Abstract
Peroxisome proliferator-activated receptors (PPARs) not only play a key role in regulating metabolic pathways but also modulate inflammatory processes, pointing to a functional interaction between PPAR and cytokine signaling pathways. In this study, we show by genome-wide transcriptional profiling that PPARβ/δ and transforming growth factor-β (TGFβ) pathways functionally interact in human myofibroblasts and that a subset of these genes is cooperatively activated by TGFβ and PPARβ/δ. Using the angiopoietin-like 4 (ANGPTL4) gene as a model, we demonstrate that two enhancer regions cooperate to mediate the observed synergistic response. A TGFβ-responsive enhancer located ∼8 kb upstream of the transcriptional start site is regulated by a mechanism involving SMAD3, ETS1, RUNX, and AP-1 transcription factors that interact with multiple contiguous binding sites. A second enhancer (PPAR-E) consisting of three juxtaposed PPAR response elements is located in the third intron ∼3.5 kb downstream of the transcriptional start site. The PPAR-E is strongly activated by all three PPAR subtypes, with a novel type of PPAR response element motif playing a central role. Although the PPAR-E is not regulated by TGFβ, it interacts with SMAD3, ETS1, RUNX2, and AP-1 in vivo, providing a possible mechanistic explanation for the observed synergism.
Keywords: AP-1 Transcription Factor, Ets Family Transcription Factor, PPAR, SMAD Transcription Factor, Transcription Regulation, Transforming Growth Factor Beta (TGFbeta), GW501516, RUNX Family Transcription Factor
Introduction
The three members of the peroxisome proliferator-activated receptor (PPAR)2 family, PPARα, PPARβ/δ and PPARγ, are nuclear receptors that modulate gene expression in response to lipid ligands, suggesting a metabolic role for PPARs as intracellular lipid sensors (1–3). PPAR ligands include various arachidonic and linoleic acid derived metabolites of the cyclooxygenase and lipoxygenase biosynthetic pathways, pointing to a role for PPARs in signaling pathways triggered by inflammatory mediators (2, 4). Although PPARs possess a high degree of structural similarities, they have distinct and non-interchangeable functions in energy metabolism. Whereas PPARα is the master regulator of fatty acid oxidation in liver, PPARγ promotes adipogenesis and lipid storage in fat cells. PPARβ/δ is ubiquitously expressed and has an essential function in catabolic pathways and energy metabolism in extrahepatic tissues. In addition to their regulatory functions in metabolism and inflammation, PPARs play roles in development, differentiation, and cell proliferation (5–7). PPARs therefore represent highly relevant drug targets, which has led to the development of several synthetic drug agonists with high subtype selectivity and high affinity binding (8).
Our own studies have revealed an essential function for PPARβ/δ in tumor stroma cells (9). Ppard deletion results in an inhibition of syngeneic tumor growth, concomitant with a severely altered, hyperplastic tumor stroma with an abnormal proportion of myofibroblasts and a lack of mature tumor microvessels. Our studies suggest a specific function for PPARβ/δ in the tumor stroma because no effects on physiological angiogenesis or related processes are detectable. We therefore believe that one of the functions of PPARβ/δ is to modulate signals triggered by tumor cytokines. PPARβ/δ has indeed been shown to modulate the expression of cytokines, adhesion molecules, and extracellular matrix proteins in immune cells (10) and to regulate macrophage polarization (11, 12). Consistent with these observations, PPARβ/δ activates genes not only by the canonical mechanism (i.e. binding as a heterodimer with retinoic acid X receptor (RXR) to a PPAR response element (PPRE)) but also regulates a number of transcription factors with functions in inflammatory signaling pathways (6), either by modulating their expression or by direct physical interactions. The former include AP-1 (13) and ATF3 (14), whereas the latter group encompasses NFκB (15–19), KLF5 (20), STAT3 (21), and BCL6 (22).
A cytokine with a pivotal role in tumor growth and tumor-stroma interactions is TGFβ (23). We therefore hypothesized that PPARβ/δ and TGFβ signaling pathways may functionally interact. In the present study, we support this hypothesis by microarray analyses of myofibroblast-like cells treated with PPARβ/δ ligands and TGFβ, which reveal an extensive cross-talk of the transcriptional pathways triggered by PPARβ/δ and TGFβ. We also identify human ANGPTL4 (angiopoietin-like 4) as a gene showing a particularly strong synergistic response to TGFβ and PPAR ligands. The ANGPTL4 gene encodes angiopoietin-like 4, an important adipokine and a putative mediator of metastatic tumor spread promoted by TGFβ (24).
The canonical TGFβ signaling pathway involves the receptor-mediated phosphorylation of regulatory SMAD (R-SMAD) proteins (SMAD2 and SMAD3), their subsequent association with the common SMAD (Co-SMAD) SMAD4, and the binding of R-SMAD·Co-SMAD complexes to specific target genes (25). SMADs recognize the consensus sequence AGAC with low affinity and therefore require an interaction with other DNA-binding proteins for selective binding to TGFβ-responsive enhancer regions. These interacting factors may serve as adaptors for DNA binding only, as exemplified by the forkhead activin signal transducer (26, 27), but frequently also contribute other functions, such as the recruitment of transcriptional coregulators. Examples of the latter category include JUN (28, 29), cAMP-response element-binding protein (30, 31), ETS (32–34), and RUNX (35, 36). Many of these transcription factors are themselves regulated by extracellular signals, thereby establishing a network of pathways interacting with TGFβ signaling.
The issue is further complicated by the fact that TGFβ also activates other signal transduction cascades, including MAPKs and AKT (25, 37), which provides a potential explanation for the observation that many genes are regulated by TGFβ through non-canonical pathways not involving SMAD4. One example is the regulation of the Mad1 gene by TGFβ through a SMAD binding element (SBE)-bound complex containing nuclear IκB kinase α and SMAD3 but lacking SMAD4 (38). Other SMAD4-independent TGFβ pathways have also been described but are not known in detail (39–41).
In this study, we identify a novel TGFβ-responsive upstream enhancer (TGF-E) in the human ANGPTL4 gene that is regulated by a non-canonical mechanism involving SMAD3, ETS1, RUNX2, and AP-1. This enhancer region cooperates with an unusual PPAR-responsive enhancer (PPAR-E) in the third intron to mediate the observed synergistic response to TGFβ and PPAR ligands.
EXPERIMENTAL PROCEDURES
Chemicals
TGFβ2 and SB431542 were purchased from Sigma-Aldrich (Karlsruhe, Germany). The PPAR ligands GW501516, GW1929, and GW7647 were obtained from Axxora (Lörrach, Germany).
Cell Culture
human keratinocyte, WPMY-1, WI-38, NIH3T3, and 2-H11 cells were obtained from ATCC. All cell lines were maintained as described (9). Human umbilical cord endothelial cells were established and cultured as described (42).
Plasmids
pcDNA-hPPARδ was constructed by inserting full-length human PPARβ/δ into pcDNA3.1. pSG5-hRXRa containing the full-length RXRa cDNA was kindly provided by Dr. A. Baniahmad (Jena, Germany). PPRE and PPAR-E luciferase reporters were constructed by inserting 36-bp oligonucleotides or a <1-kb fragment of the third intron of the human ANGPTL4 gene (43) into TATAi-pGL3 (44). The TGF-E plasmid was cloned by inserting an ANGPTL4 fragment from −9000 to −8000 into TATAi-pGL3. Mutation of transcription factor binding sites of the −8401/−8170 plasmid were generated by site-directed mutagenesis (Stratagene). Primers are listed in supplemental Table S1. The pcDNA3.1 plasmid was supplied by Invitrogen (Karlsruhe, Germany).
Luciferase Reporter Assay
Transfections were performed with polyethyleneimine (average Mr 25,000; Sigma-Aldrich). Cells were transfected on 12-well plates at 70–80% confluence in DMEM plus 2% FCS with 2.5 μg of plasmid DNA and 2.5 μl of PEI (1:1000 dilution, adjusted to pH 7.0 and preincubated for 15 min in 100 μl of phosphate-buffered saline for complex formation). Four hours after transfection, the medium was changed, and cells were incubated for 48 h in normal growth medium or serum-reduced medium when monitoring TGFβ induction. Luciferase assays were performed as described (45). Values from three biological replicates were combined to calculate averages and S.D. values.
siRNA Transfections
Cells were seeded at a density of 5 × 105 cells/6-cm dish in 4 ml of DMEM with 10% FCS and cultured for 2 h. 1280 ng of siRNA in 100 μl of Opti-MEM (Invitrogen) and 20 μl of HiPerfect (Qiagen, Hilden, Germany) were mixed and incubated for 5–10 min at room temperature prior to transfection. The cells were replated 24 h post-transfection at a density of 5 × 105 cells/6-cm dish. Transfection was repeated 48 h after start of the experiment, and cells were passaged after another 24 h. Forty-eight hours following the last transfection, cells were stimulated and harvested after another 6 h. siRNA sequences are listed in supplemental Table S2.
Quantitative RT-PCR
cDNA was synthesized from 0.1–1 μg of RNA using oligo(dT) primers and the Omniscript kit (Qiagen). qPCR was performed in a Mx3000P real-time PCR system (Stratagene, La Jolla, CA) for 40 cycles at an annealing temperature of 60 °C. PCRs were carried out using the Absolute QPCR SYBR Green mix (Abgene, Hamburg, Germany) and a primer concentration of 0.2 μm, following the manufacturer's instructions. L27 was used as normalizer. Comparative expression analyses were statistically analyzed by Student's t test (two-tailed, equal variance) and corrected for multiple-hypothesis testing via the Bonferroni method. The sequences of the primers are listed in supplemental Table S4.
Electrophoretic Mobility Shift Assays (EMSAs)
Each oligonucleotide pair (sequences in supplemental Table S3) was annealed and labeled with [γ-32P]ATP by T4 polynucleotide kinase (Fermentas). Nuclear receptor proteins were synthesized from mammalian expression vectors using the TNT T7 quick coupled transcription/translation system (Promega). Three microliters of in vitro translated proteins were mixed with 3 μg of poly(dI-dC) and 1 μg of pUC18 in 25 μl of binding buffer (20 mm Tris-HCl, 50 mm NaCl, 1 mm MgCl2, 10% glycerol, 3 mm DTT, 0,2 mm PMSF, 20 μm ZnCl2) and preincubated for 30 min at 30 °C. After adding 5 μl of 32P- labeled double-stranded probes, the samples were incubated for another 15 min at 30 °C and resolved on a 4% native polyacrylamide gel in 1× RA-buffer (6.7 mm Tris-HCl, pH 7.9, 3.3 mm sodium acetate, 1 mm EDTA, 2.5% glycerol) and visualized by autoradiography.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described (46), except that nuclei were resuspended at 2.5 × 107/ml, 60–70 pulses were applied during sonification, and chromatin from 8 × 106 nuclei was used per sample. The following antibodies were used: IgG pool, I5006 (Sigma-Aldrich); α-RNA polymerase II, sc-899; α-PPARβ/δ, sc-7197; α-RXRα, sc-553; α-CBP, sc-369; α-JUN, sc-44; α-FOS, sc-253; α-ETS1, sc-350 (all from Santa Cruz (Heidelberg, Germany)); α-SMAD3, ab28379 (Abcam, Cambridge, UK). Comparative binding analyses were statistically analyzed by Student's t test (two-tailed, equal variance) and corrected for multiple-hypothesis testing via the Bonferroni method. Primer sequences are listed in supplemental Table S5.
Microarrays
RNA was isolated using the Nucleospin RNA II kit (Macherey-Nagel, Düren, Germany). RNA quality was assessed using the Experion automated electrophoresis station with RNA StdSens chips (Bio-Rad, Munich, Germany). For microarray studies, total RNA samples were amplified and labeled using the Agilent Quick Amp labeling kit (Agilent, Santa Clara, CA) according to the manufacturer's instructions. The amplification procedure consists of reverse transcription of total RNA, including spike-in with an oligo(dT) primer bearing a T7 promoter followed by in vitro transcription of the resulting cDNA with T7 RNA polymerase in the presence of dye-labeled CTP to generate multiple fluorescence-labeled copies of each mRNA. After purification, the labeled amplified RNA was quantified, and hybridization samples were prepared according to the manufacturer's instructions. Human Agilent 4-plex Array 44K was used for the analysis of the gene expression of the different samples in a reference design assay. For the reference, a pool of all samples to be analyzed was used. This reference probe was labeled with Cy3, whereas the samples were labeled with Cy5 dye. The hybridization assembly was performed as described (59). After a 17-h hybridization at 65 °C, Slides were washed as described by the manufacturer and subsequently scanned using an Agilent DNA microarray scanner G2505C (scan software: Agilent Scan Control version A.8.1.3; quantification software: Agilent Feature Extraction version 10.5.1.1 (FE Protocol GE_105_Dec08)).
Bioinformatics
Raw microarray data were normalized using the “loess” method implemented within the marray package of R/BioConductor (available on the World Wide Web). Regulated probes were selected on the basis that the logarithmic (base 2) average intensity value was ≥6%, and that the fluctuation between replicates was ≤50%.
Normalized expression values for ligand-treated cells were determined by calculating the ratio of signals in the presence and absence of ligands. The induction by TGFβ in the presence of GW501516 was calculated as (normalized expression with GW501516 + TGFβ)/(normalized expression with GW501516), and the induction by GW501516 in the presence of TGFβ was calculated as (normalized expression with GW501516 + TGFβ)/(normalized expression with TGFβ). The theoretical additive induction by both ligands was calculated by the formula, (normalized expression with GW501516) + (normalized expression with TGFβ) − 1.
RESULTS
Cross-talk of TGFβ and PPARβ/δ Pathways Determined by Transcriptional Profiling
To assess possible interactions of the TGFβ and PPARβ/δ signaling pathways in myofibroblasts, we performed microarray analyses of WPMY-1 cells, either untreated (solvent) or treated with the PPARβ/δ-selective agonist GW501516 (0.3 μm), TGFβ2 (10 ng/ml), or both ligands for 6 h (EMBL-EBI ArrayExpress accession number E-MEXP-2748). As illustrated by the Venn diagram in Fig. 1A, 617 probes showed induction by TGFβ, and 91 probes showed induction by GW501516 (≥20% change) with an overlap of 20 probes, representing 12 annotated genes and six transcripts of unknown function induced by both ligands. Next, we sought to determine the fraction of genes that is cooperatively regulated by TGFβ and GW501516 in WPMY-1 cells. To address this question, we identified those probes showing a ≥50% difference in signal intensity after co-treatment with TGFβ plus GW501516 compared with treatment with either ligand alone (Fig. 1B). This analysis identified 165 probes, representing 34 annotated genes and 124 transcripts of unknown function that were induced by both ligands, whereas three probes indicated repression. This number of cooperatively induced genes is substantially bigger than the overlap in Fig. 1A (15 genes and 20 probes), indicating that 140 of theses genes are not responsive to a single ligand.
FIGURE 1.
Genome-wide transcriptional response of human myofibroblasts to treatment with TGFB, PPARβ/δ agonist or both ligands. A, Venn diagram of probes showing induction by TGFβ or GW501516 in WPMY-1 myofibroblasts (≥20% change). The overlap represents the probes indicating induction by both ligands. B, graphic representation of probe sets showing cooperative regulation by TGFβ and GW501516 in WPMY-1 cells. The number of probes showing a ≥50% difference in expression after co-treatment with TGFβ plus GW501516 compared with treatment with either ligand alone. The plot includes both activated (left) and repressed sequences (right). C, dot plot depicting cooperative effects for individual probes. Relative expression levels measured after co-treatment of WPMY-1 cells with TGFβ plus GW501516 were plotted against the calculated sum of expression levels observed after treatment with either ligand alone. The shaded area indicates purely additive effects (threshold ±50%); data points located above this area show synergistic activation, and data points below the shaded area synergistic repression by TGFβ and GW501516. Circles mark the data points representing the ANGPTL4 gene (two independent probes of the microarray). D, differential responses of target genes to PPARβ/δ activation and TGFβ verified by RT-qPCR. WPMY-1 cells were treated with the indicated ligands (300 nm GW501516, 10 ng/ml TGFβ2, or both) or solvent for 6 h, and the relative expression levels of ANGPTL4, THBS1, CYP24A1, LIPG, ABCA1, PAI1, PDGFA, ADRP, and SLC25A20 were determined by RT-qPCR. ***, **, and *, significant difference from untreated sample (p < 0.001 by t test, p < 0.01, and p < 0.05, respectively). # and ## induction by both ligands significantly higher than induction by either ligand alone (p < 0.05 and p < 0.01, respectively). Error bars, S.D.
To obtain a more detailed picture of the observed cooperative effects, we plotted for individual probes the experimentally measured cooperativity against the calculated cooperative induction that would be expected if the effects of both ligands were simply additive. The data in Fig. 1C show that the cotreatment with both ligands resulted in effects that were more than additive for 3.2% of the probes (n = 200; ≥50%; data points above the shaded area). Furthermore, 0.1% of the probes (n = 4) indicated repression by both ligands that was more than additive (≥50%; data points below the shaded area in Fig. 1C). These findings clearly point to an extensive cross-talk between the two pathways.
The synergistic induction of five genes discovered by microarray analyses was confirmed by RT-qPCR (i.e. ANGPTL4 coding for angiopoietin-like 4, THBS1 encoding thrombospondin-1, CYP24A1 coding for cytochrome P450 24A1, LIPG encoding endothelial lipase G, and ABCA1 coding for the cholesterol efflux-regulating ATP-binding cassette subfamily A protein) (Fig. 1D). Cooperative induction by TGFβ and GW501516 for each of these genes was significantly higher than additive (98% for ANGPTL4, 48% for THBS1, 363% for CYP24A1, 46% for LIPG, and 10% for ABCA1). In contrast, PAI1 and PDGFA were selectively induced by TGFβ, whereas ADRP and SLC25A20 were responsive only to GW5101516, as expected (Fig. 1D). A particular strong induction (>20-fold) in addition to a strong synergistic effect was observed with ANGPTL4 (represented by two probes on the array; circled data points in Fig. 1C). We therefore focused our further studies on the regulation of this gene.
Synergistic Activation of ANGPTL4 Transcription by PPAR Ligands and TGFβ in Human Cells
Synergistic activation of ANGPTL4 by GW501516 plus TGFβ was not restricted to WPMY-1 cells because clear cooperative effects were also observed with human umbilical cord endothelial cells (54% higher than additive) and immortalized human keratinocytes (208%; Fig. 2A). Treatment of WPMY-1 cells with a pool of four verified PPARD siRNAs (supplemental Table S2 and Figs. S1–S4) abolished both the induction by GW501516 and the cooperative effect with TGFβ (Fig. 2B). As expected, the response to TGFβ alone was not affected. ChIP analysis showed an enhanced occupation by RNA polymerase II of a transcribed region of the ANGPTL4 gene (intron 3) in response to either GW501516 or TGFβ, and this was potentiated by cotreatment with both ligands (Fig. 2C). These data clearly show that the observed synergism of GW501516 plus TGFβ operates at the level of transcription. Finally, we tested the synergistic response of the ANGPTL4 gene to TGFβ and different PPAR subtype-specific ligands (Fig. 2D) and found similar induction values with the PPARα ligand GW7647 (14.6-fold), the PPARβ/δ ligand GW501516 (26.7-fold), and the PPARγ ligand GW1929 (11.5-fold).
FIGURE 2.
ANGPTL4 transcription is synergistically activated by PPAR ligands and TGFβ in human cells. A, cooperative transcriptional activation of ANGPTL4 expression by PPARβ/δ and TGFβ in different cell types. ANGPTL4 expression levels in GW501516- and/or TGFβ-treated WPMY-1, human umbilical cord endothelial, and human keratinocyte cells were measured as in Fig. 1D. B, verification of a function for PPARβ/δ in the observed synergistic activation of ANGPTL4 expression by TGFβ and GW501516. WPMY-1 cells were transfected with PPARD or control siRNA and exposed to 300 nm GW501516, 2 ng/ml TGFβ2, or both for 6 h, and ANGPTL4 mRNA levels were measured by RT-qPCR. C, PPARβ/δ and TGFβ synergize at the level of ANGPTL4 transcription. WPMY-1 cells were treated with 300 nm GW501516, 2 ng/ml TGFβ2, or both for 1 h, and ChIP was carried out with antibodies against RNA polymerase II (PolII) or a nonspecific IgG pool, and a genomic DNA fragment at +3500 bp relative to the TSS of ANGPTL4 was amplified by qPCR. The signals were calculated relative to 1% of input DNA. D, synergistic transcriptional activation of ANGPTL4 by TGFβ and different PPAR subtype-specific ligands. WPMY-1 cells were treated with combinations of PPAR ligands (GW7647, PPARα; GW501516, PPARβ/δ; GW1929, PPARγ; each at 300 nm) and 2 ng/ml TGFβ2 for 6 h, and relative ANGPTL4 expression levels were measured by RT-qPCR. ***, **, and *, significant difference from untreated sample (p < 0.001 by t test, p < 0.01, p < 0.05); # and ##, induction by both ligands significantly higher than induction by either ligand alone (p < 0.05, p < 0.01). Error bars, S.D.
Identification of a PPAR-responsive Enhancer (PPAR-E) in the Third Intron of the ANGPTL4 Gene Harboring Three Functional PPREs
To identify PPARβ/δ binding sites in the ANGPTL4 gene, we performed ChIP with GW501516-treated WPMY-1 cells using 42 primer pairs spanning a region from −10 to +10.5 kb relative to the ANGPTL4 transcription start site (TSS) (Fig. 3A). This analysis revealed a single PPARβ/δ-occupied region located ∼3.5 kb downstream of the TSS in the third intron. This signal was largely abolished after treatment with PPARD siRNAs, verifying the occupation of this site by PPARβ/δ (Fig. 3B). The ChIP result was confirmed by ChIP-Seq analysis,3 which also identified a single peak at the ANGPTL4 locus (within 600 kb of the TSS). Interestingly, the ChIP-Seq peak observed at the ANGPTL4 gene was considerably broader than other peaks, reflecting single PPREs, as in the CPT1A gene (Fig. 3C), suggesting that the ANGPTL4 site might comprise multiple binding sites.
FIGURE 3.
The third intron of ANGPTL4 harbors a PPAR-responsive enhancer with three functional PPREs. A, identification of PPARβ/δ binding sites in the ANGPTL4 locus in vivo. Chromatin was immunoprecipitated from GW501516 (GW)-treated (1 h) WPMY-1 cells with a PPARβ/δ-specific antibody, and genomic fragments were amplified with primer pairs covering the ANGPTL4 locus from −10 to +10.5 kb relative to the TSS located with a spacing of ∼500 bp between adjacent primer pairs. B, verification of PPARβ/δ binding to the PPAR-responsive enhancer in the third ANGPTL4 intron. WPMY-1 cells were transfected with PPARD or control siRNA. ChIP was carried out with the indicated antibodies. The efficiency and specificity of the PPARD siRNAs are shown in supplemental Figs. S1–S4. C, signals derived from a ChIP-sequencing experiment in WPMY-1 cells with PPARβ/δ-specific antibodies and a nonspecific IgG pool. The genomic regions around the ANGPTL4 PPREs and the single CPT1A PPRE are shown. D, in vitro binding of PPAR·RXR heterodimers to the three putative PPRE sequences identified in silico shown at the top. PPRE3 represents the published ANGPTL4 PPRE (43). EMSA was carried out with corresponding double-stranded oligonucleotides and recombinant PPARβ/δ and RXRα. The arrow points to the band representing the PPRE-bound PPARβ/δ·RXRα heterodimer. *, a nonspecific background band. E, in vivo binding of PPARβ/δ and RXRα to the three putative PPRE sequences. After ChIP with antibodies against PPARβ/δ, RXRα, or a nonspecific IgG pool, DNA was amplified with primers encompassing each of the putative PPREs or an intergenic control region. F, transcriptional activity and GW501516 inducibility of the three PPREs in a transient luciferase reporter assay. WPMY-1 cells were co-transfected with luciferase reporter plasmids and expression vectors encoding for PPARβ/δ and RXRα (or the empty expression vector). Four hours after transfection, the cells were treated with 300 nm GW501516, and luciferase activity was measured 44 h later. G, response of PPRE2 and PPRE3 reporter constructs to PPARβ/δ and RXR ligands. WPMY-1 cells were co-transfected with luciferase reporter plasmids and expression vectors coding for PPARβ/δ or RXRα, both vectors, or the empty expression vector. Four hours after transfection, the cells were treated with 300 nm GW501516, 300 nm 9-cis-RA, or both ligands, and luciferase activity was measured 44 h later. ***, **, and *, significant difference from untreated sample (p < 0.001 by t test, p < 0.01, and p < 0.05, respectively). Error bars, S.D.
Inspection of the nucleotide sequence in this region revealed three motifs resembling PPREs (Fig. 3D), the most 3′ element (PPRE3) representing the published ANGPTL4 PPRE (43). Electrophoretic mobility shift assays confirmed the binding of in vitro synthesized PPARβ/δ·RXRα heterodimers to all three PPREs with similar efficiency (Fig. 3E). Likewise, PPARβ/δ ChIP analysis with primers specifically amplifying fragments harboring either PPRE1, PPRE2, or PPRE3 showed binding to all three regions (Fig. 3E). A higher intensity was seen with PPRE2, which may in part be due to the binding of PPARβ/δ to the adjacent PPREs.
We also tested the three isolated PPREs in transient luciferase reporter assays. As depicted in Fig. 3F, PPRE2 clearly showed the strongest response both to GW501516 and to cotransfected PPARβ/δ and RXRα. PPRE3 showed a clearly reduced, albeit readily detectable induction, whereas the response of PPRE1 was comparably modest. Finally, we analyzed the response of PPRE2 and PPRE3 luciferase reporter constructs to 9-cis-retinoic acid and/or GW501516. As shown in Fig. 3G, PPRE3 was cooperatively induced by both ligands in cells cotransfected with expression vectors for PPARD and RXRA, whereas PPRE2 was responsive only to GW501516. We conclude that the intronic PPAR-responsive enhancer (PPAR-E) consists of three PPREs that are functionally not equivalent.
Identification of TGF-E, an Upstream TGFβ-responsive, SMAD3-dependent Enhancer
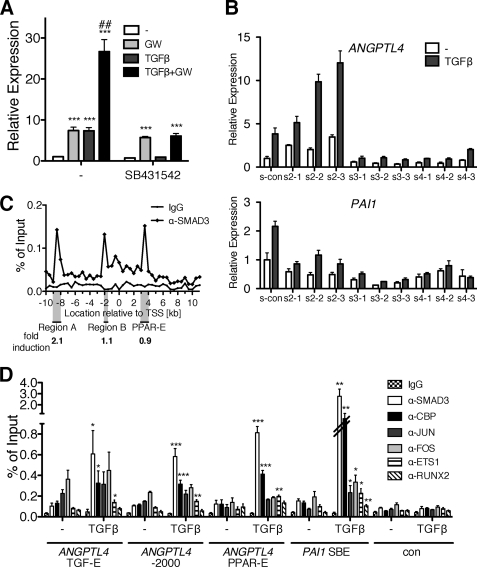
We next addressed the mechanism of the TGFβ-mediated induction of the ANGPTL4 gene. Fig. 4A shows that both the TGFβ response and the synergism with GW501516 were blocked by SB431542, indicating an essential role for the classical TGFβ receptor ALK5 (47). siRNA-mediated knockdown of individual SMAD family members (supplemental Figs. S5 and S6) strongly suggests that not all components of the canonical pathway play a central role in mediating ANGPTL4 induction by TGFβ. Although SMAD3 and SMAD4 siRNA clearly inhibited the TGFβ response, SMAD2 siRNA increased the basal level activity of the ANGPTL4 gene (Fig. 4B). The same experiment was performed with the PAI1 gene, a bona fide TGFβ target gene induced through the canonical pathway, which showed the expected requirements for all three SMADs. These data suggest that SMAD3 and SMAD4 are necessary for a full induction of the ANGPTL4 gene by TGFβ, whereas SMAD2 is not required and may rather have an inhibitory effect.
FIGURE 4.
Identification of an upstream TGFβ-responsive, SMAD3-dependent enhancer interacting with multiple transcription factors. A, ANGPTL4 induction is blocked by a TGFβ kinase inhibitor. WPMY-1 cells were treated with 300 nm GW501516, 10 ng/ml TGFβ2, a 10 μm concentration of the ALK4/5/7 inhibitor SB431542, or combinations for 6 h. Relative ANGPTL4 expression levels were measured by RT-qPCR. B, effects of siRNA-mediated knockdown of SMAD2, SMAD3, or SMAD4 on TGFβ induction of the ANGPTL4 gene. Forty-eight hours after transfection of the indicated siRNAs (three siRNAs for each gene), cells were treated with 2 ng/ml TGFβ2 or solvent control, and the relative ANGPTL4 expression was determined 6 h later. PAI1 was included in this experiment as a positive control. The efficiencies of the siRNAs are shown in supplemental Figs. S3 and S4. C, identification of SMAD3 binding sites at the ANGPTL4 locus in vivo. WPMY-1 cells were treated with 2 ng/ml TGFβ2 for 1 h and analyzed by ChIP with antibodies against SMAD3 or a control IgG pool. The experiment was performed as described in the legend to Fig. 3A for PPARβ/δ. The three indicated genomic regions (Region A, Region B, and PPAR-E) were cloned into luciferase reporter vectors. Four hours after transfection, WPMY-1 cells were treated with TGFβ2 (2 ng/ml) in serum-reduced medium, and luciferase activity was measured 44 h later. Results are expressed as -fold induction relative to untreated cells. This analysis identifies Region A as the TGFβ-responsive enhancer of the ANGPTL4 gene (referred to as TGF-E hereafter). D, recruitment of transcription factors to the TGF-E, Region B, PPAR-E, TGFβ-inducible region of PAI1 (positive control), and an irrelevant genomic fragment (negative control). The ChIP analysis of serum-starved WPMY-1 cells shows TGFβ-induced recruitment of SMAD3, CBP, and ETS1; constitutive binding of JUN and FOS family members to all three regions; and constitutive binding of RUNX2 to the TGF-E and PPAR-E. In addition, the data show TGFβ-inducible recruitment of RUNX2 to the PPAR-E. ***, **, and *, significant difference from untreated sample (p < 0.001 by t test, p < 0.01, and p < 0.05, respectively). ##, induction by both ligands significantly higher than induction by either ligand alone (p < 0.01). Error bars, S.D.
To identify the TGFβ-responsive enhancer in the ANGPTL4 gene, we performed ChIP for SMAD3 with untreated and TGFβ-treated WPMY-1 cells using the same 42-primer set as in Fig. 3A. This analysis clearly identified three SMAD3-occupied regions located at approximately −8.5 kb (Region A), −2.0 kb (Region B), and +3.5 kb (Fig. 4C), the last region overlapping the PPAR-E (Fig. 3A). To clarify which of these regions is inducible by TGFβ, we cloned each region as a genomic fragment of ∼1 kb (Region A, PPAR-E) or ∼0.5 kb (Region B) into a luciferase vector and tested the TGFβ response in transiently transfected WPMY-1 cells. Although Region A showed a clear induction of 2.1-fold, Region B and the PPAR-E were unresponsive to TGFβ.
Interspecies alignment of Region A showed a strong conservation among human, cow, horse, dolphin, and many other vertebrate species but a surprising deviation from the corresponding murine sequences, where this conserved region is largely absent. In agreement with this observation, ANGPTL4 expression was induced by TGFβ in bovine endothelial cells but not in three different mouse cell lines (supplemental Fig. S7). These data strongly support the conclusion that Region A harbors the TGFβ-responsive enhancer (TGF-E).
In silico analysis of the TGF-E by Genomatix MatInspector predicted the presence of multiple transcription factor binding sites, including putative recognition motifs for regulatory proteins previously connected to SMAD-induced transcription. We therefore performed ChIP analyses, including the published TGFβ-responsive fragment of the PAI1 gene for comparison. As shown in Fig. 4D, a clear TGFβ-inducible recruitment of SMAD3, CBP, and ETS1 to both genes was detectable. Transcription factors identified by pan-JUN and pan-FOS antibodies were also present on both genomic regions, but a clear TGFβ dependence was seen only with PAI1. Surprisingly, we also observed JUN/FOS binding and inducible recruitment of SMAD3, CBP, and ETS1 to Region B and the PPAR-E (Fig. 4D), although the corresponding fragments of the ANGPTL4 locus were not TGFβ-responsive in transient luciferase assays. Furthermore, the ANGPTL4 TGF-E and PPAR-E and the PAI1 SBE regions showed a weak but readily detectable enrichment of RUNX2.
Identification of Functional Transcription Factor Binding Sites in the TGF-E
In order to define the TGF-E enhancer more precisely, we constructed a number of deletion mutants of the genomic fragment identified in Fig. 4C and tested their inducibility by TGFβ in transient transfection assays. Deletions from both ends resulting in a construct spanning positions −8401 to −8170 led to a decrease in overall transcriptional activity without impairing inducibility by TGFβ (Fig. 5A). Further 5′ deletions completely impaired the TGFβ response (Fig. 5A). Importantly, induction of the minimal −8401/−8170 construct by TGFβ was strictly dependent on SMAD3, as demonstrated by siRNA-mediated knockdown (Fig. 5B). This fragment harbors several potential transcription factor binding sites (Fig. 5A), including an AP-1 site, an ETS binding site (EBS), a RUNX binding element (RBE), an SP1/GC-box, and three SBEs.
FIGURE 5.
Delineation of functional binding sites in the TGFβ-responsive fragment of the ANGPTL4 TGF-enhancer in WPMY-1 cells. A, delineation of a minimal TGFβ-responsive fragment of the ANGPTL4 upstream region. The indicated fragments of the TGF-E region were linked to the firefly luciferase gene and transfected into WPMY-1 cells. Luciferase activity was measured 48 h after transfection and treatment with 2 ng/ml TGFβ2 or solvent control. Potential transcription factor binding sites are indicated as colored boxes. SBE, SMAD-binding element; AP1, AP-1 site; RBE, RUNX-binding element; EBS, ETS binding site; SP, SP1/GC-box. B, dependence of the minimal TGFβ-responsive fragment (−8401 to −8170) on SMAD3. After siRNA-mediated knockdown of SMAD3 (si-S3) or control siRNA (si-con), WPMY-1 cells were transfected with the −8401/−8170 reporter construct. Luciferase activity was determined as in A. C, identification of functional elements in the minimal TGFβ-responsive fragment (−8401 to −8170). Putative transcription factor binding sites were verified by site-directed mutagenesis (mutations are indicated by diagonal lines). Luciferase activity was measured as in A. D, effects of siRNA-mediated knockdown of ETS1 and RUNX2 on TGFβ induction of the ANGPTL4 gene in WPMY-1 cells. Two siRNAs were used for each gene. Experimental details were as in Fig. 4B. The efficiencies of the siRNAs are shown in supplemental Fig. S8. ***, **, and *, significant difference from untreated sample (p < 0.001 by t test, p < 0.01, and p < 0.05, respectively). Error bars, S.D.
To identify the functionally relevant elements, we performed site-directed mutagenesis and determined the functionality of the resulting constructs in transient luciferase assays (Fig. 5C). The data show that mutation of the RBE and SBE2 completely abrogated TGFβ induction. A clearly diminished TGFβ response was also observed with mutated versions of the AP-1 site, the EBS, SBE1, and SBE3. In contrast, inducibility was not significantly affected by mutation of the SP1/GC-box. Mutations of the EBS, the RBE, and SBE3 also significantly decreased basal transcriptional activity. Taken together, our results suggest that multiple sites contribute to the function of the TGF-E, with a predominant role for the EBS, the RBE, SBE2, and SBE3.
This conclusion is supported by loss-of-function experiments, which show a clearly decreased inducibility of the endogenous ANGPTL4 gene by TGFβ upon siRNA-mediated knockdown (supplemental Fig. S8) of ETS1 or RUNX2 (from 5.3-fold down to 1.8–3.2-fold) and a dramatic decrease in overall TGFβ-induced transcription (Fig. 5D). No effect was seen with RUNX1 siRNA (data not shown). The third RUNX family member, RUNX3, is expressed at extremely low levels in WPMY-1 cells (not shown).
SMAD3 Interaction with the PPAR-E Enhancer
As demonstrated by the qPCR analyses in Fig. 2, TGFβ and GW501516 synergistically induce ANGPTL4 transcription. The results of the luciferase assay in Fig. 6A show that the synergistic effect of the two ligands could be recapitulated by combining the TGF-E and PPAR-E enhancers in a reporter construct. Although no cooperative effects were seen with either enhancer alone, an induction that was clearly more than additive was seen in a construct harboring both the TGF-E and PPAR-E (6.6-fold compared with a calculated additive induction of 3.9-fold).
FIGURE 6.
Functional interaction of the TGF-E and PPAR-E enhancers in the ANGPTL4 gene. A, cooperative induction of an ANGPTL4 reporter construct by TGFβ and PPARβ/δ. Luciferase reporter assays with vectors containing the TGF-E, the PPAR-E, or both were transfected into WPMY-1 cells in serum-reduced medium, treated as indicated (300 nm GW501516, 2 ng/ml TGFβ2), and harvested 48 h later for determination of luciferase activity. B, effect of TGFβ and GW501516 on the interaction of SMAD3 with the PPAR-E. WPMY-1 cells were treated with solvent, 300 nm GW501516, 2 ng/ml TGFβ2, or both ligands for 1 h, and analyzed by ChIP for SMAD3/PPAR-E interactions. IgG, negative antibody control. C, PPARβ/δ-independent cross-linking of SMAD3 to the PPAR-E. WPMY-1 cells were treated with either control siRNA (si-con) or PPARD siRNA, stimulated with 2 ng/ml TGFβ2 for 1 h, and analyzed by ChIP for cross-linking of SMAD3 and PPARβ/δ to the PPAR-E. D, effect of a triple knockdown of all PPAR subtypes on the interaction of SMAD3 with the PPAR-E. WPMY-1 cells were treated with either control siRNA (si-con) or three siRNA pools against PPARα, PPARβ/δ, or PPARγ; stimulated with 2 ng/ml TGFβ2 for 1 h; and analyzed by ChIP for interaction of SMAD3 and the three PPAR subtypes with the PPAR-E. ***, **, and *, significant difference (p < 0.001 by t test, p < 0.01, and p < 0.05, respectively) compared with untreated cells (A), solvent-treated sample (B), or control siRNA (C and D). #, induction by both ligands significantly higher than highest induction by a single ligand (p < 0.05) (A). Error bars, S.D.
The data in Fig. 3, C and D, indicate that SMAD3 interacts with the PPAR-E in a TGFβ-dependent fashion. Fig. 6B extends this observation by showing that this interaction is influenced neither by GW501516 (Fig. 6B) nor by the siRNA-mediated silencing of PPARβ/δ expression (Fig. 6C). Likewise, a triple knockdown of PPARα, PPARβ/δ, and PPARγ did not diminish the PPAR-E/SMAD3 interaction, although the recruitment of all PPAR subtypes to the PPAR-E was reduced (Fig. 6D). These findings provide strong evidence for a PPAR-independent mechanism mediating the recruitment of SMAD3 to the PPAR-E.
DISCUSSION
Cross-talk of the TGFβ and PPARβ/δ Pathways
The tumor stroma phenotype of Ppard knock-out mice led to the hypothesis that PPARβ/δ may interact with cytokine signaling. We therefore analyzed in the present study whether the transcriptional outcome of TGFβ signaling pathways is influenced by the PPARβ/δ agonist GW501516 in the myofibroblastic cell line WPMY-1. Our microarray data clearly suggest that this is indeed the case. We identified 34 annotated genes and 124 transcripts of unknown function that showed a cooperative induction by both ligands, whereas cooperative repression was observed for only three probes (Fig. 1B). Remarkably, many of these genes were not responsive to a single ligand, as indicated by the relatively small overlap in Fig. 1A (15 genes), pointing to a true sensitization by one ligand to stimulation by the other.
Synergistic induction by TGFβ and GW501516 was confirmed for a number of genes by RT-qPCR. These included several genes with potentially interesting functions in the biological context of TGFβ and PPARβ/δ because they point to a cross-talk of both pathways in biological processes related to tumor stroma function, tumor progression, and metabolism. Angiopoietin-like 4 encoded by the ANGPTL4 gene plays a crucial role in peripheral triglyceride metabolism and has been connected to tumor progression and metastasis (43, 48–50). LIPG encodes endothelial lipase, an enzyme with an important role in the metabolism of plasma lipoproteins and a putative function in modulating atherosclerosis (51). The THBS1 gene codes for thrombospondin-1, which is an essential regulator of angiogenesis (52). The CYP24A1-encoded cytochrome P450 24A1 initiates the degradation of 1,25-dihydroxyvitamin D3 and thereby plays a role not only in calcium homeostasis but probably also in tumorigenesis by abrogating local anti-cancer effects of 1,25-dihydroxyvitamin D3 (53).
PPAR-E, a PPAR-inducible Intronic Enhancer of the ANGPTL4 Gene
Of all PPAR target genes, ANGPTL4 shows by far the strongest response to GW501516 (7-fold in the microarray described in Fig. 1), followed by ADRP (2.5-fold). We made similar observations with other cell lines, including WI-38 (diploid human fibroblasts) and C2C12 cells (mouse myoblasts), where genome-wide transcriptional profiling identified ANGPTL4 as the best PPARβ/δ target gene.4 Our data demonstrate that the PPAR-E represents the only region of the ANGPTL4 gene occupied by PPARβ/δ in vivo within an area of several hundred kb in both directions of the TSS (Fig. 3A).3 Furthermore, the PPAR-E is sufficient to recapitulate both the strong ANGPTL4 response to GW501516 and, in concert with the TGF-E, the synergistic activation by TGFβ and PPARβ/δ in luciferase assays (Fig. 3E). These findings clearly point to specific features of the PPAR-E that determine the unusual response to PPARβ/δ ligands, as discussed below.
First, The PPAR-E harbors at least three contiguous PPREs, which extends a previous study suggesting that a single PPRE (PPRE3 in Fig. 3D) mediates ANGPTL4 induction by PPAR ligands (43). Our data show that each of the three PPREs interacts with PPARβ/δ·RXRα complexes in vitro (Fig. 3D), is most likely occupied in vivo (Fig. 3, C and E), and is able to confer inducibility by GW501516 (Fig. 3F). To our knowledge, ANGPTL4 is the first PPARβ/δ target gene containing more than two contiguous functional PPREs.
Second, PPRE2, which shows the strongest response to GW501516 (Fig. 3F), is structurally different from classical PPREs (i.e. direct repeats of similar motifs spaced by one nucleotide) (DR1 elements; AGGNCA A AGGTCA) (54, 55). Although PPRE1 and PPRE3 resemble the consensus DR1 motif (RGGNCA A AGGTCA; Genomatix MatInspector), the PPRE2 sequence substantially deviates from the consensus in the 3′ half-site (GG instead of TC) and thus appears to represent a novel type of PPRE with two identical half-sites (AGGGGA A AGGGGA). It has been shown that the classical DR1 element is functionally asymmetrical with the PPAR partner of the interacting heterodimer bound to the 5′ half-site and RXR contacting the 3′ half-site (56). Our EMSA data indicate that PPARβ/δ·RXRα heterodimers efficiently interact with PPRE2. This raises the intriguing question of whether PPAR complex binding to the unusual PPRE2 motif induces a specific conformational change and consequently an altered binding of coregulators in vivo. The increased recruitment of PPAR·RXR complexes in vivo, which is not seen in in vitro binding assays, would be consistent with this hypothesis.
Our third observation relevant in this context (i.e. the lack of inducibility of PPRE2 by the RXR agonist 9-cis retinoic acid) (Fig. 3G) could also be explained by an altered conformation and/or complex composition that makes the ligand binding pocket of RXR non-accessible to ligands.
TGF-E, a TGFβ-inducible Upstream Enhancer of the ANGPTL4 Gene
As shown by siRNA-mediated silencing, SMAD3 is indispensable for full transcriptional activation by TGFβ (Fig. 4B). We therefore searched by ChIP analyses for SMAD3 binding sites within a region of ∼10 kb in both directions of the ANGPTL4 transcriptional start site and identified three regions that are occupied by SMAD3 in vivo (Fig. 4C). Region A turned out to be a bona fide TGFβ-regulated enhancer because a 1-kb fragment encompassing this SMAD3 binding site was able to confer inducibility on a luciferase reporter construct (Fig. 4C), SMAD3 recruitment was inducible by TGFβ (Fig. 4D), and TGFβ triggered the recruitment of the SMAD coactivator CBP to the same region (Fig. 4C). In contrast, Region B and the PPAR-E did not respond to TGFβ in transient luciferase assays. We therefore concluded that Region A (TGF-E) is required and sufficient for induction of the ANGPTL4 gene by TGFβ.
The second R-SMAD, SMAD2, was dispensable (Fig. 4B). One SMAD2 siRNA (s2-2) even increased the TGFβ response, which may be related to the existence of inhibitory SMAD2 splice variants (57, 58). This is clearly different from the “classical” target gene PAI1, where SMAD2 is required for full induction (Fig. 4B).
siRNA-mediated interference with SMAD4 expression, on the other hand, had an inhibitory effect on TGFβ inducibility, similar to the knockdown of SMAD3 (Fig. 4B). In agreement with this finding, SMAD4 has been reported to be required for ANGPTL4 induction by TGFβ in a human breast cancer cell line (24). It appears, however, that the presence of SMAD4 is not an absolute requirement because the SMAD4 knockdown inhibited ANGPTL4 induction only partially, whereas PAI1 induction was completely blocked (Fig. 4B). Likewise, we observed ANGPTL4 induction by TGFβ in pancreatic and breast cancer cells lacking functional SMAD4 (Capan-1, Capan-2, and MDA-MB468; data not shown), supporting the view that SMAD4 is involved in regulating the ANGPTL4 gene but is not absolutely essential.
In addition to SMADs, other transcription factors play essential roles. Mutation of an RBE or an SBE in the TGF-E completely abolished TGFβ inducibility in luciferase assays (Fig. 5C). Mutational inactivation of either of two other SBEs, an EBS or an AP-1 site, within the same enhancer fragment also diminished TGFβ induction by nearly 50%. Of note, recruitment of ETS1 to the TGF-E was inducible by TGFβ (Fig. 4D). These transcription factors have been implicated in the induction of other genes by TGFβ, where they most likely function by anchoring SMAD3 to SBEs (28, 29, 32–36). These data can be corroborated into a model where TGFβ triggers the formation of a multiprotein complex on the TGF-E consisting of multiple DNA-binding transcription factors. These include members of the ETS, RUNX, and FOS/JUN families that anchor SMAD3 or SMAD3/4 to adjacent SBEs. The fact that multiple transcription factors bind to the TGF-E in concert with SMADs could also explain the observation that SMAD4 expression is not an absolute requirement for the TGFβ-mediated induction of ANGPTL4.
Synergistic Regulation of the ANGPTL4 Gene by TGFβ and PPARβ/δ
Clearly, TGFβ and PPARβ/δ inducibility is determined by functionally and spatially different regions of the ANGPTL4 gene, the TGF-E and PPAR-E, separated by ∼12 kb. These two enhancers cooperate in transcriptional regulation in a synergistic fashion, pointing to a functional interaction of these regions, which could be explained by different models. Both models accommodate the observation that all three PPAR subtypes are able to activate the PPAR-E and synergize with TGF (Fig. 2D).
The first model postulates a direct physical interaction of the TGF-E and PPAR-E that results in increased transcriptional activity (Fig. 7). Despite our finding that SMAD3 recruitment is not modulated by GW501516 and that PPARβ/δ binding is not affected by TGFβ, it is conceivable that the protein complex formed on one of the enhancer regions influences the recruitment of cofactors to the other region, resulting in an altered chromatin structure favoring activated transcription. If looping between the TGF-E and PPAR-E indeed occurs, how is this established? Our data show that TGFβ induces the recruitment of SMAD3 and ETS1 to the PPAR-E, which also interacts with AP-1 and RUNX2. Likewise, the TGF-E and Region B are also bound by SMAD3, ETS1, and AP-1. It is therefore conceivable that these transcription factors are directly involved in establishing a loop between the two enhancer regions and probably also with Region B.
FIGURE 7.
Model of putative interactions between different functional regions of the ANGPTL4 gene. The model postulates a physical interaction between the TGF-E and the PPAR-E, which may also involve Region B (-2000). These interactions may be established by TGF-E-bound transcription factors making contacts with SMAD3 and other transcription factors recruited to the PPAR-E and possibly Region B, in turn promoting contacts with the basal machinery. Interaction of the TGF-E and PPAR-R may involve currently unidentified transcription factor(s) binding to both enhancer regions (indicated by a question mark).
In an alternative model, the TGF-E and PPAR-E functionally interact in the absence of mutual physical contacts. It is possible that the chromatin modifications at and/or the remodeling proteins recruited to different enhancer regions complement each other in a synergistic way (e.g. by exerting complementary effects on preinitiation complex formation). It may also be conceivable that the protein complexes interacting with the two enhancer regions affect different stages of transcription (i.e. preinitiation complex formation and promoter clearance).
Clarification of the precise mechanism mediating the synergistic activation of the ANGPTL4 gene by TGF-E and PPAR-E enhancers and perhaps Region B will require extensive chromatin conformation capture and ChIP studies investigating DNA looping, histone modifications, recruitment of chromatin-modifying enzymes, chromatin structure, and RNA polymerase II positioning. The identification of the TGF-E and PPAR-E and their interacting transcription factors in the present study provides the basis for these complex future investigations.
Supplementary Material
Acknowledgments
We thank Dr. A. Baniahmad (Jena, Germany) for the RXRα expression plasmid, Drs. M. Krause and Birgit Samans for help with microarray analyses, and Margitta Alt for excellent technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft Grant Mu601/12-1, the core facility of the SFB-TR17, and the LOEWE-Schwerpunkt “Tumor and Inflammation.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S5 and Figs. S1–S8.
T. Adhikary, K. Kaddatz, F. Finkernagel, A. Grahovac, W. Meissner, M. Scharfe, M. Jarek, H. Blöcker, S. Müller-Brüsselbach, and R. Müller, manuscript in preparation.
K. Kaddatz, T. Adhikary, F. Finkernagel, W. Meissner, S. Müller-Brüsselbach, and R. Müller, unpublished observations.
- PPAR
- peroxisome proliferator-activated receptor
- ChIP-Seq
- ChIP sequencing
- PPAR-E
- PPAR-responsive ANGPTL4 intronic enhancer
- PPRE
- PPAR response element
- qPCR
- quantitative PCR
- RXR
- retinoic acid X receptor
- TGF-E
- TGFβ-responsive ANGPTL4 upstream enhancer
- SBE
- SMAD binding element
- RBE
- RUNX binding element
- R-SMAD and Co-SMAD
- regulatory and common SMAD, respectively.
REFERENCES
- 1.Feige J. N., Gelman L., Michalik L., Desvergne B., Wahli W. (2006) Prog. Lipid Res. 45, 120–159 [DOI] [PubMed] [Google Scholar]
- 2.Desvergne B., Michalik L., Wahli W. (2006) Physiol. Rev. 86, 465–514 [DOI] [PubMed] [Google Scholar]
- 3.Barish G. D., Narkar V. A., Evans R. M. (2006) J. Clin. Investig. 116, 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forman B. M., Chen J., Evans R. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michalik L., Wahli W. (2007) Biochim. Biophys. Acta 1771, 991–998 [DOI] [PubMed] [Google Scholar]
- 6.Ricote M., Glass C. K. (2007) Biochim. Biophys. Acta 1771, 926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters J. M., Gonzalez F. J. (2009) Biochim. Biophys. Acta 1796, 230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peraza M. A., Burdick A. D., Marin H. E., Gonzalez F. J., Peters J. M. (2006) Toxicol. Sci. 90, 269–295 [DOI] [PubMed] [Google Scholar]
- 9.Müller-Brüsselbach S., Kömhoff M., Rieck M., Meissner W., Kaddatz K., Adamkiewicz J., Keil B., Klose K. J., Moll R., Burdick A. D., Peters J. M., Müller R. (2007) EMBO J. 26, 3686–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilgore K. S., Billin A. N. (2008) Curr. Opin. Investig. Drugs 9, 463–469 [PubMed] [Google Scholar]
- 11.Kang K., Reilly S. M., Karabacak V., Gangl M. R., Fitzgerald K., Hatano B., Lee C. H. (2008) Cell Metab. 7, 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odegaard J. I., Ricardo-Gonzalez R. R., Red Eagle A., Vats D., Morel C. R., Goforth M. H., Subramanian V., Mukundan L., Ferrante A. W., Chawla A. (2008) Cell Metab. 7, 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan N. S., Michalik L., Di-Poï N., Ng C. Y., Mermod N., Roberts A. B., Desvergne B., Wahli W. (2004) EMBO J. 23, 4211–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nawa T., Nawa M. T., Cai Y., Zhang C., Uchimura I., Narumi S., Numano F., Kitajima S. (2000) Biochem. Biophys. Res. Commun. 275, 406–411 [DOI] [PubMed] [Google Scholar]
- 15.Rival Y., Benéteau N., Taillandier T., Pezet M., Dupont-Passelaigue E., Patoiseau J. F., Junquéro D., Colpaert F. C., Delhon A. (2002) Eur. J. Pharmacol. 435, 143–151 [DOI] [PubMed] [Google Scholar]
- 16.Inoue I., Itoh F., Aoyagi S., Tazawa S., Kusama H., Akahane M., Mastunaga T., Hayashi K., Awata T., Komoda T., Katayama S. (2002) Biochem. Biophys. Res. Commun. 290, 131–139 [DOI] [PubMed] [Google Scholar]
- 17.Planavila A., Rodríguez-Calvo R., Jové M., Michalik L., Wahli W., Laguna J. C., Vázquez-Carrera M. (2005) Cardiovasc. Res. 65, 832–841 [DOI] [PubMed] [Google Scholar]
- 18.Ding G., Cheng L., Qin Q., Frontin S., Yang Q. (2006) J. Mol. Cell Cardiol. 40, 821–828 [DOI] [PubMed] [Google Scholar]
- 19.Coll T., Alvarez-Guardia D., Barroso E., Gómez-Foix A. M., Palomer X., Laguna J. C., Vázquez-Carrera M. (2010) Endocrinology 151, 1560–1569 [DOI] [PubMed] [Google Scholar]
- 20.Oishi Y., Manabe I., Tobe K., Tsushima K., Shindo T., Fujiu K., Nishimura G., Maemura K., Yamauchi T., Kubota N., Suzuki R., Kitamura T., Akira S., Kadowaki T., Nagai R. (2005) Cell Metab. 1, 27–39 [DOI] [PubMed] [Google Scholar]
- 21.Kino T., Rice K. C., Chrousos G. P. (2007) Eur. J. Clin. Invest. 37, 425–433 [DOI] [PubMed] [Google Scholar]
- 22.Lee C. H., Chawla A., Urbiztondo N., Liao D., Boisvert W. A., Evans R. M., Curtiss L. K. (2003) Science 302, 453–457 [DOI] [PubMed] [Google Scholar]
- 23.Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padua D., Zhang X. H., Wang Q., Nadal C., Gerald W. L., Gomis R. R., Massagué J. (2008) Cell 133, 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massagué J. (2000) Nat. Rev. Mol. Cell Biol. 1, 169–178 [DOI] [PubMed] [Google Scholar]
- 26.Liu F., Pouponnot C., Massagué J. (1997) Genes Dev. 11, 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou S., Zawel L., Lengauer C., Kinzler K. W., Vogelstein B. (1998) Mol. Cell 2, 121–127 [DOI] [PubMed] [Google Scholar]
- 28.Yingling J. M., Datto M. B., Wong C., Frederick J. P., Liberati N. T., Wang X. F. (1997) Mol. Cell. Biol. 17, 7019–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Feng X. H., Derynck R. (1998) Nature 394, 909–913 [DOI] [PubMed] [Google Scholar]
- 30.Sano Y., Harada J., Tashiro S., Gotoh-Mandeville R., Maekawa T., Ishii S. (1999) J. Biol. Chem. 274, 8949–8957 [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Derynck R. (2000) J. Biol. Chem. 275, 16979–16985 [DOI] [PubMed] [Google Scholar]
- 32.Koinuma D., Tsutsumi S., Kamimura N., Taniguchi H., Miyazawa K., Sunamura M., Imamura T., Miyazono K., Aburatani H. (2009) Mol. Cell. Biol. 29, 172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jinnin M., Ihn H., Asano Y., Yamane K., Trojanowska M., Tamaki K. (2004) Oncogene 23, 1656–1667 [DOI] [PubMed] [Google Scholar]
- 34.Lindemann R. K., Ballschmieter P., Nordheim A., Dittmer J. (2001) J. Biol. Chem. 276, 46661–46670 [DOI] [PubMed] [Google Scholar]
- 35.Leboy P., Grasso-Knight G., D'Angelo M., Volk S. W., Lian J. V., Drissi H., Stein G. S., Adams S. L. (2001) J. Bone Joint Surg. Am. 83, Suppl. 1, S15–S22 [PubMed] [Google Scholar]
- 36.Javed A., Bae J. S., Afzal F., Gutierrez S., Pratap J., Zaidi S. K., Lou Y., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2008) J. Biol. Chem. 283, 8412–8422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giehl K., Imamichi Y., Menke A. (2007) Cells Tissues Organs 185, 123–130 [DOI] [PubMed] [Google Scholar]
- 38.Descargues P., Sil A. K., Sano Y., Korchynskyi O., Han G., Owens P., Wang X. J., Karin M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2487–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy L., Hill C. S. (2005) Mol. Cell. Biol. 25, 8108–8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He W., Dorn D. C., Erdjument-Bromage H., Tempst P., Moore M. A., Massagué J. (2006) Cell 125, 929–941 [DOI] [PubMed] [Google Scholar]
- 41.Furumatsu T., Ozaki T., Asahara H. (2009) Int J Biochem. Cell Biol. 41, 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graulich W., Nettelbeck D. M., Fischer D., Kissel T., Müller R. (1999) Gene 227, 55–62 [DOI] [PubMed] [Google Scholar]
- 43.Mandard S., Zandbergen F., Tan N. S., Escher P., Patsouris D., Koenig W., Kleemann R., Bakker A., Veenman F., Wahli W., Müller M., Kersten S. (2004) J. Biol. Chem. 279, 34411–34420 [DOI] [PubMed] [Google Scholar]
- 44.Jérôme V., Müller R. (1998) Hum. Gene Ther. 9, 2653–2659 [DOI] [PubMed] [Google Scholar]
- 45.Gehrke S., Jérôme V., Müller R. (2003) Gene 322, 137–143 [DOI] [PubMed] [Google Scholar]
- 46.Naruhn S., Meissner W., Adhikary T., Kaddatz K., Klein T., Watzer B., Müller-Brüsselbach S., Müller R. (2010) Mol. Pharmacol. 77, 171–184 [DOI] [PubMed] [Google Scholar]
- 47.Inman G. J., Nicolás F. J., Callahan J. F., Harling J. D., Gaster L. M., Reith A. D., Laping N. J., Hill C. S. (2002) Mol. Pharmacol. 62, 65–74 [DOI] [PubMed] [Google Scholar]
- 48.Cazes A., Galaup A., Chomel C., Bignon M., Bréchot N., Le Jan S., Weber H., Corvol P., Muller L., Germain S., Monnot C. (2006) Circ. Res. 99, 1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito Y., Oike Y., Yasunaga K., Hamada K., Miyata K., Matsumoto S., Sugano S., Tanihara H., Masuho Y., Suda T. (2003) Cancer Res. 63, 6651–6657 [PubMed] [Google Scholar]
- 50.Lichtenstein L., Kersten S.Biochim. Biophys. Acta 1801, 415–420 [DOI] [PubMed] [Google Scholar]
- 51.Brown R. J., Rader D. J. (2007) Curr. Drug Targets 8, 1307–1319 [DOI] [PubMed] [Google Scholar]
- 52.Moserle L., Amadori A., Indraccolo S. (2009) Curr. Mol. Med. 9, 935–941 [DOI] [PubMed] [Google Scholar]
- 53.King A. N., Beer D. G., Christensen P. J., Simpson R. U., Ramnath N. (2010) Anticancer Agents Med. Chem. 10, 213–224 [DOI] [PubMed] [Google Scholar]
- 54.Palmer C. N., Hsu M. H., Griffin H. J., Johnson E. F. (1995) J. Biol. Chem. 270, 16114–16121 [DOI] [PubMed] [Google Scholar]
- 55.Heinäniemi M., Uski J. O., Degenhardt T., Carlberg C. (2007) Genome Biol. 8, R147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gampe R. T., Jr., Montana V. G., Lambert M. H., Miller A. B., Bledsoe R. K., Milburn M. V., Kliewer S. A., Willson T. M., Xu H. E. (2000) Mol. Cell 5, 545–555 [DOI] [PubMed] [Google Scholar]
- 57.Yagi K., Goto D., Hamamoto T., Takenoshita S., Kato M., Miyazono K. (1999) J. Biol. Chem. 274, 703–709 [DOI] [PubMed] [Google Scholar]
- 58.Ueberham U., Lange P., Ueberham E., Brückner M. K., Hartlage-Rübsamen M., Pannicke T., Rohn S., Cross M., Arendt T. (2009) Int. J. Dev. Neurosci. 27, 501–510 [DOI] [PubMed] [Google Scholar]
- 59.Agilent Technologies (2006) Agilent Microarray Hybridization Chamber User Guide, Version 2.0, Agilent Technologies, Santa Clara, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.