Abstract
The assembly of the β-barrel proteins present in the outer membrane (OM) of Gram-negative bacteria is poorly characterized. After translocation across the inner membrane, unfolded β-barrel proteins are escorted across the periplasm by chaperones that reside within this compartment. Two partially redundant chaperones, SurA and Skp, are considered to transport the bulk mass of β-barrel proteins. We found that the periplasmic disulfide isomerase DsbC cooperates with SurA and the thiol oxidase DsbA in the folding of the essential β-barrel protein LptD. LptD inserts lipopolysaccharides in the OM. It is also the only β-barrel protein with more than two cysteine residues. We found that surAdsbC mutants, but not skpdsbC mutants, exhibit a synthetic phenotype. They have a decreased OM integrity, which is due to the lack of the isomerase activity of DsbC. We also isolated DsbC in a mixed disulfide complex with LptD. As such, LptD is identified as the first substrate of DsbC that is localized in the OM. Thus, electrons flowing from the cytoplasmic thioredoxin system maintain the integrity of the OM by assisting the folding of one of the most important β-barrel proteins.
Keywords: Bacteria, Disulfide, Electron Transfer, Membrane Proteins, Protein Folding, Thiol, Periplasm, Thioredoxin
Introduction
The outer membrane (OM)4 is a complex macromolecular structure that is essential for the viability of Gram-negative bacteria and serves as a permeability barrier against hydrophobic antibiotics. It is a unique asymmetric lipid bilayer with phospholipids forming the inner leaflet and lipopolysaccharides (LPS) forming the outer leaflet. The proteins that are present in the OM are either lipoproteins that are anchored to the inner leaflet via a lipid moiety or β-barrel proteins that are integral membrane proteins. The mechanisms that govern the assembly of the OM are poorly understood (1–3). In particular, we have a poor understanding of how the β-barrel proteins, which are synthesized in the cytoplasm and translocated unfolded across the inner membrane by the Sec translocon, are then transported across the periplasm and inserted in the OM. According to the most widely accepted model, unfolded β-barrel proteins are escorted across the periplasm by soluble periplasmic chaperones that deliver them to an OM multiprotein complex involving the β-barrel protein BamA (3, 4). Two partially redundant periplasmic chaperone pathways have been described in the Escherichia coli periplasm (5, 6). The first one involves SurA, a periplasmic prolyl cis-trans-isomerase, which is considered as the primary chaperone in the periplasm. SurA has been shown to be involved in the biogenesis of the most abundant β-barrel proteins, such as OmpA, OmpF, and OmpC (6, 7), and is the preferred chaperone for the essential protein LptD (previously Imp) (8). The second pathway involves the chaperone Skp and the protease/chaperone protein DegP. This pathway has been proposed to serve as a backup for SurA, assisting the biogenesis of the proteins that fell off the SurA pathway (6). The fact that surAskp and surAdegP mutants are not viable (5) indicates that cells cannot survive without at least one of the two chaperone pathways.
In addition to SurA, Skp, and DegP, other periplasmic proteins have been shown to exhibit a chaperone activity in vitro (reviewed in Ref. 2), but the real significance of this chaperone activity in vivo is not known. One of the proteins displaying a chaperone activity in vitro is DsbC, a protein-disulfide isomerase involved in the oxidative folding of periplasmic proteins with multiple cysteine residues (9, 10).
DsbC is a V-shaped dimeric protein. Each subunit of DsbC possesses a catalytic domain with a CXXC motif present in a thioredoxin fold as well as an N-terminal dimerization domain (11). The main function of DsbC is to correct the non-native disulfides that are introduced by the disulfide bond forming protein DsbA in proteins in which disulfides need to be formed between nonconsecutive cysteine residues (9). The first cysteine of the CXXC catalytic motif of DsbC performs a nucleophilic attack on a non-native disulfide present in a substrate protein, which results in the formation of an unstable mixed disulfide complex between DsbC and the substrate. This mixed disulfide will be resolved either by attack of another cysteine of the misfolded protein, resulting in the formation of a more stable disulfide in the substrate and the release of reduced DsbC, or by attack of the other cysteine of the CXXC motif. In this latter case, DsbC functions as a reductase, reducing the non-native disulfide and giving DsbA a new chance to form a correct disulfide. Direct evidence that this second mechanism functions in vivo has been recently reported (12). To maintain DsbC as functional in the oxidizing environment of the periplasm, its two catalytic cysteine residues are kept reduced by DsbD, an inner membrane protein that transfers reducing equivalents from the cytoplasmic thioredoxin system to the periplasm (13). Four DsbC substrates with multiple cysteines have been identified so far; they include the penicillin-insensitive endopeptidase MepA (14, 15), the ribonuclease RNase I (14, 15), the endonuclease End 1 (15), and the acid phosphatase AppA (16). Recently, we discovered that DsbC, together with another thioredoxin-related protein DsbG, also protects single cysteine residues from oxidation by reactive oxygen species present in the periplasm (17). Moreover, as stated above, DsbC exhibits a chaperone activity in vitro and can assist the refolding of denaturated proteins like lysozyme or glyceraldehyde-3-phosphate dehydrogenase (18). The active site cysteine residues are not required for this chaperone activity (19).
The pathways that govern OM biogenesis are not well characterized, and it is likely that all of the players have not yet been identified. Our objective was to find out whether DsbC, either via its isomerase or its chaperone activity, participates in the assembly of β-barrel proteins. We show here that DsbC is involved in the biogenesis of LptD. LptD is an essential β-barrel protein that is required for LPS insertion in the OM (20, 21) and that has recently been shown to be the cellular target of a new class of peptidomimetic antibiotics in Pseudomonas aeruginosa (22). Thus, the disulfide isomerization pathway that shuttles electrons from the cytoplasmic pool of NADPH to the cell envelope is involved in the folding of one of the most important β-barrel proteins present in the OM.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. All of the strains are derivatives of the E. coli K-12 strain MC4100 (23). Unless otherwise indicated, the bacteria were grown aerobically at 37 °C in Luria-Bertani (LB) medium. When necessary, growth media were supplemented with either chloramphenicol (25 μg/ml), kanamycin (50 μg/ml), or ampicillin (200 μg/ml). All of the alleles were moved by bacteriophage P1 transduction using standard procedures (24).
TABLE 1.
Strains and plasmids used in this study
| Strains and plasmids | Relevant genotype or features | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | F−, araD139, Δ(arg F-lac)U169, ptsF25, relA1, flb5301, rpsL 150·λ− | Ref. 23 |
| JAS27 | MC4100 ΔaraBAD | Received from T. Silhavy |
| JAS29 | MC4100 ΔaraBAD surA | Received from T. Silhavy |
| KD26 | JAS27 dsbA::kan | This study |
| KD27 | JAS29 dsbA::kan | This study |
| KD41 | JAS29 dsbD::kan | This study |
| KD42 | JAS29 dsbC::kan | This study |
| KD44 | JAS27 dsbC::kan | This study |
| KD45 | JAS27 dsbD::kan | This study |
| JFC470 | MC4100 Zae-502::Tn10 skp | Received from T. Silhavy |
| KD102 | JFC470 dsbC::kan | This study |
| Plasmids | ||
| pMD35 | pBAD33a DsbCCXXS-His6 | Ref. 15 |
| pKD69 | pQE60 | Qiagen |
| pMD21 | pQE60 LptD | This study |
| pCB21 | pQE60 LptD-His6 | This study |
| pJFC282 | pBAD33a | Ref. 35 |
| pJFC355 | pBAD33a DsbC | Received from J. Bardwell |
| pJFC369 | pBAD33a DsbCSXXS | Received from J. Bardwell |
Plasmid Construction
The LptD expression vector was constructed as followed: the region encoding the LptD protein was amplified from the chromosome using primers Imp N-term (5′-GCATCCATGGAAAAACGTATCCCCACTCTCCTGGCC-3′) and Imp C-term (5′-GCGCGGATCCTCACAAAGTGTTTTGATACG-3′) and cloned into pQE60 to yield plasmid pMD21 (Table 1). The stop codon at the 3′ end of the LptD gene was removed by site-directed mutagenesis using primers IMPSTOP_Fw (5′-CGTATCAAAACACTTTGGGAGGATCCAGATCTC-3′) and IMPSTOP_Rv (5′-GAGATCTGGATCCTCCCAAAGTGTTTTGATACG-3′) to yield plasmid pCB21 (Table 1). The sequence was verified.
Temperature Sensitivity
Spot titrations were performed to study the growth properties of the mutants at low temperatures. Briefly, the strains were first grown overnight at 37 °C in LB medium without selective antibiotic. After standardization to an A600 of 1.5, the cultures were serially diluted 105-fold in 10-fold increments into LB. Five microliters of each dilution were then spotted on LB plates and grown overnight at 21 °C. All of the spot titrations were performed in triplicate.
Antibiotics Sensitivity
BBL Sensi-Disc antimicrobial susceptibility test discs were used to test strain sensitivity to erythromycin (15 μg), rifampin (25 μg), vancomycin (30 μg), novobiocin (5 μg), and bacitracin (10 IU). The strains were first grown overnight at 37 °C in LB medium without selective antibiotic. One hundred microliters from these LB broth cultures were mixed with 3 ml of molten LB top agar and poured over an LB agar plate. The discs containing antibiotics were placed on top of the LB top agar, and the plates were incubated overnight at 37 °C. The diameter of the zone of inhibition of growth observed after overnight incubation was measured. The discs used were 6 mm in diameter.
Trapping and Purification of the DsbC-LptD Complex
A 1-liter LB culture of the dsbC strain carrying pBAD33 DsbCCXXS was grown at 25 °C to an A600 of 0.6. Expression was induced for 5 h by the addition of 0.2% l-arabinose. After precipitation with trichloroacetic acid (10%), the proteins were resuspended in 25 ml of 100 mm NaPi, pH 8, 300 mm NaCl, 0.3% SDS, 8 m urea, and 100 mm iodoacetamide to prevent any further disulfide bond rearrangement. The lysate was centrifuged at 14,000 rpm for 45 min. 2 ml of Ni-NTA resin were then added to the cleared mixture and incubated overnight at room temperature. A 1-ml column was packed with the protein-Ni-NTA resin complex and was washed thoroughly using NaPi, pH 8, 100 mm, NaCl 300 mm, and SDS 0.3%. The complexes involving DsbCCXXS were eluted with a step gradient from 30 to 300 mm imidazole. Only one fraction eluted from the column. It was concentrated 10-fold and analyzed by SDS-PAGE with or without DTT. After electrophoresis, the proteins were first transferred to a nitrocellulose membrane and probed with an anti-LptD antibody (1/5000) produced from a rabbit immunized with the purified protein (Eurogentec, Liège, Belgium). Anti-rabbit IgG (Sigma) was used as the secondary antibody at a concentration of 1/5000. Thermo Scientific Pierce ECL Western blotting substrate and Amersham Biosciences HyperfilmTM (GE Healthcare) were used to visualize the protein bands. In a complementary approach, the purified proteins were concentrated and analyzed by SDS-PAGE with or without DTT, and the gel was stained with Coomassie Blue.
Identification of Proteins by Mass Spectrometry
The bands of interest were cut out from the gel and digested with chymotrypsin or trypsin. The peptides were analyzed by capillary LC-tandem mass spectrometry in a LTQ XL ion trap mass spectrometer (ThermoScientific, San Jose, CA) fitted with a microelectrospray probe. The scan routine was a triple play experiment consisting of a full MS scan (400–2000 m/z) followed by a Zoom and MS/MS scan for the three most abundant ions. Dynamic exclusion allowed fragmentation of co-eluting peptides. The data were analyzed with the ProteomeDiscoverer software (ThermoScientific, version 1.1.0.263), and the proteins were identified with SEQUEST against a target decoy nonredundant E. coli protein database obtained from Uniprot. The false discovery rate was set below 5%. A search for disulfide linking was performed by DBond, using 3.5-Da precursor mass tolerance, 0.8-Da fragment ion tolerance, methionine oxidation, and carbamidomethylation on cysteine as variable modifications.
RESULTS
The surAdsbC Mutants Have a Synthetic Phenotype
We set out to determine whether DsbC is involved in the mechanisms that govern OM biogenesis. We hypothesized that the chaperone activity exhibited by this protein in vitro could play a role in one of the two major chaperone networks that govern the assembly of β-barrel proteins in vivo (the SurA and the Skp/DegP pathways). To circumvent the redundancy of these chaperone systems, we generated double skpdsbC and surAdsbC mutants using bacteriophage P1 transduction (24), and we studied their phenotypes. Whereas skpdsbC mutants were generated easily and had no particular phenotype, surAdsbC transductants grew poorly and formed very small colonies. We then studied the growth properties of the double mutants at various temperatures. The growth of the surAdsbC mutant was similar to that of the other single and double mutants at 37 and 42 °C (not shown). However, we found that the surAdsbC mutant exhibits a serious growth defect at lower temperatures, in contrast to the skpdsbC mutant (Fig. 1A).
FIGURE 1.
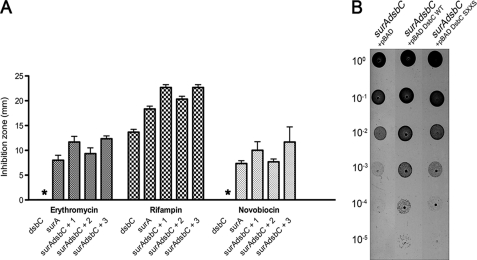
surAdsbC mutants have a growth defect at low temperatures. A, spot titers of WT MC4100, surA, dsbC, dsbD, surAdsbD, surAdsbC, skp, and skpdsbC mutants. After standardization, the cultures were serially diluted 105-fold in 10-fold increments. Five microliters of each dilution were then spotted on LB plates and grown overnight at 21 °C. B, spot titers of surAdsbC mutants harboring either an empty plasmid (pQE60) or a plasmid expressing LptD (pQE60 LptD).
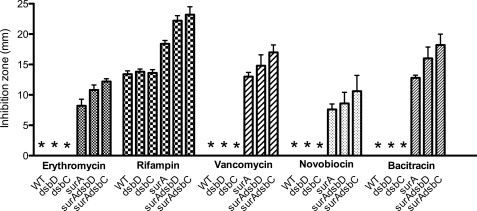
The function of SurA is clearly linked to OM biogenesis. For instance, surA mutants have decreased levels of β-barrel proteins in the OM. They also have an increased sensitivity to hydrophobic antibiotics caused by a decreased OM integrity. Therefore, we tested whether deletion of dsbC further decreases the resistance of surA mutants to those compounds. We compared the sensitivity of surA, dsbC, skp, surAdsbC, and skpdsbC mutants toward hydrophobic antibiotics, such as rifampin, novobiocin, bacitracin, vancomycin, and erythromycin. As shown in Fig. 2, we found that deletion of dsbC further increases the sensitivity of surA mutants. In contrast, the sensitivity of skpdsbC mutants was comparable with that of skp single mutants (not shown).
FIGURE 2.
surAdsbC mutants have an increased sensitivity to hydrophobic antibiotics. Antibiotics sensitivity of WT MC4100, dsbD, dsbC, surA, surAdsbD, and surAdsbC mutants at 37 °C is shown. Discs containing various hydrophobic antibiotics were used in disc diffusion assays as described under “Experimental Procedures.” The diameter of the zone of inhibition of growth is shown in mm. The strains for which no zone of inhibition of growth was observed are indicated by asterisks. The data shown are the averages of five experiments. The error bars indicate S.D., which was calculated using the following formula: S.D. = √(Σ(X − M)2/(n − 1)), where X refers to the individual data points, M is the mean, and n is the number of data points.
The data obtained so far indicate that surAdsbC mutants exhibit a synthetic phenotype. In particular, deletion of dsbC further increases the sensitivity of the surA strain to antibiotics. This strongly suggests that DsbC plays a role in the mechanisms that govern the integrity of the OM. At this point, we need to consider two different hypotheses. First, the synthetic phenotype of surAdsbC mutants could result from the absence of the potential chaperone activity of DsbC, which would place DsbC in the Skp/DegP pathway (remember that the surAskp and surAdegP mutants are synthetically lethal). Alternatively, the phenotype of surAdsbC mutants could result from the absence of the isomerase activity of DsbC, which would suggest that DsbC cooperates with SurA in the folding of some OM proteins.
The Isomerase Activity of DsbC Is Important for OM Integrity
To test these two hypotheses, we first compared the ability of wild-type DsbC and of a DsbC mutant in which both catalytic cysteine residues are replaced by serine (DsbCSXXS) to complement the phenotype of the surAdsbC mutant. DsbCSXXS lacks the isomerase activity of DsbC but keeps the chaperone activity of the protein (18). As shown in Fig. 3, we observed that only the expression of wild-type DsbC was able to complement the sensitivity of the surAdsbC mutant to antibiotics (Fig. 3A) as well as the growth defect exhibited by this strain at low temperatures (Fig. 3B). This indicates that the function of DsbC in OM biogenesis depends on the presence of the catalytic cysteine residues and, therefore, that the chaperone activity is not involved.
FIGURE 3.
Expression of wild-type DsbC complements the synthetic phenotype of surAdsbC mutants. A, antibiotics sensitivity of dsbC, surA, and surAdsbC strains harboring either an empty pBAD33a plasmid (+1), pBAD33a expressing wild-type DsbC (+2), or pBAD33a expressing DsbCSXXS (+3) at 37 °C. Discs containing erythromycin, rifampin, and novobiocin were used in disc diffusion assays as described under “Experimental Procedures.” The strains for which no zone of inhibition of growth was observed are indicated by an asterisk. The data shown are the averages of five experiments. The error bars indicate S.D., which was calculated using the following formula: S.D. = √(Σ(X − M)2/(n − 1)), where X refers to the individual data points, M is the mean, and n is the number of data points. B, spot titers of the surAdsbC mutant harboring either pBAD33, pBAD33 DsbC, or pBAD33 DsbCSXXS. The strains were grown on LB plates at 21 °C as described in the legend to Fig. 1A.
We found recently that DsbC cooperates with DsbA in the oxidation of secreted proteins, serving as a backup for DsbA (15). However, this activity does not depend on the presence of DsbD, in contrast to the isomerase activity of DsbC. Thus, to determine whether it is the absence of the isomerase activity that causes the phenotype of the surAdsbC mutant, we studied the phenotype of surAdsbD mutants. We found that surAdsbD mutants are more sensitive than surA mutants and have a phenotype similar to that of surAdsbC mutants (Fig. 1A). These results indicate therefore that it is the absence of the isomerase activity of DsbC that causes the synthetic phenotype of surAdsbC double mutants.
All of the DsbC substrates that have been identified so far are periplasmic soluble enzymes (2) that degrade substrates present in the periplasm. It is unlikely that the misfolding of any of these soluble proteins could explain the decreased OM integrity of the surAdsbC mutant. The results presented above suggest instead that DsbC is involved in the folding of at least one β-barrel protein and that misfolding of this protein leads to the observed decrease in OM integrity. We searched the ECHOlocation database (25) to identify the β-barrel proteins possessing more than two cysteine residues because only those proteins can form incorrect disulfides requiring the isomerase activity of DsbC. LptD, with four cysteine residues, turned out to be the only β-barrel protein with more than two cysteines. Interestingly, LptD has been shown to contain disulfide bonds (26) and to interact with DsbA (27). LptD is also a true SurA substrate (8). Moreover, LptD, which inserts LPS in the outer leaflet of the OM, is an essential player in envelope biogenesis, and cells expressing LptD mutants have a decreased OM integrity (28). LptD was therefore a prime candidate.
LptD Is a New DsbC Substrate
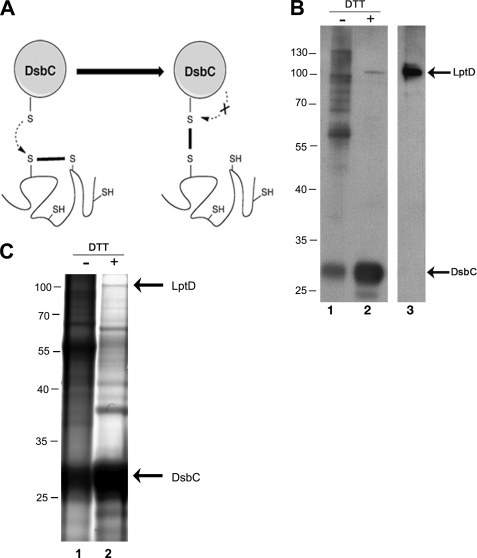
To test whether DsbC interacts with LptD in vivo, we expressed a mutated version of DsbC presenting a CXXS catalytic site in a dsbC strain. Proteins from the thioredoxin family, including DsbC, are characterized by a CXXC catalytic motif. When these proteins function as reductases or isomerases, the reaction takes off with a nucleophilic attack of the N-terminal cysteine of the conserved CXXC motif on a target disulfide (29). As a result, an intermediate mixed disulfide complex is formed between the thioredoxin protein and the substrate protein. This mixed disulfide in turn is reduced by a nucleophilic attack of the C-terminal cysteine of the CXXC motif. Thus, to trap the substrates linked to DsbC, we replaced the C-terminal cysteine by a serine. This mutant protein is still able to react with non-native disulfides but lacks the second cysteine of the catalytic motif required to resolve the mixed disulfide complex (Fig. 4A). This approach has previously been used to trap the substrates of DsbG, a reductase present in the E. coli periplasm (17). After expression of the mutant protein in the periplasm of a dsbC deletion strain, the proteins were precipitated with trichloroacetic acid, and all free cysteines were alkylated with iodoacetamide to prevent any further disulfide bond rearrangement. The proteins were resuspended in a buffer containing 0.3% SDS and no reductant to maintain DsbCCXXS covalently bound to its potential substrate proteins. DsbCCXXS was then purified in the presence of SDS using a His tag present at the C terminus of the protein. Only one peak eluted from the affinity column when an imidazole gradient was applied. DsbC was the most abundant protein in that peak, but other proteins of higher molecular weight were also present (Fig. 4B). Because the purification was performed under denaturing conditions, nonspecific binding to the column is minimal, and the bands co-eluting with DsbC likely correspond to complexes between DsbC and other proteins. To determine whether LptD was trapped in complex with DsbC, the fractions were analyzed by Western blot using an anti-LptD antibody. We observed that the addition of DTT led to the appearance of a band detected by the anti-LptD antibody and migrating at the size expected for LptD (∼100 kDa) (26) (Fig. 4B). Thus, this result strongly suggests that LptD was trapped in complex with DsbC. LptD is released from the mixed disulfide complex with DsbC upon DTT addition.
FIGURE 4.
Identification of LptD trapped in complex with DsbC. A, the first cysteine residue of the CXXC motif of DsbC performs a nucleophilic attack on an incorrect disulfide, which results in the formation of a mixed disulfide complex. To trap the proteins linked to DsbC, we replaced the C-terminal cysteine with a serine. This mutant protein is still able to react with non-native disulfides but lacks the second cysteine of the catalytic motif required to resolve the mixed disulfide complex. B, the complex formed between DsbCCXXS and LptD was purified using Ni-NTA resin. It was concentrated 10-fold and analyzed by SDS-PAGE without (lane 1) or with DTT (lane 2). Two bands were detected by Western blot using the anti-LptD antibody after DTT treatment. Whereas the upper band migrates at the same size as the purified LptD protein (lane 3), the lower band corresponds to DsbCCXXS. The anti-LptD antibody was obtained using a His-tagged LptD protein and reacts with His-tagged DsbC. The positions of the relevant molecular weight markers are indicated in kDa. C, the fraction that eluted from the Ni-NTA resin was concentrated 10-fold and subjected to SDS-PAGE without (lane 1) or with DTT (lane 2). The protein bands were visualized by staining with Coomassie Blue. After DTT treatment, three major bands appeared in addition to DsbC. The band migrating at ∼100 kDa (the expected mass for LptD) (26) was cut out of the gel and digested with chymotrypsin. Tandem mass spectrometry analysis indicated that this band corresponds to LptD. The positions of the relevant molecular weight markers are indicated in kDa.
We then decided to unambiguously confirm the identity of this band as LptD using mass spectrometry. After further concentration, the sample was analyzed by SDS-PAGE, and the gel was stained with Coomassie Blue. Upon DTT treatment, three major bands appeared in addition to DsbC, including one migrating near 100 kDa (Fig. 4C). This latter band was cut out of the gel and digested. Analysis by MS/MS confirmed that this band corresponds to LptD. Thus, we can conclude that LptD forms disulfide-linked complexes with DsbC in vivo.
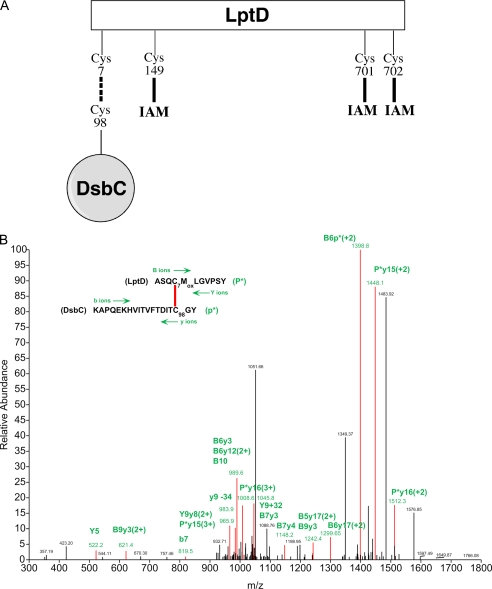
As stated above, LptD has four cysteine residues. We sought to identify the cysteines of LptD that are able to form a mixed disulfide with Cys98 of DsbC, the nucleophilic cysteine of the catalytic motif. The proteins present in the sample containing the covalent complex between LptD and DsbC were digested with chymotrypsin, and the pool of peptides was analyzed by MS/MS. To identify mixed disulfides between LptD and Cys98 of DsbC, we analyzed the tandem mass spectrometry data with DBond, a new algorithm for the identification of intact disulfide linkages (30). We found one peptide whose fragmentation pattern likely corresponds to that of a mixed disulfide involving Cys7 of LptD and Cys98 of LptD (Fig. 5B). Remarkably, when we searched the MS/MS data for cysteine-containing peptides derived from LptD, we found only three LptD peptides containing Cys149, Cys701, and Cys702, respectively, but we did not find the peptide containing Cys7. This further suggests that Cys7 is involved in a mixed disulfide with DsbC. Moreover, we observed that Cys149, Cys701, and Cys702 were all modified by iodoacetamide (Fig. 5A). As explained above, iodoacetamide was added after precipitation of the proteins, before the purification of the DsbC complexes, to prevent any further disulfide bond rearrangement. Because iodoacetamide does not modify cysteines that are disulfide-bonded, this indicates that Cys149, Cys701, and Cys702 of LptD were reduced in the DsbC-LptD complex in vivo and, therefore, that LptD bound to DsbC is not folded.
FIGURE 5.
Mass spectrometry analysis of the LptD-DsbC complex. A, we found that Cys149, Cys701, and Cys702 of LptD are modified by iodoacetamide (IAM). This indicates that these three cysteines are reduced in the DsbC-LptD complex and, therefore, that LptD is not folded. The peptide containing Cys7 was not identified in the proteolytic digest. This cysteine is likely to be involved in a mixed disulfide, as suggested by the analysis with DBond (30). B, a triply charged parent ion of [M+3H]3+ = 1106.7 Da shows fragmentation characteristics of a disulfide linkage between Cys7 of LptD and Cys98 of DsbC, as determined by the DBond software (30). P*, one strand of a dipeptide; p*, the other strand of a dipeptide; capital letters, fragment ions from peptide P*; lowercase letters, fragment ions from peptide p*. The loss of −34 or the addition of +32 represents formation of dehydroalanine or persulfide from C–S bond fragmentation.
LptD Overexpression Complements the Phenotype of surAdsbC Mutants
Our results indicate that DsbC assists the folding of LptD and suggest that the phenotype exhibited by the surAdsbC mutant results from an impaired assembly of LptD. To confirm this, we expressed LptD from a plasmid in a surAdsbC mutant to test whether LptD overexpression rescues the growth defect of this mutant at low temperatures. The idea is that by increasing LptD synthesis, we also increase the levels of correctly folded LptD in the OM. In agreement with this hypothesis, we previously showed that LptD overexpression results in higher amounts of LptD in the OM of surA strains, although LptD depends on SurA for folding (8). Remarkably, we observed that surAdsbC mutants overexpressing LptD are able to grow at low temperatures, whereas cells harboring an empty plasmid cannot (Fig. 1B). This confirms that the synthetic phenotype of the surAdsbC mutant results from decreased levels of LptD in the OM.
DsbA Cooperates with SurA in the Folding of LptD
LptD contains four cysteine residues that have previously been shown to form disulfide bonds (26). Moreover, LptD was trapped in complex with the thiol oxidase DsbA (27), the protein that is responsible for disulfide bond formation in the periplasm. We tested whether deletion of dsbA would further decrease the integrity of the OM of surA strains. We observed that surAdsbA double mutants also have an increased sensitivity to hydrophobic antibiotics (Fig. 6). This sensitivity is most probably due to a decreased disulfide formation in LptD. Thus, our results indicate that an impairment of the disulfide bond formation process in LptD, either by deletion of dsbC or dsbA, results in a decreased integrity of the OM.
FIGURE 6.
surAdsbA mutants are more sensitive to antibiotics. Antibiotics sensitivity of WT MC4100, dsbA, surA, and surAdsbA mutants at 37 °C. Discs containing various hydrophobic antibiotics were used in disc diffusion assays as described under “Experimental Procedures.” The diameter of the zone of inhibition of growth is shown in mm. The strains for which no zone of inhibition of growth was observed are indicated by asterisks. The data shown are the averages of five experiments. The error bars indicate S.D., which was calculated using the following formula: S.D. = √(Σ(X − M)2/(n − 1)), where X refers to the individual data points, M is the mean, and n is the number of data points.
DISCUSSION
The periplasm contains several chaperones and protein folding catalysts that assist the folding of OM proteins. However, partly because of the redundancy between these folding factors, the role and function of these proteins are not yet fully understood.
We found that the periplasmic protein-disulfide isomerase DsbC is involved in OM assembly. We identified LptD as a protein whose correct folding was affected by the absence of DsbC. The synthetic phenotype of the surAdsbC mutant fits well with the identification of LptD as a DsbC substrate, because strains expressing low levels or mutated versions of LptD also are sensitive to antibiotics (28, 31). Regarding the growth defect exhibited by the surAdsbC mutant at low temperatures, we propose that it results from perturbations in the lipid content of the OM caused by problems in the LPS insertion process. Cells are indeed known to react to cold shock conditions by modifying membrane fluidity and the proportion of unsaturated lipids in the membranes (32). Low levels of LptD could directly or indirectly have an impact on this adaptability mechanism.
LptD has four cysteine residues that are oxidized by DsbA (27). Here, we show that LptD interacts also with DsbC. Our results indicate that DsbC functions as an isomerase/reductase in LptD folding. We found indeed that strains lacking SurA and DsbD, the protein that allows DsbC to function as an isomerase, have a synthetic phenotype similar to that of surAdsbC strains. The involvement of the isomerase activity of DsbC in LptD assembly strongly suggests that LptD contains at least one disulfide between two nonconsecutive cysteines. The fact that we identified a likely mixed disulfide between LptD and DsbC involving the first cysteine of LptD (Cys7) fits well with this hypothesis. If LptD indeed contains a disulfide between two nonconsecutive cysteines, then this disulfide involves either Cys7 or Cys149 bound to Cys701 or Cys702. Thus, if DsbA introduces a disulfide between Cys7 and Cys149, which are consecutive in the sequence, this disulfide will need to be corrected by DsbC. If the nucleophilic cysteine of DsbC attacks Cys7 of LptD, this will lead to the formation of a mixed disulfide with Cys7, as supported by our data.
How can we reconcile that LptD is essential with the fact that dsbA and dsbC mutants are viable? We will first consider the case of the dsbA mutants. The fact that dsbA mutants are viable whereas LptD is essential indicates that either disulfide bond formation is not essential for the function of LptD or the levels of LptD containing disulfides spontaneously formed by small oxidants present in the periplasm are sufficient for survival. We favor the latter hypothesis because most proteins containing disulfides depend strictly on the formation of the correct disulfide bond pattern to be stable and functional. For instance, periplasmic RNase I, a protein with four disulfides including one formed between two nonconsecutive cysteines, becomes active only after formation of the native disulfide bonds in the protein (33). If we assume that the formation of native disulfide bonds is important for the function of LptD, then the fact that dsbC mutants are viable indicates that DsbA catalyzes sufficient correct disulfides in LptD for viability. This is in agreement with our previous work on RNase I, in which we showed that DsbA is a sufficient catalyst for correct disulfide bond formation in this protein (33). Thus, although DsbC is involved in the biogenesis of LptD, it is not required for viability because DsbA produces sufficient amounts of correctly oxidized LptD. The fact that DsbA does a good job in oxidizing LptD suggests that it has a more prominent role than DsbC in LptD biogenesis.
How can we explain the phenotype of the surAdsbC mutant? From previous studies, it is known that the assembly of LptD involves the chaperone SurA and that LptD levels are significantly decreased in surA mutants (8). We propose that under conditions in which LptD biogenesis is compromised, the role of DsbC becomes more important; in the absence of SurA, DsbC is needed to correctly fold LptD in levels sufficient to maintain cellular viability. A similar model can be proposed to explain the increased sensitivity of double surAdsbA mutants; when less LptD is correctly inserted in the membrane because of the absence of SurA, spontaneous oxidation is not enough to fold sufficient amounts of this β-barrel protein. Furthermore, we recently observed that double dsbAdsbC mutants have a synthetic phenotype and present defects in the OM that are more severe than those exhibited by the single mutants (15). For instance, they are sensitive to hydrophobic antibiotics and detergents. The phenotype of the dsbAdsbC mutant could result, at least in part, from defects in the LptD folding pathway; in the absence of DsbA, LptD relies upon the spontaneous formation of its disulfides, a process that is less efficient than the oxidation catalyzed by DsbA. Deletion of the gene coding for DsbC in a dsbA mutant further compromises LptD biogenesis, decreasing the integrity of the OM of the double mutant.
CONCLUSIONS
Our initial objective was to find out whether the protein-disulfide isomerase DsbC is involved in OM assembly, possibly by cooperating with one of the chaperone systems that have been identified in the periplasm. Our results show that DsbC is involved in the biogenesis of LptD, an essential protein that inserts LPS in the OM. Thus, the correct assembly of LptD depends on the interplay between three periplasmic folding factors: the thiol oxidase DsbA, which introduces disulfides into LptD; the disulfide isomerase DsbC, which corrects the non-native disulfides introduced by DsbA; and the chaperone SurA, which most likely escorts LptD across the periplasm and delivers it to the BamA complex. Of all these proteins, SurA seems to be the most crucial. Interestingly, while this paper was under revision, it has been shown that the assembly of LptD in the OM also depends on the lipoprotein LptE (34). Thus, future work should be done to decipher the complex folding pathway of LptD and understand how disulfides are introduced into this essential β-barrel protein.
LptD is the first substrate of DsbC that is localized in the OM. DsbC is kept reduced in the periplasm by DsbD, which transports electrons received from the thioredoxin system across the inner membrane. Thus, our results expand the role of the disulfide isomerization pathway, showing that electrons transported from the cytoplasmic pool of NADPH to the periplasm play a role in maintaining the integrity of the OM.
Acknowledgments
We thank Asma Boujtat and Steve Calberson for technical assistance and Joris Messens, Matthieu Depuydt, Celine Lafaye, and Pauline Leverrier for critical reading of the manuscript.
This work was supported by funds from Interuniversity Attraction Pole Programme-Belgian Science Policy Network P6/05 (to J.-F. C. and K. D.), and Pb/28 (to D. V.) 21C Frontier Functional Proteomics Project Grant FPR-08-A1-020 from the Korean Ministry of Education, Science and Technology, National Core Research Center Program Grant R15-2006-020 through the Center for Cell Signaling Research and Drug Discovery Research (to E. P.), and grants from the Fonds de la Recherche Scientifique–FNRS (to J.-F. C.).
- OM
- outer membrane
- LPS
- lipopolysaccharide(s)
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Ruiz N., Kahne D., Silhavy T. J. (2006) Nat. Rev. Microbiol. 4, 57–66 [DOI] [PubMed] [Google Scholar]
- 2.Leverrier P., Vertommen D., Collet J. F. (2010) Proteomics 10, 771–784 [DOI] [PubMed] [Google Scholar]
- 3.Knowles T. J., Scott-Tucker A., Overduin M., Henderson I. R. (2009) Nat. Rev. Microbiol. 7, 206–214 [DOI] [PubMed] [Google Scholar]
- 4.Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., Kahne D. (2005) Cell 121, 235–245 [DOI] [PubMed] [Google Scholar]
- 5.Rizzitello A. E., Harper J. R., Silhavy T. J. (2001) J. Bacteriol. 183, 6794–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sklar J. G., Wu T., Kahne D., Silhavy T. J. (2007) Genes Dev. 21, 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouvière P. E., Gross C. A. (1996) Genes Dev. 10, 3170–3182 [DOI] [PubMed] [Google Scholar]
- 8.Vertommen D., Ruiz N., Leverrier P., Silhavy T. J., Collet J. F. (2009) Proteomics 9, 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Messens J., Collet J. F. (2006) Int. J. Biochem. Cell Biol. 38, 1050–1062 [DOI] [PubMed] [Google Scholar]
- 10.Gleiter S., Bardwell J. C. (2008) Biochim. Biophys. Acta. 1783, 530–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haebel P. W., Goldstone D., Katzen F., Beckwith J., Metcalf P. (2002) EMBO J. 21, 4774–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shouldice S. R., Cho S. H., Boyd D., Heras B., Eser M., Beckwith J., Riggs P., Martin J. L., Berkmen M. (2010) Mol. Microbiol. 75, 13–28 [DOI] [PubMed] [Google Scholar]
- 13.Katzen F., Beckwith J. (2000) Cell 103, 769–779 [DOI] [PubMed] [Google Scholar]
- 14.Hiniker A., Bardwell J. C. (2004) J. Biol. Chem. 279, 12967–12973 [DOI] [PubMed] [Google Scholar]
- 15.Vertommen D., Depuydt M., Pan J., Leverrier P., Knoops L., Szikora J. P., Messens J., Bardwell J. C., Collet J. F. (2008) Mol. Microbiol. 67, 336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berkmen M., Boyd D., Beckwith J. (2005) J. Biol. Chem. 280, 11387–11394 [DOI] [PubMed] [Google Scholar]
- 17.Depuydt M., Leonard S. E., Vertommen D., Denoncin K., Morsomme P., Wahni K., Messens J., Carroll K. S., Collet J. F. (2009) Science 326, 1109–1111 [DOI] [PubMed] [Google Scholar]
- 18.Chen J., Song J. L., Zhang S., Wang Y., Cui D. F., Wang C. C. (1999) J. Biol. Chem. 274, 19601–19605 [DOI] [PubMed] [Google Scholar]
- 19.Liu X., Wang C. C. (2001) J. Biol. Chem. 276, 1146–1151 [DOI] [PubMed] [Google Scholar]
- 20.Wu T., McCandlish A. C., Gronenberg L. S., Chng S. S., Silhavy T. J., Kahne D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11754–11759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz N., Kahne D., Silhavy T. J. (2009) Nat. Rev. Microbiol. 7, 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srinivas N., Jetter P., Ueberbacher B. J., Werneburg M., Zerbe K., Steinmann J., Van der Meijden B., Bernardini F., Lederer A., Dias R. L., Misson P. E., Henze H., Zumbrunn J., Gombert F. O., Obrecht D., Hunziker P., Schauer S., Ziegler U., Käch A., Eberl L., Riedel K., DeMarco S. J., Robinson J. A. (2010) Science 327, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 23.Casadaban M. J. (1976) J. Mol. Biol. 104, 541–555 [DOI] [PubMed] [Google Scholar]
- 24.Miller J. H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25.Misra R. V., Horler R. S., Reindl W., Goryanin I. I., Thomas G. H. (2005) Nucleic Acids Res. 33, D329–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun M., Silhavy T. J. (2002) Mol. Microbiol. 45, 1289–1302 [DOI] [PubMed] [Google Scholar]
- 27.Kadokura H., Tian H., Zander T., Bardwell J. C., Beckwith J. (2004) Science 303, 534–537 [DOI] [PubMed] [Google Scholar]
- 28.Ruiz N., Falcone B., Kahne D., Silhavy T. J. (2005) Cell 121, 307–317 [DOI] [PubMed] [Google Scholar]
- 29.Collet J. F., Messens J. (2010) Antioxid. Redox. Signal., in press [Google Scholar]
- 30.Choi S., Jeong J., Na S., Lee H. S., Kim H. Y., Lee K. J., Paek E. (2010) J. Proteome Res. 9, 626–635 [DOI] [PubMed] [Google Scholar]
- 31.Abe S., Okutsu T., Nakajima H., Kakuda N., Ohtsu I., Aono R. (2003) Microbiology 149, 1265–1273 [DOI] [PubMed] [Google Scholar]
- 32.Yamanaka K. (1999) J. Mol. Microbiol. Biotechnol. 1, 193–202 [PubMed] [Google Scholar]
- 33.Messens J., Collet J. F., Van Belle K., Brosens E., Loris R., Wyns L. (2007) J. Biol. Chem. 282, 31302–31307 [DOI] [PubMed] [Google Scholar]
- 34.Chng S. S., Ruiz N., Chimalakonda G., Silhavy T. J., Kahne D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5363–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]