Abstract
Airway smooth muscle hypertrophy is one of the hallmarks of airway remodeling in severe asthma. Several human diseases have been now associated with dysregulated microRNA (miRNA) expression. miRNAs are a class of small non-coding RNAs, which negatively regulate gene expression at the post-transcriptional level. Here, we identify miR-26a as a hypertrophic miRNA of human airway smooth muscle cells (HASMCs). We show that stretch selectively induces the transcription of miR-26a located in the locus 3p21.3 of human chromosome 3. The transcription factor CCAAT enhancer-binding protein α (C/EBPα) directly activates miR-26a expression through the transcriptional machinery upon stretch. Furthermore, stretch or enforced expression of miR-26a induces HASMC hypertrophy, and miR-26 knockdown reverses this effect, suggesting that miR-26a is a hypertrophic gene. We identify glycogen synthase kinase-3β (GSK-3β), an anti-hypertrophic protein, as a target gene of miR-26a. Luciferase reporter assays demonstrate that miR-26a directly interact with the 3′-untranslated repeat of the GSK-3β mRNA. Stretch or enforced expression of miR-26a attenuates the endogenous GSK-3β protein levels followed by the induction of HASMC hypertrophy. miR-26 knockdown reverses this effect, suggesting that miR-26a-induced hypertrophy occurs via its target gene GSK-3β. Overall, as a first time, our study unveils that miR-26a is a mechanosensitive gene, and it plays an important role in the regulation of HASMC hypertrophy.
Keywords: C/EBP Transcription Factor, Glycogen Synthase Kinase 3, MicroRNA, Signal Transduction, Smooth Muscle
Introduction
MicroRNAs (miRNAs)2 are an evolutionarily conserved novel class of small non-coding RNAs that have achieved status as potent regulators of gene expression. According to their genomic location relative to protein-coding gene locus, miRNAs can be divided into intergenic miRNAs and intragenic miRNAs. The former have independent transcriptional units, including promoter, transcript sequence, and terminator; therefore, they do not overlap with other genes (1, 2). The later are found in the introns of protein-coding host genes, and they generally share the same regulatory motifs with their host genes (3–5). Most of the miRNAs are transcribed by RNA polymerase II as primary miRNAs (1, 2) and processed by the RNase III enzymes Drosha and Dicer to produce 21- to 23-nucleotide double-stranded RNA duplexes (6, 7). These smaller RNAs are then exported to the cytoplasm by Exportin 5 (8, 9), where they are subsequently processed into mature miRNAs by Dicer. The mature miRNAs are loaded into the miRNA-induced silencing complex (7), where they recognize their target protein-coding mRNAs to inhibit mostly mRNA translation or degradation (10) by base pairing to complementary sequences within the 3′-untranslated region (3′ UTR). With respect to miRNAs functions, they play pivotal roles in the pathophysiological processes such as apoptosis, cell differentiation, cell proliferation, and organ development (4, 11). Functionally speaking, several human diseases have now been associated with dysregulated miRNAs expression.
Airway remodeling is a characteristic feature observed in the airways of patients having severe asthma and chronic obstructive pulmonary disease. Clinical studies have shown that both hypertrophy and hyperplasia of human airway smooth muscle cells (HASMCs) play key roles in airway remodeling (12–16). Many lines of evidence have demonstrated that hypertrophy in cardiomyocytes, skeletal myotubes, and smooth muscle cells is induced by various hypertrophic stimuli, including mechanical stretch (17–27). Recently, it has been shown that miRNAs play important roles in the induction of cardiac hypertrophy as well as response to hypertrophic stimuli (28–32). However, the role of miRNAs in the regulation of smooth muscle hypertrophy is completely unknown. Furthermore, the induction of specific miRNAs in response to hypertrophic stimuli, including stretch, is also lacking in these cells.
The present study was aimed to investigate whether stretch can induce miRNAs expression, and whether miRNAs are involved in the regulation of airway smooth muscle hypertrophy. Our results show that stretch induces HASMC hypertrophy through miR-26a up-regulation. Promoter analysis of the miR-26a gene reveals that C/EBPα directly binds to the promoter of miR-26a and activates its expression. In addition, we identify glycogen synthase kinase-3β (GSK-3β) as a target gene of miR-26a. Luciferase reporter assay demonstrate that miR-26a directly interacts with the 3′UTR of the GSK-3β mRNA. Consequently, miR-26a is able to convey the hypertrophic signal by suppressing the translation of GSK-3β mRNA. These findings reveal that miR-26a is a novel regulator in airway smooth muscle hypertrophy.
EXPERIMENTAL PROCEDURES
Cell Culture and Stretch
Primary HASMCs (obtained from Lonza, Walkersville, MD) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1× non-essential amino acids, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator containing 5% CO2 at 37 °C. Prior to experiments, 2 × 105 cells at passage 7 were plated on type I collagen-coated Bioflex 6-well plates or normal cell culture 6-well plates in the above growth medium. All experiments were conducted after the cells were serum-deprived for 24 h. For stretch stimulation, cells grown on Bioflex plates were stimulated with a 1-h cyclic strain at 1 Hz (0.5 s of deformation alternating with 0.5 s of relaxation) for every 12 h or unless otherwise stated using a computer-controlled vacuum strain apparatus (Flexercell Strain Unit, FlexCell International, Hillsborough, NC) with a vacuum pressure that is sufficient to generate 12% strain. Cells grow in normal cell culture plates were used for non-stretch experiments.
miRNA Microarray Analysis
After stretch, total RNA samples were isolated by TRIzol reagent according to the manufacturer's protocol (Invitrogen). Ten micrograms of total RNA was size-fractionated with YM-100 Microcon centrifugal filter (Millipore, Billerica, MA), and then used for miRNA expression analysis with miRNA microarray (LC Sciences, Houston, TX).
Construction of Expression Plasmids
miR-26a precursor DNA containing 77-bp stem-loop sequence and 100-bp native flank sequence to both upstream and downstream of the stem loop (as shown in Fig. 4A) was synthesized with miR-26a-C-F and miR-26a-C-R primers, and cloned into pSilencer 4.1-CMV vector (Ambion, Austin, TX) according to the manufacturer's instructions. GSK-3β cDNA with or without 3′UTR was synthesized and cloned into pcDNA 3.1D/V5-His-TOPO vector (Invitrogen) according to the manufacturer's instructions. GSK-3β-F1 and GSK-3β-R1 primers were used to synthesize GSK-3β cDNA with 3′UTR. GSK-3β cDNA without the 3′UTR was synthesized using GSK-3β-F1 and GSK-3β-R2 primer sets. To generate reporter vector bearing miR-23a-binding sites, a 650-bp human GSK-3β 3′UTR sequence was synthesized and cloned into pmirGLO vector (Promega, Madison, WI) according to the manufacturer's instructions. GSK-3β-3′UTR-F and GSK-3β-3′UTR-R primers were used. To generate reporter vector bearing C/EBPα-binding elements, a 750-bp and a 1000-bp human CTDSPL/miR-26a 5′UTR DNA (as shown in Fig. 4) were synthesized and cloned into pGL4.1 luciferase reporter vector (Promega). The following primer pairs were used: miR-26a 5′UTR-F1 and miR-26a 5′UTR-R1 (for 1000 bp), and miR-26a 5′UTR-F2 and miR-26a 5′UTR-R2 (for 750 bp). PCRs were performed to synthesis inserts with AccuPrime Pfx DNA polymerase according to the manufacturer's protocols (Invitrogen). Primers information was detailed in Table 1. Constructs were sequenced by the DNA Sequence Core Facility of Baylor College of Medicine to verify insert identities.
FIGURE 4.
Enforced expression of miR-26a induces hypertrophy. A, structure of pSilencer-miR-26a construct contains pre-miR-26a, cytomegalovirus promoter (CMV), simian virus 40 (SV40), and neomycin. The blue letters represent two restriction sites BamHI and HindIII. The red letters indicate 22 bases of mature miR-26a sequence. The underlined letters represent 77-bp stem-loop sequence, and the green letters indicate 100-bp native flank sequence to both upstream and downstream of the stem-loop sequence. B–E, cells were transfected with pSilencer or pSilencer-miR-26a and/or anti-miR-26a for 24 h followed by a 1-h stretch for every 12 h. B, 36 h after transfection, the overexpression of pre-miR-26a and mature miR-26a was confirmed by RT-qPCR (upper panel) and solution hybridization methods (lower panel), respectively. Cell size, DNA synthesis, protein synthesis, and cell count (C), and α-actin, SM22, and MHC expressions (D) were analyzed 72 h after transfection. Protein/DNA ratio (E) was also analyzed. β-Actin served as a loading control. Gel pictures are representative of three separate experiments. δ, p < 0.05 versus control and *, p < 0.05 versus pSilencer-miR-26a. Each bar indicates mean ± S.E. (n = 3).
TABLE 1.
Primers used in PCR
| Primer name | Sequence 5′-3′ | Underline | Purpose |
|---|---|---|---|
| miR-26a-C-F | GGATCCGTGATATCACAAGGTCCCAG | BamHI | Cloning |
| miR-26a-C-R | AAGCTTCTACATGCAAAGGGCAGGAG | HindIII | Cloning |
| GSK-3β-F1 | CACCGGTGATTCGCGAAGAGAGTG | Cloning | |
| GSK-3β-R1 | TCATGGAGTTGGAAGCTGATGCA | Cloning | |
| GSK-3β-F1 | CACCGGTGATTCGCGAAGAGAGTG | Cloning | |
| GSK-3β-R2 | TGTGCAGCTGGCTGCTCGGG | Cloning | |
| GSK-3β-3′UTR-F | GCTAGCTGCTGGAGTATACACCAACTGCCC | SacI | Cloning |
| GSK-3β-3′UTR-R | CTCGAGGCATGAGGCAGGAGTCCTGTTTTT | XhoI | Cloning |
| miR-26a 5′UTR-F1 | GGTACCCCTGTTTGGCCTCGCCTGCT | KpnI | Cloning |
| miR-26a 5′UTR-R1 | GAGCTCGGAGGCAGGAGCGAGGAAGG | SacI | Cloning |
| miR-26a 5′UTR-F2 | GGTACCCCTTCCAGTGCCCCCCACGG | KpnI | Cloning |
| miR-26a 5′UTR-R2 | GAGCTCTGGGTGCGCGGCGCGGCGGC | SacI | Cloning |
| ChIP-F1000 | CTGTTTGGCCTCGCCTGCTG | qPCR | |
| ChIP-R1000 | GGAGGCAGGAGCGAGGAAGG | qPCR | |
| ChIP-F750 | CTTCCAGTGCCCCCCACGGG | qPCR | |
| ChIP-R750 | GGGTGCGCGGCGCGGCGGCC | qPCR | |
| CTDSPL-F | TGCTGAGGGAGGGGAGTGAG | qPCR | |
| CTDSPL-R | GCAGCATGCCACAGGTTGTC | qPCR | |
| CTDSP2-F | CCAGCTGGCCGTGAAGAGGC | qPCR | |
| CTDSP2-R | GGACACGGGGCGGTTTCCAG | qPCR | |
| GSK-3β-F2 | GGCCCAGAACCACCTCCTTT | qPCR | |
| GSK-3β-R3 | CCTTGCTGCCGTCCTTGTCT | qPCR | |
| GAPDH-F | GAAGGTGAAGGTCGGAGTCA | qPCR | |
| GAPDH-R | TGGAAGATGGTGATGGGATT | qPCR |
Transfection and Luciferase Assays
Cells were grown in Opti-MEM I medium (Invitrogen) for 24 h before transfection. Cells were transfected with 2.2 μg of expression vector bearing has-miR-26a precursor, GSK-3β cDNA, or GSK-3β cDNA without 3′UTR, or 2.4 μg of reporter vector bearing GSK-3β 3′UTR or C/EBPα-binding elements by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. The Renilla luciferase vector pGL4.74 was co-transfected with firefly luciferase vectors as a normalizer. For miR-26a inhibitor assays, cells were transfected with 400 ng of has-miR-26a miRCURY LNA knockdown probe (antagomir) or scrambled probe (Exiqon, Woburn, MA). For small interference RNA-mediated knockdown studies, 500 pmol of small interference RNA specific for human C/EBPα or nonspecific small interference RNA (Santa Cruz Biotechnology, Santa Cruz, CA) was used to transfect HASMCs. RNA transfection studies were performed with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. After 8 h, the transfection medium was replaced with the growth medium. Subsequent assays were made after 24- to 48-h transfection. Luciferase activity was measured with the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer's protocol.
ChIP Assays
Cells were fixed with 1% formaldehyde at room temperature for 10 min and washed with ice-cold 1x phosphate-buffered saline. After cells were scraped off in buffer I (0.25% Triton X-100/10 mm EDTA/0.5 mm EGTA/10 mm Hepes, pH 6.5), cell were pelleted by centrifugation and washed in buffer II (200 mm NaCl/1 mm EDTA/0.5 mm EGTA/10 mm Hepes, pH 6.5). Two hundred-microliter cell pellets were resuspended in 1 ml of lysis buffer (0.5% SDS/10 mm EDTA/50 mm Tris, pH 8.1/1× protease inhibitor mixture (Roche Applied Science, Indianapolis, IN)/1 mg/ml 4-(2-aminoethyl)benzenesulfonyl fluoride) and sonicated four times for a 30-s interval of 0.5-s pulses (Fisher, model 550 Sonic Dismembrator). Cell debris was removed by centrifugation, and the chromatin solutions were diluted 5× with dilution buffer (1% Triton X-100/2 mm EDTA/150 mm NaCl/20 mm Tris, pH 8.1/1× protease inhibitor mixture). Chromatin fragments were immunoprecipitated with specific antibodies overnight at 4 °C. For a 5-ml diluted chromatin solution, the following amount of antibodies was used: 5 μg of C/EBPα (Upstate Biotechnology, Lake Placid, NY), 5 μg of RNA polymerase II or 4 μg of non-immune serum. The purified DNA was used as template and quantified using qPCR system with ChIP-F1000 and ChI-R1000, and ChIP-F750 and ChIP-R750 primer sets (Table 1). The amplified PCR products were resolved by using 1% agarose gel.
Solution Hybridization Detection Analysis
The expression levels of mature miRNAs were measured by solution hybridization detection method with mirVana miRNA and BrightStar BioDetect Kits (Ambion) according to the manufacturer's protocol. Briefly, 5 μg of total RNA was incubated with 5′-biotin end-labeled miR-26a probes, which were obtained from Exiqon, at 42 °C for overnight. Unhybridized RNA and excess probe were removed by a rapid ribonuclease digestion step. The hybridized RNA fragments were resolved by 15% denaturing polyacrylamide gel, transferred to positively charged nylon membrane, and detected by using a chemiluminescent detection system.
RT-qPCR Expression Analysis
All RNAs were treated with TURBO DNase (Ambion). RT-qPCRs for miRNA expression were performed by using the mirVana RT-qPCR miRNA detection kit (Ambion) following the manufacturer's instructions. Fifty nanograms of RNA and 10 μm of miRCURY LNA PCR Primer Sets (Exiqon) specific for hsa-miR-26a or U6 small nuclear RNA (Ambion) as a sample normalizer were used in each reaction. RT-qPCRs for mRNA expression were performed by using the following method. One microgram of total RNA was reverse-transcribed by using SuperScript III First-Strand Synthesis Super Mix according to the manufacturer's protocols (Invitrogen). PCRs were performed by using Brilliant II SYBR Green QPCR Master Mix (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The following primer sets were used in PCRs: human GAPDH primers were used as a sample normalizer. RT-qPCRs were performed on an Mx 3005p Real Time PCR system (Stratagene). The temperature cycle profile for the qPCR reactions was 95 °C for 15 min and 40 cycles of 94 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. Melting curve analysis was also included at one cycle of 95 °C for 1 min, 55 °C for 30 s, and 95 °C for 30 s to verify the specificity of the amplified PCR products. The amount of amplified transcripts (2 − ΔCT) was estimated by the comparative CT (ΔCT) method and normalized to an endogenous reference (GAPDH) relative to a calibrator. All PCR products were verified on agarose gel stained with ethidium bromide to discriminate between the correct amplification products and the potential primer dimers.
Western Blot
Cell lysates were isolated by using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) according to the manufacturer's instructions. Forty micrograms of proteins was resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Membrane was blocked with 5% fat-free milk for 1 h and probed with mouse anti-α-smooth muscle (SM) actin, anti-SM22, anti-smMHC (myosin heavy chain), anti-GSK-3β (glycogen synthase kinase-3β), or anti-tubulin. Antibody binding was detected with a peroxidase-conjugated goat anti-mouse IgG and chemiluminescence (Pierce).
DNA and Protein Synthesis Analyses
We used Click-iT EdU (5-ethynyl-2′-deoxyuridine, a thymidine analog) and Clic-iT HPG (l-homopropargylglycine, a glycine analog) according to the manufacturer's instructions (Invitrogen). Briefly, after treatments, cells were incubated with 10 μm EdU or HPG for the indicated periods, harvested, washed, and fixed with Click-iT fixative for 15 min. Cells were permeabilized with 1× saponin-based permeabilization and wash buffer for 30 min. After washing, the incorporation of EdU or HPG was detected by using Click-iT Cell Reaction Buffer Kit (Invitrogen) according to the manufacturer's instructions. Flow cytometry was used to estimate the fluorescence intensity of Alexa Fluor-488 bound EdU or HPG.
Cell Size Analysis
Cells were stained for α-actin fibers, and cell size was determined by computer-assisted planimetry. 100–200 cells in 20–30 fields were examined in each experiment.
Statistical analysis
The results are expressed as means ± S.E. of at least three independent experiments. The comparison among different groups was performed by one-way analysis of variance followed by Bonferroni test using SigmaStat 3.5 software. Paired data were evaluated by Student's t test. p < 0.05 was considered statistically significant.
RESULTS
Mechanical Stretch Up-regulates miRNAs Expression
Others and we have previously shown that stretch can induce HASMCs gene expressions through mechanosignal transduction pathways (33–38). To explore whether stretch can induce the expression of miRNAs in HASMCs, we performed a microRNA array screen using total RNA isolated from HASMCs 12 h after stretch. Among 837 mRNAs, the array uncovered the induction of 50 differentially regulated mechanosensitive miRNAs, and this was based on a p value of 0.01. Among those, 28 miRNAs were up-regulated, including the highly up-regulated miRNAs miR-16, miR-26a, and miR-140 (Fig. 1A), and 22 miRNAs were down-regulated. To confirm the validity of miR-16, miR-26a, and miR-140 up-regulations by stretch, a portion of the RNA used for the microarray was converted into cDNA and subjected to qPCR or solution hybridization analysis. Consistent with the microarray findings, miR-16, miR-26a, and miR-140 were strongly up-regulated by stretch (Fig. 1, B and C). The small nuclear RNA U6, a control and normalizer for miRNAs, was relatively unchanged by stretch. These results indicate that HASMCs respond to stretch by strongly up-regulating miR-16, miR-26a, and miR-140, and the genes that transcribe these miRNAs are mechanosensitive.
FIGURE 1.
Stretch alters micro-RNAs expression in HASMCs. A, HASMCs were stimulated with stretch for 1 h. Total RNA was isolated after 12 h and used to conduct a microarray analysis to determine the expression levels of human miRNAs. Data are presented on a scatter plot showing log 10-transformed signal intensities for each probe on both channels for the Cy3-labeled controls (no stretch) and Cy5-labeled stretch stimulated samples. Each dot represents one miRNA probe. B and C, RNA used in A was analyzed by solution hybridization technique with 5′-biotin-labeled miR-16, miR-26a, and miR-140 and small nuclear RNA U6 (B) and, in a separate experiment, by qPCR to assay the expression of miR-16, miR-26a, miR-140, and U6 under the same conditions (C). U6 served as both loading control and normalizer. *, p < 0.05 versus corresponding control without stretch (white bar). Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (n = 3).
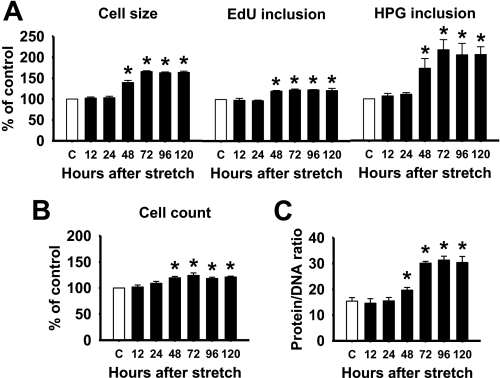
Stretch Induces HASMCs Hypertrophy
Accumulating evidences have demonstrated that stretch can induce cardiac, skeletal, and smooth muscle cell hypertrophy (17–27). To test this in HASMCs, we stimulated the cells with a 1-h stretch for every 12 h up to 3 days. Our data showed that cells stimulated with stretch displayed an increase in cell size (Fig. 2A). Changes in cell size were accompanied by an increase in protein synthesis, because the HPG incorporation was enhanced. Stretching of HASMCs also increased DNA synthesis as evidenced by higher incorporation of EdU (Fig. 2A). The increase in DNA synthesis was accompanied by an increase in proliferation (Fig. 2B). The protein synthesis was much higher than the DNA synthesis as evidenced by the increased protein/DNA ratio (Fig. 2C). All these changes were effective from 48 h after initiation of stretch. These data provide experimental evidences demonstrating that stretch can induce both hypertrophy and hyperplasia of HASMCs, but the former is more dominant response than the later one.
FIGURE 2.
Stretch induces HASMC hypertrophy and hyperplasia. HASMCs were stimulated with a 1-h stretch for every 12-h period, and then cell size, DNA synthesis, protein synthesis (A), protein/DNA ratio (B), and cell count (C) were analyzed for the indicated periods. *, p < 0.05 versus control without stretch (white bar) for the indicated time point. Each bar indicates mean ± S.E. (n = 3).
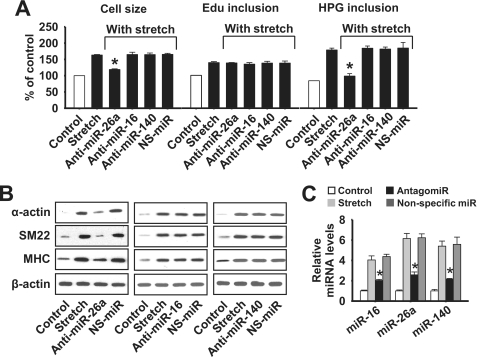
Stretch-induced miR-26a Participates in Initiating Hypertrophy
Recent studies have shown that miRNAs play important roles in cardiac hypertrophy (28–32). Induction of miRNAs and hypertrophy by stretch led us to consider whether the above stretch-induced miRNAs may participate in conveying the hypertrophic effect of stretch. To this end, we used miR-16, miR-26a, and miR-140 antagomirs (anti-miR) to knock down their endogenous expressions in the stretch-induced hypertrophic cascade. Interestingly, miR-26a knockdown attenuated the stretch-induced hypertrophic responses as evidenced by decrease in cell size, HPG incorporation (Fig. 3A), and contractile proteins such as α-smooth muscle actin, SM22, and smooth muscle MHC expressions (Fig. 3B). In contrast, neither miR-16 nor miR-140 had a role in any of the above stretch-induced hypertrophic responses. These results indicate that miR-26a unlike miR-16 and miR-140 is involved in mediating the hypertrophic effects of stretch. In addition, miR-26a, miR-16, and miR-140 did not attenuate the EdU incorporation, which was increased by stretch (Fig. 3A), suggesting that these miRNAs are unable to inhibit the stretch-induced hyperplasia. We tested whether transfection of HASMCs with the antagomir of miR-16, miR-26a, or miR-140 could influence their endogenous levels of expression. As expected, miR-16, miR-26a, and miR-140 antagomirs inhibited their endogenous levels of expressions, suggesting the specificity of the antagomirs (Fig. 3C).
FIGURE 3.
Stretch-induced miR-26a is involved in initiating hypertrophy. HASMCs were transfected with anti-miR-26a, anti-miR-16, anti-miR-140, or nonspecific miRNA (NS-miR). Twenty-four hours after transfection cells were stimulated with a 1-h stretch for every 12 h. A, cell size, DNA synthesis, and protein synthesis were analyzed after 72 h. B, a portion of cells used in A was analyzed to show contractile proteins such as α-actin, smooth muscle 22 (SM22), and myosin heavy chain (MHC) expressions by Western blot. C, cells were transfected with anti-miR-26a, anti-miR-16, anti-miR-140, or NS-miR for 24 h followed by a 1-h stretch. Total RNA was isolated after 36 h, and the levels of miRNAs were quantified by RT-qPCR. Gel pictures are representative of three separate experiments. *, p < 0.05 versus stretch. Each bar indicates mean ± S.E. (n = 3).
Enforced Expression of miR-26a Induces Hypertrophy
We overexpressed miR-26a in HASMCs to evaluate whether miR-26a itself induces hypertrophy in the absence of stretch. We engineered an miR-26a-expressing construct in an expression vector (Fig. 4A). Expression of the pre-miR-26a and mature miR-26a were observed in HASMCs stably transfected with the construct (Fig. 4B). Our results showed that miR-26a alone could induce HASMC hypertrophy as evidenced by increased cell size, HPG incorporation (Fig. 4C), contractile proteins expressions (Fig. 4D), and protein-DNA ratio (Fig. 4E). Introduction of anti-miR-26a blocked the miR-26a-induced hypertrophic effects. Surprisingly, miR-26a inhibited cell proliferation, because the Edu incorporation and total number of cells were reduced (Fig. 4C). Overall, regardless of stretch, miR-26a up-regulation induces HASMC hypertrophy. Furthermore, miR-26a inhibits the proliferation of HASMCs, and such inhibitory effect occurs only in the absence of stretch.
GSK-3β Is a Target of miR-26a
To find out the molecular target of miR-26a, we searched for predicted miR-26a targets, focusing our attention on the regulators of hypertrophy. In agreement with this, the public data base of animal miRNA miRGen (available on-line) has listed GSK-3β as one of the potential targets of miR-26a. GSK-3β has a conservative miR-26a seed sequence in its 3′UTR (Fig. 5A). Recent studies have shown that GSK-3β is an anti-hypertrophic molecule and negatively regulates hypertrophy in airway smooth muscle, cardiac, and skeletal muscles (39–47). These data provided a strong rationale to test our hypothesis of whether GSK-3β is functionally a downstream target of miR-26a in the hypertrophic pathway of HASMCs. First, we detected the expression levels of GSK-3β in the hypertrophic model of stretch. Both total and phosphorylated GSK-3β protein levels were decreased in a time-dependent manner upon stretch (Fig. 5B). Second, we tested whether miR-26a transcriptionally or post-transcriptionally suppresses the endogenous GSK-3β expression. To test this, we overexpressed miR-26a in HASMCs using the pSilencer-miR-26a construct. The enforced expression of miR-26a significantly decreased GSK-3β protein but not mRNA levels, (Fig. 5C), suggesting that miR-26a predominantly suppresses GSK-3β translation. To confirm whether miR-26a influences the protein translation of GSK-3β, we analyzed the protein levels of GSK-3β in cells that overexpress both miR-26a and GSK-3β. We cloned GSK-3β full-length cDNA in the mammalian expression vector pcDNA 3.1. Expression of GSK-3β mRNA and protein were observed in HASMCs stably transfected with the construct (Fig. 5D). GSK-3β expression was suppressed by miR-26a in a dose-dependent manner (Fig. 5E).
FIGURE 5.
GSK-3β is a target of miR-26a. A, sequence alignment of putative miR-26a targeting site in 3′UTR of GSK-3β shows a high level of complementarily and sequence conservation. B, HASMCs were stimulated with a 1-h stretch. Total and phosphorylated (p) GSK-3β protein levels were analyzed by Western blot for the indicated periods. C, HASMCs were transfected with pSilencer or pSilencer-miR-26a expression construct. GSK-3β mRNA levels were analyzed 24 h after transfection by RT-qPCR (left panel), and total GSK-3β protein levels were analyzed 48 h after transfection by Western blot (right panel). D, cells were transfected with pcDNA-GSK-3β construct, and overexpressions of GSK-3β mRNA (after 36 h) and protein (after 48 h) were confirmed by RT-PCR (upper panel) and Western blot (lower panel) methods, respectively. E, cells were transfected with pcDNA-GSK-3β construct, along with indicated amount of pSilencer-miR-26a construct. GSK-3β protein expression was analyzed by Western blot 48 h after transfection. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (n = 3).
Subsequently, we analyzed the effect of miR-26a on GSK-3β translation. To do this, a reporter construct containing the luciferase gene fused to the GSK-3β-3′UTR (luc-GSK-3β-3′UTR) was transfected into HASMCs with or without miR-26a overexpression vector. As shown in Fig. 6A, cells transfected with luc-GSK-3β-3′UTR alone had luciferase activity, and cells co-transfected with pSilencer-miR-26a significantly reduced the luciferase activity. Introduction of miR-26a antagomir was reverted to the inhibitory activity of miR-26a. SC-miR-26a antagomir was also able to rescue luc-GSK-3β-3′UTR luciferase activity (Fig. 6A). Finally, we tested whether miR-26a suppresses GSK-3β by base pairing on the 3′UTR of GSK-3β mRNA. To do this, we cloned GSK-3β cDNA without 3′UTR in the pcDNA 3.1 vector, and the expression of GSK-3β mRNA without the 3′UTR in HASMCs stably transfected with the construct was confirmed by RT-qPCR and Western blot (Fig. 6B). As shown in Fig. 6C, miR-26a was unable to influence the expression of GSK-3β without its 3′UTR, suggesting that the GSK-3β 3′UTR contains an active seed of miR-26a. Overall, these data provided experimental evidences demonstrating that GSK-3β is a target gene of miR-26a.
FIGURE 6.
miR-26a directly binds on the 3′UTR of the GSK-3β mRNA. A, HASMCs were transfected with the GSK-3β-3′UTR-luciferase construct, along with pSilencer-miR-26a construct. Forty-eight hours after transfection, cells were collected and then firefly luciferase activities were estimated and normalized to Renilla luciferase activities. *, p < 0.05 versus GSK-3β-3′UTR-luciferase construct alone. B, cells were transfected with pcDNA or pcDNA-GSK-3β without 3′ UTR. Forty-eight hours after transfection, cells were collected for the analysis of GSK-3β mRNA and protein expressions by RT-PCR (upper panel) and Western blot (lower panel) methods, respectively. C, cells were co-transfected with pcDNA or pcDNA-GSK-3β without 3′ UTR and/or pSilencer-miR-26a. Forty-eight hours after transfection, cells were collected for the analysis of GSK-3β protein expression by Western blot method. D, cells were transfected with pcDNA or pcDNA-GSK-3β construct (with 3′UTR), and cell size and protein synthesis were determined 72 h after transfection. *, p < 0.05 versus miR-26a alone. E, a portion of cells used in C was stimulated with a 1-h stretch every 12 h. Cell size and protein synthesis were measured 72 h after transfection. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (n = 3).
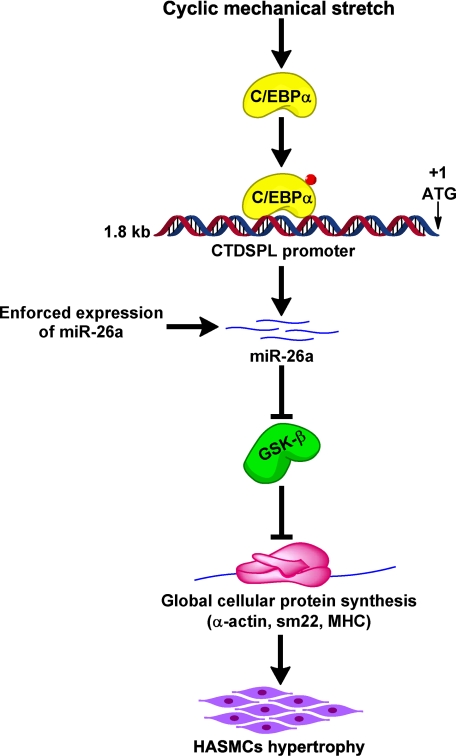
To understand if enforced expression of GSK-3β can influence the hypertrophic cascades induced by stretch or miR-26a we used cells that overexpress GSK-3β from Fig. 5D. Transfection of miR-26a induced HASMC hypertrophy, and such hypertrophy was inhibited by the ectopic expression of GSK-3β (Fig. 6D). Ectopic expression of GSK-3β also was able to attenuate the stretch-induced hypertrophy (Fig. 6E). These data suggest that miR-26a and GSK-3β are both involved in the regulation of HASMC hypertrophy. Overall, our data support a model in which miR-26a participates in HASMC hypertrophy by suppressing GSK-3β protein expression, which in turn triggers the expression of smooth muscle-specific markers and hypertrophy (see Fig. 10).
FIGURE 10.
miR-26a pathway in the initiation of HASMC hypertrophy. Up-regulation of miR-26a by stretching of HASMCs or enforced expression of mir-26a suppresses GSK-3β protein expression that triggers global cellular protein synthesis. As a result, smooth muscle hypertrophic marker proteins (α-actin, SM22, and MHC) are up-regulated, which, in turn, initiates hypertrophy.
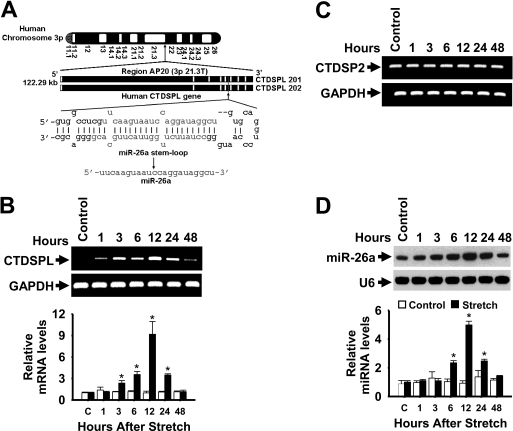
Stretch Selectively Transcribes miR-26a
Two loci align with miR-26a in the human genome (48). miR-26a-1 is located in an intron of C-terminal domain RNA polymerase II polypeptide A small phosphatase-like (CTDSPL) at chromosome 3 (Fig. 7A), and miR-26a-2 is located in an intron of C-terminal domain RNA polymerase II polypeptide A small phosphatase 2 (CTDSP2) at chromosome 12 (figure not shown). To understand whether the stretch-induced miR-26a transcription occurs via the activation of CTDSPL and/or CTDSP2 gene(s) we performed qPCR with primers specific for CTDSPL and CTDSP2. Surprisingly, stretch increased the mRNA levels of CTDSPL (Fig. 7B), but not CTDSP2 (Fig. 7C), in a time-dependent manner. More precisely, CTDSPL mRNA was detectable at 1 h, elevated at 12 h, suddenly reduced at 24 h, and was reduced at 48 h after stretch. To monitor the kinetics of miR-26a induction, mature miR-26a was assayed over a 48-h time course after stretch. miR-26a induction by stretch followed a similar pattern of expression as CTDSPL, reaching its highest levels at 12 h and slowly decreasing at 24 and 48 h after stretch (Fig. 7D). In these experiments, the levels of U6 and GAPDH, a control and normalizer for miR-26a and CTDSPL, respectively, were relatively unchanged by stretch. These findings provide evidence that 1) human CTDSPL is expressed at lower levels, and its levels increases by stretch, 2) most of the stretch-induced miR-26a transcribes from the miR located on chromosome 3, and 3) the expression of miR-26a is not correlated with the expression of CTDSP2 in response to stretch.
FIGURE 7.
Kinetics of stretch induction of CTDSPL mRNA and mature miR-26a. A, depiction of the genomic structure of the human CTDSPL coding RNA genes in chromosome 3p and location of miR-26a in introns 5 (CTDSPL 201) and 4 (CTDSPL 202). B and D, HASMCs were stimulated with a 1-h stretch, and total RNA was isolated over a 48-h period. The levels of CTDSPL (B) and CTDSP2 (C) mRNAs were analyzed by RT-qPCR (data for CTDSPL mRNA levels by qPCR are not shown) and resolved in 2% agarose gel electrophoresis. Primers were designed to target CTDSPL (identical for CTDSPL 201 and 202) and CTDSP2 sequences extending outside of miR-26a. GAPDH served as both loading control and normalizer. The levels of CTDSPL were presented relatively to that of GAPDH level. *, p < 0.05 versus corresponding control without stretch for the indicated time points. RNA used in the above experiments was also analyzed by solution hybridization technique with 5′-biotin-labeled miR-26a and small nuclear RNA U6 and in a separate experiment by RT-qPCR to assay expression of miR-26a and U6 under the same conditions (D). U6 served as both loading control and normalizer. *, p < 0.05 versus corresponding control without stretch for the indicated time points. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (n = 3).
miR-26a Is under the Control of C/EBPα in Stretch-induced Hypertrophy
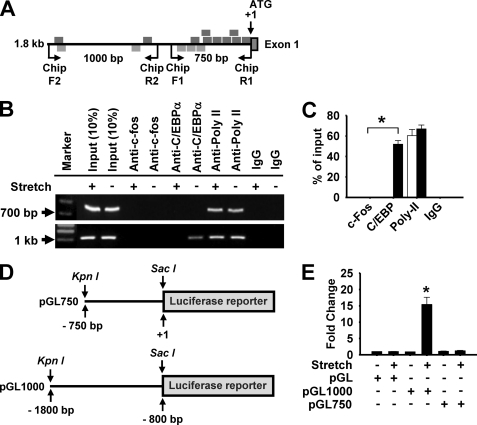
To determine the specific transcription factor by which primary miR-26a was induced by stretch we first analyzed the miR-26a promoter region using the public software PATCH (available on-line), focusing on mechanosensitive AP-1 and C/EBPα transcription factors that are activated by stretch in HASMCs (34). A scan of 1.8 kb genomic sequence located upstream of the ATG of CTDSPL gene identified eight putative AP-1 and three C/EBPα consensus binding sites in the first 750 bp. In addition to this, two AP-1 and C/EBPα-binding sites were identified between 750 and 1800 bp upstream of the ATG (Fig. 8A). The presence of AP-1 and C/EBPα binding elements on the CTDSPL 5′UTR (miR-26a promoter) led us to consider whether miR-26a is a transcriptional target of AP-1 and/or C/EBPα. First, using ChIP assays, we tested whether these transcription factors could directly bind to the promoter region of the miR-26a. As shown in Fig. 8 (B and C), activation of miR-26a promoter by stretch was completely dependent on the C/EBPα-binding sites as evidenced by qPCR (Fig. 8B) and the visualization of PCR products on 1% agarose gel (Fig. 8C). In contrast, stretch had no effect on c-fos activity on the miR-26a promoter. Second, we tested whether C/EBPα could influence the promoter activity of miR-26a. We generated two promoter constructs by cloning the first 750 bp, and a 1000 bp region (from −800 to −1800) upstream of the ATG (Fig. 8D) into luciferase reporter plasmid, named pGL750 and pGL1000, and tested them in a luciferase reporter assay 48 h after transfection. Cells transfected with the pGL1000 construct had higher luciferase activities than control cells upon stretch (Fig. 8E). In contrast, stretch did not activate the luciferase gene in cells transfected with the pGL750 construct. Finally, we tested whether the endogenous C/EBPα is able to influence the promoter activity of miR-26a. To achieve this, C/EBPα RNAi was produced, and they could decrease their endogenous expression levels both in total and phosphorylated states (Fig. 9A). The promoter of miR-26a was activated upon stretch, but knockdown of C/EBPα by RNAi abolished the stretch-induced miR-26 promoter activation (Fig. 9B). We next explored the relationship between C/EBPα and miR-26a in the hypertrophic cascades of stretch. To do this, we carried out the above RNAi strategy. Knockdown of C/EBPα inhibited the hypertrophic responses (Fig. 9C) of stretch. Taken together, these data suggest that miR-26a is a direct transcriptional target of C/EBPα during stretch-induced HASMC hypertrophy.
FIGURE 8.
miR-26a is a C/EBPα-dependent gene. A, schematic representation of the 5′UTR of the human CTDSPL gene (miR-26a promoter). Region between −1800 and +1 bp contains putative binding elements for AP-1 (bottom) and C/EBPα (top). B and C, HASMCs were stimulated with a 1-h stretch, and then the chromatin was isolated and precipitated with anti-c-Fos, anti-C/EBPα, anti-RNA Poly II, or nonspecific IgG. qPCRs were performed with two sets of primers, as shown in A, specific for miR-26a promoter to identify the specific transcription factor and its region of binding to the miR-26a promoter (B) and resolved in 1% agarose gel (C). D and E, a 750-bp (pGL750) and 1000-bp (pGL1000) promoter region was synthesized and linked to luciferase (Luc) reporter gene (D). Cells were transfected with empty vector or the pGL750 or pGL1000 miR-26a promoter region. Forty-eight hours after transfection, cells were stimulated with a 1-h stretch, and then firefly luciferase activities were estimated and normalized to Renilla luciferase activities (E). *, p < 0.05 versus pGL alone. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (n = 3).
FIGURE 9.
Knockdown of endogenous C/EBPα expression reduces miR-26a promoter activity. A, HASMCs were transfected with C/EBPα RNAi or its scramble form (SC-C/EBPα RNAi). Forty-eight hours after transfection, total and phosphorylated (p) C/EBPα levels were detected by Western blots. B, in a separate experiment, forty-eight hours after RNAi transfection, cells were transfected with the empty vector (pGL) or the constructs of the pGL1000 vector. Forty-eight hours after transfection, cells were stimulated with a 1-h stretch, and then firefly luciferase activities were estimated and normalized to Renilla luciferase activities. C, cells were transfected as shown in A. Twenty-four hours after transfection, cells were stimulated with a 1-h stretch for every 12 h. Cell size and protein synthesis were measured 72 h after transfection. *, p < 0.05 versus stretch alone. Gel pictures are representative of three separate experiments. Each bar indicates mean ± S.E. (n = 3).
DISCUSSION
Many lines of evidence demonstrate that physical forces elicit a number of biologically relevant signals in the human body. The best characterized system responses to mechanical stimuli are the cardiovascular (17–21), musculoskeletal (22–24), and pulmonary physiology (25, 33–38). Structural changes to airway smooth muscle, due to hypertrophy and/or hyperplasia, are a well known feature of chronic airway diseases, such as asthma, chronic obstructive pulmonary disease, and cystic fibrosis (12–16). Alterations in the regulation of gene expression by mechanical stimuli play an important role in the thickening and remodeling of the airway wall in subjects with the above airway diseases (25). In the last few years, miRNAs have been recognized as potent regulators of gene expression, and as a key modulator of variety of biological processes (4, 11). More recently, several studies have demonstrated the role of miRNAs in cardiac hypertrophy (28–32). Thus, it is plausible that miRNAs may be involved in the regulation of airway smooth muscle hypertrophy. In this study, we have investigated the potential involvement of a specific miRNA in the regulation of HASMC hypertrophy by evaluating the expression profile of miRNAs in response to stretch. Our microarray and RT-qPCR data showed that miR-16, miR-26a, and miR-140 were the only miRNAs highly up-regulated in HASMCs (more than 6-fold) by stretch. Recent studies have shown that these miRNAs inhibit proliferation of tumor cells by targeting the regulators of the cell cycle (49–61), suggesting that these miRNAs may have a role in the regulation of HASMC hypertrophy rather than hyperplasia. Consistent with these findings, our results showed that miR-26a, but neither miR-16 nor miR-140, induced HASMC hypertrophy regardless of the hypertrophic stimuli stretch. Our results also demonstrated that these miRNAs could not inhibit HASMCs proliferation, which was induced by stretch. In contrast, enforced expression of miR-26a in HASMCs inhibited cell proliferation, and then transforms them to hypertrophic phenotype in the absence of stretch. One possible explanation is that some other signaling pathway(s) that might regulate the stretch-induced proliferation may overdominate the inhibitory role of miR-26a in cell proliferation. To our knowledge, the identification of miR-26a as a stretch-responsive or mechanosensitive miRNA in primary HASMCs represents the first evidence linking miRNA to airway smooth muscle hypertrophy.
Like miR-26a, many proteins have emerged to regulate hypertrophy and promote disease upon their dysregulated expression. Among those, one of the potential proteins that regulates hypertrophy is GSK-3β. GSK-3β is constitutively active in unstimulated cells and inactivated upon phosphorylation at Ser9 (62). Although initially described as an inhibitor of glycogen synthesis through phosphorylation of glycogen synthase (63), GSK-3β was later revealed as a key signaling molecule regulating many aspects of cellular function, including protein synthesis, cytoskeletal integrity, and gene expression (64). More importantly, GSK-3β negatively regulates cardiac (39–43), skeletal (44, 45), and airway smooth muscle (44, 47) hypertrophy as evidenced by the finding that GSK-3β overexpression inhibits the hypertrophic phenotype, protein synthesis, and hypertrophic genes expression. In this study, we determined the expression of α-actin, SM22, and smMHC genes as the indicators of airway smooth muscle hypertrophy. Many studies have shown the up-regulation of these proteins occurs when airway smooth muscle cells enter into the hypertrophic cascade upon GSK-3β activation (46–47). Interestingly, our results showed that stretch decreased GSK-3β phosphorylation due to the inhibition of total protein production in a time-dependent manner, suggesting that the stretch-induced hypertrophy in HASMCs could be mediated through the suppression of GSK-3β gene expression. Interestingly, our data show that GSK-3β is a downstream target of miR-26a, which down-regulates GSK-3β expression. The following bioinformatics and experimental evidences confirm the molecular mechanism by which miR-26a suppresses GSK-3β gene expression. 1) GSK-3β has a conservative miR26a seed sequence in its 3′UTR, suggesting that miR-26a may inhibit GSK-3β expression; 2) overexpression of miR-26a significantly suppressed GSK-3β protein but not mRNA levels, indicating that miR-26a predominantly suppresses GSK-3β translation; 3) miR-26a suppressed GSK-3β expression in a dose-dependent manner; 4) miR-26a could not inhibit GSK-3β expression when the GSK-3β was overexpressed without its 3′UTR, indicating that GSK-3β 3′UTR contains an active seed of miR-26a; and 5) HASMCs transfected with luc-GSK-3β-3′UTR construct had higher luciferase activity than when those cells were co-transfected with miR-26a expression vector, and introduction of miR-26a antagomir could revert this effect, confirming the presence of miR-26a active seed on the GSK-3β 3′UTR.
In addition, miR-26a overexpression abolished the hypertrophic effect of GSK-3β, and enforced expression of GSK-3β inhibited the stretch-induced hypertrophy. This provides a molecular link between miR-26a and GSK-3β in HASMC hypertrophy. In this study, the GSK-3β-miR-26a-induced hypertrophy was documented by the increased cell size and relative expression of specific contractile proteins as well as a global increase in protein synthesis. A similar finding has been reported in HASMCs, in which suppression of GSK-3β by RNAi increased cell size and protein synthesis (46). In addition, we found that the miR-26a-GSK-3β-mediated signaling pathway was not involved in the stretch-induced proliferation of HASMCs. This is also consistent with the previous finding demonstrating that GSK-3β does not participate in HASMC hyperplasia (46). Our search for GSK-3β targets provided only five miRNAs (has-miR-199a, -199b, -26a, -26b, and -302a*). Because miR-199a, miR-199b, miR-26b, and miR-302a* were not induced by stretch we cannot rule out the possibility that these miRNAs are not upstream targets of GSK-3β. It is well recognized that miRNAs may function according to a “combinational circuitry model,” whereby a single miRNA targets multiple mRNAs and several miRNAs may target a single mRNA (65). Thus, further studies are required to elucidate specifically whether hypertrophic stimuli other than stretch can induce the expression of these predicted miRNAs, and whether miR-26a can regulate proteins that are involved in the hypertrophic cascade of HASMCs other than GSK-3β.
Gene expression-modulating miRNAs are encoded in diverse genomic locations, including intergenic regions, introns of protein-coding genes, and introns/exons of noncoding RNA genes (66). Most human miRNAs lie between protein-coding genes, whereas about one-third is within the introns of annotated mRNAs (3, 67). In the human genome, two distinct genes, CTDSPL and CTDSP2, encode different pri-miR-26a (miR-26a-1 and miR-26a-2) leading to the generation of identical mature miR-26a. miR-26a-1 is located in an intron of CTDSPL at chromosome 3, and miR-26a-2 is located in an intron of CTDSP2 at chromosome 12. It is likely that the intronic miRNAs are processed from the same primary transcript as the precursor mRNAs, and thus, their expression levels are regulated by the expression of the host mRNA (68). We examined the correlation between the miR-26a expression profile and the expression profile of the host genes by RT-qPCR in response to stretch. Surprisingly, in a time-dependent manner, stretch increased up-regulation of CTDSPL mRNA levels, but had no effect on CTDSP2 mRNA levels. As anticipated, miR-26a induction by stretch followed a similar pattern of the expression of CTDSPL levels. These results indicate that most of the miR-26a in HASMCs is miR-26a-1 in response to stretch, and is processed from the same primary transcript as its host gene CTDSPL. In contrast, in the absence of stretch, most of the miR-26a transcripts are miR-26a-2 that occurs by the activation of the host gene CTDSP2. Moreover, our study also showed that, in the absence of stretch, the basal CTDSPL mRNA levels were barely detectable, unlike CTDSP2 mRNA levels, and the basal miR-26a levels were high. This suggests the existence of a strong correlation between the expression of miR-26a and CTDSP2 mRNA, consistent with previous finding (48). We also determined the specific transcription factor by which pri-miR-26a is induced by stretch. Using ChIP assay, we found that activation of miR-26a promoter by stretch was completely dependent on the C/EBPα-binding sites. The luciferase reporter assays demonstrated that the stretch-induced activation of miR-26a promoter by C/EBPα was effective only in the presence of the distal miR-26a promoter region (1000 bp), which contains two C/EBPα-binding sites. Moreover, the small interference RNA-mediated knockdown of C/EBPα experiment confirmed the requirement of C/EBPα for the activation of miR-26a promoter by stretch. The present study also corroborated the necessity of C/EBPα for the induction of HASMC hypertrophy by stretch. Our study indicates that C/EBPα plays a key role in the induction of HASMC hypertrophy via miR-26a up-regulation in response to stretch.
Taken together, our data demonstrate that miR-26a is a mechanosensitive and hypertrophic miRNA of HASMCs. Stretch activates the miR-26a promoter through C/EBPα transcription factor, which leads to the up-regulation of miR-26a. In addition, stretch selectively induces the transcription of miR-26a located in the locus 3p21.3 of human chromosome 3. Molecular functional analyses indicate that miR-26a-induced hypertrophy is mediated through the suppression of its target gene GSK-3β expression (Fig. 10). We anticipate our study can serve as a model system for studying gene regulation by miRNAs in the development of gene therapy for airway diseases such as asthma and chronic obstructive pulmonary disease.
This work was supported, in whole or in part, by National Institutes of Health Grants HL63134 and HL-072839 from NHLBI. This work was also supported by a grant from the National Science Foundation.
- miRNA
- microRNA
- HASMC
- human airway smooth muscle cell
- C/EBP-α
- CCAAT enhancer-binding protein α
- GSK-3β
- glycogen synthase kinase-3β
- UTR
- untranslated region
- RT
- reverse transcription
- CTDSPL
- C-terminal domain, RNA polymerase II, polypeptide A small phosphatase-like
- CTDSP2
- C-terminal domain, RNA polymerase II, polypeptide A small phosphatase 2
- ChIP
- chromatin immunoprecipitation
- GAPDH
- glyceraldehydes-3-phosphate dehydrogenase
- MHC
- myosin heavy chain
- CMV
- cytomegalovirus
- qPCR
- quantitative PCR
- RNAi
- RNA interference
- EdU
- 5-ethynyl-2′-deoxyuridine
- HPG
- l-homopropargylglycine.
REFERENCES
- 1.Cai X., Hagedorn C. H., Cullen B. R. (2004) RNA 10, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., Kim V. N. (2004) EMBO J. 23, 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez A., Griffiths-Jones S., Ashurst J. L., Bradley A. (2004) Genome Res. 14, 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 5.Lin S. L., Miller J. D., Ying S. Y. (2006) J. Biomed. Biotechnol. 4, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. (2004) Nature 432, 235–240 [DOI] [PubMed] [Google Scholar]
- 7.Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. (2005) Nature 436, 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yi R., Qin Y., Macara I. G., Cullen B. R. (2003) Genes Dev. 17, 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J., Lee Y., Yeom K. H., Nam J. W., Heo I., Rhee J. K., Sohn S. Y., Cho Y., Zhang B. T., Kim V. N. (2006) Cell 125, 887–901 [DOI] [PubMed] [Google Scholar]
- 10.Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. (2006) Genes Dev. 20, 515–524 [DOI] [PubMed] [Google Scholar]
- 11.Plasterk R. H. (2006) Cell 124, 877–881 [DOI] [PubMed] [Google Scholar]
- 12.Ebina M., Takahashi T., Chiba T., Motomiya M. (1993) Am. Rev. Respir. Dis. 148, 720–726 [DOI] [PubMed] [Google Scholar]
- 13.Jeffery P. K. (2001) Am. J. Respir. Crit. Care Med. 164, S28–S38 [DOI] [PubMed] [Google Scholar]
- 14.Benayoun L., Druilhe A., Dombret M. C., Aubier M., Pretolani M. (2003) Am. J. Respir. Crit. Care Med. 167, 1360–1368 [DOI] [PubMed] [Google Scholar]
- 15.Hays S. R., Ferrando R. E., Carter R., Wong H. H., Woodruff P. G. (2005) Thorax 60, 226–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regamey N., Ochs M., Hilliard T. N., Mühlfeld C., Cornish N., Fleming L., Saglani S., Alton E. W., Bush A., Jeffery P. K., Davies J. C. (2008) Am. J. Respir. Crit. Care Med. 177, 837–843 [DOI] [PubMed] [Google Scholar]
- 17.Komuro I. (2000) Jpn. Heart J. 41, 117–129 [DOI] [PubMed] [Google Scholar]
- 18.Komuro I., Katoh Y., Kaida T., Shibazaki Y., Kurabayashi M., Hoh E., Takaku F., Yazaki Y. (1991) J. Biol. Chem. 266, 1265–1268 [PubMed] [Google Scholar]
- 19.Sadoshima J., Izumo S. (1993) J. Recept. Res. 13, 777–794 [DOI] [PubMed] [Google Scholar]
- 20.Ruwhof C., van der Laarse A. (2000) Cardiovasc. Res. 47, 23–37 [DOI] [PubMed] [Google Scholar]
- 21.Sugden P. H. (2001) Circulation 103, 1375–1377 [DOI] [PubMed] [Google Scholar]
- 22.Adachi R., Yabusaki K., Obinata T. (2003) Zool Sci. 20, 557–565 [DOI] [PubMed] [Google Scholar]
- 23.Vandenburgh H., Kaufman S. (1979) Science 203, 265–268 [DOI] [PubMed] [Google Scholar]
- 24.Sasai N., Agata N., Inoue-Miyazu M., Kawakami K., Kobayashi K., Sokabe M., Hayakawa K. (2010) Muscle Nerve 41, 100–1406 [DOI] [PubMed] [Google Scholar]
- 25.Tschumperlin D. J., Drazen J. M. (2001) Am. J. Respir. Crit. Care Med. 164, S90–94 [DOI] [PubMed] [Google Scholar]
- 26.Galvin D. J., Watson R. W., Gillespie J. I., Brady H., Fitzpatrick J. M. (2002) Am. J. Physiol. Renal Physiol. 283, F1192–1199 [DOI] [PubMed] [Google Scholar]
- 27.Hellstrand P., Albinsson S. (2005) Can J. Physiol. Pharmacol. 83, 869–875 [DOI] [PubMed] [Google Scholar]
- 28.van Rooij E., Sutherland L. B., Liu N., Williams A. H., McAnally J., Gerard R. D., Richardson J. A., Olson E. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M. L., Segnalini P., Gu Y., Dalton N. D., Elia L., Latronico M. V., Høydal M., Autore C., Russo M. A., Dorn G. W., 2nd., Ellingsen O., Ruiz-Lozano P., Peterson K. L., Croce C. M., Peschle C., Condorelli G. (2007) Nat. Med. 13, 613–618 [DOI] [PubMed] [Google Scholar]
- 30.Sayed D., Hong C., Chen I. Y., Lypowy J., Abdellatif M. (2007) Circ. Res. 100, 416–424 [DOI] [PubMed] [Google Scholar]
- 31.Lin Z., Murtaza I., Wang K., Jiao J., Gao J., Li P. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12103–12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callis T. E., Pandya K., Seok H. Y., Tang R. H., Tatsuguchi M., Huang Z. P., Chen J. F., Deng Z., Gunn B., Shumate J., Willis M. S., Selzman C. H., Wang D. Z. (2009) J. Clin. Invest. 119, 2772–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith P. G., Moreno R., Ikebe M. (1997) Am. J. Physiol. Lung Cell Mol. Physiol. 272, L20–L27 [DOI] [PubMed] [Google Scholar]
- 34.Kumar A., Knox A. J., Boriek A. M. (2003) J. Biol. Chem. 278, 18868–18876 [DOI] [PubMed] [Google Scholar]
- 35.Hasaneen N. A., Zucker S., Cao J., Chiarelli C., Panettieri R. A., Foda H. D. (2005) FASEB J. 04–3350fje [DOI] [PubMed] [Google Scholar]
- 36.Ito S., Kume H., Oguma T., Ito Y., Kondo M., Shimokata K., Suki B., Naruse K. (2006) Eur. J. Pharmacol. 552, 135–142 [DOI] [PubMed] [Google Scholar]
- 37.Kanefsky J., Lenburg M., Hai C. M. (2006) Am. J. Respir. Cell Mol. Biol. 34, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito S., Kume H., Naruse K., Kondo M., Takeda N., Iwata S., Hasegawa Y., Sokabe M. (2008) Am. J. Respir. Cell Mol. Biol. 38, 407–413 [DOI] [PubMed] [Google Scholar]
- 39.Haq S., Choukroun G., Kang Z. B., Ranu H., Matsui T., Rosenzweig A., Molkentin J. D., Alessandrini A., Woodgett J., Hajjar R., Michael A., Force T. (2000) J. Cell Biol. 151, 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morisco C., Seta K., Hardt S. E., Lee Y., Vatner S. F., Sadoshima J. (2001) J. Biol. Chem. 276, 28586–28597 [DOI] [PubMed] [Google Scholar]
- 41.Antos C. L., McKinsey T. A., Frey N., Kutschke W., McAnally J., Shelton J. M., Richardson J. A., Hill J. A., Olson E. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badorff C., Ruetten H., Mueller S., Stahmer M., Gehring D., Jung F., Ihling C., Zeiher A. M., Dimmeler S. (2002) J. Clin. Invest. 109, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hardt S. E., Sadoshima J. (2002) Circ. Res. 90, 1055–1063 [DOI] [PubMed] [Google Scholar]
- 44.Vyas D. R., Spangenburg E. E., Abraha T. W., Childs T. E., Booth F. W. (2002) Am. J. Physiol. Cell Physiol. 283, C545–551 [DOI] [PubMed] [Google Scholar]
- 45.Rochat A., Fernandez A., Vandromme M., Molès J. P., Bouschet T., Carnac G., Lamb N. J. (2004) Mol. Biol. Cell 15, 4544–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng H., Dokshin G. A., Lei J., Goldsmith A. M., Bitar K. N., Fingar D. C., Hershenson M. B., Bentley J. K. (2008) J. Biol. Chem. 283, 10198–10207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bentley J. K., Deng H., Linn M. J., Lei J., Dokshin G. A., Fingar D. C., Bitar K. N., Henderson W. R., Jr., Hershenson M. B. (2009) Am. J. Physiol. Lung Cell Mol. Physiol. 296, L176–L184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kashuba V. I., Li J., Wang F., Senchenko V. N., Protopopov A., Malyukova A., Kutsenko A. S., Kadyrova E., Zabarovska V. I., Muravenko O. V., Zelenin A. V., Kisselev L. L., Kuzmin I., Minna J. D., Winberg G., Ernberg I., Braga E., Lerman M. I., Klein G., Zabarovsky E. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4906–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bottoni A., Piccin D., Tagliati F., Luchin A., Zatelli M. C., degli Uberti E. C. (2005) J. Cell Physiol. 204, 280–285 [DOI] [PubMed] [Google Scholar]
- 51.Tuddenham L., Wheeler G., Ntounia-Fousara S., Waters J., Hajihosseini M. K., Clark I., Dalmay T. (2006) FEBS Lett. 580, 4214–4217 [DOI] [PubMed] [Google Scholar]
- 52.Xi Y., Formentini A., Chien M., Weir D. B., Russo J. J., Ju J., Kornmann M., Ju J. (2006) Bio. Insight 1, 113–121 [PMC free article] [PubMed] [Google Scholar]
- 53.Chang T. C., Yu D., Lee Y. S., Wentzel E. A., Arking D. E., West K. M., Dang C. V., Thomas-Tikhonenko A., Mendell J. T. (2008) Nat. Genet. 40, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Linsley P. S., Schelter J., Burchard J., Kibukawa M., Martin M. M., Bartz S. R., Johnson J. M., Cummins J. M., Raymond C. K., Dai H., Chau N., Cleary M., Jackson A. L., Carleton M., Lim L. (2007) Mol. Cell Biol. 27, 2240–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Visone R., Pallante P., Vecchione A., Cirombella R., Ferracin M., Ferraro A., Volinia S., Coluzzi S., Leone V., Borbone E., Liu C. G., Petrocca F., Troncone G., Calin G. A., Scarpa A., Colato C., Tallini G., Santoro M., Croce C. M., Fusco A. (2007) Oncogene 26, 7590–7595 [DOI] [PubMed] [Google Scholar]
- 56.Bonci D., Coppola V., Musumeci M., Addario A., Giuffrida R., Memeo L., D'Urso L., Pagliuca A., Biffoni M., Labbaye C., Bartucci M., Muto G., Peschle C., De Maria R. (2008) Nat. Med. 14, 1271–1277 [DOI] [PubMed] [Google Scholar]
- 57.Nicolas F. E., Pais H., Schwach F., Lindow M., Kauppinen S., Moulton V., Dalmay T. (2008) RNA 14, 2513–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bandi N., Zbinden S., Gugger M., Arnold M., Kocher V., Hasan L., Kappeler A., Brunner T., Vassella E. (2009) Cancer Res. 69, 5553–5559 [DOI] [PubMed] [Google Scholar]
- 59.Guo C. J., Pan Q., Jiang B., Chen G. Y., Li D. G. (2009) Apoptosis 14, 1331–1340 [DOI] [PubMed] [Google Scholar]
- 60.Kota J., Chivukula R. R., O'Donnell K. A., Wentzel E. A., Montgomery C. L., Hwang H. W., Chang T. C., Vivekanandan P., Torbenson M., Clark K. R., Mendell J. R., Mendell J. T. (2009) Cell 137, 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song B., Wang Y., Xi Y., Kudo K., Bruheim S., Botchkina G. I., Gavin E., Wan Y., Formentini A., Kornmann M., Fodstad O., Ju J. (2009) Oncogene 28, 4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohen P., Frame S. (2001) Nat. Rev. Mol. Cell Biol. 2, 769–776 [DOI] [PubMed] [Google Scholar]
- 63.Parker P. J., Caudwell F. B., Cohen P. (1983) Eur. J. Biochem. 130, 227–234 [DOI] [PubMed] [Google Scholar]
- 64.Grimes C. A., Jope R. S. (2001) Prog. Neurobiol. 65, 391–426 [DOI] [PubMed] [Google Scholar]
- 65.He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. (2005) Nature 435, 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J. (2008) Nucleic Acids Res. 36, D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saini H. K., Griffiths-Jones S., Enright A. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17719–17724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cullen B. R. (2004) Mol. Cell 16, 861–865 [DOI] [PubMed] [Google Scholar]