Abstract
The cg1324 gene (rosR) of Corynebacterium glutamicum encodes a MarR-type transcriptional regulator. By a comparative transcriptome analysis with DNA microarrays of a ΔrosR mutant and the wild type and subsequent EMSAs with purified RosR protein, direct target genes of RosR were identified. The narKGHJI operon, which encodes a nitrate/nitrite transporter and the dissimilatory nitrate reductase complex, was activated by RosR. All other target genes were repressed by RosR. They encode four putative monooxygenases, two putative FMN reductases, a protein of the glutathione S-transferase family, a putative polyisoprenoid-binding protein, and RosR itself. The DNA binding site of RosR was characterized as an 18-bp inverted repeat with the consensus sequence TTGTTGAYRYRTCAACWA. The in vitro DNA binding activity of RosR was reversibly inhibited by the oxidant H2O2. Mutational analysis of the three cysteine residues present in RosR (Cys-64, Cys-92, and Cys-151) showed that these are responsible for the inhibition of DNA binding by H2O2. A deletion mutant (Δcg1322) lacking the putative polyisoprenoid-binding protein showed an increased sensitivity to H2O2, supporting the role of RosR in the oxidative stress response of C. glutamicum.
Keywords: Bacterial Transcription, DNA-binding Protein, DNA-Protein Interaction, Oxidation Reduction, Oxidative Stress, Transcription Regulation, Corynebacterium glutamicum, MarR-type Regulator
Introduction
Corynebacterium glutamicum is a predominantly aerobic, nonpathogenic, biotin-auxotrophic, Gram-positive soil bacterium that was isolated by Kinoshita et al. (1) in a screen for bacteria that excrete l-glutamate, which is used as a flavor enhancer in the form of monosodium glutamate. Over the past 45 years, different strains of this species have been used for the industrial production of l-glutamate (2) and several other amino acids, in particular the feed additive l-lysine. Today, more than 2 million tons of amino acids are produced annually with C. glutamicum.
Because of its importance in industrial biotechnology, the genome sequence of C. glutamicum has been determined independently several times (3, 4), and the species has become a model organism for systems biology (5). In this context, we and others started to study the function of transcriptional regulators in this species. Based on the genome sequence, 127 one-component DNA-binding transcriptional regulators, 13 two-component signal transduction systems, and 7 sigma factors were annotated (6, 7). Within the past years, we analyzed five of the one-component regulators, i.e. ClgR (8–10), AcnR (11), RipA (12), DtxR (13), GntR1, and GntR2 (14), and three of the two-component systems, MtrAB (15, 16), PhoRS (7, 17), and CitAB (18).
In the present study, we performed a comprehensive analysis of a previously uncharacterized MarR-type regulator of C. glutamicum. With nine thus far uncharacterized representatives, the MarR family is one of the most prevalent families of transcriptional regulators in C. glutamicum (6). The MarR (multiple antibiotic resistance regulator) family includes proteins that are involved in the control of virulence factor production, the response to environmental stresses, or the regulation of catabolic pathways for aromatic compounds (19). In general, these regulators exist as homodimers in either a free or DNA-bound state. Most members of the MarR family are transcriptional repressors and often bind to the −10 or −35 region in the promoter causing a steric inhibition of RNA polymerase binding (19).
The MarR-type regulator studied here is encoded by the cg1324 gene. Our results indicate that the Cg1324 protein is involved in the response of C. glutamicum to oxidative stress, and therefore we named the corresponding protein RosR, an acronym for regulator of oxidative stress response.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Media, and Growth Conditions
All strains and plasmids used in this work are listed in Table 1. C. glutamicum type strain ATCC 13032 (1) was used as the wild type. The strains ΔrosR and Δcg1322 are derivatives with in-frame deletions of the rosR gene and the cg1322 gene, respectively. All C. glutamicum strains were routinely grown at 30 °C. For growth experiments, chloramphenicol acetyltransferase (CAT)2 assays, and RNA isolations, 5 ml of BHI (brain-heart infusion) medium (Difco Laboratories, Detroit, MI) was inoculated with colonies from a fresh BHI agar plate and incubated for 6 h on an orbital shaker at 120 rpm. After washing, the cells of this first preculture were used to inoculate a 500-ml baffled shake flask containing 60 ml of CGXII minimal medium (20) with 4% (w/v) glucose. This second preculture was incubated overnight at 120 rpm and then used to inoculate the main culture (CGXII minimal medium with 4% (w/v) glucose) to an optical density at 600 nm (A600) of ∼1. The trace elements were always added after autoclaving. If appropriate, kanamycin was added to a final concentration of 25 μg/ml. For anaerobic cultivation of C. glutamicum, 5 ml of LB medium (22) was inoculated with cells from a fresh BHI agar plate and cultivated aerobically overnight at 120 rpm. After washing with BT medium (21), cells were used to inoculate 100 ml of anoxic BT medium containing 0.5% (w/v) glucose and 30 mm potassium nitrate as the terminal electron acceptor and 1 μg/ml resazurin as an oxygen indicator. The medium was prepared in a 200-ml bottle, and oxygen was removed by bubbling it with 100% nitrogen gas for 2 h. Subsequently, the bottle was closed with a rubber stopper. After inoculation with a syringe, the bottles were shaken at 120 rpm and 30 °C. For all cloning purposes, Escherichia coli DH5α was used. Overproduction of RosR with plasmid pET16b-rosR or pET-TEV-rosR was performed with E. coli BL21(DE3)/pLysS. The E. coli strains were cultivated in LB medium at 37 °C or at room temperature for RosR overproduction. If appropriate, ampicillin (100 μg/ml) or kanamycin (50 μg/ml) was added.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− f80lacZDM15 D(lacZYA-argF) U169 endA1 recA1 hsdR17 (rk−, mk+) supE44 thi-1 gyrA96 relA1 phoA, host for cloning purposes | Invitrogen |
| E. coli BL21(DE3) with pLysS | F−ompT hsdSB (rB−, mB−) gal dcm (DE3); host for overproduction of RosR | Ref. 60 |
| C. glutamicum ATCC 13032 | Biotin-auxotrophic wild-type strain | American Type Culture Collection |
| C. glutamicum ΔrosR | Strain with an in-frame deletion of the rosR (cg1324) gene | This work |
| C. glutamicum Δcg1322 | Strain with an in-frame deletion of the cg1322 gene encoding a putative polyisoprenoid-binding protein | This work |
| Plasmids | ||
| pK19mobsacB | KanR; vector for allelic exchange in C. glutamicum (pK18 oriVE.c., sacB, lacZa) | Ref. 61 |
| pK19mobsacB-ΔrosR | KanR; pK19mobsacB derivative containing an overlap extension PCR product composed of the up- and downstream regions of rosR | This work |
| pK19mobsacB Δcg1322 | KanR; pK19mobsacB derivative containing an overlap extension PCR product composed of the up- and downstream regions of cg1322 | This work |
| pET16b | AmpR; plasmid for overexpression of genes in E. coli, adding an N-terminal decahistidine tag and a factor Xa cleavage site to the target protein (pBR322 oriVE.c., PT7, lacI) | Novagen |
| pET16b-rosR | KanR; pET16b derivative for overproduction of RosR with an N-terminal decahistidine tag | This work |
| pET16b-rosRC64S | pET16b-rosR derivative coding for a RosR protein with a Cys-64 → Ser exchange | This work |
| pET16b-rosRC92S | pET16b-rosR derivative coding for a RosR protein with a Cys-92 → Ser exchange | This work |
| pET16b-rosRC151S | pET16b-rosR derivative coding for a RosR protein with a Cys-151 → Ser exchange | This work |
| pET16b-rosRC64S,C92S | pET16b-rosR derivative coding for a RosR protein with Cys-64,Cys-92 → Ser exchanges | This work |
| pET16b-rosRC64S,C151S | pET16b-rosR derivative coding for a RosR protein with Cys-64,Cys-151 → Ser exchanges | This work |
| pET16b-rosRC92S,C151S | pET16b-rosR derivative coding for a RosR protein with Cys-92,Cys-151 → Ser exchanges | This work |
| pET16b-rosRC64S,C92S,C151S | pET16b-rosR derivative coding for a RosR protein with Cys-64,Cys-92,Cys-151 → Ser exchanges | This work |
| pET28b | KanR; plasmid for overexpression of genes in E. coli, adding an N-terminal hexahistdine tag and a thrombin cleavage site to the target protein (pBR322 oriVE.c., PT7, lacI) | Novagen |
| pET-TEV | KanR; pET28b derivative for overexpression of genes in E. coli, adding an N-terminal decahistdine tag and a TEV protease cleavage site to the target protein (pBR322 oriVE.c., PT7, lacI) | This work |
| pET-TEV-rosR | KanR; pET-TEV derivative for overproduction of RosR with an N-terminal decahistidine tag, which can be cleaved off using TEV protease | This work |
| pAN6 | KanR; C. glutamicum/E. coli shuttle vector for regulated gene expression; derivative of pEKEx2 (Ptac, lacIq, pBL1 oriVC.g., pUC18 oriVE.c.) | Ref. 14 |
| pAN6-rosR | KanR; pAN6 derivative carrying the rosR gene under the control of an IPTGa-inducible tac promoter | This work |
| pET2 | KanR; promoter probe plasmid for C. glutamicum | Ref. 26 |
| pET2-rosR | KanR; pET2-derivative with a 284-bp fragment covering the C. glutamicum rosR promoter | This work |
a Isopropyl 1-thio-β-d-galactopyranoside.
General DNA Techniques and Sequence Analysis
Standard methods such as PCR, restriction digestion, or ligation were carried out according to established protocols (22). E. coli was transformed as described (23). DNA sequencing was performed with a 3100-Avant genetic analyzer (Applied Biosystems, Darmstadt, Germany). Sequencing reactions were carried out with the ABI PRISMTM Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems). Alternatively, sequencing was performed by Agowa (Berlin, Germany). All oligonucleotides used in this study are listed in supplemental Table S1.
Mapping of Transcriptional Start Sites by Primer Extension Analysis
To determine the transcriptional start site of the cg1322, cg1426, and cg3084 genes, primer extension analysis was performed as described previously (8) using the IRD800-labeled oligonucleotides cg1322-1, cg1322-2, cg1426-1, cg1426-2, cg3084-1, and cg3084-2 listed in supplemental Table S1. Total RNA was isolated as described previously (24) from C. glutamicum ΔrosR cells grown in glucose minimal medium up to the exponential growth phase (A600, 5–6). The length of the primer extension products was determined by running the four lanes of a DNA sequencing reaction set up using the same IRD800-labeled oligonucleotide as for reverse transcription alongside the primer extension products. As templates for the sequencing reactions PCR products were used that had been obtained with the oligonucleotide pairs cg1322-A/cg1322-B, cg1426-A/cg1426-B, and cg3084-A/cg3084-B (supplemental Table S1) and chromosomal DNA of C. glutamicum wild type as template.
Mapping of Transcriptional Start Sites by Rapid Amplification of cDNA Ends (RACE)
To determine the transcriptional start sites of the rosR (cg1324), cg1150, cg2329, and cg1848 genes, 5′- RACE analysis was performed using the gene-specific primers cg1324-sp1, cg1324-sp2, cg2329-sp1, cg2329-sp2, cg1150-sp1, cg1150-sp2, cg1848-sp1, and cg1848-sp2 (supplemental Table S1) and the 5′/3′ RACE Kit, 2nd Generation (Roche Diagnostics). Total RNA was isolated as described previously (24) from C. glutamicum ΔrosR cells grown in glucose minimal medium up to the exponential growth phase (A600, 5–6). Subsequent PCR amplification of the 5′-junction and sequence analysis identified the transcriptional start sites.
Construction of Strains and Plasmids
In-frame deletion mutants of C. glutamicum lacking either the rosR gene or the gene cg1322 were constructed via a two-step homologous recombination procedure as described previously (25). The oligonucleotides used for this purpose (D1–1124, D2–1124, D3–1124, and D4–1124 for the rosR deletion and D1-cg1322, D2-cg1322, D3-cg1322, and D4-cg1322 for the cg1322 deletion) are listed in supplemental Table S1. The chromosomal deletions of rosR and cg1322 were confirmed by PCR using oligonucleotide pairs D1324-fw/D1324-rv and Dcg1322-fw/Dcg1322-rv, respectively.
For construction of the expression plasmid pET16b-rosR, which codes for a RosR derivative with an N-terminal decahistidine tag, the rosR coding region was amplified by PCR using oligonucleotides 1124-NdeN and 1124-XhoC, which included NdeI and XhoI restriction sites for cloning purposes (supplemental Table S1). After digestion with NdeI and XhoI, the PCR product was cloned into the expression vector pET16b (Novagen). The PCR-derived part of the resulting pET16b-rosR plasmid and the ligation sites were sequenced in order to exclude unwanted mutations. The RosR protein encoded by this plasmid contained 21 additional amino acids (MGHHHHHHHHHHSSGHIEGRH) at the N terminus, including a factor Xa cleavage site (SGHIEGR). Plasmids pET16b-rosR-C64S, -C92S, -C151S, -C64S,C92S, -C64S,C151S, -C92S,C151S, and -C64S,C92S,C151S, which code for RosR derivatives where the three cysteine residues Cys-64, Cys-92, and Cys-151 were replaced individually or in combination by serine residues, were constructed using the QuikChange XL site-directed mutagenesis kit (Stratagene) using pET16b-rosR as template and the oligonucleotides c1-for, c1-rev, c2-for, c2-rev, c3-for, and c3-rev (supplemental Table S1). All recombinant plasmids were sequenced to exclude unwanted mutations in the rosR gene.
For construction of plasmid pET-TEV, a DNA fragment encoding a decahistidine tag, a linker region, and a TEV (Tobacco Etch Virus) protease cleavage site (ENLYFQG) was generated using the partially complementary oligonucleotides pET28–10HisTEV-fw and pET28–10HisTEV-rv and Taq DNA polymerase to fill in the overhanging single strands of the annealed oligonucleotides and to amplify the double-stranded DNA fragment. The resulting product was digested with NcoI and NdeI and cloned into a pET28b plasmid digested with the same restriction enzymes, resulting in the expression vector pET-TEV. The PCR-derived fragment of this vector was sequenced to exclude unwanted mutations. An open reading frame, ligated into this vector by using the NdeI site, contained 29 additional codons and at the protein level 29 additional N-terminal amino acids (MGSSHHHHHHHHHHDYDIPTTENLYFQGH). After cleavage of the resulting protein with TEV protease, only two amino acids (GH) remain at the N terminus of the target protein. The plasmid pET-TEV-rosR was constructed analogously to pET16b-rosR and sequenced to exclude unwanted mutations.
For construction of the pET2-rosR plasmid, which was used for CAT assays, a 282-bp promoter fragment of rosR (extending from position +10 to −272 with respect to the rosR transcriptional start site) was amplified by PCR with the oligonucleotides pET2–1324-for and pET2–1324-rev (supplemental Table S1) containing BamHI and SacI restriction sites for cloning. After digestion with BamHI and SacI, the PCR product was cloned into the promoter probe plasmid pET2 (26). The PCR-derived part of the resulting pET2-rosR plasmid and the ligation sites were sequenced to exclude unwanted mutations.
Overproduction and Purification of RosR
For overproduction of RosR and its mutated derivatives, E. coli BL21(DE3)/pLysS was transformed with expression plasmid pET16b-rosR or pET-TEV-rosR and cultivated in LB medium at 37 °C to an A600 of 0.5. Then, expression of the target gene was induced by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside, and the culture was incubated for another 3 h at room temperature. After the cells were harvested, purification of RosR by Ni2+-NTA affinity chromatography was performed essentially as described previously for the RipA protein (12). The RosR-containing elution fractions were desalted by size exclusion chromatography using PD10 columns (GE Healthcare) and buffer containing 20 mm Tris/HCl, pH 7.5, and 10% (v/v) glycerol. For determination of apparent KD values, tag-free RosR was prepared as follows. After Ni2+-NTA affinity chromatography of cell extracts from the pET-TEV-rosR carrying expression host, the elution fractions were diluted 2× into buffer T (50 mm Tris/HCl, pH 8.0, 0.5 mm EDTA, 1 mm DTT), and TEV protease was added at a ratio ranging from 1:50 to 1:100 (w/w). Cleavage was performed at 4 °C overnight. On the following day the protein solution was concentrated to 1 ml and the buffer exchanged to TNI5 using an Amicon Ultra-15 centrifugal filter unit with a cut-off of 10 kDa. A second Ni2+-NTA affinity chromatography was performed to remove TEV protease and uncleaved RosR. This was followed by a size exclusion chromatography using a HiLoadTM 16/60 SuperdexTM 200 column with binding buffer (20 mm Tris/HCl, pH 8.0, 50 mm KCl, 10 mm MgCl2, 5% (v/v) glycerol, 0.5 mm EDTA) to get rid of any aggregated protein. After gel filtration the protein was flash-frozen in liquid nitrogen in 60-μl aliquots and stored at −70 °C until used for further studies. A fresh aliquot was used for each series of electrophoretic mobility shift assays (EMSAs).
Global Gene Expression Analysis Using DNA Microarrays
Preparation of RNA and synthesis of fluorescently labeled cDNA were carried out as described (35). Custom-made DNA microarrays for C. glutamicum ATCC 13032 printed with 70mer oligonucleotides, obtained from Operon (Cologne, Germany), are based on the genome sequence entry NC_006958 (21). Hybridization and stringent washing of the microarrays were performed according to the instructions of the supplier. Hybridization was carried out for 16–18 h at 42 °C using a MAUI hybridization system (BioMicro Systems, Salt Lake City, UT). After washing, the microarrays were dried by centrifugation (5 min, 520 × g), and fluorescence was determined at 532 nm (Cy3-dUTP) and 635 nm (Cy5-dUTP) with 10 μm resolution using an Axon GenePix 6000 laser scanner (Axon Instruments, Sunnyvale, CA). Quantitative image analysis was carried out using GenePix image analysis software, and the results were saved as a GPR file (GenePix Pro 6.0, Axon Instruments). For data normalization, GPR files were processed using the BioConductor/R packages limma and marray. Processed and normalized data as well as experimental details (MIAME (10)) were stored in the in-house microarray database for further analysis (40).
Chloramphenicol Acetyltransferase Assay
For analyzing the expression of the rosR gene, C. glutamicum wild type and the deletion mutant ΔrosR were transformed with plasmid pET2-rosR, which is based on the corynebacterial promoter probe vector pET2 (26) and contains the rosR promoter region in front of a promoterless cat (chloramphenicol acetyltransferase) gene. The promoter activity test was performed as described previously by measuring the formation of 5-thio-2-nitrobenzoate photometrically at 412 nm and 37 °C (27, 28). One unit of enzyme activity is defined as the turnover of 2 μmol of acetyl-CoA/min.
Gene Expression Analysis Using Real-time PCR
500 ng of RNA was transcribed into cDNA using specific primers for the genes of interest. The product was quantified via real-time PCR using a LightCycler instrument 1.0 (Roche Diagnostics) with SYBR Green I as the fluorescence dye following the instructions of the supplier (Qiagen, Hilden, Germany). The accuracy of the PCR product was checked by melting curve analysis. To quantify the amount of cDNA, a calibration curve was generated from known concentrations of template DNA (eight for each gene) that were processed in parallel via real-time PCR. These reference samples were generated by PCR using chromosomal DNA of C. glutamicum as a template and KOD DNA polymerase (Invitrogen). Expression values are given in relation to the ddh gene (cg2900) encoding diaminopimelate dehydrogenase, which was used as a reference gene with constitutive expression.
Electrophoretic Mobility Shift Assay
The binding of RosR protein or its mutated derivatives to putative target promoters was tested as described previously for RipA (12). Purified protein was incubated with DNA fragments (100–700 bp, final concentration 8–20 nm) in a total volume of 20 μl. The binding buffer contained 20 mm Tris/HCl, pH 8.0, 50 mm KCl, 10 mm MgCl2, 5% (v/v) glycerol, and 0.5 mm EDTA. 1 mm DTT was freshly added to the reaction mixture before incubation. Promoter fragments of putative non-target genes (∼13 nm) of RosR were used as negative controls. The reaction mixtures were incubated at room temperature for 20 min and then loaded onto 10 or 15% native polyacrylamide gels. Electrophoresis was performed at room temperature and 170 V using 1× TBE (89 mm Tris base, 89 mm boric acid, 2 mm EDTA) as the electrophoresis buffer. The gels were subsequently stained either with SYBR Green I or GelRedTM and photographed. All PCR products used in the gel shift assays were purified with a PCR purification kit (Qiagen) and eluted in water.
The reversibility of the loss of binding due to oxidation was tested as follows. RosR was used in three different concentrations (100, 200, and 400 nm), and aliquots were taken for EMSA (final RosR concentrations 25, 50, and 100 nm). Afterward, H2O2 was added to each RosR solution to a final concentration of 10 mm, and immediately aliquots were taken for EMSA. In the next step, DTT was added to the H2O2-treated RosR solutions to a final concentration of 50 mm, and again aliquots were taken for EMSA. The final RosR concentrations in the EMSAs were 25, 50, and 100 nm. All nine aliquots were incubated in binding buffer with 80 ng of DNA covering the promoter region of cg2329 for 20 min at room temperature and separated on a 10% native polyacrylamide gel at 180 V for 1.5 h on ice. The gel was stained using GelRedTM.
For the determination of apparent KD values, increasing concentrations of the RosR dimer (0.5–100 nm) were incubated for 20 min at room temperature with 80 ng of DNA covering the promoter regions of the RosR target genes. The samples were applied onto a 10% native polyacrylamide gel and separated at 180 V for 1–1.5 h on ice. The gels were stained with GelRedTM and photographed. The bands were quantified using ImageQuant software (GE Healthcare), and the percentage of shifted DNA was calculated. These values were plotted against the RosR concentration in log10 scale, and a sigmoidal fit was performed using Origin 7 software (OriginLab Corp., Northampton, MA), considering the error bars as well as 0 and 100% shifted DNA as asymptotes, The turning point of the curve was defined as the apparent KD value. All determinations were performed at least in triplicate.
Size Exclusion Chromatography
The native molecular mass of purified RosR was estimated by size exclusion chromatography using a HiLoadTM 16/60 SuperdexTM 200 prep grade column (Amersham Biosciences) integrated into an ÄKTATM FPLC system (Amersham Biosciences). The column was equilibrated with 20 mm HEPES buffer, pH 8.0, containing 500 mm NaCl. After the application of 0.5–1 mg of purified protein, chromatography was performed at 15 °C with a flow rate of 1 ml/min. The column was calibrated with protein molecular mass markers (MWGF-200, Sigma).
Two-dimensional Gel Electrophoresis and Protein Identification by Mass Spectrometry
Two-dimensional gel electrophoresis was performed as described previously (29). 300 μg of protein extract of the wild type and the deletion mutant ΔrosR were used for each two-dimensional gel. Extracts were prepared from cells cultivated in glucose minimal medium and harvested in the exponential growth phase (A600, 5–6). Identification of proteins from Coomassie-stained two-dimensional gels was performed by peptide mass fingerprinting of tryptic digests as described (29), except that peptides were extracted by the addition of 0.2% (v/v) trifluoroacetic acid in 30% (v/v) acetonitrile. MALDI-TOF-MS was performed with an Ultraflex III TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). MASCOT software (30) was used to compare the peptide mass patterns obtained with those of all proteins from the theoretical C. glutamicum proteome. The molecular weight search (MOWSE) scoring scheme (31) with a cut-off value of 50 was used for unequivocal identification of proteins.
RESULTS
Influence of a rosR Deletion on Global Gene Expression
The cg1324 gene of C. glutamicum ATCC 13032 encodes a protein of 162 amino acids (mass, 18,641 Da), which has been annotated as a putative transcriptional regulator of the MarR family (4). The role of this protein, which we named RosR, has not been studied hitherto. To get clues on the function, we searched for putative RosR target genes by analyzing the influence of a deletion of the rosR gene on global gene expression. For this purpose, strain C. glutamicum ΔrosR was constructed, which contains an in-frame deletion of the rosR gene. Global gene expression of the ΔrosR mutant was compared with that of the wild type by transcriptome analysis using DNA microarrays. The two strains were cultivated in minimal medium with 4% (w/v) glucose as carbon and energy source. No differences in growth behavior were observed under these conditions: both strains grew at a growth rate of 0.39 h−1 and reached a final A600 of ∼60. For the transcriptome comparisons, cells were harvested in the exponential growth phase (A600, ∼5), and total RNA was isolated. Three DNA microarray experiments were performed, each starting from an independent culture. In total, 55 genes showed an at least 2-fold change in the mRNA ratio with a p value of ≤0.05; these are listed in Table 2. Table 2 also includes a few genes that do not meet the criteria but are part of operons containing genes that met the criteria.
TABLE 2.
Genome-wide comparison of mRNA levels in C. glutamicum wild type and the ΔrosR mutant using DNA microarrays
The mRNA ratios represent mean values from three independent microarray experiments starting from independent cultures (see “Experimental Procedures”). The strains were cultivated in CGXII minimal medium with 4% (w/v) glucose, and mRNA was isolated in the exponential growth phase. The table includes those genes that showed a ≥2-fold changed mRNA level (increased or decreased) in at least two of the three experiments and that had a p value ≤ 0.05 (exception is gluD; here only one experiment was evaluable). The genes are ordered according to their position on the genome. The mRNA ratios for the genes narJ and gluD were marked with an asterisk, as the mRNA ratio (narJ) or the p value (gluD) were not within the defined range. However, these genes were included, as they are part of operons of which all other genes fulfilled the selected criteria.
| Accession no. | Gene | Annotation | mRNA ratio DrosR/WT |
|---|---|---|---|
| Genes with a decreased mRNA level in strain DrosR | |||
| cg0416 | Secreted protein carrying a eukaryotic domain | 0.24 | |
| cg0415 | ptpA2 | Low molecular weight phosphotyrosine protein phosphatase | 0.32 |
| cg1109 | porB | Anion-specific porin precursor | 0.46 |
| cg1324 | rosR | Transcriptional regulator of MarR family | 0.00 |
| cg1345 | narK | Putative nitrate/nitrite antiporter | 0.34 |
| cg1344 | narG | Respiratory nitrate reductase, α-subunit | 0.31 |
| cg1343 | narH | Respiratory nitrate reductase, β-subunit | 0.43 |
| cg1342 | narJ | Respiratory nitrate reductase, δ-subunit | 0.55* |
| cg1341 | narI | Respiratory nitrate reductase, γ-subunit | 0.42 |
| cg1382 | glgE | Putative α-amylase | 0.49 |
| cg1671 | Putative membrane-associated GTPase | 0.42 | |
| cg2136 | gluA | ABC transporter for glutamate, ATP-binding protein | 0.39 |
| cg2137 | gluB | ABC transporter for glutamate, secreted glutamate-binding protein | 0.36 |
| cg2138 | gluC | ABC transporter for glutamate, permease | 0.37 |
| cg2139 | gluD | ABC transporter for glutamate, permease | 0.29* |
| cg2649 | Secreted penicillin-binding protein | 0.39 | |
| cg2773 | Uncharacterized protein with SCP/PR1 domain | 0.32 | |
| Genes with an increased mRNA level in strain DrosR | |||
| cg0072 | Membrane protein of unknown function | 189.00 | |
| cg0144 | rbtT | Putative arabinitol or ribitol transporter | 2.25 |
| cg0292 | tnp16a | Transposase (ISCg16a) | 2.17 |
| cg0364 | Membrane protein | 3.51 | |
| cg0384 | rluC1 | Ribosomal large subunit, pseudouridine synthase C | 2.57 |
| cg0408 | Membrane protein | 2.82 | |
| cg0823 | Putative monooxygenase | 69.95 | |
| cg0824 | tnp5a | Transposase (ISCg5a) | 3.10 |
| cg1001 | mscL | Large conductance mechanosensitive channel | 2.24 |
| cg1150 | Putative NAD(P)H-dependent FMN reductase | 169.08 | |
| cg1255 | HNH endonuclease | 3.48 | |
| cg1322 | Putative polyisoprenoid-binding protein | 20.66 | |
| cg1426 | Protein of glutathione S-transferase family | 26.02 | |
| cg1848 | Luciferase-like monooxygenase | 22.24 | |
| cg1850 | Putative FMN reductase | 419.73 | |
| cg2329 | Luciferase-like monooxygenase | 140.65 | |
| cg2426 | tnp2d | Transposase (ISCg2d) | 4.14 |
| cg2485 | phoD | Secreted alkaline phosphatase precursor | 2.31 |
| cg2504 | Protein of unknown function | 4.93 | |
| cg2599 | Pirin-related protein | 3.68 | |
| cg2759 | tnp15b | Transposase (ISCg15b) | 2.87 |
| cg2844 | pstA | ABC transporter for phosphate uptake, permease component | 3.51 |
| cg2914 | tnp5b | Transposase (ISCg5b) | 3.51 |
| cg2953 | xylC | Putative benzaldehyde dehydrogenase | 2.13 |
| cg3004 | gabD1 | Putative succinate semialdehyde dehydrogenase | 2.34 |
| cg3084 | Putative flavin-containing monooxygenase | 40.64 | |
| cg3085 | Luciferase-like monooxygenase | 2.46 | |
| cg3258 | rluC2 | Putative pseudouridine synthase | 2.38 |
| cg3272 | Membrane protein (putative Fe2+/Mn2+ transporter) | 2.67 | |
| cg3273 | Protein of unknown function | 2.38 | |
| cg3274 | Site-specific recombinase (resolvase) fragment | 3.69 | |
| cg3284 | cgtS9 | Two-component histidine kinase CgtS9 | 198.14 |
| cg3288 | Hypothetical protein predicted by Glimmer | 2.58 | |
| cg3289 | Putative thiol-disulfide isomerase | 2.46 | |
| cg3291 | Putative transcriptional regulator of Crp/FNR family | 95.66 | |
| cg3293 | Protein of unknown function | 3.17 | |
| cg3295 | Probable cation-transporting P-type ATPase | 4.19 | |
| cg3329 | Member of uncharacterized protein family UPF0027 (homolog of E. coli RtcB) | 2.06 | |
| cg3413 | azlC | Putative branched chain amino acid permease (azaleucine resistance) | 2.21 |
| cg3418 | Putative secreted protein | 3.21 | |
17 genes showed a 2–4-fold decreased mRNA concentration in the ΔrosR mutant. This group included, for example, the narKGHJI operon coding for a nitrate/nitrite transporter (NarK) and the dissimilatory nitrate reductase (NarGHJI), which allows restricted anaerobic growth of C. glutamicum by nitrate respiration (21, 32), or the gluABCD operon coding for an ABC-type glutamate uptake system (33).
40 genes were found to have a ≥2-fold increased mRNA concentration in the ΔrosR mutant, with 11 of them showing increases between 20- and 420-fold. The genes with a strongly increased expression code for a membrane protein of unknown function (cg0072), a putative polyisoprenoid-binding protein (cg1322, gene upstream of and divergent from rosR), a sensory histidine kinase (cgtS9), a putative transcriptional regulator of the Crp/FNR family (cg3291), a protein of the glutathione S-transferase family (cg1426), two putative FMN reductases (cg1150 and cg1850), and four putative monooxygenases (cg0823, cg1848, cg2329, and cg3084). Besides the multiplicity of monooxygenase genes, another peculiar feature was that many genes of the HGC1 (high G + C content) region of the C. glutamicum genome, which extends from cg3267 to cg3295 and is flanked by defective insertion sequences (34), showed an increased mRNA level in the ΔrosR mutant.
To verify the results obtained by the DNA microarray experiments, quantitative real-time PCR was performed for some of the genes with altered mRNA levels in the ΔrosR mutant, namely narK and narG as examples of down-regulated genes and cg1426 and cg2329 as representatives of up-regulated genes. The mRNA ratios in relation to the reference gene ddh were 0.35 for narK, 0.63 for narG, 27.7 for cg1426, and 58.6 for cg2329, thus confirming the results of the DNA microarray experiments (Table 2).
Purification of RosR and Binding to the Promoter Regions of narKGHJI, cg1426, and cg2329
To determine which of the genes showing a different expression level in the DNA microarray experiments were direct target genes of the transcriptional regulator RosR, EMSAs were performed with purified RosR (supplemental Fig. S1A). An N-terminal His tag did not interfere with DNA binding by RosR, but the His-tagged protein showed a stronger tendency for aggregation and precipitation. Therefore, the tag was cleaved off for experiments in which it was crucial to keep the protein stable over some time to get reproducible results, such as the determination of the apparent KD values or the reversibility of the inhibition of DNA binding by H2O2. In addition, it was necessary for those experiments to remove aggregates because these would contribute to the measured protein concentration, although they probably no longer bound DNA. We found the protein to be most stable without the His tag and when it was stored frozen at −70 °C. The native mass of RosR-His as determined by size exclusion chromatography was found to be ∼40 kDa (supplemental Fig. S1B). This indicates that RosR forms a homodimer, similar to other MarR-type regulators such as MarR or MexR (19).
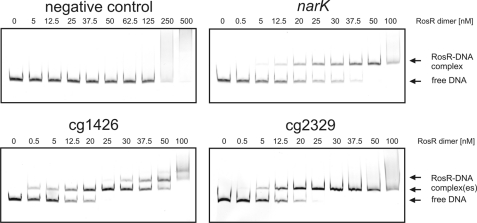
In the first series of EMSAs, DNA fragments covering the promoter regions of the narKGHJI operon (−500 to +86 bp), of cg1426 (−500 to +120 bp), and of cg2329 (−500 to +89 bp) were amplified by PCR (all distances are with respect to the translational start site). As shown in Fig. 1, the promoter fragments of narKGHJI, cg1426, and cg2329 were completely shifted with 50 nm RosR dimer. In contrast, no specific shift was observed with a DNA fragment covering parts of the coding regions of narK and narG (+855 to +1545 bp with respect to the translational start site of narK) and with the promoter region of the sdhCAB operon (−490 to +110 bp). The sdhCAB genes showed unaltered mRNA levels in the ΔrosR mutant (data not shown). According to these results, narKGHIJ, cg1426, and cg2329 presumably represent direct target genes of RosR. In further EMSAs the promoter regions of the gluABCD operon and of cg3291 were tested, but no binding of RosR was observed (data not shown).
FIGURE 1.
EMSAs with purified RosR protein and DNA fragments (80 ng each) covering the promoter regions of narKGHJI, cg1426, and cg2329. A DNA fragment containing parts of the coding regions of narK and narG was used as the negative control. The DNA fragments were incubated either without RosR or with the indicated concentrations of RosR for 20 min at room temperature. Then the samples were separated by native polyacrylamide gel electrophoresis (10%), and the DNA was stained with GelRedTM.
Identification of the RosR Binding Motif
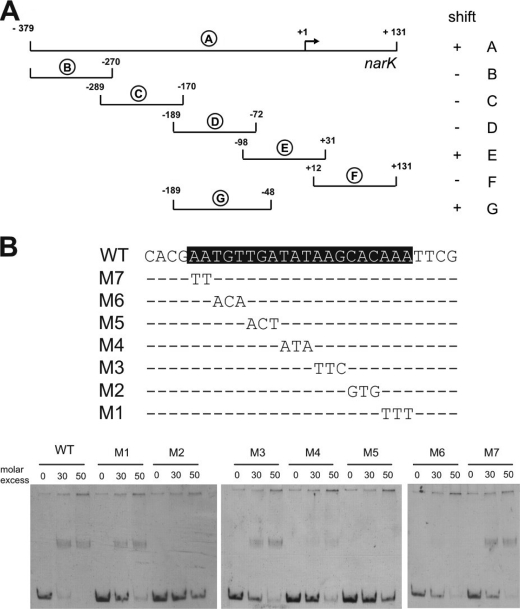
To locate the binding site(s) of RosR in the promoter region of the narKGHJI operon, the 510-bp fragment A used for the initial binding studies was split into several smaller fragments, which were synthesized by PCR and analyzed in EMSAs for binding of RosR. As shown in Fig. 2A, only fragments E and G were bound by RosR. As fragment G differs from fragment D only by 24 additional bp at the promoter-proximal site, the most important part of the RosR binding site was assumed to be located within these 24 bp, which extends from position −71 to −48 with respect to the narK transcriptional start site determined previously (21). The relevance of this region was analyzed by mutational analysis. Seven mutated fragments were amplified by PCR (Fig. 2B), each with two or three nucleotides exchanged, and tested again in EMSAs. In the case of fragments M2, M4, M5, and M6, the mutations prevented the binding of RosR, indicating that the corresponding nucleotides were required for binding (Fig. 2B).
FIGURE 2.
Identification of the RosR binding site in the promoter region of the narKGHJI operon. A, localization of the RosR binding site using narKGHJI promoter fragments (designated A–G) and purified RosR in EMSAs. The numbers show the position of the fragments relative to the transcriptional start site (+1). ‘+’ and ‘−’ signs indicate whether the fragment was shifted by RosR. B, mutational analysis of the RosR binding site (shaded in black) in the narKGHJI promoter region. The mutations M1–M7 were introduced by PCR and are shown below the wild-type sequence. The corresponding DNA fragments were analyzed by EMSAs with RosR.
DNA sequence motifs similar to the one identified in the narK promoter region (ATGTTGATATAAGCACAA) were also identified in the promoter regions of cg1426 (TTGTTGACATATCATCTA) and cg2329 (TTGTTGATATATCTACAA). To confirm their relevance for RosR binding, promoter fragments including or lacking the proposed binding sites (cg1426, −49 to +120 bp and −20 to +120 bp; cg2329, −64 to +89 bp and −36 to +89 bp with respect to the translational start sites) were synthesized by PCR. EMSAs showed that only the DNA fragments containing the motif were shifted by RosR (supplemental Fig. S2).
Identification of Further RosR Target Genes
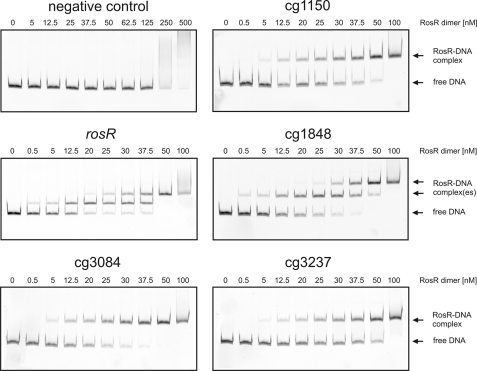
The identification of the RosR binding sites in the promoter regions of narKGHJI, cg1426, and cg2329 allowed us to derive a first generation RosR consensus binding motif (ANTGTTGANATANNNNCNAA), which was used for a genome-wide in silico search (allowing two mismatches) for similar sequences in the genome of C. glutamicum ATCC 13032 using ERGO software (Integrated Genomics Inc.). In total 67 matches were obtained, 17 of which were located between open reading frames and thus within putative promoter regions. This group included, of course, the promoters of narKGHJI, cg1426, and cg2329. The remaining 14 promoter regions were amplified and tested for binding of RosR in EMSAs. A shift in the presence of purified RosR was observed for five fragments, which represent the promoter regions of cg1150 (putative NADPH-dependent FMN reductase), rosR itself, cg1848 (luciferase-like monooxygenase), cg3084 (putative flavin-containing monooxygenase), and sodA (cg3237, superoxide dismutase) (Fig. 3). The latter result indicates that RosR is subject to negative autoregulation, a feature known for many MarR-type transcriptional regulators (19).
FIGURE 3.
EMSAs with purified RosR protein and the promoter regions of putative target genes predicted from an in silico search. DNA fragments (80 ng) covering the promoter regions of cg3237 (superoxide dismutase), cg1150 (putative FMN reductase), cg1426 (putative protein of the glutathione S-transferase family), cg2329 (putative monooxygenase), cg1848 (putative monooxygenase), rosR, and cg3084 (putative monooxygenase) were incubated for 20 min at room temperature with the given concentrations of purified RosR. The negative control contained the narK promoter without the predicted RosR binding site. Samples were separated by native PAGE (10%) and stained with GelRedTM.
In the case of the promoter fragments of rosR, cg1426, and cg1848, two RosR-DNA complexes were formed, indicating the presence of two binding motifs in these regions (Figs. 1 and 4). Inspection of the cg1848 promoter sequence revealed the presence of two RosR binding motifs in close proximity. Also in the intergenic region between rosR and the divergent cg1322 gene, two motifs could be identified, one of which overlaps with the proposed translational start site of rosR (Fig. 4). The promoter regions of rosR and cg1848 were separated between the two predicted binding sites and each fragment was analyzed separately by EMSAs. All four fragments led to the formation of a single RosR-DNA complex (Fig. 5 and supplemental Fig. S3). The fact that cg1322, which codes for a putative polyisoprenoid-binding protein, was strongly up-regulated in the ΔrosR mutant (Table 2) indicates that this gene is also repressed by RosR. In the case of cg1426, no obvious second RosR binding site could be identified. Therefore the promoter region used in the EMSA shown in Fig. 1 was divided into two smaller fragments, and each was tested by EMSAs. The fragment containing the predicted RosR binding site was shifted, yielding a single RosR-DNA complex. The other fragment located upstream of the predicted binding site was not shifted. Hence we assumed that the second binding site can be bound only after the primary site has been occupied by RosR, maybe due to DNA bending. The downstream fragment with the primary binding site was elongated stepwise in the upstream direction and tested by EMSAs. None of the longer fragments showed a double shift, and therefore the origin of this second shift is still unclear (supplemental Fig. S4).
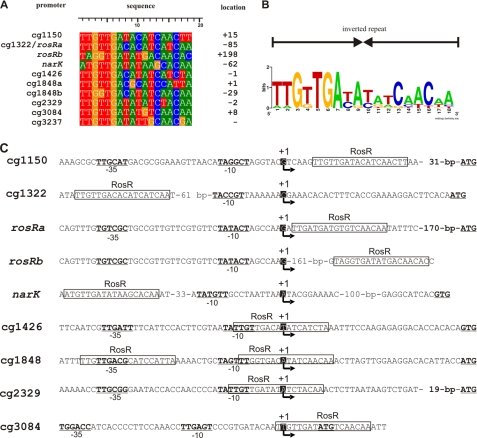
FIGURE 4.
Definition of a RosR consensus DNA binding site and distance of RosR binding sites to the determined transcriptional start sites. A, alignment of the experimentally identified RosR binding sites. The distance of the 11th nucleotide of the 20-bp binding sites to the transcriptional start site of the target genes is indicated (location). B, DNA sequence logo representing the RosR binding site in C. glutamicum. The verified binding sites were used as input for WebLogo (59). The sequence logo represents the information content of the alignment of RosR DNA binding sites, showing the sequence conservation (overall height at each position) and the relative frequency of each nucleotide at each position (nucleotide height). C, transcriptional start sites of RosR target genes. Sequences show the transcriptional start sites (+1 and arrow) identified by primer extension or 5′-RACE experiments. The RosR binding sites are indicated as boxes, and the −10 and −35 regions are underlined and printed in bold characters. The RosR binding site in front of cg1322 is the same as the one shown for rosRa.
FIGURE 5.
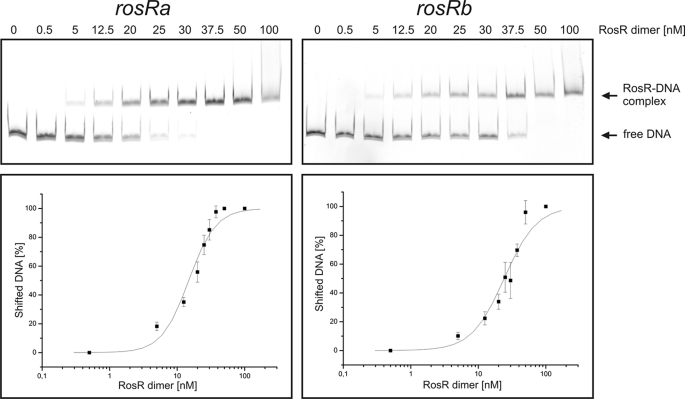
Determination of the apparent KD values for the two RosR binding sites in the rosR promoter region. Two DNA fragments (80 ng), each covering a single binding site, were incubated with increasing RosR concentrations, resolved on a 10% native polyacrylamide gel, and stained with GelRedTM At least three independent gels were performed for each binding site. The bands were quantified using ImageQuant software (GE Healthcare), and the percentage of shifted DNA was calculated. These values were plotted against the RosR concentration in log10 scale, and a sigmoidal fit was performed. The turning point of the curve was defined as the apparent KD value.
Except for the sodA gene, all other newly identified RosR target genes showed significantly increased mRNA ratios in the ΔrosR/wt transcriptome comparison (cg1150, 169.1; cg1322, 20.7; cg1848, 22.2; cg3084, 40.6). Inspection of the corresponding genomic regions indicated that the cg1848 gene presumably forms an operon with cg1849 and cg1850 (putative FMN reductase), the latter of which also showed a 420-fold increased mRNA level in the ΔrosR mutant. The cg1849 gene, coding for a hypothetical protein, also showed an increased mRNA level in all three experiments, but in this case the p value was above the threshold of 0.05. The cg3084 gene is presumably co-transcribed with cg3085 (luciferase-like monooxygenase), which showed a 2.5-fold increased expression in the ΔrosR mutant. Alignment of the DNA binding sites of RosR in the promoter regions of narKGHJI, cg1150, rosR, cg1426, cg1848, cg2329, cg3084, and sodA led to the definition of a second generation consensus binding motif of 18 bp, which represents an inverted repeat (Fig. 4, A and B).
Determination of the Apparent KD Values for the Different RosR Binding Sites
The determination of the apparent KD is exemplarily shown in Fig. 5 for the two binding sites of the rosR promoter and described under “Experimental Procedures.” The apparent KD values for all tested targets varied between 15 and 40 nm RosR dimer (Table 3), which is within the range found for other transcriptional regulators. The KD for the narK promoter, which is activated by RosR, is in the same range as those for the promoters of the repressed genes.
TABLE 3.
Apparent KD values determined for the different RosR binding sites by EMSAs
| RosR binding site | Apparent KDa |
|---|---|
| nm | |
| cg1150 | 27.7 ± 0.9 |
| cg1322/rosR-a | 14.8 ± 1.2 |
| rosR-b | 24.1 ± 1.8 |
| nark | 16.3 ± 1.3 |
| cg1426 | 39.8 ± 2.1 |
| cg1848-a | 27.8 ± 0.7 |
| cg1848-b | 33.4 ± 2.0 |
| cg2329 | 35.1 ± 2.8 |
| cg3084 | 21.8 ± 1.4 |
| cg3237 | 21.3 ± 1.3 |
a Calculated for dimeric RosR.
Identification of Transcriptional Start Sites of RosR Target Genes
According to the results described above, RosR acts as a transcriptional activator for the narKGHJI operon and as a transcriptional repressor for all other known target genes. In many cases, the function as transcriptional activator or repressor correlates with a binding site upstream of the −35 region or within or downstream of the −10/35 regions of the promoter, respectively. To analyze whether such a correlation is also true in the case of RosR, we determined the transcriptional start sites (TSS) of all known target genes except for the narKGHJI operon. Based on the fact that the narK upstream sequence of C. glutamicum ATCC 13032 is completely identical to that of C. glutamicum strain R, it was assumed that also the TSS of narK in strain ATCC 13032 is identical to the one determined previously for strain R (21). In the narK promoter the RosR binding motif, which is also completely conserved in strain R, stretches from position −131 to −112 with respect to the TSS (position +1).
The TSS of cg1322, cg1426, and cg3084 were determined by primer extension using two different primers for each gene (see supplemental Table S1). As shown in supplemental Fig. S5, the TSS were found to be located 32, 34, and 7 bp upstream of the proposed translational start sites of cg1322, cg1426, and cg3084, respectively. The TSS of rosR, cg1150, cg2329, and cg1848 were determined by 5′-RACE PCR (for primers see supplemental Table S1). In each case, 10 clones containing the amplified cDNA were sequenced to determine the 5′-ends of the mRNA. A transcriptional start site was assumed when at least three of the 10 clones showed the same 5′-end and when the other clones contained non-identical shorter transcripts. The TSS were found to be located 57, 196, 32, and 45 bp upstream of the proposed translational start sites of cg1150, rosR, cg1848, and cg2329, respectively.
In Fig. 4C the promoter sequences based on the newly identified TSS are shown. In all cases except for cg1322, the RosR binding site either overlaps with the −35 or −10 regions or is located at or immediately downstream of the TSS, indicating that repression is achieved by inhibition of RNA polymerase binding or of initiation complex formation. In the case of cg1322, the RosR binding sites are centered 86 and 247 bp upstream of the TSS. In this case, the mechanism of repression is unclear. It might involve blockage of RNA polymerase binding to the cg1322 promoter by the stalled RNA polymerase at the rosR promoter, as the −35 regions of the two divergent promoters are separated by only 5 bp.
Negative Autoregulation of rosR
The EMSAs showed binding of RosR to its own promoter region. The two RosR binding sites were centered at +11 (immediately downstream of the TSS) and +172 (overlapping the proposed translational start site) (Fig. 4C), indicating a negative autoregulation of RosR. To investigate this possibility, the rosR promoter region (extending from position −272 to +12 with respect to TSS) was cloned in front of a promoterless CAT gene in the promoter probe vector pET2. The resulting plasmid pET2-rosR was transferred into C. glutamicum wild type and the ΔrosR mutant. CAT activity was measured for cells that were cultivated in CGXII minimal medium with 4% (w/v) glucose and harvested in the exponential growth phase. The experiments were performed in triplicate and revealed a specific CAT activity of 52 ± 2 nmol/min/mg protein for wild-type cells and 318 ± 15 nmol/min/mg protein for ΔrosR cells. This result confirmed that rosR expression is negatively autoregulated.
DNA Binding of RosR Is Reversibly Inhibited by H2O2 and Dependent on the Oxidation Status of Cysteine Residues
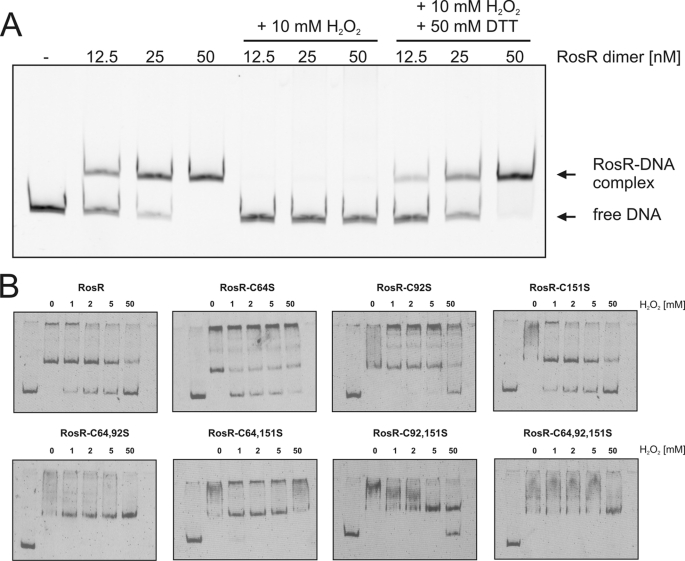
Based on the fact that several target genes encode putative monooxygenases and FMN reductases that might act in concert, we considered the possibility that some kind of oxidative stress could be sensed by RosR. To test this hypothesis, the effects of the oxidant H2O2 on the DNA binding properties of RosR were tested by EMSAs. As shown in Fig. 6A, binding of RosR to the cg2329 promoter fragment was completely inhibited by 10 mm H2O2. A similar effect was observed when cumene hydroperoxide was added in concentrations of 5 and 25 mm (data not shown). In a control experiment we tested the effect of up to 50 mm H2O2 on the binding of the response regulator MtrA of C. glutamicum to its target promoter, nlpC (16) and found no inhibition (data not shown). This indicates that there is no general inhibition of protein-DNA interaction by H2O2. Importantly, the inhibition of DNA binding by H2O2 was reversible, as the addition of 50 mm DTT to H2O2-treated RosR samples restored the DNA binding ability of the protein (Fig. 6A).
FIGURE 6.
Reversible inhibition of the DNA binding activity of RosR by H2O2 and role of cysteine residues. A, inhibition of the DNA binding activity of RosR by H2O2 and reversal of the inhibition by DTT. RosR was prepared in three different concentrations, and aliquots were taken for EMSAs (control). Then H2O2 was added to each RosR sample to a final concentration of 10 mm, and again aliquots were taken for EMSAs. In the next step DTT was added to a final concentration of 50 mm, and aliquots were taken for EMSAs. The nine aliquots were incubated in binding buffer, pH 8.0, with 80 ng of DNA covering the promoter region of cg2329 for 20 min at room temperature and then separated on a 10% native polyacrylamide gel at 180 V for 1.5 h on ice. The gel was stained using GelRedTM. B, influence of the cysteine residues in RosR on its DNA binding activity. Shown is the binding of wild-type RosR and the mutated derivatives RosR-C64S, RosR-C92S, RosR-C151S, RosR-C64S,C92S, RosR-C64S,C151S, RosR-C92S,C151S, and RosR-C64S,C92S,C151S (each 450 nm) to the promoter region of cg2329 (15 nm) in the absence and presence of H2O2 (1, 2, 5, and 50 mm) as oxidant. The samples were incubated for 20 min at room temperature and then separated by native PAGE (15%) and stained with SYBR Green I.
Inspection of the RosR amino acid sequence revealed three cysteine residues located at positions 64, 92, and 151, which might be responsible for the inhibitory effect of H2O2 on DNA binding. To test this assumption, the cysteine residues were exchanged with serine residues by site-directed mutagenesis either individually or in all possible combinations. The resulting seven RosR variants were overproduced in E. coli, purified, and used for EMSAs. As shown in Fig. 6B, all of the RosR variants were still binding-competent in the absence of H2O2, as the cg2329 promoter fragment was completely shifted at a 30-fold molar excess of these proteins. Thus, none of the cysteine residues is essential for DNA binding or dimerization of RosR. In the presence of increasing H2O2 concentrations, the DNA binding properties of the RosR derivatives varied. The mutated proteins RosR-C64S and RosR-C151S behaved similarly to wild-type RosR, meaning that DNA binding was inhibited already in the presence of 1 mm H2O2. In contrast, the DNA binding of RosR-C92S remained unaffected up to 5 mm H2O2. This indicates that oxidation of Cys-92 is important for inhibition of DNA binding by H2O2. The mutated protein RosR-C92S,C151S showed DNA binding properties similar to RosR-C92S, whereas in the case of RosR-C64S,C92S even 50 mm H2O2 did not prevent DNA binding. The same was true for the triple mutated protein RosR-C64S,C92S,C151S. Surprisingly, RosR-C64S,C151S also showed no inhibition of DNA binding by up to 50 mm H2O2, although the single mutated proteins RosR-C64S and RosR-C151S were sensitive to H2O2. Thus, the combined mutation of both residues can also lead to H2O2 insensitivity. The above results show that the inhibition of DNA binding by H2O2 is caused by the oxidation of cysteine residues, in particular Cys-92, and that H2O2-insensitive, but still binding-competent, RosR derivatives can be obtained by exchange of the cysteine residues with serines.
Evidence for a Role for Cg1322 in H2O2 Resistance
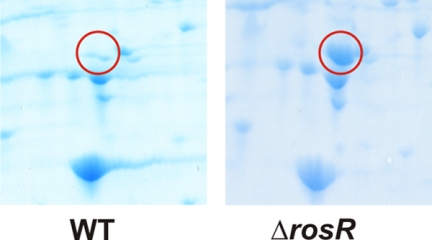
In a proteome comparison of the C. glutamicum wild type and the ΔrosR mutant using two-dimensional gel electrophoresis, Cg1322 was the only protein that showed a highly increased abundance in the ΔrosR mutant (Fig. 7). To investigate the role of the Cg1322 protein in the response of C. glutamicum to oxidative stress, the in-frame deletion mutant Δcg1322 was constructed and analyzed in agar diffusion assays. A paper disc containing 1 or 2 m H2O2 was placed on a freshly plated lawn of Δcg1322 and wild-type cells (supplemental Fig. S6). The inhibition zone around the paper disc was about 2-fold larger in the case of Δcg1322 cells (2.6 ± 1 mm for 1 m H2O2 and 4.75 ± 0.43 mm for 2 m H2O2) than in the case of wild-type cells (1.25 ± 0.82 mm for 1 m H2O2 and 2.8 ± 1.4 mm for 2 m H2O2), indicating that the absence of Cg1322 causes a significantly increased H2O2 sensitivity.
FIGURE 7.
Section of Coomassie-stained two-dimensional gels, pH 4–7, showing the strong induction of the Cg1322 protein (encircled) in the ΔrosR mutant compared with the wild type (WT). The cells were grown in glucose minimal medium and used in the exponential growth phase for preparation of cell extracts. Cg1322 was identified by peptide mass fingerprinting.
DISCUSSION
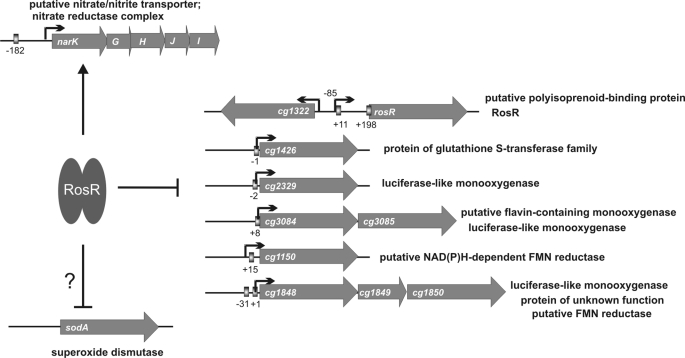
In this work we have analyzed the function of the as yet uncharacterized MarR-type transcriptional regulator RosR (Cg1324) of C. glutamicum. By combining a comparative transcriptome analysis of C. glutamicum wild type and a ΔrosR mutant and DNA interaction studies with purified RosR, direct target genes of RosR could be identified and an 18-bp consensus DNA binding site could be defined. On the basis of these data, we determined that RosR functions as an activator of the narKGHJI operon and as a repressor of at least nine genes.
The narKGHJI operon codes for a nitrate-nitrite antiporter (NarK) and dissimilatory nitrate reductase (NarGHJI), allowing C. glutamicum to reduce nitrate to nitrite and to grow under anaerobic conditions using nitrate as a final electron acceptor (2, 21). As genes for nitrite reductase or denitrification are absent in C. glutamicum (48), nitrite cannot be further metabolized and thus accumulates (2, 21). Although the activation of narKGHJI expression by RosR is missing in the ΔrosR mutant, this strain showed the same growth behavior under anaerobic conditions with nitrate as a terminal electron acceptor as did the wild type (data not shown). This might be explainable by the assumption that activation of the narKGHJI operon by RosR is not strong enough to effect growth or occurs primarily under aerobic conditions. The reduced expression of the nar operon under stress conditions that inactivate RosR might help to avoid additional stress by the accumulation of nitrite or products derived from nitrite. The nar operon of C. glutamicum was shown previously to be transcriptionally regulated also by ArnR, GlxR, and RipA (12, 35, 36).
Although none of the proteins repressed by RosR (except for RosR itself) has been studied experimentally, their bioinformatic analysis revealed some noticeable features. Four of the repressed proteins (Cg1848, Cg2329, Cg3084, and Cg3085) might be active as monooxygenases, and three of them (Cg1848, Cg2329, and Cg3085) belong to PFAM family PF00296, which harbors luciferase-like monooxygenases (37). These are flavin monooxygenases that catalyze the oxidation of long-chain aldehydes using reduced flavin as second substrate: RCHO + FMNH2 + O2 → RCOO− + H+ + FMN + H2O + light. This family also includes nonfluorescent flavoproteins such as alkanesulphonate monooxygenase, which catalyze the following reaction: R-CH2-SO3H + FMNH2 + O2 → R-CHO + FMN + sulfite + H2O (38). In agreement with the requirement of these enzymes for reduced FMN, two of the proteins repressed by RosR encode putative FMN reductases (Cg1150 and Cg1850). A better knowledge of the substrates of the putative monooxygenases would help our understanding of the function of RosR.
The protein encoded by cg1426 belongs to the glutathione S-transferase family (PFAM family PF00043). The members of this family have a variety of functions, such as detoxification of electrophilic compounds by catalyzing their conjugation to glutathione or biodegradative metabolism (39, 40). Corynebacteria, mycobacteria, and related genera contain mycothiol instead of glutathione (41, 42). Therefore, Cg1426 might form a conjugate of mycothiol with compounds such as cumene hydroperoxide.
Cg1322, which is encoded by the gene next to rosR (Fig. 8), is a member of the YceI family (PFAM family PF04264). In E. coli, YceI is induced by alkaline pH and salt stress (43, 44) and is proposed to be a secreted protein. Recently, the crystal structure of a member of the YceI protein family, TT1927b (TTHA0802) from Thermus thermophilus HB8, has been solved (45). The structure consists of an extended, eight-stranded, antiparallel β-barrel. In the hydrophobic pore of the barrel, the ligand octaprenyl pyrophosphate was identified, which was unintentionally co-purified with the protein from E. coli. The polyisoprenoid chain was bound by hydrophobic interactions. As the amino acid residues that contact the ligand are largely conserved in C. glutamicum Cg1322, it is likely that this protein also binds octaprenyl phosphate or a closely related polyisoprenoid. In E. coli, octaprenyl phosphate is synthesized by octaprenyl-pyrophosphate synthase IspB (46) and forms the side chain of the respiratory quinones menaquinone-8 (MK-8), demethylmenaquinone-8 (DMK-8), and ubiquinone-8 (UQ-8). It is attached to 1,4-dihydroxy-2-naphthoic acid, resulting in the formation of DMK-8, which is then methylated to MK-8. The former step is catalyzed by the 1,4-dihydroxy-2-naphthoic acid octaprenyltransferase encoded by the menA gene (47) and the latter by the DMK methyltransferase MenG using S-adenosyl-methionine as the methyl group donor. In C. glutamicum, a similar pathway for the synthesis of menaquinone is present (48). Interestingly, a C. glutamicum Δcg1322 mutant showed an increased sensitivity to hydrogen peroxide, indicating that the Cg1322 protein is involved in the response to oxidative stress. The mechanism of this involvement is as yet unknown, but it may be related to the observation that the adventitious autoxidation of reduced menaquinone in the cytoplasmic membrane releases a steady flux of superoxide into the periplasm of E. coli (49). Superoxide is detoxified by superoxide dismutase, which leads to the formation of hydrogen peroxide. The C. glutamicum sodA gene encoding superoxide dismutase was identified as a putative RosR target in the in silico search based on the first generation consensus binding site. Although a specific DNA binding of RosR to the sodA promoter region could be demonstrated (Fig. 3), the effect of RosR on sodA expression remains unclear. In the DNA microarray studies comparing the wild type and ΔrosR mutant, the sodA mRNA level was not changed. In additional DNA microarray experiments comparing a RosR overproducing strain with the wild type, the sodA transcript level was decreased 2-fold in the overproducing strain, suggesting a repressing effect of RosR (data not shown). Further studies are required to clarify the role of RosR in sodA expression.
FIGURE 8.
Overview of the RosR regulon. Under reducing conditions, RosR is active and binds to the 18-bp sequence motifs present in the target promoters (indicated as boxes); the numbers show the distance to the TSS (black arrow). Under oxidizing conditions, RosR is inactivated, causing the derepression of the repressed genes and loss of narKGHJI activation.
Hydrogen peroxide was found to inhibit the DNA binding ability of RosR, indicating that this regulator is redox-active. This inhibition could be reversed by the addition of excess DTT. In addition, the inhibition by H2O2 could be partially or completely prevented by exchange of one (Cys-92), two (Cys-64 and Cys-92; Cys-64 and Cys-151; and Cys-92 and Cys-151) or all three cysteine residues (Cys-64, Cys-92, and Cys-151) present in the protein to serine residues. None of the three cysteine residues per se is required for DNA binding. Therefore it is assumed that H2O2 causes the formation of intra- or intermolecular disulfide bridges in the RosR dimer, which is accompanied by conformational changes that attenuate or prohibit DNA binding. The formation of a disulfide bond is well known to serve as a mechanism for activation or inactivation of redox-responsive transcriptional regulators. The best studied example is the LysR-type regulator OxyR of E. coli, which is activated by H2O2 through the formation of a disulfide bond between Cys-199 and Cys-208 and then activates the transcription of genes necessary for defense against this type of oxidative stress (50, 51). An example of inactivation by disulfide bond formation is provided by the transcriptional regulator CprK of Desulfitobacterium dehalogenans, which controls genes involved in dehalorespiration (52, 53). Although the inactivation of RosR by the formation of disulfide bonds is an attractive model, other mechanisms such as S-thiolation cannot be excluded.
Some members of the MarR family were previously shown to be involved in the response to oxidative stresses. Bacillus subtilis OhrR is an organic peroxide sensor that represses expression of an inducible peroxiredoxin, OhrA. The DNA binding activity of OhrR is regulated by the oxidation status of its sole cysteine residue, Cys-15. After oxidation to a sulfenic acid intermediate, which retains DNA binding activity, further reactions generate either a mixed disulfide or a protein sulfenamide, both of which prevent DNA binding and cause derepession of ohrA (54). MhqR (YkvE) of B. subtilis represses genes encoding multiple dioxygenases/glyoxylases, oxidoreductases, and an azoreductase (55). MhqR target proteins confer resistance to 2-methylhydroquinone and catechol, as shown by the fact that a ΔmhqR mutant is hyper-resistant to these compounds, whereas mutants lacking target genes such as azoR2 are sensitive to these compounds. Binding of MhqR to its target genes is not affected by thiol-reactive compounds such as 2-methylhydroquinone, catechol, diamide, or methylglyoxal and hydrogen peroxide. YodB of B. subtilis represses the yocJ (azoR1) gene encoding an azoreductase, and its DNA binding activity is directly inhibited by thiol-reactive compounds and H2O2 (56).
In C. glutamicum strain R, the redox-sensing transcriptional regulator QorR has been described recently (57, 58). QorR represses the qor2 gene, located upstream and divergent from qorR and its own structural gene. The qor2 gene encodes a quinone oxidoreductase, which is involved in diamide resistance. The DNA binding activity of QorR is inhibited by oxidants such as diamide, H2O2, and cumene hydroperoxide in vitro; its only cysteine residue, Cys-17, is essential for redox-responsive regulation of QorR. The qor2 transcript level does not change in the presence of hydrogen peroxide stress but under diamide stress (58). This might be because C. glutamicum is highly resistant to hydrogen peroxide.
In summary, we identified RosR as a novel transcriptional regulator in C. glutamicum, in which DNA binding activity can be inhibited by hydrogen peroxide in vitro. Except for the narKGHJI operon, all other currently known target genes, such as cg1322, are repressed by RosR (Fig. 8). Deletion of cg1322, which encodes a putative polyisoprenoid-binding protein, caused increased sensitivity to hydrogen peroxide, supporting the role of Cg1322 and RosR in the oxidative stress response. Homologs of RosR and Cg1322 are found in other members of the Corynebacteriaceae (supplemental Fig. S7A) and some other species of the order Actinomycetales, where the corresponding genes show the same divergent organization as in C. glutamicum (supplemental Fig. S7B). Amino acid sequence alignments of the RosR homologs revealed the conservation of the cysteine residue Cys-92, found to be particularly sensitive to hydrogen peroxide.
Supplementary Material
This work was supported by the Health & Nutrition business unit of Evonik Degussa GmbH and the German Ministry of Education and Research (BMBF).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Table S1.
- CAT
- chloramphenicol acetyltransferase
- RACE
- rapid amplification of cDNA ends
- NTA
- nitrilotriacetic acid
- TSS
- transcriptional start site
- MK
- menaquinone
- DMK
- demethylmenaquinone
- TEV
- tobacco etch virus
- EMSA
- electrophoretic mobility shift assay.
REFERENCES
- 1.Kinoshita S., Udaka S., Shimono M. (1957) J. Gen. Appl. Microbiol. 3, 193–205 [PubMed] [Google Scholar]
- 2.Liebl W. (2006) Prokaryotes 3, 796–818 [Google Scholar]
- 3.Ikeda M., Nakagawa S. (2003) Appl. Microbiol. Biotechnol. 62, 99–109 [DOI] [PubMed] [Google Scholar]
- 4.Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A., Dusch N., Eggeling L., Eikmanns B. J., Gaigalat L., Goesmann A., Hartmann M., Huthmacher K., Krämer R., Linke B., McHardy A. C., Meyer F., Möckel B., Pfefferle W., Pühler A., Rey D. A., Rückert C., Rupp O., Sahm H., Wendisch V. F., Wiegräbe I., Tauch A. (2003) J. Biotechnol. 104, 5–25 [DOI] [PubMed] [Google Scholar]
- 5.Wendisch V. F., Bott M., Kalinowski J., Oldiges M., Wiechert W. (2006) J. Biotechnol. 124, 74–92 [DOI] [PubMed] [Google Scholar]
- 6.Brune I., Brinkrolf K., Kalinowski J., Pühler A., Tauch A. (2005) BMC Genomics 6, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kocan M., Schaffer S., Ishige T., Sorger-Herrmann U., Wendisch V. F., Bott M. (2006) J. Bacteriol. 188, 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engels S., Schweitzer J. E., Ludwig C., Bott M., Schaffer S. (2004) Mol. Microbiol. 52, 285–302 [DOI] [PubMed] [Google Scholar]
- 9.Engels S., Ludwig C., Schweitzer J. E., Mack C., Bott M., Schaffer S. (2005) Mol. Microbiol. 57, 576–591 [DOI] [PubMed] [Google Scholar]
- 10.Russo S., Schweitzer J. E., Polen T., Bott M., Pohl E. (2009) J. Biol. Chem. 284, 5208–5216 [DOI] [PubMed] [Google Scholar]
- 11.Krug A., Wendisch V. F., Bott M. (2005) J. Biol. Chem. 280, 585–595 [DOI] [PubMed] [Google Scholar]
- 12.Wennerhold J., Krug A., Bott M. (2005) J. Biol. Chem. 280, 40500–40508 [DOI] [PubMed] [Google Scholar]
- 13.Wennerhold J., Bott M. (2006) J. Bacteriol. 188, 2907–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frunzke J., Engels V., Hasenbein S., Gätgens C., Bott M. (2008) Mol. Microbiol. 67, 305–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Möker N., Brocker M., Schaffer S., Krämer R., Morbach S., Bott M. (2004) Mol. Microbiol. 54, 420–438 [DOI] [PubMed] [Google Scholar]
- 16.Brocker M., Bott M. (2006) FEMS Microbiol. Lett. 264, 205–212 [DOI] [PubMed] [Google Scholar]
- 17.Schaaf S., Bott M. (2007) J. Bacteriol. 189, 5002–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brocker M., Schaffer S., Mack C., Bott M. (2009) J. Bacteriol. 191, 3869–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson S. P., Grove A. (2006) Curr. Issues Mol. Biol. 8, 51–62 [PubMed] [Google Scholar]
- 20.Keilhauer C., Eggeling L., Sahm H. (1993) J. Bacteriol. 175, 5595–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura T., Vertès A. A., Shinoda Y., Inui M., Yukawa H. (2007) Appl. Microbiol. Biotechnol. 75, 889–897 [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J., MacCallum P., Russell D. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23.Inoue H., Nojima H., Okayama H. (1990) Gene 96, 23–28 [DOI] [PubMed] [Google Scholar]
- 24.Lange C., Rittmann D., Wendisch V. F., Bott M., Sahm H. (2003) Appl. Environ. Microbiol. 69, 2521–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niebisch A., Bott M. (2001) Arch. Microbiol. 175, 282–294 [DOI] [PubMed] [Google Scholar]
- 26.Vasicova P., Abrhamova Z., Nesvera J., Patek M., Sahm H., Eikmanns B. (1998) Biotechnol. Tech. 12, 743–746 [Google Scholar]
- 27.Gerstmeir R., Wendisch V. F., Schnicke S., Ruan H., Farwick M., Reinscheid D., Eikmanns B. J. (2003) J. Biotechnol. 104, 99–122 [DOI] [PubMed] [Google Scholar]
- 28.Engels V., Lindner S. N., Wendisch V. F. (2008) J. Bacteriol. 190, 8033–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaffer S., Weil B., Nguyen V. D., Dongmann G., Günther K., Nickolaus M., Hermann T., Bott M. (2001) Electrophoresis 22, 4404–4422 [DOI] [PubMed] [Google Scholar]
- 30.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 31.Pappin D. J., Hojrup P., Bleasby A. J. (1993) Curr. Biol. 3, 327–332 [DOI] [PubMed] [Google Scholar]
- 32.Takeno S., Ohnishi J., Komatsu T., Masaki T., Sen K., Ikeda M. (2007) Appl. Microbiol. Biotechnol. 75, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 33.Kronemeyer W., Peekhaus N., Krämer R., Sahm H., Eggeling L. (1995) J. Bacteriol. 177, 1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalinowski J. (2005) in Handbook of Corynebacterium glutamicum (Eggeling L., Bott M. eds) pp. 37–56, CRC Press, Boca Raton, FL [Google Scholar]
- 35.Nishimura T., Teramoto H., Vertès A. A., Inui M., Yukawa H. (2008) J. Bacteriol. 190, 3264–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohl T. A., Baumbach J., Jungwirth B., Pühler A., Tauch A. (2008) J. Biotechnol. 135, 340–350 [DOI] [PubMed] [Google Scholar]
- 37.Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) Nucleic Acids Res. 36, D281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kertesz M. A., Schmidt-Larbig K., Wüest T. (1999) J. Bacteriol. 181, 1464–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vuilleumier S., Pagni M. (2002) Appl. Microbiol. Biotechnol. 58, 138–146 [DOI] [PubMed] [Google Scholar]
- 40.Vuilleumier S. (1997) J. Bacteriol. 179, 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.den Hengst D., Buttner M. J. (2008) Biochim. Biophys. Acta 1780, 1201–1216 [DOI] [PubMed] [Google Scholar]
- 42.Newton G. L., Fahey R. C. (2002) Arch. Microbiol. 178, 388–394 [DOI] [PubMed] [Google Scholar]
- 43.Weber A., Kögl S. A., Jung K. (2006) J. Bacteriol. 188, 7165–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stancik L. M., Stancik D. M., Schmidt B., Barnhart D. M., Yoncheva Y. N., Slonczewski J. L. (2002) J. Bacteriol. 184, 4246–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Handa N., Terada T., Doi-Katayama Y., Hirota H., Tame J. R., Park S. Y., Kuramitsu S., Shirouzu M., Yokoyama S. (2005) Protein Sci. 14, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okada K., Suzuki K., Kamiya Y., Zhu X., Fujisaki S., Nishimura Y., Nishino T., Nakagawa T., Kawamukai M., Matsuda H. (1996) Biochim. Biophys. Acta 1302, 217–223 [DOI] [PubMed] [Google Scholar]
- 47.Suvarna K., Stevenson D., Meganathan R., Hudspeth M. E. (1998) J. Bacteriol. 180, 2782–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bott M., Niebisch A. (2003) J. Biotechnol. 104, 129–153 [DOI] [PubMed] [Google Scholar]
- 49.Korshunov S., Imlay J. A. (2006) J. Bacteriol. 188, 6326–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng M., Aslund F., Storz G. (1998) Science 279, 1718–1721 [DOI] [PubMed] [Google Scholar]
- 51.Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. (2001) J. Bacteriol. 183, 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pop S. M., Gupta N., Raza A. S., Ragsdale S. W. (2006) J. Biol. Chem. 281, 26382–26390 [DOI] [PubMed] [Google Scholar]
- 53.Pop S. M., Kolarik R. J., Ragsdale S. W. (2004) J. Biol. Chem. 279, 49910–49918 [DOI] [PubMed] [Google Scholar]
- 54.Lee J. W., Soonsanga S., Helmann J. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8743–8748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Töwe S., Leelakriangsak M., Kobayashi K., Van Duy N., Hecker M., Zuber P., Antelmann H. (2007) Mol. Microbiol. 66, 40–54 [DOI] [PubMed] [Google Scholar]
- 56.Leelakriangsak M., Huyen N. T., Töwe S., van Duy N., Becher D., Hecker M., Antelmann H., Zuber P. (2008) Mol. Microbiol. 67, 1108–1124 [DOI] [PubMed] [Google Scholar]
- 57.Nakunst D., Larisch C., Hüser A. T., Tauch A., Pühler A., Kalinowski J. (2007) J. Bacteriol. 189, 4696–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ehira S., Ogino H., Teramoto H., Inui M., Yukawa H. (2009) J. Biol. Chem. 284, 16736–16742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Studier F. W., Moffatt B. A. (1986) J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 61.Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994) Gene 145, 69–73 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.