is retained in internal membranes. We also know from these experiments that TREK1 cannot act as a chaperone to allow TREK1ΔEx4 to traffic to the plasma membrane. On the contrary, TREK1ΔEx4 appears to retain TREK1 in internal membranes. Consequently, it appears most likely that TREK1ΔEx4 reduces surface expression of TREK1 rather than modulating its biophysical properties. At present, our knowledge about transcriptional and translational regulation as well as trafficking of two-pore domain K+ channels is in its infancy (27, 47–49), and we can only speculate whether unmasking of an ER retention signal would cause the reduced surface expression. A sequence of negative charges (EDE) in the proximal C terminus has been identified as a positive signal crucial for TASK3 channel trafficking to the surface membrane in Xenopus oocytes (48). However, such a sequence is not present in the C terminus of TREK1. In contrast, the intracellular C-terminal sequence of TREK1 contains several stretches of positive charges (RRR, KRK), which might act as ER retention signals (50). Alternatively, by analogy to what has been found for KATP channels, dimerization might be required to mask this retention signal and allow trafficking to the plasma membrane (51). Alternatively, a different protein partner might bind to this part of TREK1 and thus mask the retention signal and allow trafficking to the plasma membrane. A recent study by Sandoz et al. (27) suggests that the microtubule-associated protein Mtap2 does exactly this for TREK1 and TREK2 but not TRAAK channels.
COP1 (coatamer protein complex 1) has been suggested to promote the retention of TASK channels in the ER by binding to the N terminus dibasic motif ((M)KR; see Refs. 52 and 53, but also see Ref. 54), whereas the chaperone protein 14-3-3 binds to the extreme C terminus of the channel, causing COP1 to dissociate from it (54, 55). Thus, the presence of the N terminus of TASK channels without the C terminus will promote retention in the ER. In contrast, the N-terminal intracellular sequence retained in TREK1ΔEx4 contains no such potential ER retention motif; nor does it contain the EDE trafficking signal. This suggests that a different mechanism retains this protein in internal membranes. It also seems unlikely that the frame-shifted sequence after Gln104 of TREK1ΔEx4 is essential for the observed suppression of current because expression of the simply truncated protein TREK1_Q105X had a similar inhibitory effect. However, the slightly reduced effect of TREK1_Q105X as compared with TREK1ΔEx4 suggests that a larger size construct might be more effective at inhibiting TREK1. We cannot tell whether TREK1ΔEx4 interacts with a fully formed TREK1 dimer and prevents its surface expression or whether TREK1ΔEx4 binds to individual TREK1 subunits that then fail to dimerize to fully functional channels. At present, it is also impossible for us to tell which intracellular compartment TREK1 is trapped in by co-expression with TREK1ΔEx4. Further analysis of this splice variant might shed more light onto these issues.
Significance for ATI
Our results very clearly show that TREK1ΔEx4 has no effect on N-terminally truncated TREK channels derived from the alternative translation initiation described by Thomas et al. (28). Thus, in cells where both isoforms are present, TREK1ΔEx4 would only down-regulate the full-length channel. Interestingly, Thomas et al. (28) describe a reduced selectivity for potassium for the N-terminal truncated channel when expressed in Xenopus oocytes. In our hands, when expressed in tsA201 cells, especially after enhancement with DEPC, no difference between TREK1Δ1-41 and the full-length channels TREK1 and TREK1_M42I was seen when comparing reversal potentials at different external K+ concentrations (supplemental material). The more positive reversal potential observed in the absence of DEPC seems to reflect the larger contribution of the tsA201 cell intrinsic background conductance to the small TREK1Δ1–41 currents rather than a reduced K+ selectivity of the channel itself. Whether the discrepancy with the study in Xenopus oocytes (28) is due to expression of the channel in mammalian instead of amphibian cells in our experiments remains to be determined.
On a molecular level, the only difference between the long and short forms of TREK1 derived from ATI is the lack of the first 41 amino acids at the N terminus of the short form. The large difference in current density between the two forms suggests that this region influences either the functional or trafficking properties of the channel (or both; see also Refs. 28 and 29). The large increase in current density of the short form in the presence of DEPC does, however, suggest that a considerable number of channels are trafficked to the membrane. The lack of effect of TREK1ΔEx4 on the short form suggests that these first 41 amino acids are necessary for the interaction with TREK1ΔEx4, either because this region interacts directly with TREK1ΔEx4 or because the presence of this region alters the conformation of the long form to allow interaction with TREK1ΔEx4.
CONCLUSIONS
The results presented here demonstrate that a heavily truncated potassium channel subunit that shows no ion conductance itself can still be translated and have unexpected regulatory effect on related proteins. The differential regulatory role of TREK1ΔEx4 in the long and short form of TREK1 suggests that expression of this splice variant will alter the functional profile of TREK1 current in neurons where they are expressed, thereby regulating the functional properties and excitability of these cells.
Supplementary Material
Acknowledgments
We thank Helen Meadows (Glaxo SmithKline) for the gift of TREK1 cDNA and Frances Ashcroft (Oxford University) for the gift of Kir6.2 cDNA. We also thank Ian Brown, Naina Bajaria, and Catherine Clear for contributions to experiments.
This work was supported by grants from the Medical Research Council (Ref. G0600928 (to S. T.) and Ref. G0200370 ( to E. L. V. and A. M.)).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- ATI
- alternative translation initiation
- pF
- picofarad
- DEPC
- diethylpyrocarbonate
- TM
- transmembrane domain
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Talley E. M., Lei Q., Sirois J. E., Bayliss D. A. (2000) Neuron 25, 399–410 [DOI] [PubMed] [Google Scholar]
- 2.Millar J. A., Barratt L., Southan A. P., Page K. M., Fyffe R. E., Robertson B., Mathie A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3614–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein S. A., Bayliss D. A., Kim D., Lesage F., Plant L. D., Rajan S. (2005) Pharmacol. Rev. 57, 527–540 [DOI] [PubMed] [Google Scholar]
- 4.Talley E. M., Solorzano G., Lei Q., Kim D., Bayliss D. A. (2001) J. Neurosci. 21, 7491–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talley E. M., Sirois J. E., Lei Q., Bayliss D. A. (2003) Neuroscientist 9, 46–56 [DOI] [PubMed] [Google Scholar]
- 6.Aller M. I., Wisden W. (2008) Neuroscience 151, 1154–1172 [DOI] [PubMed] [Google Scholar]
- 7.Franks N. P., Lieb W. R. (1999) Nat. Neurosci. 2, 395–396 [DOI] [PubMed] [Google Scholar]
- 8.Lesage F. (2003) Neuropharmacology 44, 1–7 [DOI] [PubMed] [Google Scholar]
- 9.Chen X., Talley E. M., Patel N., Gomis A., McIntire W. E., Dong B., Viana F., Garrison J. C., Bayliss D. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3422–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathie A., Veale E. L. (2007) Curr. Opin. Investig. Drugs 8, 555–562 [PubMed] [Google Scholar]
- 11.Mathie A. (2007) J. Physiol. 578, 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trapp S., Aller M. I., Wisden W., Gourine A. V. (2008) J. Neurosci. 28, 8844–8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maingret F., Lauritzen I., Patel A. J., Heurteaux C., Reyes R., Lesage F., Lazdunski M., Honoré E. (2000) EMBO J. 19, 2483–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heurteaux C., Guy N., Laigle C., Blondeau N., Duprat F., Mazzuca M., Lang-Lazdunski L., Widmann C., Zanzouri M., Romey G., Lazdunski M. (2004) EMBO J. 23, 2684–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alloui A., Zimmermann K., Mamet J., Duprat F., Noël J., Chemin J., Guy N., Blondeau N., Voilley N., Rubat-Coudert C., Borsotto M., Romey G., Heurteaux C., Reeh P., Eschalier A., Lazdunski M. (2006) EMBO J. 25, 2368–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linden A. M., Aller M. I., Leppä E., Vekovischeva O., Aitta-Aho T., Veale E. L., Mathie A., Rosenberg P., Wisden W., Korpi E. R. (2006) J. Pharmacol. Exp. Ther. 317, 615–626 [DOI] [PubMed] [Google Scholar]
- 17.Noël J., Zimmermann K., Busserolles J., Deval E., Alloui A., Diochot S., Guy N., Borsotto M., Reeh P., Eschalier A., Lazdunski M. (2009) EMBO J. 28, 1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang D., Choe C., Kim D. (2005) J. Physiol. 564, 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D. (2003) Trends Pharmacol. Sci. 24, 648–654 [DOI] [PubMed] [Google Scholar]
- 20.Franks N. P., Honoré E. (2004) Trends Pharmacol. Sci. 25, 601–608 [DOI] [PubMed] [Google Scholar]
- 21.Honoré E. (2007) Nat. Rev. Neurosci. 8, 251–261 [DOI] [PubMed] [Google Scholar]
- 22.Heurteaux C., Lucas G., Guy N., El Yacoubi M., Thümmler S., Peng X. D., Noble F., Blondeau N., Widmann C., Borsotto M., Gobbi G., Vaugeois J. M., Debonnel G., Lazdunski M. (2006) Nat. Neurosci. 9, 1134–1141 [DOI] [PubMed] [Google Scholar]
- 23.Fink M., Duprat F., Lesage F., Reyes R., Romey G., Heurteaux C., Lazdunski M. (1996) EMBO J. 15, 6854–6862 [PMC free article] [PubMed] [Google Scholar]
- 24.Honoré E., Maingret F., Lazdunski M., Patel A. J. (2002) EMBO J. 21, 2968–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chemin J., Patel A. J., Duprat F., Lauritzen I., Lazdunski M., Honoré E. (2005) EMBO J. 24, 44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandoz G., Thümmler S., Duprat F., Feliciangeli S., Vinh J., Escoubas P., Guy N., Lazdunski M., Lesage F. (2006) EMBO J. 25, 5864–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandoz G., Tardy M. P., Thümmler S., Feliciangeli S., Lazdunski M., Lesage F. (2008) J. Neurosci. 28, 8545–8552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas D., Plant L. D., Wilkens C. M., McCrossan Z. A., Goldstein S. A. (2008) Neuron 58, 859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simkin D., Cavanaugh E. J., Kim D. (2008) J. Physiol. 586, 5651–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X. T., Dyachenko V., Zuzarte M., Putzke C., Preisig-Müller R., Isenberg G., Daut J. (2006) Cardiovasc. Res. 69, 86–97 [DOI] [PubMed] [Google Scholar]
- 31.Hentze M. W., Kulozik A. E. (1999) Cell 96, 307–310 [DOI] [PubMed] [Google Scholar]
- 32.Chen C., Okayama H. (1987) Mol. Cell Biol. 7, 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Downing J. E., Role L. W. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 7739–7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meadows H. J., Benham C. D., Cairns W., Gloger I., Jennings C., Medhurst A. D., Murdock P., Chapman C. G. (2000) Pflugers Arch. 439, 714–722 [DOI] [PubMed] [Google Scholar]
- 35.Chapman C. G., Meadows H. J., Godden R. J., Campbell D. A., Duckworth M., Kelsell R. E., Murdock P. R., Randall A. D., Rennie G. I., Gloger I. S. (2000) Brain Res. Mol. Brain Res. 82, 74–83 [DOI] [PubMed] [Google Scholar]
- 36.Sakura H., Ammälä C., Smith P. A., Gribble F. M., Ashcroft F. M. (1995) FEBS Lett. 377, 338–344 [DOI] [PubMed] [Google Scholar]
- 37.Inagaki N., Gonoi T., Clement J. P., 4th, Namba N., Inazawa J., Gonzalez G., Aguilar-Bryan L., Seino S., Bryan J. (1995) Science 270, 1166–1170 [DOI] [PubMed] [Google Scholar]
- 38.Tucker S. J., Gribble F. M., Zhao C., Trapp S., Ashcroft F. M. (1997) Nature 387, 179–183 [DOI] [PubMed] [Google Scholar]
- 39.Lauritzen I., Zanzouri M., Honoré E., Duprat F., Ehrengruber M. U., Lazdunski M., Patel A. J. (2003) J. Biol. Chem. 278, 32068–32076 [DOI] [PubMed] [Google Scholar]
- 40.Brenner T., O'Shaughnessy K. M. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E1480–E1486 [DOI] [PubMed] [Google Scholar]
- 41.Karschin C., Wischmeyer E., Preisig-Müller R., Rajan S., Derst C., Grzeschik K. H., Daut J., Karschin A. (2001) Mol. Cell Neurosci. 18, 632–648 [DOI] [PubMed] [Google Scholar]
- 42.Kang D., Han J., Talley E. M., Bayliss D. A., Kim D. (2004) J. Physiol. 554, 64–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei L., Wiser O., Slavin A., Mu D., Powers S., Jan L. Y., Hoey T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7803–7807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandoz G., Douguet D., Chatelain F., Lazdunski M., Lesage F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu W., Schlichthörl G., Hirsch J. R., Engels H., Karschin C., Karschin A., Derst C., Steinlein O. K., Daut J. (2002) J. Physiol. 539, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu K., Seharaseyon J., Dong P., Bour S., Marbán E. (2004) Mol. Cell 14, 259–267 [DOI] [PubMed] [Google Scholar]
- 47.Renigunta V., Yuan H., Zuzarte M., Rinné S., Koch A., Wischmeyer E., Schlichthörl G., Gao Y., Karschin A., Jacob R., Schwappach B., Daut J., Preisig-Müller R. (2006) Traffic 7, 168–181 [DOI] [PubMed] [Google Scholar]
- 48.Zuzarte M., Rinné S., Schlichthörl G., Schubert A., Daut J., Preisig-Müller R. (2007) Traffic 8, 1093–1100 [DOI] [PubMed] [Google Scholar]
- 49.Zanzouri M., Lauritzen I., Lazdunski M., Patel A. (2006) Biochem. Biophys. Res. Commun. 348, 1350–1357 [DOI] [PubMed] [Google Scholar]
- 50.Michelsen K., Yuan H., Schwappach B. (2005) EMBO Rep. 6, 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. (1999) Neuron 22, 537–548 [DOI] [PubMed] [Google Scholar]
- 52.Shikano S., Coblitz B., Sun H., Li M. (2005) Nat. Cell Biol. 7, 985–992 [DOI] [PubMed] [Google Scholar]
- 53.O'Kelly I., Butler M. H., Zilberberg N., Goldstein S. A. (2002) Cell 111, 577–588 [DOI] [PubMed] [Google Scholar]
- 54.Zuzarte M., Heusser K., Renigunta V., Schlichthörl G., Rinné S., Wischmeyer E., Daut J., Schwappach B., Preisig-Müller R. (2009) J. Physiol. 587, 929–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Kelly I., Goldstein S. A. (2008) Traffic 9, 72–78 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.