Abstract
In vascular smooth muscle cells, exposed to hyperglycemia and insulin-like growth factor-I (IGF-I), SHPS-1 functions as a scaffold protein, and a signaling complex is assembled that leads to AKT activation. However, the underlying mechanism by which formation of this complex activates the kinase that phosphorylates AKT (Thr308) is unknown. Therefore, we investigated the mechanism of PDK1 recruitment to the SHPS-1 signaling complex and the consequences of disrupting PDK1 recruitment for downstream signaling. Our results show that following IGF-I stimulation, PDK1 is recruited to SHPS-1, and its recruitment is mediated by Grb2, which associates with SHPS-1 via its interaction with Pyk2, a component of the SHPS-1-associated complex. A proline-rich sequence in PDK1 bound to an Src homology 3 domain in Grb2 in response to IGF-I. Disruption of Grb2-PDK1 by expression of either a Grb2 Src homology 3 domain or a PDK1 proline to alanine mutant inhibited PDK1 recruitment to SHPS-1, leading to impaired IGF-I-stimulated AKT Thr308 phosphorylation. Following its recruitment to SHPS-1, PDK1 was further activated via Tyr373/376 phosphorylation, and this was required for a maximal increase in PDK1 kinase activity and AKT-mediated FOXO3a Thr32 phosphorylation. PDK1 recruitment was also required for IGF-I to prevent apoptosis that occurred in response to hyperglycemia. Assembly of the Grb2-PDK1 complex on SHPS-1 was specific for IGF-I signaling because inhibiting PDK1 recruitment to SHPS-1 had no effect on EGF-stimulated AKT Thr308 phosphorylation. These findings reveal a novel mechanism for recruitment of PDK1 to the SHPS-1 signaling complex, which is required for IGF-I-stimulated AKT Thr308 phosphorylation and inhibition of apoptosis.
Keywords: Adaptor Proteins, AKT PKB, Apoptosis, Insulin-like Growth Factor (IGF), Phosphatidylinositol-dependent Kinase-1 (PDK1), Smooth Muscle, Membrane Localization, SHPS-1, Cell Survival
Introduction
AKT is phosphorylated at two key regulatory sites, Thr308 in the activation loop of the catalytic domain and Ser473 in the COOH-terminal domain. PDK1 (3-phosphoinositide-dependent kinase 1) is the kinase responsible for phosphorylation of Thr308 (1), and this is required for AKT-mediated inhibition of apoptosis (2–5). Although constitutive autophosphorylation of PDK1 Ser241 is required for PDK1 kinase activity, it is not regulated by growth factors (1, 6). However, PDK1 activity is regulated by changes in its conformation, tyrosine phosphorylation, and subcellular localization (7–12). PDK1 undergoes tyrosine phosphorylation in response to insulin, angiotensin II, high glucose, and insulin-like growth factor-I (IGF-I)2 (9, 11–17). Previous studies indicate that optimal activation of PDK1 requires phosphorylation of Tyr373/376 (11, 12, 14, 17), and growth factor receptor activation leads to PDK1 recruitment to the plasma membrane, followed by sequential phosphorylation of Tyr9 and then Tyr373/376 (14, 17).
A previous study suggested that Pyk2 (proline-rich tyrosine kinase 2) plays an important role in PDK1 activation. Tanniyama et al. (9) showed that Pyk2 was required for optimal PDK1 tyrosine phosphorylation in response to angiotensin II and that Pyk2 and PDK1 were co-localized to the focal adhesions. Pyk2 facilitates PDK1 interaction with Src, and Tyr373/376 phosphorylation is increased in response to Src activation (9, 12, 14, 18), but the mechanism by which these signaling components interact, the involvement of other signaling components, the exact sites of protein/protein interaction, and the mechanism of PDK1 membrane recruitment leading to AKT Thr308 phosphorylation have not been determined.
IGF-I has diverse biological actions, including regulation of cellular proliferation, differentiation, migration, and survival (19). The biological effects of IGF-I are mediated through its receptor tyrosine kinase, which phosphorylates specific substrates to activate downstream signaling (20, 21). SHPS-1 (SH2 domain-containing protein tyrosine phosphatase substrate 1) is an integral membrane protein that acts as a scaffold for multiprotein signaling complexes that are assembled in response to IGF-I in vascular smooth muscle cells or endothelial cells in response to hyperglycemia. The SHPS-1 cytoplasmic domain (SHPS-1/CD) contains four tyrosines that are phosphorylated by the IGF-I receptor (22). This leads to recruitment of the SH2 domain containing phosphatase SHP2. Subsequently, c-Src, p52Shc/Grb2 (growth factor receptor-bound 2), and the p85 subunit of PI3K are recruited and activated, leading to stimulation of both the PI3K and mitogen-activated protein kinase (MAPK) signaling pathways (23–25). Truncation of the cytoplasmic domain (CD) of SHPS-1 significantly impairs IGF-I-stimulated AKT activation (26). Recently, we reported that IGF-I stimulates Pyk2 recruitment to the SHPS-1 signaling complex via Src-Pyk2 association and that Src phosphorylates Pyk2 Tyr881, creating a binding site for Grb2 (27). Because, following hyperglycemic stress, IGF-I-stimulated SHPS-1 phosphorylation results in a significant increase in AKT phosphorylation, we wished to study whether PDK1 is recruited to SHPS-1 signaling complex and, if so, to determine the role of Grb2 in PDK1 recruitment in response to IGF-I. Our findings demonstrate that Grb2 mediates the recruitment of PDK1 to the SHPS-1 signaling complex, that these signaling components interact through an SH3 domain-polyproline motif interaction, and that PDK1 recruitment is required for AKT Thr308 phosphorylation and cell survival in response to IGF-I.
EXPERIMENTAL PROCEDURES
Human IGF-I was a gift from Genentech (South San Francisco, CA). Dulbecco's modified Eagle's medium (DMEM) containing 4,500 mg of glucose/liter (25 mm) was purchased from Invitrogen, and penicillin and streptomycin were purchased from Invitrogen. Blasticidin was obtained from Invitrogen. Phosphatidylinositol substrate was purchased from Avanti Polar Lipids (Alabaster, AL). [γ-32P]ATP was from GE Healthcare. Antibodies against phospho-AKT (Ser473), phospho-AKT (Thr308), AKT, PDK1, cleaved caspase-3, β-actin, and HA were from Cell Signaling Technologies (Danvers, MA). An antibody that detected pPDK1 (Tyr373/376) was from Abcam (Cambridge, MA). The anti-Grb2 (rabbit), anti-p27 (rabbit), and the monoclonal anti-phosphotyrosine antibodies (Tyr(P)99) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The Grb2, caveolin, and Pyk2 monoclonal antibodies (mouse) were purchased from BD Biosciences. The anti-Pyk2 (rabbit) and anti-p85α subunit antibodies were purchased from Upstate Cell Signaling Solutions/Millipore (Lake Placid, NY). The anti-Src antibody was purchased from Calbiochem. A polyclonal antibody for the extracellular domain of SHPS-1 was generated in our laboratory (26). The PI3K inhibitor (LY294002) was purchased from Calbiochem. The DeadEnd TUNEL kit was purchased from Promega (Madison, WI). All other reagents were purchased from Sigma unless otherwise stated.
Cell Culture
Vascular smooth muscle cells were prepared from porcine aortas and maintained in culture as described previously (28). All experiments except those shown in Fig. 6 (C and D) were conducted in hyperglycemic conditions (25 mm glucose). For the experiments in Fig. 6 (C and D), the cells were grown in 10% FBS and DMEM containing 5 mm glucose and then starved overnight in serum-free DMEM containing 25 mm glucose before the treatments were added.
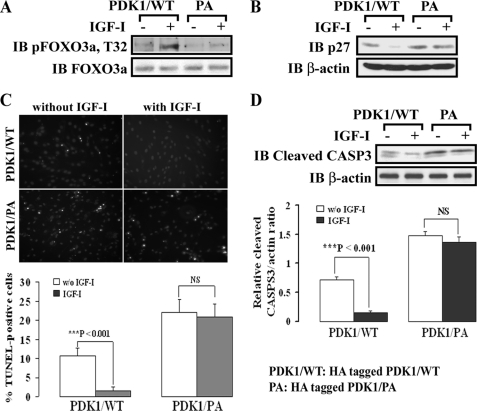
FIGURE 6.
Grb2-PDK1 association regulates AKT-dependent FOXO3a phosphorylation, p27 protein expression, and cell survival in response to IGF-I. A, quiescent PDK1/WT- and PDK1/PA-expressing SMCs were stimulated with IGF-I for 5 min. The cell lysates were directly immunoblotted (IB) for phospho-FOXO3a (Thr32). The blot was stripped and reprobed with anti-FOXO3a as a loading control. B, quiescent PDK1/WT- and PDK1/PA-expressing SMCs were treated with or without IGF-I overnight. The cell lysates were directly immunoblotted for p27. The blot was stripped and reprobed with anti-β-actin as a loading control. C and D, apoptotic cell death and caspase-3 activity are attenuated by IGF-I treatment, which requires PDK1 activation. The TUNEL assay was conducted as described under “Experimental Procedures.” SMCs expressing PDK1/WT or PDK1/PA were grown to 80% confluence in normal glucose (5 mm) with 10% FBS in 6-well culture plates and then starved overnight in serum-free DMEM-H containing 25 mm glucose. 48 h after the addition of IGF-I, the apoptotic cell number was determined by counting apoptotic nuclei using fluorescence microscopy (C, top, photomicrograph; bottom, quantitation of apoptotic cells). ***, p < 0.001, a significant difference between two different treatments. Additional cultures were lysed and analyzed by immunoblotting using anti-cleaved caspase-3 (cleaved CASP3) (D). The blots were stripped and reprobed with anti-β-actin as a loading control. The protein levels were quantified using scanning densitometry. The graph shows the mean result from three independent experiments expressed as the cleaved CASP3/β-actin ratio (D, bottom). ***, p < 0.001, a significant difference between two different treatments; NS, not significant. Error bars represent S.E.
pLenti-HA-SHPS-1/Wild Type (SHPS-1/WT) and pLenti-HA-SHPS-1/−CD (MT-CD)
The full-length SHPS-1/WT and the truncated cytoplasmic domain of SHPS-1 mutant (deleting amino acids from 399 to 503) were PCR-amplified with HA-containing primers using previously generated pcDNA-SHPS-1 as a template (23). That the construct contained the correct sequence was confirmed by DNA sequencing, and then the cDNAs were cloned into pLenti viral transduction system as described previously (23, 26).
pLenti-HA-Grb2 Wild Type (Grb2/WT), pLenti-HA-Grb2 mutant (Grb2/WA), pLenti-HA-Pyk2 wild type (Pyk2/WT), and pLenti-HA-Pyk2/Y881F (Pyk2/Y881F)
Grb2/WT, Grb2/WA, Pyk2/WT, and Pyk2/Y881F were prepared as described previously (25, 27).
Generation of pLenti Expression Vectors pLenti-HA-PDK1 Wild Type (PDK1/WT) and pLenti-HA-PDK1 Mutant (PDK1/PA)
The full-length PDK1/WT cDNA was prepared by PCR using a pcDNA-myc-PDK1/WT construct kindly provided by Dr. Brian Hemmings (Friedrich Miescher Institute, Basel, Switzerland) and cloned into the pENTR/D-TOPO Gateway entry vector according to the manufacturer's instructions (Invitrogen). The forward and reverse primers used to generate the PCR product were as follows: forward primer, 5′-CACC ATG ATG GCC AGG ACC ACC AGC CAG CTG-3′; reverse primer, 5′-TTA AGC GTA ATC TGG AAC ATC GTA TGG GTA TTA CTG CAC AGC GGC GTC CGG G-3′. The forward primer includes an ATG (underlined) start site. The reverse primer contained the sequence encoding HA epitope (underlined) followed by the stop codon (boldface type).
Using the pENTR-PDK1/WT as template, the prolines at positions 71 and 74 were charged to alanine (PDK1/PA) using double-stranded mutagenesis. PCR amplification was carried out using forward primer 5′- gcatgcccagcctGcgccgcagGctcggaagaagcggc-3′ and reverse primer 5′-gccgcttcttccgagCctgcggcgCaggctgggcatgc-3′, where capitalized bases indicate the substitutions. After selection of correct clones as determined by sequencing, the cDNAs encoding the wild type and mutant proteins were transferred from the entry vector into pLenti6/V5-DEST Gateway vector using the LR Clonase reaction following the manufacturer's instructions (Invitrogen).
Construction of a Plasmid Containing Short Hairpin RNA (shRNA) Template for Pyk2 Silencing
The construction of the plasmid containing shRNA template for Pyk2 was prepared as described previously (27). The expression vector of shRNA template for LacZ was used as a control. After confirmation of the sequences, the plasmid DNAs were prepared and purified using the Plasmid Midi kit (Promega, Madison, WI) according to the manufacturer's instructions.
Generation of Virus Stocks and Establishment of SMCs Expressing pLenti Constructs
293FT cells (Invitrogen) were utilized for generation of virus stocks that were prepared, purified, and transfected into vascular smooth muscle cells to obtain cells expressing SHPS-1/WT, SHPS-1/−CD, Pyk2/WT, Pyk2/Y881F, Grb2/WT, Grb2/WA, PDK1/WT, PDK1/PA, si-Pyk2, and si-LacZ. These cell lines were established using procedures that have been described previously (23, 25, 27). The expression of the HA-tagged SHPS-1/WT, SHPS-1/−CD, Pyk2/WT, Pyk2/Y881F, Grb2/WT, Grb2/WA, PDK1/WT, or PDK1/PA protein was detected by immunoblotting with an anti-HA antibody (1:1,000); The effectiveness of shRNA for inhibiting Pyk2 expression was determined by immunoblotting using an anti-Pyk2 antibody and comparing the results to SMCs expressing the LacZ shRNA. The immune complexes were detected using either a horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibody and developed with enhanced chemiluminescence following the manufacturer's instructions (Pierce).
Immunoprecipitation and Immunoblotting
The immunoprecipitation and immunoblotting procedures were performed as described previously (26).
In Vitro Binding Assay
The in vitro [35S]methionine (MP Biomedicals, Solon, OH)-labeled PDK1/WT and PDK1/PA mutant were generated by a coupled in vitro transcription and translation (TNT) system (Promega) according to the manufacturer's instructions. For the in vitro binding assay using agarose bead-conjugated Grb2 protein (Santa Cruz Biotechnology, Inc.), a 10-μl aliquot of the individual TNT mixture was incubated with 5 μg of Grb2 beads in binding buffer (10 mm Hepes, pH 7.4, 150 mm NaCl, 0.1% Tween 20, 1 mm dithiothreitol) for 4 h at 4 °C. The beads were then washed three times with binding buffer. The precipitated proteins were eluted in 30 μl of SDS sample buffer, boiled for 5 min, and subjected to SDS-polyacrylamide gel. The images were developed and analyzed using a Storm860 PhosphorImager (Amersham Biosciences).
Isolation Cytoplasmic and Membrane Fraction Proteins
Isolation of cytoplasmic and membrane fraction proteins was performed as described previously (29).
PI3K Assay
The immunoprecipitates containing the p85-p110 complex were washed once with phosphate-buffered saline, twice with lysis buffer, and once with the reaction buffer (30) and resuspended in 50 μl of the PI3K reaction buffer containing 0.1 mg/ml phosphoinositide (Avanti Polar Lipids). The reactions were performed, and the phosphorylated lipids were separated by TLC as described previously (25, 31).
In Vitro Kinase Assay for PDK1
In vitro PDK1 activity was measured as described previously (14, 16). In brief, cells were incubated in serum-free medium overnight and then exposed to 0 or 50 ng/ml IGF-I for 5 min. The cells were lysed in radioimmune precipitation buffer, and the PDK1-containing immune complexes were prepared by immunoprecipitating 0.5 mg of total protein with a 1:250 dilution of anti-PDK1 antibody. The immune complexes were washed once with lysis buffer containing 500 mm NaCl, followed by lysis buffer and finally with kinase assay buffer (50 mm Tris-HCl, pH 7.5, 0.1% (v/v) 2-mercaptoethanol). In vitro kinase assays were performed for 45 min at 30 °C in 50 μl of kinase assay buffer containing 30 μl of immunoprecipitate, 0.1 mm Suntide (RRKDGATMKTFCGTPE) as substrate, 10 mm MgCl2, 1 μm protein kinase A inhibitor peptide (Millipore), and 10 μCi of [γ-32P] ATP. Reactions were stopped by adding EDTA to a final concentration of 50 mm. The amount of 32P that was incorporated into the substrate peptide, which reflects the activity of PDK1 kinase, was quantified by liquid scintillation counting (Packard, Meriden, CT).
TUNEL Assay
Apoptosis was determined by a TUNEL assay using the DeadEnd fluorometric TUNEL system according to the manufacturer's instructions (Promega, Madison, WI). SMCs were washed once with PBS and then fixed with 4% paraformaldehyde solution in PBS (pH 7.4) for 25 min at 4 °C. Samples were permeabilized with 0.2% Triton X-100 solution in PBS for 5 min and then incubated with recombinant terminal deoxynucleotidyltransferase for 1 h at 37 °C in a humidified chamber. The slides were mounted in VECTASHIELD with DAPI (Vector Laboratories, catalog no. H-1200) to stain nuclei and detect localized green fluorescence of apoptotic cells on a blue background using fluorescence microscopy. The number of apoptotic cells in each sample was quantified by counting the number of TUNEL-positive cells in three randomly selected, non-overlapping regions. Each treatment was analyzed in triplicate. The results represent mean values of three independent experiments.
Statistical Analysis
Student's t test was used to compare differences between treatments. The results that are shown in all experiments are the representative of at least three independent experiments and expressed as the mean ± S.E.
RESULTS
IGF-I Stimulates PDK1 Recruitment to SHPS-1 and Loss of PDK1 Recruitment Attenuates AKT Thr308 Phosphorylation
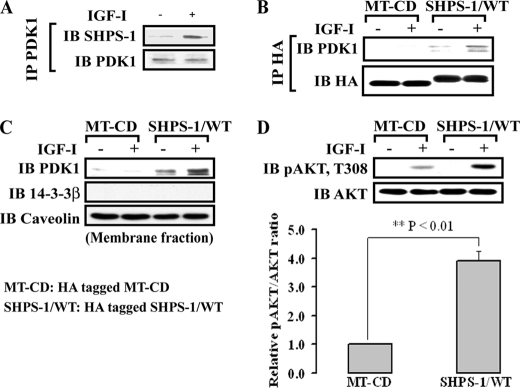
Because PDK1 is a kinase that phosphorylates AKT on Thr308 and its membrane localization is essential for PDK1 activation (1, 32), we determined whether PDK1 recruitment to SHPS-1 was stimulated by IGF-I. Untransfected SMCs exposed to hyperglycemia and IGF-I showed a 4.6 ± 1.1 (mean ± S.E.)-fold increase in PDK1-SHPS-1 association (Fig. 1A). In addition, IGF-I stimulated a 2.1 ± 0.5-fold increase in PDK1 recruitment to the membrane fraction in untransfected cells (supplemental Fig. 1). In contrast, cells expressing a truncated form of SHPS-1 in which the cytoplasmic domain was deleted (SHPS-1/−CD; MT-CD) showed no detectable PDK1-SHPS-1 association after IGF-I treatment (Fig. 1B). The levels of HA were confirmed to be the same when SHPS-1/WT- and MT-CD-expressing cells were compared (supplemental Fig. 2A). Further, analysis of the PDK1 content in the membrane fraction showed that it was significantly reduced in cells expressing MT-CD, suggesting that SHPS-1 association was required for its recruitment to the membrane fraction (Fig. 1C). Following IGF-I stimulation, there was a significant reduction (74.5 ± 10.1%, n = 3, p < 0.01) in AKT Thr308 phosphorylation in cells that expressed MT-CD compared with cells expressing SHPS-1/WT (Fig. 1D). Because deletion of the SHPS-1/CD would be predicted to alter the activation of multiple signaling elements, we wished to establish a method for selectively inhibiting PDK1 association with the SHPS-1 signaling complex. Therefore, we conducted studies to determine the mechanism of PDK1 recruitment to SHPS-1.
FIGURE 1.
PDK1 is recruited to SHPS-1 in response to IGF-I, resulting in enhanced AKT phosphorylation. A, nontransfected, confluent SMC cultures were serum-starved overnight and then stimulated with IGF-I (50 ng/ml). The cells were lysed, and immunoprecipitation was performed using an anti-PDK1 antibody, followed by immunoblotting with anti-SHPS-1. To control for loading, the blot was stripped and reprobed with anti-PDK1. B, cells that were expressing HA-tagged SHPS-1/WT or HA-tagged MT-CD were serum-starved and stimulated with IGF-I. Lysates were immunoprecipitated (IP) with anti-HA, followed by immunoblotting with an anti-PDK1. The blot was stripped and reprobed with anti-HA as a loading control. C, membrane proteins were isolated as described under “Experimental Procedures” and immunoblotted with anti-PDK1. The blots were stripped and reprobed with anti-caveolin as a membrane fraction marker and anti-14-3-3β as a cytoplasmic fraction marker. D, both SHPS-1 WT and MT-CD cells were exposed to IGF-I, and cell lysates were analyzed by direct immunoblotting for phospho-AKT (Thr308). The blot was stripped and reprobed with anti-AKT as a loading control. The protein levels were quantified using scanning densitometry. The graph shows the mean result from three independent experiments expressed as relative phospho-AKT/AKT ratio, which was calculated using arbitrary scanning units (D, bottom). **, p < 0.01 indicates the significant difference between the two cell types. Error bars represent S.E.
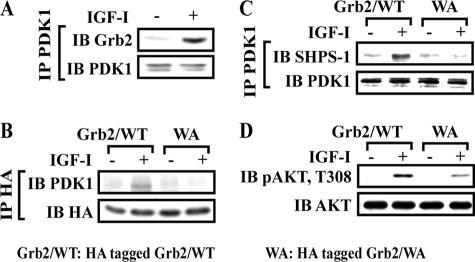
Grb2 Mediates PDK1 Recruitment to SHPS-1
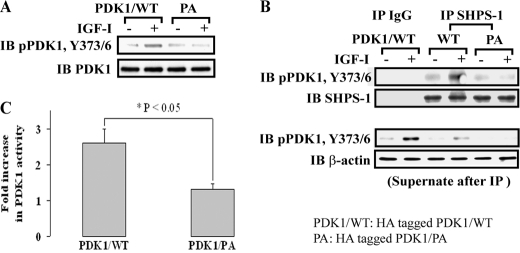
PDK1 does not contain SH2 domains and does not bind directly to phospho-SHPS-1. Therefore, we searched the PDK1 sequence for motifs that might bind to known SHPS-1 binding partners. PDK1 does contain a polyproline motif that has the potential to bind to the SH3 domain in Grb2, a component of the SHPS-1 complex. Following IGF-I stimulation, PDK1-Grb2 association was increased 5.4 ± 1.2-fold (p < 0.01) (Fig. 2A). To determine if this was mediated through the Grb2 SH3 domain, we utilized a mutant in which both SH3 domains in Grb2 were altered by substituting tryptophans at positions 36 and 193 with alanine (Grb2/WA). These substitutions had been shown to alter Grb2 SH3 domain-polyproline motif interactions (25, 33). Grb2 binding to PDK1 was significantly inhibited in cells expressing the Grb2/WA mutant compared with Grb2/WT-expressing cells (Fig. 2B) (0.9 ± 0.2 versus 4.5 ± 0.9-fold, n = 3, p < 0.05). Importantly, PDK1 association with SHPS-1 was also reduced in response to IGF-I in cells expressing Grb2/WA (1.0 ± 0.2- versus 5.2 ± 1.1-fold, n = 3, p < 0.01) (Fig. 2C). To control for nonspecific conformational changes in Grb2, we measured Pyk2-Grb2 association because this interaction occurs through an SH2 domain in Grb2 binding to phosphotyrosine 881 in Pyk2 (27, 34–37). The results showed that IGF-I-stimulated Pyk2-Grb2 association was unchanged (supplemental Fig. 3). SMCs expressing Grb2/WA mutant also showed decreased AKT Thr308 phosphorylation in response to IGF-I (67 ± 11% reduction in PDK1/PA mutant cells, n = 3, p < 0.01) (Fig. 2D).
FIGURE 2.
Grb2/WA mutant impairs PDK1 recruitment to SHPS-1 in response to IGF-I. A, nontransfected, confluent cultures were serum-starved overnight and then stimulated with IGF-I. Cell lysates were immunoprecipitated (IP) with anti-PDK1 and immunoblotted (IB) for Grb2. To control the loading, the blot was stripped and reprobed with anti-PDK1. B and C, quiescent HA-tagged Grb2/WT or HA-tagged Grb2/WA-expressing SMCs were stimulated with IGF-I. Cell lysates were immunoprecipitated with anti-HA (B) or PDK1 (C) and immunoblotted for the protein of interest. To control the loading, the blots were stripped and reprobed with anti-HA (B) or anti-PDK1 antibodies (C). D, 20 μl of cell lysate from the same experiment was used for detection of phospho-AKT (Thr308). The blot was stripped and reprobed with anti-AKT as a loading control.
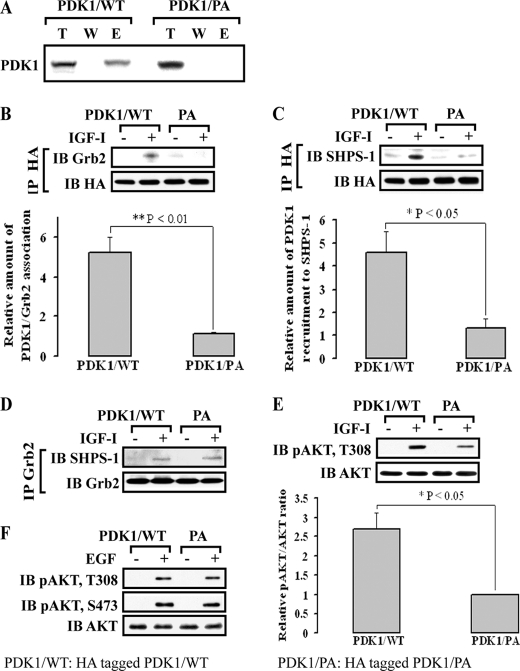
Because it was likely that the SH3 domain in Grb2 was binding to a proline-rich sequence in PDK1, a mutant in which prolines 71 and 74 in PDK1 were substituted with alanine (PDK1/PA) was constructed and expressed in vitro and in living cells. In vitro binding assays clearly showed that wild-type PDK1 (PDK1/WT) was able to directly bind to Grb2, whereas the PDK1/PA mutant did not bind to Grb2 in vitro (Fig. 3A). Following transfection, the levels of HA and PDK1 were confirmed to be the same when PDK1/WT and PDK1/PA mutant-expressing cells were compared (supplemental Fig. 2B). The cells expressing the PDK1/PA mutant had significantly reduced PDK1-Grb2 association in response to IGF-I compared with PDK1/WT-expressing cells (Fig. 3B) (1.1 ± 0.1- versus 5.2 ± 0.8-fold, n = 3, p < 0.01). More importantly, substitution for these prolines in PDK1 resulted in a major reduction in PDK1 recruitment to SHPS-1 compared with the PDK1/WT-expressing cells (1.3 ± 0.4- versus 4.6 ± 0.9-fold, n = 3, p < 0.05) (Fig. 3C). However, expression of this mutant did not alter Grb2 association with SHPS-1 (Fig. 3D). AKT Thr308 phosphorylation was also significantly decreased in cells expressing the PDK1/PA mutant (63 ± 9% reduction in PDK1/PA mutant cells, n = 3, p < 0.05) (Fig. 3E). To determine whether Grb2-mediated recruitment of PDK1 to SHPS-1 was specific for regulating IGF-I-stimulated AKT Thr308 phosphorylation, EGF was used in a similar experiment. In contrast to IGF-I, expression of the PDK1/PA mutant did not alter EGF-stimulated phosphorylation of AKT Thr308 (Fig. 3F).
FIGURE 3.
PDK1 polyproline motif provides a binding site for the Grb2 SH3 domain, which mediates IGF-I-induced PDK1 recruitment to SHPS-1. A, PDK1 directly binds to Grb2 through a polyproline-SH3 domain interaction in vitro. The interaction of [35S]methionine-labeled PDK1/WT or PDK1/PA mutant with Grb2 was determined using in vitro binding assays as described under “Experimental Procedures.” Images were obtained using a Storm 870 PhosphorImager. T, the expressed protein; W, the last wash; E, the eluate (supernatant after elution from the Grb2 beads). B–D, quiescent HA-tagged PDK1/WT or HA-tagged PDK1/PA-expressing SMCs were stimulated with IGF-I. Cell lysates were immunoprecipitated (IP) with anti-HA (B and C) or anti-Grb2 (D) and immunoblotted (IB) for the protein of interest. To control the loading, the blots were stripped and reprobed with anti-HA (B and C) or anti-Grb2 (D). The graphs show the mean result from three independent experiments expressed as relative amount of Grb2 associated with PDK1/total PDK1 that was immunoprecipitated or PDK1 associated with SHPS-1/total PDK1 in response to IGF-I (B and C, bottom). **, p < 0.01; *, p < 0.05; significant difference in two cell types. E, quiescent HA-tagged PDK1/WT- and PDK1/PA-expressing SMCs were stimulated with IGF-I. The cell lysates were directly immunoblotted for detection of phospho-AKT (Thr308). The blots were stripped and reprobed with anti-AKT as a loading control. The graph shows the mean result from three independent experiments expressed as relative phospho-AKT/AKT ratio for phospho-Thr308 (bottom). *, p < 0.05, significant difference in two cell types. F, EGF-induced AKT Thr308 or Ser473 phosphorylation in the PDK1/PA mutant or PDK1/WT cells. Both PDK1/WT and PDK1/PA cells were exposed to EGF (50 ng/ml, 5 min), and cell lysates were analyzed by direct immunoblotting for phospho-AKT (Thr308 or Ser473). The blots were stripped and reprobed with anti-AKT as a loading control. Error bars represent S.E.
Prior studies had shown that Pyk2 binding to Grb2 occurred through Pyk2 phosphotyrosine 881 binding to a Grb2 SH2 domain (34–37), and we have shown that tyrosine phosphorylation of Pyk2 mediates Grb2 recruitment to the SHPS-1 signaling complex (27). To confirm the importance of Grb2 association with SHPS-1 for PDK1 recruitment and to control for PDK1 overexpression, we utilized Pyk2/Y881F mutant-expressing cells that we had shown previously had attenuated Grb2 association with SHPS-1 (27). Mutation of Pyk2/Y881F significantly inhibited Pyk2-PDK1 association (1.0 ± 0.1 versus 4.7 ± 0.8-fold, n = 3, p < 0.01) and PDK1 recruitment to SHPS-1 (1.1 ± 0.1 versus 5.0 ± 1.1-fold, n = 3, p < 0.01) (supplemental Fig. 4A), leading to impaired AKT Thr308 phosphorylation (supplemental Fig. 4C).
IGF-I-stimulated PDK1 Recruitment to SHPS-1 Is Required for AKT Thr308 Phosphorylation
To determine the functional significance of loss of PDK1 recruitment to SHPS-1, the PKD1/PA and Pyk2/Y881F mutants were utilized. The PDK1/PA mutant cells had impaired phosphorylation of PDK1 on Tyr373/376 in response to IGF-I compared with PDK1/WT cells (0.9 ± 0.3 versus 3.6 ± 0.5-fold, n = 3, p < 0.05) (Fig. 4A). Similarly, cells expressing Pyk2/Y881F had impaired Tyr373/376 phosphorylation in response to IGF-I (1.1 ± 0.1- versus 3.9 ± 0.6-fold, n = 3, p < 0.01) (supplemental Fig. 4B). To confirm the importance of SHPS-1 localization for PDK1 activation, Tyr373/376 phosphorylation was determined following SHPS-1 immunoprecipitation. As shown in Fig. 4B, the PDK1 that was immunoprecipitated with SHPS-1 from cells expressing PDK1/WT showed a major increase in Tyr373/376 phosphorylation, whereas the same immunoprecipitate from cells expressing the PDK1/PA mutant showed minimal tyrosine phosphorylation. More importantly, the mutation of PDK1/PA inhibited the ability of IGF-I to enhance PDK1 kinase activity compared with cells expressing PDK1/WT (1.38 ± 0.16- versus 2.8 ± 0.4-fold, n = 5, p < 0.05) (Fig. 4C).
FIGURE 4.
Impairment of Grb2-PDK1 association leads to reduced tyrosine phosphorylation and activation of PDK1 in response to IGF-I. A, quiescent HA-tagged PDK1/WT and PDK1/PA-expressing SMCs were stimulated with IGF-I. The cell lysates were directly immunoblotted for phospho-PDK1 (Tyr373/376). The blot was stripped and reprobed with anti-PDK1 as a loading control. B, confluent HA-tagged PDK1/WT and PDK1/PA cell cultures were serum-starved overnight and treated with or without IGF-I for 5 min. Cell lysates were immunoprecipitated (IP) with anti-SHPS-1 or control IgG and immunoblotted (IB) for pPDK1 (Tyr373/376). To control the loading, the blot was stripped and reprobed with anti-SHPS-1. The supernatants were immunoblotted for detection of phospho-PDK1 (Tyr373/376) (bottom). The blot was stripped and reprobed with anti-β-actin as a loading control. C, confluent HA-tagged PDK1/WT and PDK1/PA cell cultures were serum-starved overnight and treated with IGF-I. Equivalent amounts of protein were immunoprecipitated with anti-PDK1, and the immunoprecipitates were assayed for PDK1 activity as described under “Experimental Procedures.” The values are the mean ± S.E. (error bars) from three independent experiments. *, p < 0.05, significant difference between the two cell types.
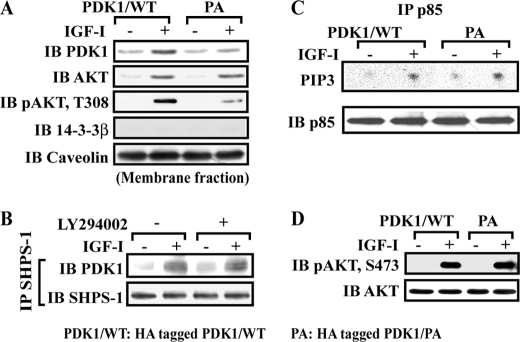
AKT membrane localization (38) has been shown to be essential for its activation, including Thr308 phosphorylation (1, 39). To exclude the possibility that decreased AKT Thr308 phosphorylation was due to altered AKT membrane localization, AKT membrane recruitment in response to IGF-I stimulation was analyzed. SMC expressing PDK1/PA mutant had the same amount of AKT localized in the membrane fraction compared with cells expressing PDK1/WT, whereas localization of PDK1 in the membrane fraction was impaired in response to IGF-I (1.4 ± 0.3- versus 3.3 ± 0.5-fold, n = 3, p < 0.05) (Fig. 5A). Similarly, AKT Thr308 phosphorylation in the membrane fraction was significantly decreased in cells expressing the PDK1/PA mutant (74 ± 12% reduction in PDK1/PA mutant cells, n = 3, p < 0.05) (Fig. 5A). To exclude the possibility that changes in PIP3 generation by PI3K accounted for the change in PDK1 recruitment to SHPS-1 in response to IGF-I, SMCs were exposed to IGF-I in the presence of a PI3K inhibitor, and SHPS-1-PDK1 association was determined. Inhibition of PI3K had no effect on PDK1-SHPS-1 association (Fig. 5B). Additionally, there was a comparable increase in PI3K activity in cells expressing the PDK1/PA mutant compared with cells expressing PDK1/WT (Fig. 5C). Similarly, when we quantified the AKT Ser473 phosphorylation response in cells expressing this mutant, there was no difference compared with SMCs expressing PDK1/WT (Fig. 5D).
FIGURE 5.
Expression of the PDK1/PA mutant reduces PDK1 membrane localization but not PI3K activity. A, membrane proteins were isolated as described under “Experimental Procedures” and immunoblotted (IB) with anti-PDK1, anti-AKT, or anti-phospho-AKT (Thr308). The blots were stripped and reprobed with anti-caveolin as a membrane fraction marker and anti-14-3-3β as a cytoplasmic fraction marker. B, confluent cultures were serum-starved overnight and incubated with or without LY294002 (10 μm) for 1 h. IGF-I was added for 5 min. The cell lysates were immunoprecipitated (IP) with anti-SHPS-1, followed by immunoblotting for PDK1. The blot was stripped and reprobed with anti-SHPS-1 as a loading control. C, confluent HA-tagged PDK1/WT and PDK1/PA cell cultures were serum-starved overnight and treated with or without IGF-I for 5 min. The anti-p85 immunoprecipitates were washed, and the pellets were incubated with PIP3 and [γ-32P]ATP, as described under “Experimental Procedures.” Radiolabeled PIP3 was separated using thin layer chromatography. A representative autoradiogram is shown. D, quiescent PDK1/WT- and PDK1/PA-expressing SMCs were stimulated with IGF-I. The cell lysates were directly immunoblotted for detection of phospho-AKT (Ser473). The blots were stripped and reprobed with anti-AKT as a loading control.
Disruption of PDK1-mediated Signaling Increases Cell Apoptosis
AKT enhances cell survival by phosphorylating Thr32 in the forkhead transcription factor FOXO3a, which leads to its nuclear exclusion (40). Members of the FOXO family of transcription factors exert their apoptosis-inducing or cell cycle-inhibitory functions by promoting the transcription of various proteins, including p27Kip1 (p27) (41). In cells expressing the PDK1/PA mutant, phosphorylation of Thr32 in FOXO3a was significantly decreased compared with PDK1/WT-expressing cells in response to IGF-I (Fig. 6A) (1.0 ± 0.1- versus 2.5 ± 0.3-fold, n = 3, p < 0.01). SMCs expressing PDK1/PA had impaired IGF-I-dependent down-regulation of p27 compared with SMC expressing PDK1/WT (Fig. 6B) (3 ± 2% versus 73 ± 8% in IGF-I-dependent down-regulation of p27, n = 3, p < 0.01). Because this signaling event is linked to apoptosis, we investigated apoptotic cell death in SMC expressing the mutant PDK1/PA following acute exposure to high glucose (26, 42). Using this paradigm, cells expressing PDK1/WT had an 87 ± 14% reduction in the number of TUNEL-positive cells in response to IGF-I, whereas IGF-I conferred no protection from apoptotic cell death in the PDK1/PA mutant-expressing cells (3.5% reduction in the number of TUNEL-positive cells) (Fig. 6C). To further characterize cellular apoptosis, we measured cleaved caspase-3, which indicates caspase-3 activity in this cell type. In the PDK1/WT cells, IGF-I was more effective in preventing caspase-3 cleavage than in PDK1/PA mutant cells (79.1% reduction versus 6.5% reduction, n = 3, p < 0.001) (Fig. 6D). In addition, PDK1/PA mutant-expressing cells had increased basal cleaved caspase-3, which is consistent with increased TUNEL-positive cells compared with PDK1/WT (Fig. 6, C and D). These results indicate that impairment of PDK1 recruitment to SHPS-1 in cells expressing PDK1/PA mutant inhibits IGF-I-induced AKT activation, leading to an increase in apoptotic cell death.
DISCUSSION
Previous studies have shown that in response to hyperglycemic stress, SHPS-1 functions as a scaffold protein for IGF-I signaling and that recruitment of signaling proteins to this scaffold is required for IGF-I-stimulated PI3K and MAPK activation as well as cell migration and proliferation (23–25). Recently, we reported that IGF-I stimulates the recruitment of Pyk2 to the SHPS-1 signaling complex via its direct binding to Src. Src then phosphorylates Pyk2 Tyr881, which creates a binding site for Grb2, resulting in recruitment of Grb2 to the SHPS-1 complex (27). Grb2 also recruits the p85 subunit of PI3K, leading to PI3K activation and enhanced AKT Ser473 phosphorylation (25). Although membrane recruitment of PDK1 is known to be essential for AKT Thr308 phosphorylation (1, 32), the underlying mechanism that mediates PDK1 membrane recruitment and the specific adaptor proteins that are required have not been delineated. Recently, Nakamura et al. (43) demonstrated that Freund-1/Akil, a cytoplasmic protein, bound to PDK1 and, in conjunction with the epidermal growth factor (EGF) receptor, modulated PDK1 translocation to the plasma membrane following EGF stimulation. This interaction was required for EGF-stimulated PDK1 activation. In contrast, they showed that IGF-I-stimulated PDK1 activation and AKT Thr308 phosphorylation did not depend upon Fruend-1-PDK1 association (43). This finding led those authors to speculate that other scaffold proteins might exist that mediated the effect of IGF-I receptor activation of PDK1.
The results of this study provide evidence that SHPS-1 is an important scaffold for assembly of a complex that includes Pyk2-Grb2 and that Grb2 mediates PDK1 recruitment in response to IGF-I. Assembly of this complex is required for membrane localization and tyrosine phosphorylation of PDK1, leading to its activation, which results in enhancement of AKT Thr308 phosphorylation and cell survival.
In contrast to EGFR-Fruend1-PDK1 complex assembly, the complex studied herein does not associate directly with IGF-I receptor. Furthermore, unlike EGFR-Freund-1-PDK1, there is no direct interaction between SHPS-1 and PDK1. SHP2, which is the component of this complex that binds directly to SHPS-1, recruits Src-Pyk2-Grb2, and Grb2 binds directly to PDK1. Consistent with the IGF-I-regulated mechanism being distinct from the EGF-mediated pathway is our observation that EGF-stimulated AKT Thr308 phosphorylation was not impaired in cells expressing the PDK1/PA mutant that could not bind to Grb2. This supports the conclusion that EGF and IGF-I are stimulating PDK1 membrane recruitment through different signaling pathways that involve the assembly of distinct membrane-associated scaffolds, each containing unique components.
Several proteins have been reported to interact with PDK1, including HSP90, Grb14, mAKAP α, and TUSC4. HSP90 and TUSC4 have been reported to modulate PDK1 activity (10, 44–47). However, only Freund-1-EGFR and Grb14-IR have been shown to recruit PDK1 to the cell membrane and thereby facilitate its activation. In the present study, SHPS-1-Grb2-mediated membrane recruitment of PDK1 was dependent upon a direct binding interaction between Grb2 and PDK1 (Fig. 5A). This occurred via the SH3 domain in Grb2 binding to a polyproline sequence in PDK1. Grb2 is a versatile adapter protein that interacts with multiple signaling elements through its SH2 and SH3 domains. In addition to adaptor protein-mediated membrane localization of PDK1, it is well established that phospholipid metabolites generated by PI3K modulate PDK1 membrane association and activation (48). However, in this study, cells expressing the PDK1/PA mutant showed normal PI3K activation and PIP3 production in response to IGF-I (Fig. 5C) but impaired PDK1 membrane recruitment and activation. Although previous studies have shown that PIP3 generation plays an important role in pleckstrin homology domain-mediated PDK1 localization to the membrane (32, 48, 49), our findings suggest that the generation of PIP3 alone is not sufficient for PDK1 membrane recruitment in response to IGF-I. This is consistent with a prior study that showed that a pleckstrin homology domain-deleted form of PDK1 underwent insulin-dependent membrane association (10). Therefore, we propose that PDK1 association with the adaptor proteins, which can be localized on membrane-associated scaffolds, represents an important mechanism for PDK1 recruitment.
Full activation of PDK1 requires not only membrane recruitment but also tyrosine phosphorylation. Growth factor stimulation leads to sequential phosphorylation of Tyr9 and Tyr373/376 (11, 14, 16, 17), and substitutions for these phosphotyrosines result in decreased PDK1 activity (9, 13, 14, 16, 47). Previous studies showed that Pyk2 activates PDK1, but the mechanism was not determined (9, 12, 18). Our recent study showed that IGF-I stimulates Pyk2 Tyr881 phosphorylation, and expression of Pyk2/Y881F that had impaired Grb2 binding led to failure to recruit PDK1 to SHPS-1 (supplemental Fig. 4A) (27). Conversely, overexpression of Pyk2 resulted in an increased phosphorylation of PDK1 at Tyr373/376 in response to IGF-I (supplemental Fig. 4B). We propose that this allows Src to be in close proximity to PDK1, resulting in enhanced Src-dependent PDK1 Tyr373/376 tyrosine phosphorylation. That this interaction occurred when both proteins were associated with SHPS-1 is suggested by our observation that, following SHPS-1 pull-down, the immunoprecipitate contained almost all of the tyrosine-phosphorylated PDK1 (Fig. 4B).
Prior studies have reported that IGF-I enhances PDK1 activity and increases cell survival (17, 50). Our studies show that when SMCs are exposed to hyperglycemic stress, they undergo apoptosis unless they are exposed to IGF-I (42) and that assembly of the signaling complex on SHPS-1 is required to prevent apoptosis (26). Because inhibition of PDK1 recruitment to SHPS-1 impaired IGF-I-dependent cell survival, we conclude that PDK1 recruitment to SHPS-1 scaffold is an integral component of the process by which IGF-I inhibits hyperglycemic stress-induced apoptosis. We noted a 2-fold difference in basal apoptotic cell death between PDK1/WT and PDK1/PA mutant-overexpressing cells. We postulate that a difference in wild-type PDK1 could account for this trend. The increased amount of PDK1 in PDK1/WT-overexpressing cells may amplify the effect of growth factors on cell survival. In contrast, cells overexpressing the PDK1/PA mutant may have suppressed the endogenous PDK1, which would amplify hyperglycemia-induced cell apoptosis. To support this conclusion, we also detected a difference in the basal level of FOXO3a phosphorylation, p27 expression, and cleaved caspase-3.
PDK1-mediated phosphorylation of AKT Thr308 activates its downstream signaling events that mediate PDK1-dependent cell survival (43, 50, 51). Recent studies have emphasized the importance of AKT Thr308 phosphorylation for downstream signaling and growth factor-stimulated biologic actions. Ito et al. (52) showed that selective loss of PDK1-mediated Thr308 phosphorylation in mice with retention of Ser473 phosphorylation resulted in deficient FOXO3a nuclear transport and increased cardiomyocyte apoptosis. Similarly, Padmanabhan et al. (53) showed that selective inhibition of AKT Thr308 dephosphorylation resulted in FOXO3a nuclear exclusion and decreased FOXO3a augmented gene expression in response to insulin. Other studies have shown that full activation of AKT Thr308 phosphorylation is required for phosphorylation of GSK3α and FOXO3a (51, 54, 55), which are required to inhibit apoptosis. Taken together, these results support the conclusion that selective activation of AKT Thr308 by PDK1 is an important component of IGF-I receptor-linked signaling events that stimulate FOXO3a nuclear exclusion, thus leading to down-regulation of p27 and inhibition of apoptosis.
Our prior studies have demonstrated that AKT is also recruited to SHPS-1 (26). Therefore, it is possible that the decrease in AKT Thr308 phosphorylation is due to a reduction in AKT recruitment to SHPS-1. However, our results show that in cells expressing the PDK1/PA mutant, AKT Ser473 phosphorylation and PIP3 generation in response to IGF-I stimulation are intact, and AKT recruitment to SHPS-1 is not attenuated (Fig. 5A). Therefore, AKT is appropriately recruited to the cell membrane, and this does not account for reduced AKT Thr308 phosphorylation. This highlights the importance of PDK1 recruitment to SHPS-1 for AKT Thr308 phosphorylation in response to IGF-I.
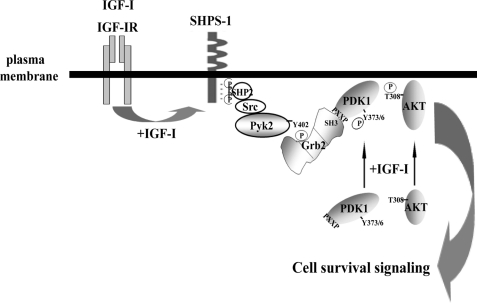
Based on these findings, we propose a model of PDK1 membrane recruitment and AKT Thr308 phosphorylation in response to IGF-I (Fig. 7). Following SHPS-1 tyrosine phosphorylation, SHP2-Src-Pyk2 is recruited to SHPS-1 complex, resulting in Pyk2 Tyr881 phosphorylation, which creates a binding site for Grb2 (27). Grb2 recruits PDK1 to the membrane via an SH3 domain-polyproline interaction, resulting in enhanced Src-Pyk2-dependent tyrosine phosphorylation of PDK1 and its optimal enzymatic activation. This leads to enhanced AKT Thr308 phosphorylation and cell survival in response to IGF-I. The localized interaction of these key signaling molecules on SHPS-1 may thus represent a novel pathway that plays a critical role in preventing hyperglycemic stress-induced apoptosis and potentially leads to enhanced SMC proliferation.
FIGURE 7.
A proposed model for PDK1 recruitment to the SHPS-1 signaling complex, which is stimulated by IGF-I and mediates cell survival during hyperglycemia. Following stimulation by IGF-I, the IGF-I receptor tyrosine kinase phosphorylates tyrosines contained in the SHPS-1 cytoplasmic domain, leading to the formation of a signaling complex that includes SHP2, Src, Pyk2, and Grb2. In response to IGF-I, a proline-rich sequence in PDK1 binds to an SH3 domain in Grb2, resulting in PDK1 recruitment to the SHPS-1 signaling complex. Following its recruitment to SHPS-1, PDK1 is further activated via Tyr373/376 phosphorylation, and this is required for maximal increase in PDK1 kinase activity and subsequently AKT Thr308 phosphorylation, a critical event for the downstream signaling events that mediate enhanced cell survival.
Supplementary Material
Acknowledgments
We thank Dr. Walker H. Busby, Jr. for help in preparing SHPS-1 antibody. We thank Drs. H. S. Earp and Lee M. Graves (University of North Carolina, Chapel Hill, NC) for providing the pcDNA-myc-Pyk2/WT and pcDNA-myc-Pyk2/Y881F constructs and Dr. Brian Hemmings (Friedrich Miescher Institute, Basel, Switzerland) for providing the pcDNA-myc-PDK1/WT construct. We thank Drs. Laura A. Maile and Lee M Graves for comments. We also thank Laura Lindsey for her help in preparing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL56850 and AG022331.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- IGF-I
- insulin-like growth factor-I
- SH2 and-3
- Src homology 2 and 3, respectively
- CD
- cytoplasmic domain
- SMC
- smooth muscle cell
- PIP3
- inositol 1,4,5-trisphosphate
- EGFR
- EGF receptor.
REFERENCES
- 1.Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 2.Belham C., Wu S., Avruch J. (1999) Curr. Biol. 9, R93–R96 [DOI] [PubMed] [Google Scholar]
- 3.Flynn P., Wongdagger M., Zavar M., Dean N. M., Stokoe D. (2000) Curr. Biol. 10, 1439–1442 [DOI] [PubMed] [Google Scholar]
- 4.Cho H., Thorvaldsen J. L., Chu Q., Feng F., Birnbaum M. J. (2001) J. Biol. Chem. 276, 38349–38352 [DOI] [PubMed] [Google Scholar]
- 5.Lawlor M. A., Alessi D. R. (2001) J. Cell Sci. 114, 2903–2910 [DOI] [PubMed] [Google Scholar]
- 6.Casamayor A., Morrice N. A., Alessi D. R. (1999) Biochem. J. 342, 287–292 [PMC free article] [PubMed] [Google Scholar]
- 7.Klarlund J. K., Guilherme A., Holik J. J., Virbasius J. V., Chawla A., Czech M. P. (1997) Science 275, 1927–1930 [DOI] [PubMed] [Google Scholar]
- 8.Biondi R. M., Komander D., Thomas C. C., Lizcano J. M., Deak M., Alessi D. R., van Aalten D. M. (2002) EMBO J. 21, 4219–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniyama Y., Weber D. S., Rocic P., Hilenski L., Akers M. L., Park J., Hemmings B. A., Alexander R. W., Griendling K. K. (2003) Mol. Cell. Biol. 23, 8019–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King C. C., Newton A. C. (2004) J. Biol. Chem. 279, 37518–37527 [DOI] [PubMed] [Google Scholar]
- 11.Fiory F., Alberobello A. T., Miele C., Oriente F., Esposito I., Corbo V., Ruvo M., Tizzano B., Rasmussen T. E., Gammeltoft S., Formisano P., Beguinot F. (2005) Mol. Cell. Biol. 25, 10803–10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Block K., Eid A., Griendling K. K., Lee D. Y., Wittrant Y., Gorin Y. (2008) J. Biol. Chem. 283, 24061–24076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grillo S., Grémeaux T., Casamayor A., Alessi D. R., Le Marchand-Brustel Y., Tanti J. F. (2000) Eur. J. Biochem. 267, 6642–6649 [DOI] [PubMed] [Google Scholar]
- 14.Park J., Hill M. M., Hess D., Brazil D. P., Hofsteenge J., Hemmings B. A. (2001) J. Biol. Chem. 276, 37459–37471 [DOI] [PubMed] [Google Scholar]
- 15.Steiler T. L., Galuska D., Leng Y., Chibalin A. V., Gilbert M., Zierath J. R. (2003) Endocrinology 144, 5259–5267 [DOI] [PubMed] [Google Scholar]
- 16.Yang K. J., Shin S., Piao L., Shin E., Li Y., Park K. A., Byun H. S., Won M., Hong J., Kweon G. R., Hur G. M., Seok J. H., Chun T., Brazil D. P., Hemmings B. A., Park J. (2008) J. Biol. Chem. 283, 1480–1491 [DOI] [PubMed] [Google Scholar]
- 17.Alberobello A. T., D'Esposito V., Marasco D., Doti N., Ruvo M., Bianco R., Tortora G., Esposito I., Fiory F., Miele C., Beguinot F., Formisano P. (2010) J. Biol. Chem. 285, 6563–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo J., Sabri A., Elouardighi H., Rybin V., Steinberg S. F. (2006) Circ. Res. 99, 1367–1375 [DOI] [PubMed] [Google Scholar]
- 19.Clemmons D. R. (2007) Nat. Rev. Drug Discov. 6, 821–833 [DOI] [PubMed] [Google Scholar]
- 20.LeRoith D., Werner H., Beitner-Johnson D., Roberts C. T., Jr. (1995) Endocr. Rev. 16, 143–163 [DOI] [PubMed] [Google Scholar]
- 21.Jones J. I., Prevette T., Gockerman A., Clemmons D. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2482–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radhakrishnan Y., Busby W. H., Jr., Shen X., Maile L. A., Clemmons D. R. (2010) J. Biol. Chem. 285, 15682–15695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling Y., Maile L. A., Lieskovska J., Badley-Clarke J., Clemmons D. R. (2005) Mol. Biol. Cell 16, 3353–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieskovska J., Ling Y., Badley-Clarke J., Clemmons D. R. (2006) J. Biol. Chem. 281, 25041–25053 [DOI] [PubMed] [Google Scholar]
- 25.Radhakrishnan Y., Maile L. A., Ling Y., Graves L. M., Clemmons D. R. (2008) J. Biol. Chem. 283, 16320–16331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen X., Xi G., Radhakrishnan Y., Clemmons D. R. (2009) Mol. Cell Proteomics 8, 1539–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen X., Xi G., Radhakrishnan Y., Clemmons D. R. (2010) Cell Mol. Life Sci., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker A., Gockerman A., Busby W. H., Clemmons D. R. (1995) Endocrinology 136, 2470–2476 [DOI] [PubMed] [Google Scholar]
- 29.Xi G., Shen X., Clemmons D. R. (2008) Mol. Endocrinol. 22, 2162–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers M. G., Jr., Sun X. J., Cheatham B., Jachna B. R., Glasheen E. M., Backer J. M., White M. F. (1993) Endocrinology 132, 1421–1430 [DOI] [PubMed] [Google Scholar]
- 31.Kwon M., Ling Y., Maile L. A., Badley-Clark J., Clemmons D. R. (2006) Endocrinology 147, 1458–1465 [DOI] [PubMed] [Google Scholar]
- 32.Anderson K. E., Coadwell J., Stephens L. R., Hawkins P. T. (1998) Curr. Biol. 8, 684–691 [DOI] [PubMed] [Google Scholar]
- 33.Pleiman C. M., Hertz W. M., Cambier J. C. (1994) Science 263, 1609–1612 [DOI] [PubMed] [Google Scholar]
- 34.Lev S., Moreno H., Martinez R., Canoll P., Peles E., Musacchio J. M., Plowman G. D., Rudy B., Schlessinger J. (1995) Nature 376, 737–745 [DOI] [PubMed] [Google Scholar]
- 35.Dikic I., Tokiwa G., Lev S., Courtneidge S. A., Schlessinger J. (1996) Nature 383, 547–550 [DOI] [PubMed] [Google Scholar]
- 36.Avraham H., Park S. Y., Schinkmann K., Avraham S. (2000) Cell. Signal. 12, 123–133 [DOI] [PubMed] [Google Scholar]
- 37.Felsch J. S., Cachero T. G., Peralta E. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5051–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toker A., Cantley L. C. (1997) Nature 387, 673–676 [DOI] [PubMed] [Google Scholar]
- 39.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 40.Brunet A., Roux D., Lenormand P., Dowd S., Keyse S., Pouysségur J. (1999) EMBO J. 18, 664–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medema R. H., Kops G. J., Bos J. L., Burgering B. M. (2000) Nature 404, 782–787 [DOI] [PubMed] [Google Scholar]
- 42.Maile L. A., Capps B. E., Ling Y., Xi G., Clemmons D. R. (2007) Endocrinology 148, 2435–2443 [DOI] [PubMed] [Google Scholar]
- 43.Nakamura A., Naito M., Tsuruo T., Fujita N. (2008) Mol. Cell. Biol. 28, 5996–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujita N., Sato S., Ishida A., Tsuruo T. (2002) J. Biol. Chem. 277, 10346–10353 [DOI] [PubMed] [Google Scholar]
- 45.Chun J., Kwon T., Lee E. J., Hyun S., Hong S. K., Kang S. S. (2005) Biochem. Biophys. Res. Commun. 326, 136–146 [DOI] [PubMed] [Google Scholar]
- 46.Michel J. J., Townley I. K., Dodge-Kafka K. L., Zhang F., Kapiloff M. S., Scott J. D. (2005) Mol. Cell 20, 661–672 [DOI] [PubMed] [Google Scholar]
- 47.Kurata A., Katayama R., Watanabe T., Tsuruo T., Fujita N. (2008) Cancer Sci. 99, 1827–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Currie R. A., Walker K. S., Gray A., Deak M., Casamayor A., Downes C. P., Cohen P., Alessi D. R., Lucocq J. (1999) Biochem. J. 337, 575–583 [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshizaki H., Mochizuki N., Gotoh Y., Matsuda M. (2007) Mol. Biol. Cell 18, 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen R. T., Krueger K. D., Dhume A., Agrawal D. K. (2005) Apoptosis 10, 525–535 [DOI] [PubMed] [Google Scholar]
- 51.Williams M. R., Arthur J. S., Balendran A., van der Kaay J., Poli V., Cohen P., Alessi D. R. (2000) Curr. Biol. 10, 439–448 [DOI] [PubMed] [Google Scholar]
- 52.Ito K., Akazawa H., Tamagawa M., Furukawa K., Ogawa W., Yasuda N., Kudo Y., Liao C. H., Yamamoto R., Sato T., Molkentin J. D., Kasuga M., Noda T., Nakaya H., Komuro I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Padmanabhan S., Mukhopadhyay A., Narasimhan S. D., Tesz G., Czech M. P., Tissenbaum H. A. (2009) Cell 136, 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rena G., Woods Y. L., Prescott A. R., Peggie M., Unterman T. G., Williams M. R., Cohen P. (2002) EMBO J. 21, 2263–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashimoto N., Kido Y., Uchida T., Asahara S., Shigeyama Y., Matsuda T., Takeda A., Tsuchihashi D., Nishizawa A., Ogawa W., Fujimoto Y., Okamura H., Arden K. C., Herrera P. L., Noda T., Kasuga M. (2006) Nat. Genet. 38, 589–593 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.