Abstract
Breast cancers that overexpress the receptor tyrosine kinase ErbB2/HER2/Neu result in poor patient outcome because of extensive metastatic progression. Herein, we delineate a molecular mechanism that may govern this malignant phenotype. ErbB2 induction of migration requires activation of the small GTPases Rac1 and Cdc42. The ability of ErbB2 to activate these small GTPases necessitated expression of p120 catenin, which is itself up-regulated by signaling through ErbB2 and the tyrosine kinase Src. Silencing p120 in ErbB2-dependent breast cancer cell lines dramatically inhibited migration and invasion as well as activation of Rac1 and Cdc42. In contrast, overexpression of constitutively active mutants of these GTPases reversed the effects of p120 silencing. Lastly, ectopic expression of p120 promoted migration and invasion and potentiated metastatic progression of a weakly metastatic, ErbB2-dependent breast cancer cell line. These results suggest that p120 acts as an obligate intermediate between ErbB2 and Rac1/Cdc42 to modulate the metastatic potential of breast cancer cells.
Keywords: Breast Cancer, Cell Adhesion, Cell Migration, Epithelial Cell, Receptor Tyrosine Kinase, Src, Tumor Metastases
Introduction
HER2/Neu/ErbB2 (epidermal growth factor receptor 2) is a member of the epidermal growth factor receptor (EGFR)2 family that is amplified and overexpressed in 20–30% of breast cancers. HER2/ErbB2-positive breast cancer yields a poor patient prognosis because of a high incidence of metastases and intrinsic resistance to endocrine and conventional chemotherapy (1). ErbB2 is the preferred heterodimerization partner for EGFR, ErbB3, and ErbB4 (2). The large range of downstream signaling targets induced by ErbB2-containing heterodimers permits this receptor and its partners to modulate a wide array of cellular processes. Of these, the nonreceptor tyrosine kinase Src, PI3K, and MAPK pathways are the most studied in mediating ErbB2 responsiveness, which includes increased proliferation, survival, motility, and invasion (3).
ErbB2-induced cell migration and invasion, and hence metastatic potential, requires activation of Src and its downstream signaling pathways (4). Src also positively provides feedback and stabilizes competent ErbB2/ErbB3 heterocomplexes to enhance PI3K/Akt signaling (5). ErbB2 overexpression in MDA-MB-435 cells enhances formation of metastases by increasing Src synthesis and decreasing its degradation by calpain (6). Similarly, signaling from ErbB2 to Akt is necessary to promote metastatic potential because attenuation of the Akt pathway by pharmacological inhibitors or RNAi decreases the formation of metastases by the ErbB2-dependent 21T breast cancer cell line (7). Together, Src and PI3K function downstream of ErbB2 to activate the RhoGEF Vav2, leading to increased activity of Rac1 and a pro-migratory phenotype (8). Although Vav2 is a key mediator of ErbB2-induced migration and invasion, it is likely that additional Src substrates also contribute to this process.
p120 catenin (p120) is a Src substrate that has been shown to be a potent activator of Rac1, Cdc42, and RhoA in addition to being a critical component of the adherens junction (9). Its purported primary function at the cell membrane is the regulation of cadherin stability (9, 10). p120 is thought to block migration and invasion by stabilizing E-cadherin, thus resulting in the inhibition of metastatic progression (11, 12). Supporting this notion is the reported loss of p120 in a subset of breast cancers (13, 14). However, p120 also resides in the cytoplasm, and its actions in this compartment regulate cellular behavior as well (15). Cytoplasmic p120 activates Rac1 and Cdc42 by directly interacting with Vav2 and p190RhoGAP, which coordinate actin rearrangement to modulate cell migration (3). In contrast to its indirect activation of Rac1 and Cdc42, p120 can directly bind to, and repress, the activity of RhoA (16, 17). In primary human esophageal squamous cells, EGFR overexpression results in a dramatic increase in cell aggregation and migration, and this is accompanied by movement of p120 from the cytoplasm to the cell membrane (18). Alterations in the subcellular localization of p120 have also been detected early in breast cancer progression with cytoplasmic p120 correlating with a loss of E-cadherin and β-catenin in atypical lobular hyperplasia and carcinomas (19). Although these studies suggest that changes in p120 expression or subcellular localization may occur in breast cancer, an analysis of these changes in relationship to molecularly defined breast cancer subtypes (i.e. luminal, HER2/ErbB2-positive, and triple-negative) has not been reported.
HER2/ErbB2-positive breast cancers have a distinct gene expression signature compared with their non-ErbB2-amplified counterparts, suggesting that the composition of this signature may be useful for identifying important mediators of ErbB2-induced tumorigenesis and metastatic progression (20, 21). We previously described an expression profile of a well established transgenic mouse (MMTV-c-Neu) model of ErbB2-positive breast cancer (22). This study revealed that p120 mRNA is induced in mammary tumors from these mice compared with normal control glands. Herein, we report the novel finding that p120 is essential for ErbB2-induced migration and invasion because of its ability to activate Rac1/Cdc42. In addition, p120 enhances the formation of experimental metastases of a weakly metastatic breast cancer cell line. These data indicate that p120 is a novel intermediate in the ErbB2 signaling cascade that potentiates metastatic progression of breast cancer cells.
EXPERIMENTAL PROCEDURES
Cell and Primary Explant Culture
Wild type FVB/N mouse mammary epithelium and MMTV-c-Neu tumor cells were isolated and cultured using procedures adapted from those of Liu et al. (12). EN cell lines from MMTV-c-Neu tumors were generated by explant culture and were maintained in Iscove's modified Dulbecco's medium, 1% FBS, 10 ng/ml EGF, 5 μg/ml insulin, 1 μg/ml hydrocortisone supplemented with penicillin/streptomycin, gentamicin, and Fungizone. MCF-10A, BT-474, and SKBR3 cells were purchased and maintained according to ATCC instructions. Phoenix, 293FT, and MCF-7 cells were cultured in DMEM with 10% FBS supplemented with penicillin/streptomycin. The cells were selected in media containing 500 μg/ml of G418, 500 μg/ml of hygromycin B, or 5 μg/ml puromycin.
Antibodies
Antibodies against Rac1, Cdc42, E-cadherin, pY228 p120 catenin, and total p120 catenin were purchased from BD Transduction Laboratories. Polyclonal antibodies against HER2/ErbB2, Src, Akt, pAkt, p42/44, pY877 ErbB2, and ZO-1 were purchased from Cell Signaling. FITC and TRITC-conjugated goat anti-mouse and goat anti-rabbit secondary antibodies were purchased from Jackson ImmunoResearch Laboratory. Antibodies to β-actin and γ-tubulin were purchased from Sigma. The monoclonal antibody for pY1248 ErbB2 was purchased from Calbiochem.
Xenograft Model of Metastasis
The luciferase gene from the Luciferase T7 control plasmid (Promega) was subcloned into LZBOB-hygro and used to generate a stable population of BT-474 cells. The cells were subsequently infected with control LZBOB-pac and LZBOB-Neo or with mp120 isoforms 1 and 3 (cloned from EN064 cells) to generate 474Luc/vec (control) or two mixed populations of cells that expressed both p120 isoforms: 474Luc/p120–1 and 474Luc/p120–2. Female NCR nu/nu mice were implanted with a 60-day release 1.5 mg/17β-estradiol pellet 2 days prior to inoculation with 3 × 105 cells via intracardiac injection into the left ventricle under 2% isoflurane. Luciferin was administered at 150 mg/ml via intraperitoneal injection (150 mg/kg). The mice were immediately imaged at the specified times on the Xenogen IVIS system (Xenogen Corporation, Hopkinton, MA).
Human Tissue Specimens and Immunohistochemistry
All of the paraffin-embedded tissues were collected from the University Hospital Department of Pathology Archives based on reported receptor (ER, PR, and HER2) status. No patient information was obtained. The samples were sectioned and de-identified. Following immunohistochemistry, all of the samples were blindly scored by two independent pathologists. Approval was obtained prior to initiating these studies from the Case Center Institutional Review Board. Immunohistochemistry for total p120 was performed utilizing the Dako Envision Plus Mouse HRP kit, as previously described (22), with minor modifications. Antigen retrieval was achieved by incubating slides in a decloaking chamber (Biocare Medical) for 15 min at 125 °C in 10 mm citrate buffer (pH 6.0). Sections were blocked with Dako peroxidase blocking reagent along with 15 μl/ml normal goat serum and then incubated with primary antibody diluted 1:50 (overnight, 4 °C). Following incubation with secondary antibody, bound antibody was detected by DAB reaction. The sections were counterstained with Gill's hematoxylin 3 (Polysciences, Inc., Washington, PA), dehydrated, cleared, and mounted in Permount.
Transwell Motility and Invasion Assays and Wound Healing Assays
Transwell® motility assays were performed as previously described (23) with minor modifications. The chemoattractant used in these experiments was complete media unless specified. Invasion assays were performed with 8-μm Matrigel-coated Transwell® filters purchased from BD Bioscience. For wound healing experiments, confluent monolayers of cells were scratched with a 200-μl pipette tip. Wound closure was monitored over a 24-h period. Cell migration into the wound was monitored in multiple wells with Image-PRO MDA version 6.0 (MediaCybernetics) and a Leica DMI 6000B microscope.
Western Blots, Pulldown Assays, and Immunofluorescence
Western, immunofluorescence, and pulldown assays were conducted in accordance with the protocols previously described (23). In brief, subconfluent cells were washed in sterile PBS and lysed in radioimmune precipitation assay buffer and cleared by centrifugation. Protein concentrations were determined by Bio-Rad protein assays. The lysates were processed for SDS-PAGE and membrane-probed with the aforementioned antibodies. In Rac1 and Cdc42 pulldown assays, fusion protein was prepared and coated onto beads. Tissue culture cells were rinsed twice with cold Tris-buffered saline (50 mm Tris-HCl, pH 7.5, 150 mm NaCl) and scraped in lysis buffer (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 10 mm MgCl2, 1 mm DTT, 1% (v/v) Nonidet P-40, 5% (v/v) glycerol, 1 mg/ml leupeptin, 1 mg/ml aprotinin, 1 mm PMSF). The mixture was incubated on ice for several min, clarified by centrifugation, and used immediately. 10% of the cell lysate was used for the total protein control, whereas the remainder was incubated with beads coated with the GST-CRIB or GST-CBD fusion proteins for 45 min to 1 h at 4 °C. The beads were centrifuged, washed three times in lysis buffer, and resuspended in 2× Laemmli sample buffer for SDS-PAGE. To assess p120, E-cadherin, and ZO-1 localization in culture cells, the cells were cultured on glass coverslips, mounted, and fixed with SlowFade Gold (Invitrogen). Confocal images were captured on a Zeiss LSM 510 META microscope with a 1.4 NA 100× oil DIC objective.
Transfections and Virus Production
The Phoenix viral packaging cell line or 293FT cells were cultured, and viruses were generated with the aforementioned vectors as previously described (23) with minor modifications. Phoenix and 293FT cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. 293FT cells were simultaneously transfected with the ViraPower Lentiviral packaging kit (Invitrogen). Transfected 293FT cells were cultured for 48 h at 37 °C before virus-containing medium was harvested. Phoenix cells were selected with puromycin to produce a stable homogenous population of virus-producing cells. Virus-containing medium was removed, filtered through a 0.45-μm filter, and supplemented with polybrene (4 mg/ml, Sigma). Target cells were plated overnight at low density to ensure isolated cells and then incubated in the virus-containing media at 32 °C overnight. Infected cells were cultured until ∼80% confluency and then selected in either 400 μg/ml G418 or 4 μg/ml puromycin.
Statistics
All of the experiments were repeated a minimum of three times. The data are expressed as the means ± S.D. Statistical analyses were performed using a one-way analysis of variance, followed by unpaired Student's t test. A minimum p value of <0.05 was considered statistically significant.
Supplemental Materials
Addition explanations for pulldown assays, immunofluorescence and for the constructs and reagents are given in the supplemental materials.
RESULTS
ErbB2 Induces Expression of p120 in Mammary Epithelial Cells
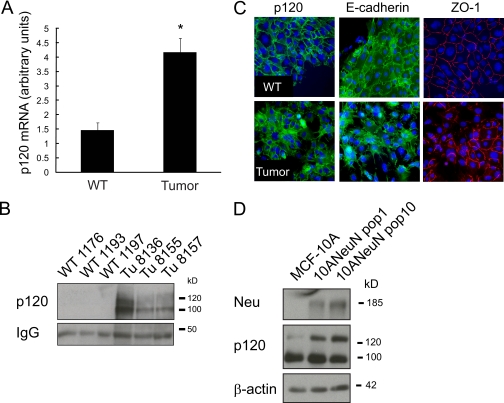
We previously characterized an ErbB2/Neu/HER2-induced mammary tumor transcriptome using a transgenic mouse model of breast cancer (MMTV-c-Neu) by comparing expression signatures of tumors to age-matched wild type (WT) mammary glands (22, 24). Further evaluation of these data revealed that p120 mRNA is increased ∼3-fold in tumors compared with wild type glands (Fig. 1A). Western blot analysis confirmed that p120 protein expression (isoforms 1 and 3) was also elevated in tumors compared with wild type glands (Fig. 1B). Given the cellular complexity of the virgin mammary fat pad, with epithelium being a minor cellular component, these Western results may not be indicative of p120 expression within the mammary epithelium. Therefore, we examined whether p120 protein expression is altered within tumor cells compared with normal mammary epithelium by isolating and culturing epithelial cells from both wild type glands and MMTV-c-Neu tumors, followed by immunocytochemistry for p120. This confirmed that p120 expression is elevated in tumor compared with normal mammary epithelial cells. In addition, the localization of p120 was distinct for the two cell types. In normal mammary epithelial cells p120, E-cadherin, and ZO-1 (zonula occludens-1) are associated with regions of cell-cell contact in a contiguous manner (Fig. 1C). Conversely, p120 expression in the tumor cells was dispersed across cell-cell contacts, cytoplasm, and the nucleus. This analysis also revealed that contiguous circumferential staining of ZO-1 was substantially reduced; suggesting disrupted intercellular contacts and increased motility. To determine whether increased p120 expression in MMTV-c-Neu tumors may be due to ErbB2 overexpression or a consequence of tumorigenesis, we ectopically expressed rat c-Neu/ErbB2 in the nontransformed human mammary epithelial MCF-10A cell line (10ANeuN). Overexpression of ErbB2 increased expression of p120 isoforms 1 and 3 (Fig. 1D), with the predominant induction being observed in the pro-migratory isoform 1 (25). Numerous studies have shown that p120 mediates its effects on cellular behavior according to physical location. Hence, these studies suggest that the up-regulation and relocalization of p120 that occurs in response to ErbB2 may modulate cellular pathways other than stabilizing the adherens junction.
FIGURE 1.
ErbB2 induces expression of p120 in mammary epithelial cells. A, Affymetrix expression array (U74Av2) analysis of age-matched wild type glands compared with MMTV-c-Neu tumors revealed a ∼4-fold increase in p120 mRNA in tumors. The values represent the means ± S.D. *, p < 0.001. B, whole cell lysates of mouse mammary tumors or WT glands were generated, and samples were processed for Western blot analysis for p120 expression. C, freshly isolated epithelial cells from WT mammary glands and MMTV-c-Neu tumors were cultured on coverslips and processed for indirect immunofluorescence. The cells were stained with antibodies against E-cadherin, p120, and ZO-1. Note the mislocalization of p120 in tumor cells compared with WT mammary epithelial cells. D, the immortalized, nontransformed human mammary epithelial cell line MCF-10A was retrovirally transduced with a NeuN/ErbB2 expression cassette, and multiple populations were generated and analyzed for p120 expression (n = minimum of three replicates for each experiment). Scale bar, 20 μm.
ErbB2-associated Migration and Invasion Requires Sustained Expression of p120
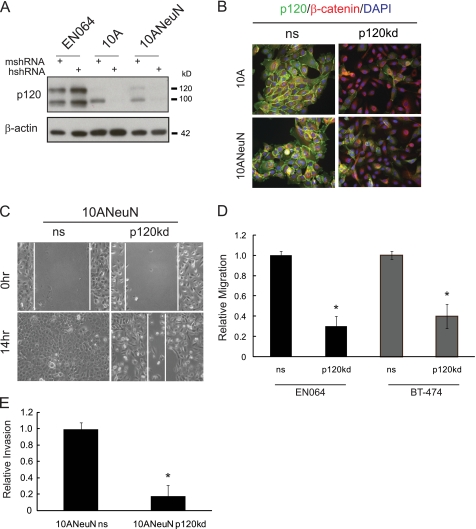
ErbB2 and p120 have independently been shown to regulate cell migration. To determine whether p120 is necessary for ErbB2-induced cell migration and invasion, we attenuated p120 expression in several human mammary epithelial cell lines that exhibit elevated expression of ErbB2 as well as cell lines that we established from MMTV-c-Neu mouse mammary tumors (EN064, EN518, and EN523). The EN cells continue to express ErbB2 and p120 (supplemental Fig. S1) and remain dependent on ErbB family signaling because treatment with the EGFR/ErbB2 selective inhibitor, PD168393, induces extensive cell death (data not shown). Species-specific p120 shRNA directed to either mouse or human mRNA sequences (10) was used to reduce p120 expression (Fig. 2A). In either case, the control and parental cells exhibited indistinguishable p120 levels. We therefore only compare knockdown cells with their nonsilencing counterpart. Consistent with previous reports, stable p120 silencing in EN064 (mouse), 10A/10ANeuN (human), and BT-474 (human) mammary epithelial cells led to a decrease in E-cadherin and β-catenin levels (10, 25) (supplemental Fig. S2A and data not shown). We refer to the nonsilencing shRNA sequence as “ns” and the silencing sequence as “p120kd.” Immunofluorescence imaging of 10A/10ANeuN cells revealed that p120 silencing (p120kd) resulted in nearly complete ablation of p120 at cell-cell borders, whereas cells expressing the nonsilencing control retained p120 at cell junctions (Fig. 2B). These results also revealed that β-catenin was shifted to the cytoplasmic pool and absent from intercellular borders. More importantly, we observed a ∼58% suppression of wound healing in 10ANeuN cells with stable silencing of p120 expression (Fig. 2C). Similar results were obtained in EN064 and BT-474 cells, with p120 silencing resulting in a decrease in serum-induced migration in Transwell® assays (Fig. 2D). In vitro invasion assays further revealed a requirement for p120 in Neu/ErbB2-induced invasion with an 80% reduction in 10ANeuN invasion upon p120 silencing compared with a nonsilencing control (Fig. 2E). These findings were further corroborated in EN064 and EN518 cells where transient p120 silencing led to a decrease in migration and invasion (supplemental Fig. S2, B and C), indicating that the inhibition observed with p120 knockdown was not cell line-specific or caused by stable suppression of p120. Together these data indicate that sustained p120 expression is required for ErbB2-induced migration and invasion of mammary epithelial cells. These results contrast with the purported anti-migratory role of p120 in breast cancer and reveal an essential action of p120 in the context of ErbB2-dependent breast cancer cell migration (19, 26, 27).
FIGURE 2.
ErbB2-associated migration and invasion requires sustained expression of p120. A, species-specific oligodeoxynucleotides were used to generate lentiviral shRNA vectors (10). Stable p120 knockdown populations were generated from EN064, MCF-10A, 10ANeuN, and BT-474 cell lines. The mshRNA targets the mouse p120 mRNA, whereas hshRNA targets the human p120 mRNA. These sequences differ by three nucleotides. B, 10A or 10ANeuN cells that express either a nonsilencing shRNA (ns) or a p120-specific shRNA (p120kd) were cultured on coverslips and processed for indirect immunofluorescence. The cells were stained with antibodies against p120 and β-catenin and counterstained with DAPI. C, migration of ErbB2-overexpressing mammary epithelial cells requires sustained p120 expression. Wound healing was assessed in 10ANeuN cells expressing a nonsilencing p120 shRNA (ns) or a p120 species-specific shRNA (p120kd) over 14 h. D, ErbB2-dependent cells require p120 expression for migratory activity. EN064 and BT474 cell migration was analyzed with or without p120 suppression over a 12-h period of chemotactic migration in modified Boyden chambers toward 10% FBS. E, the ability of p120 to regulate cell invasion was examined with 10ANeuN ns and 10ANeuN kd cells in MartigelTM-coated invasion chambers. The values represent the means ± S.D. **, p < 0.001; *, p < 0.05 compared with the same cell line not treated with inhibitor or transduced with the ns vector (n = minimum of three replicates for each experiment).
p120 Is Required for ErbB2-induced Activation of Rac1 and Cdc42
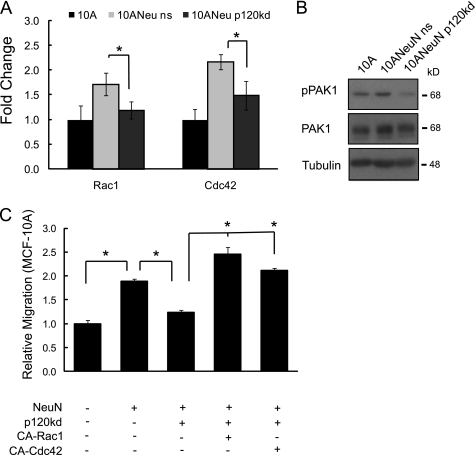
ErbB2 overexpression and/or heregulin stimulation have been shown to activate the small GTPase Rac1, which is a critical modulator of migration (28, 29). Likewise, p120 is an established regulator of Rac1, as well as Cdc42 and RhoA (17). To ascertain the impact of p120 in ErbB2-induced GTPase activity, we compared MCF-10A and 10ANeuN cells in the presence of normal or reduced levels of p120. Although RhoA was not detectable in 10A or 10ANeuN cells (data not shown), overexpression of NeuN resulted in a ∼2-fold increase in Rac1 and Cdc42 activity when compared with the parental cells (Fig. 3A). Silencing p120 in ErbB2-overexpressing 10ANeuN cells caused a ∼50% reduction in Rac1 and Cdc42 activity. Phosphorylation of p21-activated kinase (PAK1), a downstream effector of Rac1 and Cdc42 that facilitates actin cytoskeletal rearrangement and cellular locomotion (30), ErbB2 expression resulted in a 35 ± 0.5% increase in phospho-PAK1 (normalized to total PAK1), an indication of PAK1 activation. This response was mitigated by p120 knockdown, reducing the phospho-PAK level by 62 ± 2.3%. (Fig. 3B), further supporting the importance of this catenin in mediating ErbB2 activation of both GTPases. To determine whether Rac1 and Cdc42 were the primary mediators of ErbB2/p120-regulated migration, constitutively active forms of Rac1 (CA-Rac1) or Cdc42 (CA-Cdc42) were ectopically expressed in 10ANeuN cells in the presence or absence of p120 silencing. Either CA-Rac1 or CA-Cdc42 was sufficient to rescue the loss of motility observed with p120 silencing (Fig. 3C). These data suggest that p120 is necessary for ErbB2 activation of Rac1/Cdc42 and subsequent acquisition of maximal migratory potential.
FIGURE 3.
p120 is required for ErbB2-induced activation of Rac1 and Cdc42. A, ErbB2 activation of Rac1 or Cdc42 requires p120 expression. Rac1 and Cdc42 activation levels were compared between parental MCF-10A, 10ANeuN, and 10ANeuN kd cells. The values represent the means ± S.D. *, p < 0.001 (n = 3). B, silencing p120 results in reduced phosphorylation of PAK1, an established target of Rac1 and Cdc42. C, add-back cell lines were generated by retroviral transduction with expression vectors for CA-Rac1 or CA-Cdc42 into 10ANeuN kd cells. The cells were serum-starved and subjected to chemotactic migration assays toward serum over a 16-h period. The values represent the means ± S.D. *, p < 0.001 (n = 3).
Overexpression of p120 in MCF-10A Cells Mimics the Ability of ErbB2 to Induce Migration but Not Invasion
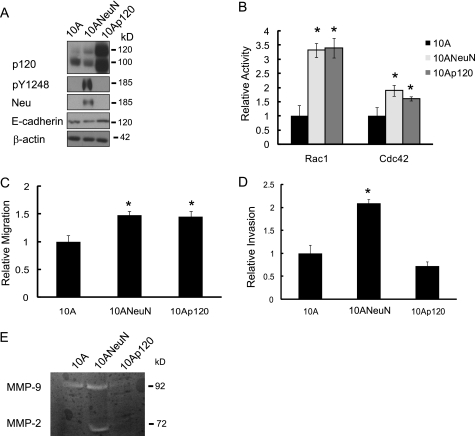
Our studies indicate that sustained expression of p120 is necessary for ErbB2-induced migration and invasion. A key question that arises is whether p120 overexpression alone is sufficient to induce migration/invasion of mammary epithelial cells in the absence of ErbB2 overexpression. We therefore retrovirally transduced both murine p120 isoforms 1 and 3 into MCF-10A cells (10Ap120) (Fig. 4). This increased both the cytosolic and nuclear pools of p120 compared with the vector control (data not shown). Protein analysis showed that p120 overexpression did not induce expression or phosphorylation of ErbB2 in these cells. However, E-cadherin expression was slightly increased as expected given the ability of p120 to stabilize this protein (10) (Fig. 4A). Overexpression of p120 leads to an increase in Rac1 and Cdc42 activity that was indistinguishable from that observed with ErbB2 overexpression (Fig. 4B). Although no obvious changes in morphology were observed with p120 overexpression (data not shown), 10Ap120 cells exhibited a 50% increase in migration compared with MCF-10A cells that was indistinguishable from the increase in migration observed with ErbB2 overexpression (Fig. 4C). A hallmark of metastatic progression is the destruction of surrounding extracellular matrix proteins and cellular invasion into the local environment. To evaluate the invasive potential conveyed by p120 overexpression, 10Ap120 cells were serum-starved, and chemotactic invasion was assessed. Unlike ErbB2 overexpression, expression of p120 was unable to increase invasion of MCF-10A cells (Fig. 4D). To further examine the basis for the differences between p120 and ErbB2 on mammary epithelial cell invasion, we assessed MMP production using gelatin-based zymography (Fig. 4E). Overexpression of ErbB2 in MCF-10A cells increased MMP-2 activity, whereas p120 overexpression alone was unable to induce the activity of MMP-2 and suppressed activity of MMP-9 compared with the vector control cells. Together, these data indicate that although p120 overexpression can mimic the migratory phenotype induced by ErbB2, it cannot completely recapitulate the signaling of this receptor to induce invasion.
FIGURE 4.
Overexpression of p120 in MCF-10A cells mimics the ability of ErbB2 to induce migration but not invasion. A, cDNA clones for p120 isoforms 1 and 3 were generated from EN064 cell mRNA. Ectopic expression was achieved by retroviral transduction of both p120 isoforms into MCF-10A cells. Protein expression was compared with 10ANeuN and parental MCF-10A cells. pY1248 is a phosphorylated form of ErbB2, indicating active signaling from this receptor. B, cells were serum-starved overnight and subjected to Rac1/Cdc42 pulldown assays. MCF-10A cells expressing p120 (10Ap120) exhibited indistinguishable levels of activated Rac1 and Cdc42 compared with ErbB2 overexpression in MCF-10A (10ANeuN) cells. C and D, MCF-10A cells overexpressing ErbB2 (10ANeuN) or p120 (10Ap120) were serum-starved overnight, and chemotactic migration (C) and invasion (D) were assessed over a 14-h period. The values represent the means ± S.D. *, p < 0.001 (n = 3). E, 10A, 10ANeuN, and 10Ap120 cells were maintained in serum-free medium for 48 h, and conditioned medium was collected. The activity of MMP2/9 in conditioned medium was analyzed by gelatin zymography (n = 2).
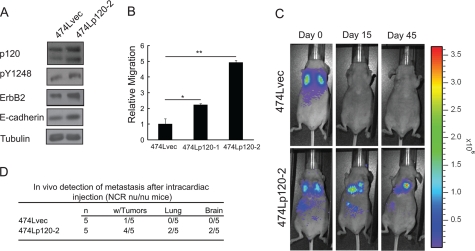
p120 Overexpression Potentiates Metastasis of the Weakly Metastatic, ErbB2-dependent BT-474 Breast Cancer Cell Line
The ability of p120 to induce migration of ErbB2-expressing cells suggested that it may promote metastatic progression of ErbB2/HER2-positive breast cancers. To assess whether p120 overexpression promotes metastatic progression, we utilized the ErbB2-dependent BT-474 cell line, which has a low tumorigenic rate in xenograft models with weak lymph node metastasis when placed in the neck of nude mice (31). Currently, no ErbB2 model of breast cancer has been reported to metastasize after orthotopic transplantation into the mammary fat pad. To circumvent the weak metastatic rate of these HER2-dependent cells, we examined the metastatic process from the point of extravasation into distal tissue sites using a cardiac injection paradigm. BT-474 cells were transduced with a luciferase expression vector to produce cells (474L) that could be noninvasively imaged in xenografted mice following intraperitoneal injection with luciferin. Luciferase expressing cells were then retrovirally transduced to express murine p120 isoforms 1 and 3 or an empty vector. Increased p120 expression was confirmed (Fig. 5A and data not shown), and two stable populations of cells (474Lp120–1 and 474Lp120–2) displayed a ∼2.5–5-fold increase in chemotactic migration compared with the vector control (Fig. 5B and data not shown). On average, p120 expression was increased 2 ± 0.5-fold in 474Lp120–2 cells. We also observed increased E-cadherin, ErbB2, and pY1248 in the 474Lp120–2 cells (60 ± 20-, 70 ± 10-, and 2.0 ± 1.5-fold, respectively) in 474Lp120–2 cells compared with 474L cells. We then assessed the metastatic potential of p120 by utilizing 474Lp120–2 cells in an experimental metastasis model. Immediately following intracardiac injection, both the vector control (474Lvec) and p120 expressing (474Lp120–2) cells were clearly dispersed throughout the vascular network, with a high concentration of cells accumulating in the lungs (Fig. 5C, top and bottom left panels). Within 15 days luciferase-expressing tumors could be detected in the lungs and brains in four of the five mice that received p120-overexpressing cells. In contrast, only one mouse that received vector control cells formed a bioluminescent focus. This occurred around the heart and was likely the result of an inaccurate injection and pooling of cells in the mediastinum (data not shown). The differential formation of tumors between 474Lp120–2 and vector control cells was sustained at 45 days after tumor cell injection (Fig. 5, C and D). Only one vector control mouse developed a detectable tumor after more than 45 days following tumor cell injection (data not shown). These data indicate that increased expression of p120 can promote metastases of ErbB2-dependent tumors.
FIGURE 5.
Ectopic expression of p120 accelerates experimental metastasis. A, 474L/vec and 474L/p120–2 cells were maintained in complete medium, and whole cell lysates were prepared in radioimmune precipitation assay buffer. Protein expression was compared by Western blot. B, evaluation of p120 overexpression on ErbB2-dependent cell migration in Transwell® assays. Migration in response to 10% serum for 16 h was examined for cells containing the empty vector (474L/vec) or 474L/p120–1 and 474L/p120–2 mixed population cell lines that overexpress isoforms 1 and 3 of p120,. *, p < 0.001 (n = 3). C, representative photographs of an 8-week-old female NCR nu/nu mice following intracardiac injections with 474L/vec or 474L/p120–2 cells and noninvasively imaged utilizing luciferase-mediated bioluminescence. The images were captured at 0, 15, and 45 days after tumor cell injection. D, extent of metastases was assessed for individual animals in the experimental metastasis model. The bioluminescence intensity above a negative tissue threshold was calculated, and any mice that had intensities higher than this threshold were considered positive for metastases (n = 7).
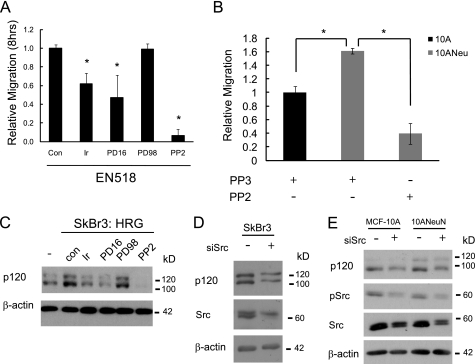
Src Activation Is Required for ErbB2 Induction of p120 Expression
Having established that p120 was up-regulated by ErbB2 and required for ErbB2-induced migration and invasion, we sought to determine the mechanism by which ErbB2 stimulates p120 expression. A panel of inhibitors that target EGFR (Iressa), EGFR/ErbB2 (PD168393), Erk1/2 (PD98059), or Src (PP2) was used to determine which may inhibit migration of EN518 cells. Inhibition of EGFR and ErbB2 (Iressa or PD168393 treatment) caused a 40–60% reduction in cell migration, whereas Erk1/2 inhibition (PD98059) had no effect (Fig. 6A). The most efficacious inhibitor of migration was PP2, blocking EN518 migration by 90%. These data suggested that Src activation is critical for ErbB2-driven motility. We further corroborated this finding by treating 10ANeuN cells with PP2 in a chemotatic migration assay. Src inhibition resulted in a 75% reduction in migration of these cells as well (Fig. 6B), and this result was nearly identical to that obtained with p120 silencing in these cells (Fig. 2C). p120 was first identified as a Src substrate; thus we anticipated that Src activity may also be required for ErbB2 induction of p120 expression (9). To test this possibility, the impact of Src suppression on p120 expression was assessed in cells undergoing transient amplification of ErbB2 activity. This was accomplished using SkBr3 cells that express endogenous ErbB2 that can be further activated by the ligand, heregulin-β1. As expected, heregulin induced phosphorylation of Akt and MAPK, two downstream targets of ErbB2 signaling, within 30 min (supplemental Fig. S3A). Heregulin also stimulated p120 levels within 5 min of treatment (∼3-fold), and induction was maintained for at least 2 h (∼2-fold at 2 h). After confirming the efficacy of the above inhibitors for preventing the activity of their respective targets (supplemental Fig. S3B), we evaluated their impact on heregulin-induced p120 expression (Fig. 6C). Mimicking the results of the migration assays with EN518 cells, inhibition of EGFR and ErbB2 prevented the increase in p120 expression, whereas inhibition of Erk1/2 had no effect. More importantly, inhibition of Src activity with PP2 prevented the stimulation of p120 expression that occurs with heregulin treatment as well as reduced the basal expression of p120 within a 2-h time course. Src regulation of p120 expression was further confirmed by transiently transfecting a siRNA pool against Src into SkBr3 (Fig. 6D). This resulted in a 42 ± 14% decrease in Src protein accompanied by a 55 ± 7% reduction in p120 levels. Transfection of the 10A and 10ANeuN cells (Fig. 6E) with the siSrc also abated Src expression by 38 ± 13 and 52 ± 9%, respectively, and this resulted in a decrease in p120 expression by 42 ± 14 and 39 ± 10%, respectively. Inhibition of Src resulted in a reduction in p120 expression in all three cell lines, indicating that sustained expression of Src is necessary for p120 expression.
FIGURE 6.
Src activation is required for ErbB2 induction of p120 expression. A, EN518- and ErbB2-expressing mouse mammary tumor cells require Src activation for migratory activity. EN518 cell migration was analyzed with or without the following inhibitors: Iressa (Ir; 1 μm), PD168393 (PD16; 1 μm), PD98059 (PD98, 20 μm), and PP2 (10 μm) over an 8-h period of chemotactic migration in modified Boyden chambers toward 10% FBS. Con, control. B, blockade of ErbB2, EGFR, and Src prevents induction of p120 expression. Heregulin (5 μg/ml) induction of p120 expression in SKBR3 cells can be attenuated by pretreatment with various inhibitors. SKBR3 cells were stimulated with heregulin (5 μg/ml) over a 2-h time course, and lysates were probed for alterations in p120 expression. Total and phosphorylated forms of each target of the inhibited proteins were assessed to confirm inhibitor efficacy (supplemental Fig. 3B). C and D, SkBr3, 10A, and 10ANeuN cells were transiently transfected with a Dharmacon siRNA SMART pool against human Src. 48 h post-transfection the cells were harvested and assessed for p120 expression. (n = minimum of three replicates for each experiment).
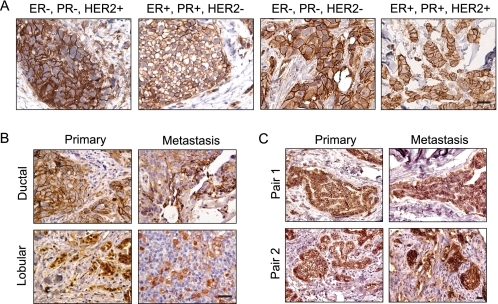
p120 Is Expressed in Most, if Not All, Primary Breast Cancers and Metastases Regardless of Receptor Status
Given the role of p120 in ErbB2/HER2-induced migration and invasion, we asked whether p120 expression is differentially expressed in primary breast tumors and whether its expression correlates with disease subtype. Breast cancers can be categorized according to their receptor expression pattern into four primary groups: 1) estrogen and progesterone receptor-positive and ErbB2/Her2-negative (ER+/PR+/HER2−); 2) estrogen and progesterone receptor-positive and ErbB2/HER2-positive (ER+/PR+/HER2+); 3) estrogen and progesterone receptor-negative and ErbB2/HER2-positive (ER−/PR−/HER2+); and 4) triple negative (ER−/PR−/HER2−) (1, 32). We immunohistochemically stained 71 human breast cancer samples (62 primary tumors and 9 metastases) for total p120 protein and examined these samples for expression levels and localization. Unexpectedly, all of the tumors examined had readily detectable p120 expression, and the level and pattern of expression was indistinguishable in tumors of distinct receptor status (representative samples shown in Fig. 7A). Hence, p120 expression is consistently maintained in the vast majority of breast cancers. In accordance with previous reports (19, 33, 34), ductal carcinomas expressed p120 primarily at the cell membrane, whereas in lobular carcinomas, p120 was localized to the cytoplasm (Fig. 7B). However, this pattern was also independent of receptor status. Of note, all of the breast cancer metastases also expressed p120. Again, the subcellular pattern of staining corresponded to tumor pathology, with lobular tumors having cytoplasmic staining and ductal tumors accumulating p120 predominantly at the cell membrane (Fig. 7B). The pattern of staining for p120 that was associated with ductal versus lobular tumors is consistent with the loss of E-cadherin that occurs in lobular carcinomas (33, 35–37). E-cadherin loss can also occur with tumor progression (27, 38–43); thus we additionally screened nine matched pairs of primary tumors and their associated metastases to determine whether p120 expression may be altered during the process of metastasis (Fig. 7C). All distal metastases maintained p120 expression that was comparable with the primary lesion. In contrast to previous suggestions that p120 may be lost during metastatic progression (12, 13), these results indicate that for breast cancer, sustained p120 occurs in most, if not all, primary tumors and is sustained in subsequent metastases. This consistent pattern of expression may be due to the common activation of Src in many breast cancer types, including those that overexpress ErbB2. Furthermore, the sustained expression in over 80 different tumor samples suggests that p120 may play a pleiotropic and essential role in breast cancer.
FIGURE 7.
Expression of p120 is maintained in most, if not all, primary human breast cancers and distal metastasis regardless of receptor status. A, representative paraffin-embedded samples of 60 primary human breast cancers were selected based on receptor composition and were immunohistochemically stained for p120 catenin. Expression of p120 was retained in all samples and differed only by intracellular location. B, the distinct patterns of subcellular localization of p120 in lobular and ductal primary tumors were preserved in unpaired samples of distal metastases. C, p120 expression is maintained in distal metastases. Representative samples of nine matched pairs of primary breast cancers and their corresponding metastases were stained for p120 catenin and evaluated for expression levels and subcellular localization. Two representative pairs are shown. Bars, 20 μm.
DISCUSSION
Activation of the pro-migratory factor Rac1 and its downstream effector PAK1 is induced by ErbB2 signaling (44), and this ultimately leads to an increase in migration. We show that ErbB2 also increases Cdc42 activity. However, the mechanism(s) by which ErbB2 activates these small GTPases is not fully understood. Mediators between ErbB2 and these GTPases likely contribute to ErbB2-induced metastases; thus their elucidation may reveal effective targets to halt disease progression. The role of p120 has predominantly been associated with the maintenance of cadherin stability, and it has only been proposed to function in a pro-metastatic capacity in cadherin-null environments or in conjunction with mesenchymal cadherins. Herein, we report the first demonstration that ErbB2-induced migration and invasion requires sustained p120 expression. The requirement of p120 to mediate ErbB2-induced migration and invasion is enigmatic to its role as a cadherin stabilizing factor because E-cadherin is an established inhibitor of metastatic progression. In general, the loss of both E-cadherin and p120 has been reported in cancer progression. However, our analyses revealed that chronic ErbB2 overexpression in MMTV-c-Neu mice results in mammary tumors with elevated p120 catenin and E-cadherin expression (Fig. 1 and data not shown). Similar to the mouse tumors, concurrent expression of ErbB2 and E-cadherin has been reported for breast cancer, with ErbB2 expression positively correlating with E-cadherin levels (45). These results contradict the commonly held premise that E-cadherin is generally lost in aggressive cancers, suggesting that sustained E-cadherin is a characteristic of ErbB2-induced tumorigenesis, possibly contributing to collective migration of these tumor cells.
The increased expression of p120 in response to ErbB2 overexpression/activation led us to hypothesize that p120 may be an important intermediary given the ability of p120 to modulate the activity of Rac1, Cdc42, and RhoA (17, 26). Supporting this postulate, we found that p120 silencing resulted in a significant reduction of Rac1 and Cdc42 activity in 10ANeuN cells that was accompanied by an attenuation of cellular migration. Conversely, the ablation of p120 in parental MCF-10A cells accelerated cell motility in wound healing and Transwell® experiments (data not shown), indicating that the effects of p120 silencing are specific to ErbB2 in this cell line. Forced expression of constitutively active forms of Rac1 and Cdc42 completely reversed the phenotype observed with p120 knockdown, indicating that the primary role of p120 in mediating ErbB2-induced migration was activation of these GTPases. Of note, we also found that p120 overexpression was sufficient to increase migration of MCF-10A cells in the absence of ErbB2, indicating that the control of p120 expression is important for determining the basal migratory rate of mammary epithelial cells. These data coupled with the ability of p120 knockdown to induce migration in parental MCF-10A cells suggest that a balance of p120 is critical in maintaining a normal phenotypic state. This may explain why elevated levels of p120 in certain models promote cell motility by increasing Rac1 and Cdc42 activity, whereas attenuating levels of p120 in other models initiates a similar phenotype (27, 46). In contrast to migration, p120 overexpression was incapable of inducing invasion of MCF-10A cells in the absence of ErbB2 overexpression. These data indicate that p120 mediates a subset of ErbB2 signaling, but not all of its properties. Indeed, although ErbB2 overexpression induces activity of MMP-2, p120 overexpression is unable to recapitulate this function.
Using BT-474 breast cancer cells, which are ErbB2-dependent but have low metastatic potential, we found that p120 overexpression increases the formation of distal metastases in an in vivo experimental metastasis model. These data indicate that p120 promotes extravasation in the context of ErbB2-positive breast cancers. Our data also suggest that p120 promotes migration and invasion but does not dismiss a possible pro-survival function. Indeed, we found that p120 silencing in BT-474 cells increases the rate of apoptosis (data not shown), suggesting a role in ErbB2-induced survival as well. Whether p120 may mediate metastases of breast cancer cells that lack constitutive ErbB2 activation is not yet known but quite possible.
We also examined the potential for p120 to control metastases of diverse breast cancer subtypes by evaluating the expression pattern of p120 in various human breast tumors and metastases. Previous immunohistological analyses of colorectal, bladder, gastric, prostate, lung, and pancreatic cancers suggested that p120 expression is decreased or lost in 20–94% cancers (41, 42, 47–49). In two separate studies p120 expression has been reported to be completely lost in 10% of breast cancer samples, whereas abnormal or decreased expression was observed in ∼50–73% of tumors (14, 40). These authors also reported that complete ablation of p120 was associated with progesterone receptor loss and rare. Sarrió et al. (19) also showed that p120 is commonly reduced (57%) in ductal carcinomas. All of these studies correlate p120 loss with increased breast cancer cell invasiveness, and this is likely due to the loss of E-cadherin in these tumors. In contrast, p120 was found to be a marker of lobular tumors, with 88% of tumors exhibiting cytoplasmic localization but not diminished expression (50, 51).
To date no study has addressed p120 expression in the context of HER2 status. Our data indicate that p120 mRNA and protein is increased in MMTV-c-Neu mouse tumors. We then sought to discern whether p120 expression was increased in HER2/Neu-positive tumors as compared with other tumors of differing receptor status (ER+/PR+/HER2+, ER−/PR−/HER+, ER−/PR−/HER2−, and ER+/PR+/HER−). To our surprise, p120 expression remained consistent in all 71 tumor samples, even in those that were triple negative, which are characteristically highly metastatic (52). These results contrast with the previous reports described above. This could be due to three possibilities. The former studies utilized a tissue microarray, which although high throughput, is limited by the amount of each tumor that can be surveyed. In addition, differences in antigen retrieval or patient populations may have contributed to the different findings. From our results, we deduce that p120 reduction is an extremely rare event, suggesting an integral role in tumorigenesis and metastatic progression. Our studies do not, however, exclude the possibility that transient attenuation may occur.
A sustained loss of p120 expression also did not occur with metastatic progression of breast cancer as seen in our matched primary/metastasis paired samples. Most importantly, the retention of p120 in all tumors and metastases suggests that p120 may play a critical role in the maintenance of these diverse pathologies. Although we observed induction of p120 expression in response to ErbB2 overexpression and activation in numerous in vitro and in vivo models, selective up-regulation of p120 was not observed in HER2-positive human breast tumors compared with other tumor subtypes. This may be due to the inherent technical limitation of immunohistochemistry, which is restricted to qualitative assessments. Alternatively, numerous mechanisms may exist for increasing p120 expression in breast cancer, with ErbB2 only being one of these. Lastly, p120 is a multi-faceted intracellular signaling protein with an extensive role in cell behavior, and it is not necessary to change its expression to have an impact on activity. Under normal conditions p120 stabilizes cadherin-mediated cell-cell adhesion, releases Kaiso-mediated transcriptional suppression, and maintains the Rac1/Cdc42/RhoA balance. Furthermore, we found that Src up-regulates the expression of p120 and that p120 is critical for mediating Src actions in breast cancer cells. The pleiotrophic activation of Src in breast cancer by a wide array of oncogenic signals, such as EGFR, HER2, VEGF, Met, IGF, and ER (53–55) among others could each result in activation of p120. Hence, tumors with very different transforming events could all be dependent upon p120 if Src is a critical factor in their transformation. If so, it would not be surprising that the majority of tumors maintain p120 expression.
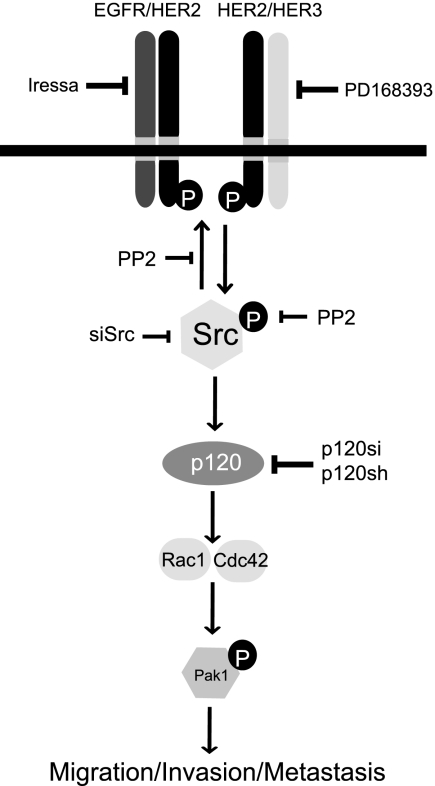
When expressed at low levels, ErbB2 regulates the proper development and growth of normal mammary epithelial cells (56, 57). However, genetic events triggering ErbB2 amplification serve as a catalyst for uncontrolled activation of downstream signals. It is this process that activates signaling pathways that accelerate cellular locomotion and invasion culminating in distal metastasis. Several studies have illuminated various signaling cascades that accentuate ErbB2 migration/invasion (58). Our studies now place p120 as an essential mediator of ErbB2-induced migration, invasion, and experimental metastasis that acts by inducing Rac1 and Cdc42 activity (see model in Fig. 8). Most cancer therapies have been developed by focusing on primary tumors without detailed analyses of the mechanisms underlying metastatic progression. Our studies have revealed that Src plays critical roles in establishing p120 levels in ErbB2 metastatic properties of tumor cells. Hence, targeting proteins such as p120 and Src may provide an avenue for the prevention of metastatic progression in ErbB2-induced tumors and potentially other breast cancer subtypes as well.
FIGURE 8.
A schematic depiction of p120 in the context of ErbB2 regulation of migration/invasion and metastasis. Oncogenic amplification and overexpression of ErbB2 initiates metastatic signals mediated by Src. Our model shows p120 to be a critical intermediary that transduces the pro-migratory signals originating from ErbB2 through Src. Other studies have shown that ErbB2-dependent cells modulate Src activity, which stabilizes functional ErbB2/ErbB3 heterodimers (5). Elevated Src activity may directly contribute to the cytoplasmic pool of p120. It is not clear whether this fraction of p120 requires phosphorylation by Src to propagate ErbB2 signaling. However, it is evident that p120 is necessary to induce Rac1 and Cdc42 activity in highly migratory, ErbB2-dependent cells.
Supplementary Material
Acknowledgments
We thank Drs. Keith R. Johnson and Margaret J. Wheelock for their gift of the LZBOB vectors, CA-Rac1, CA-Cdc42, and GST-CRIB constructs used in the pull-down assays and Dr. Susann Brady-Kalnay for the gift of anti-pY228 p120 catenin antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA090398. This work was also supported by National Institutes of Health Research Oncology Training Grant T32 CA059366, a Continuing Umbrella Research Experience (Cure) Supplement to National Institutes of Health Grant R01 CA090398, Department of Defense Grant USAMRMC W81XWH-09-1-0070, and a University Hospitals of Cleveland Pathology Research Associates Grant (to F. W. A.-K. and R. A. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S3.
- EGFR
- epidermal growth factor receptor
- p120
- p120 catenin
- TRITC
- tetramethylrhodamine isothiocyanate
- PAK
- p21-activated kinase
- ER
- estrogen receptor
- PR
- progesterone receptor.
REFERENCES
- 1.Sørlie T., Perou C. M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Thorsen T., Quist H., Matese J. C., Brown P. O., Botstein D., Eystein Lønning P., Børresen-Dale A. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moghal N., Sternberg P. W. (1999) Curr. Opin. Cell Biol. 11, 190–196 [DOI] [PubMed] [Google Scholar]
- 3.Moasser M. M. (2007) Oncogene 26, 6469–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Y., Benlimame N., Su J., He Q., Alaoui-Jamali M. A. (2009) Br. J. Cancer 100, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishizawar R. C., Miyake T., Parsons S. J. (2007) Oncogene 26, 3503–3510 [DOI] [PubMed] [Google Scholar]
- 6.Tan M., Li P., Klos K. S., Lu J., Lan K. H., Nagata Y., Fang D., Jing T., Yu D. (2005) Cancer Res. 65, 1858–1867 [DOI] [PubMed] [Google Scholar]
- 7.Qiao M., Iglehart J. D., Pardee A. B. (2007) Cancer Res. 67, 5293–5299 [DOI] [PubMed] [Google Scholar]
- 8.Arana E., Vehlow A., Harwood N. E., Vigorito E., Henderson R., Turner M., Tybulewicz V. L., Batista F. D. (2008) Immunity 28, 88–99 [DOI] [PubMed] [Google Scholar]
- 9.Reynolds A. B., Daniel J., McCrea P. D., Wheelock M. J., Wu J., Zhang Z. (1994) Mol. Cell. Biol. 14, 8333–8342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis M. A., Ireton R. C., Reynolds A. B. (2003) J. Cell Biol. 163, 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanagisawa M., Huveldt D., Kreinest P., Lohse C. M., Cheville J. C., Parker A. S., Copland J. A., Anastasiadis P. Z. (2008) J. Biol. Chem. 283, 18344–18354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Li Q. C., Miao Y., Xu H. T., Dai S. D., Wei Q., Dong Q. Z., Dong X. J., Zhao Y., Zhao C., Wang E. H. (2009) Cancer Sci. 100, 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thoreson M. A., Reynolds A. B. (2002) Differentiation 70, 583–589 [DOI] [PubMed] [Google Scholar]
- 14.Dillon D. A., D'Aquila T., Reynolds A. B., Fearon E. R., Rimm D. L. (1998) Am. J. Pathol. 152, 75–82 [PMC free article] [PubMed] [Google Scholar]
- 15.Aho S., Levänsuo L., Montonen O., Kari C., Rodeck U., Uitto J. (2002) J. Cell Sci. 115, 1391–1402 [DOI] [PubMed] [Google Scholar]
- 16.Anastasiadis P. Z., Moon S. Y., Thoreson M. A., Mariner D. J., Crawford H. C., Zheng Y., Reynolds A. B. (2000) Nat. Cell Biol. 2, 637–644 [DOI] [PubMed] [Google Scholar]
- 17.Noren N. K., Liu B. P., Burridge K., Kreft B. (2000) J. Cell Biol. 150, 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andl C. D., Mizushima T., Nakagawa H., Oyama K., Harada H., Chruma K., Herlyn M., Rustgi A. K. (2003) J. Biol. Chem. 278, 1824–1830 [DOI] [PubMed] [Google Scholar]
- 19.Sarrió D., Pérez-Mies B., Hardisson D., Moreno-Bueno G., Suárez A., Cano A., Martín-Pérez J., Gamallo C., Palacios J. (2004) Oncogene 23, 3272–3283 [DOI] [PubMed] [Google Scholar]
- 20.Alexe G., Dalgin G. S., Ganesan S., Delisi C., Bhanot G. (2007) J. Biosci. 32, 1027–1039 [DOI] [PubMed] [Google Scholar]
- 21.Dalgin G. S., Alexe G., Scanfeld D., Tamayo P., Mesirov J. P., Ganesan S., DeLisi C., Bhanot G. (2007) BMC Bioinformatics 8, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landis M. D., Seachrist D. D., Montañez-Wiscovich M. E., Danielpour D., Keri R. A. (2005) Oncogene 24, 5173–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson E., Theisen C. S., Johnson K. R., Wheelock M. J. (2004) J. Biol. Chem. 279, 31041–31049 [DOI] [PubMed] [Google Scholar]
- 24.Guy C. T., Webster M. A., Schaller M., Parsons T. J., Cardiff R. D., Muller W. J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10578–10582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagisawa M., Anastasiadis P. Z. (2006) J. Cell Biol. 174, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wildenberg G. A., Dohn M. R., Carnahan R. H., Davis M. A., Lobdell N. A., Settleman J., Reynolds A. B. (2006) Cell 127, 1027–1039 [DOI] [PubMed] [Google Scholar]
- 27.Wijnhoven B. P., Pignatelli M., Dinjens W. N., Tilanus H. W. (2005) J. Surg. Oncol. 92, 116–123 [DOI] [PubMed] [Google Scholar]
- 28.Yamauchi J., Miyamoto Y., Chan J. R., Tanoue A. (2008) J. Cell Biol. 181, 351–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang C., Liu Y., Lemmon M. A., Kazanietz M. G. (2006) Mol. Cell. Biol. 26, 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knaus U. G., Wang Y., Reilly A. M., Warnock D., Jackson J. H. (1998) J. Biol. Chem. 273, 21512–21518 [DOI] [PubMed] [Google Scholar]
- 31.van Slooten H. J., Bonsing B. A., Hiller A. J., Colbern G. T., van Dierendonck J. H., Cornelisse C. J., Smith H. S. (1995) Br. J. Cancer 72, 22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorlie T., Tibshirani R., Parker J., Hastie T., Marron J. S., Nobel A., Deng S., Johnsen H., Pesich R., Geisler S., Demeter J., Perou C. M., Lønning P. E., Brown P. O., Børresen-Dale A. L., Botstein D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mastracci T. L., Tjan S., Bane A. L., O'Malley F. P., Andrulis I. L. (2005) Mod. Pathol. 18, 741–751 [DOI] [PubMed] [Google Scholar]
- 34.Shibata T., Kokubu A., Sekine S., Kanai Y., Hirohashi S. (2004) Am J. Pathol. 164, 2269–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Da Silva L., Parry S., Reid L., Keith P., Waddell N., Kossai M., Clarke C., Lakhani S. R., Simpson P. T. (2008) Am J. Surg. Pathol. 32, 773–783 [DOI] [PubMed] [Google Scholar]
- 36.Turashvili G., Bouchal J., Ehrmann J., Fridman E., Skarda J., Kolar Z. (2007) Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech. Repub. 151, 59–64 [DOI] [PubMed] [Google Scholar]
- 37.Cleton-Jansen A. M. (2002) Breast Cancer Res. 4, 5–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreizenbeck G. M., Berger A. J., Subtil A., Rimm D. L., Gould Rothberg B. E. (2008) Cancer Epidemiol. Biomarkers Prev. 17, 949–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellovin D. I., Bates R. C., Muzikansky A., Rimm D. L., Mercurio A. M. (2005) Cancer Res. 65, 10938–10945 [DOI] [PubMed] [Google Scholar]
- 40.Nakopoulou L., Zervas A., Gakiopoulou-Givalou H., Constantinides C., Doumanis G., Davaris P., Dimopoulos C. (2000) Anticancer Res. 20, 4571–4578 [PubMed] [Google Scholar]
- 41.Karayiannakis A. J., Syrigos K. N., Alexiou D., Kalahanis N., Rosenberg T., Bastounis E., Pignatelli M. (1999) Anticancer Res. 19, 4401–4405 [PubMed] [Google Scholar]
- 42.Gold J. S., Reynolds A. B., Rimm D. L. (1998) Cancer Lett. 132, 193–201 [DOI] [PubMed] [Google Scholar]
- 43.Karayiannakis A. J., Syrigos K. N., Efstathiou J., Valizadeh A., Noda M., Playford R. J., Kmiot W., Pignatelli M. (1998) J. Pathol. 185, 413–418 [DOI] [PubMed] [Google Scholar]
- 44.Wang S. E., Shin I., Wu F. Y., Friedman D. B., Arteaga C. L. (2006) Cancer Res. 66, 9591–9600 [DOI] [PubMed] [Google Scholar]
- 45.Howard E. M., Lau S. K., Lyles R. H., Birdsong G. G., Tadros T. S., Umbreit J. N., Kochhar R. (2004) Int. J. Clin. Oncol. 9, 154–160 [DOI] [PubMed] [Google Scholar]
- 46.Macpherson I. R., Hooper S., Serrels A., McGarry L., Ozanne B. W., Harrington K., Frame M. C., Sahai E., Brunton V. G. (2007) Oncogene 26, 5214–5228 [DOI] [PubMed] [Google Scholar]
- 47.Kallakury B. V., Sheehan C. E., Ross J. S. (2001) Hum. Pathol. 32, 849–855 [DOI] [PubMed] [Google Scholar]
- 48.Karatzas G., Karayiannakis A. J., Syrigos K. N., Chatzigianni E., Papanikolaou S., Simatos G., Papanikolaou D., Bogris S. (2000) Hepatogastroenterology 47, 1465–1469 [PubMed] [Google Scholar]
- 49.Bremnes R. M., Veve R., Hirsch F. R., Franklin W. A. (2002) Lung Cancer 36, 115–124 [DOI] [PubMed] [Google Scholar]
- 50.Dabbs D. J., Bhargava R., Chivukula M. (2007) Am. J. Surg. Pathol. 31, 427–437 [DOI] [PubMed] [Google Scholar]
- 51.McGrath P. C., Holley D. T., Hamby L. S., Mattingly C. A., Freeman J. W. (1994) Surg. Oncol. 3, 69–77 [DOI] [PubMed] [Google Scholar]
- 52.Kashiwagi S., Yashiro M., Takashima T., Nomura S., Noda S., Kawajiri H., Ishikawa T., Wakasa K., Hirakawa K. (2010) Br. J. Cancer 13, 249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corsino P. E., Davis B. J., Nørgaard P. H., Parker N. N., Law M., Dunn W., Law B. K. (2008) Neoplasia 10, 1240–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X. H., Wang Q., Gerald W., Hudis C. A., Norton L., Smid M., Foekens J. A., Massagué J. (2009) Cancer Cell 16, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finn R. S., Dering J., Ginther C., Wilson C. A., Glaspy P., Tchekmedyian N., Slamon D. J. (2007) Breast Cancer Res. Treat. 105, 319–326 [DOI] [PubMed] [Google Scholar]
- 56.Stern D. F. (2003) Exp. Cell Res. 284, 89–98 [DOI] [PubMed] [Google Scholar]
- 57.Stern D. F. (2008) J. Mammary Gland Biol. Neoplasia 13, 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seton-Rogers S. E., Lu Y., Hines L. M., Koundinya M., LaBaer J., Muthuswamy S. K., Brugge J. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1257–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Zhang M., Ihrig M. M., McManus M. T., Gertler F. B., Scott M. L., Van Parijs L. (2003) Nat. Genet. 33, 401–406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.